Abstract
Objective:
To study the bacterial pathogens causing neonatal sepsis and their sensitivity pattern so that guidelines can be prepared for empirical antibiotic therapy.
Materials and Methods:
We conducted a prospective analysis of all the cases admitted to the neonatal intensive care unit (NICU) of a tertiary care hospital and studied the culture and sensitivity pattern of organisms isolated. The neonates who presented with signs and symptoms of septicemia, with/without pneumonia and/or meningitis were studied and a detailed record of the maturity, age at onset, sex, birth weight (weight on admission for home deliveries), symptoms and signs along with the maternal risk factors was made. The cases with suspect sepsis were screened using various screening markers. Blood culture was done in all the cases, while cerebrospinal fluid was analysed only in those indicated. Sensitivity of the isolated organism was tested by Kirby Bauer disc diffusion techniques and various drug resistance mechanisms were studied.
Result:
Out of the 190 neonates (M:F=1.22:1) admitted to the NICU, 60 (31.57%) shows blood culture positive. Ninety-five percent cases were due to early onset septicemia. Thirty one neonates had Gram negative, twenty seven had Gram positive septicemia and two had candidial infection. Seventy percent Gram-positive isolates were resistant to penicillin. Ninety percent Gram negative isolates were resistant to gentamycin and ampicillin. Carbapenem resistance mechanisms such as ESBL.
Conclusion:
There is an increasing trend of antibiotic resistance to the commonly used and available drugs. Continuous surveillance for antibiotic susceptibility should be done to look for resistance pattern.
Keywords: Early onset septicemia, Kirby Bauer disc diffusion, neonatal sepsis, resistance mechanisms, surveillance
INTRODUCTION
Neonatal sepsis is one of the major causes of morbidity and mortality among the newborns in the developing countries. It is a life-threatening clinical emergency that demands urgent diagnosis and treatment. High rate of antibiotic resistance against commonest bacterial pathogen has further worsened the situation. Neonatal sepsis can be defined as “a clinical syndrome characterized by systemic signs and symptoms and bacteremia during the first month of life”. Bacterial pathogens responsible for this serious condition vary with the geographical area and time.[1] Moreover, the bacterial pathogens responsible and their susceptibility pattern should be regularly monitored in a hospital setting. So, present study is conducted to determine the susceptibility pattern of pathogens causing neonatal sepsis and to provide antibiogram to pediatricians for better patient management. Neonatal sepsis occurs in two distinct patterns based on the age at onset, early onset sepsis (EOS) and late onset sepsis (LOS)[2]
MATERIALS AND METHODS
Our study is based on a prospective analysis of all the cases admitted to the neonatal intensive care unit (NICU) of a tertiary care hospital and to study the culture and sensitivity pattern of organisms isolated. We had taken the permission of ethical committee of our institute to carry out the study. The study was carried out in 2011 during July and August months. The NICU in our hospital is 30 bedded with facilities like ventilators, centralized O2 supply, warmers and phototherapy unit. The neonates who presented with signs and symptoms of septicemia, with/without pneumonia and/or meningitis were included and a detailed record of the maturity, age at onset, sex, birth weight (weight on admission for home deliveries), along with the maternal risk factors was also considered. The cases with suspected sepsis were screened using various screening markers like C-reactive protein, total leucocyte count and platelet count. Blood culture was done in all the cases, while cerebrospinal fluid (CSF) was analysed only in those indicated. The results were categorized into intramural or in-born babies, extramural or out-born babies, early onset and late onset sepsis.[3]
An automated versa trek system was used to detect bacterial growth. Positive blood cultures were subcultured onto blood, chocolate and Mac Conkey agar plates and incubated for 37°C for 24 hours. Quantitative cultures were not performed. Antimicrobial susceptibility testing was done by a Kirby-Bauer disc diffusion method following overnight incubation on Muller-Hinton agar plates. [4] Susceptibility results were interpreted as per recent CLSI guidelines.
RESULT
Total 190 patients were studied during the study period. Out of which 60 (M:F=1.22:1) shows blood culture positive, positivity rate being 31.57%. Gram-negative organisms were isolated in 31 cases (52%), Gram-positive in 27 cases (45%) and Candida spp. in 2 cases (3%). Early onset septicemia (neonates presented with sign and symptoms of septicemia within 72 hours of life)[5,6] was noted in 57 cases (95%), while late onset septicemia (neonates presented with sign and symptoms of septicemia after 72 hours of life)[5,6] was noted only in 3 cases (5%). Prematurity accounted for 70% cases in the study group. Low birth weight (1-2 kg) was associated in 70% of neonates with sepsis. C-reactive protein, one of the septic screen markers, was positive in 70% cases. Platelet counts were low in all cases of septicemia.
Among Gram-negative isolates, Escherichia coli (20%) was commonest organism isolated followed by Klesiella spp.(12%), Pseudomonas spp.(10%), Acinetobacter spp (7%), and others(3%). While in Gram-positive isolates, Coagulase-negative Staphylococci (CONS) was most common (27%) followed by Staphylococcus aureus (13%) and Enterococci spp. (5%). The antimicrobial sensitivity pattern of various organisms was also studied [Figures 1 and 2].
Figure 1.
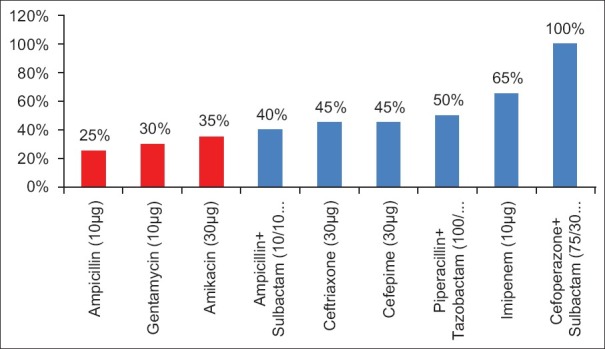
Percent susceptibility in Gram-negative organisms
Figure 2.
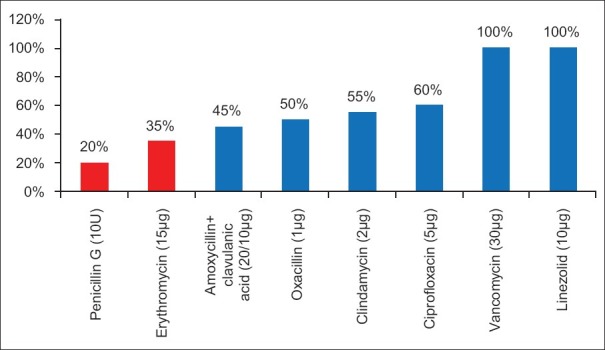
Percent susceptibility in Gram-positive organisms
In Gram-negative isolates, 14% were ESBL producers and 29% were carbapenamase producers. Methicillin resistance in S. aureus was seen in as high as 50% isolates. And methicillin resistance noted in CONS was 68%. Inducible clindamycin resistance was noted in 9%.
DISCUSSION
Sepsis is one of the important causes of neonatal morbidity and mortality.[3] The causative organisms in neonatal sepsis vary from place to place and the frequency of the causative organisms is different in different hospitals and even in the same hospital at different time.[7,8] In Western countries, Group B Streptococci (GBS) is mainly responsible for neonatal sepsis.[9,10] In present study there was not a single case of sepsis by GBS. A possible explanation might be due to the fact that GBS has a tendency to colonize the cervix and sexual partners of infected women can harbor GBS in their urethra and possibly act as reservoirs in certain societies. Therefore, ethnic and socioeconomic differences may contribute to the varying incidence of GBS infection among neonates in different populations.[11]
Thus causative organisms vary in developed and developing countries. In this study, Gram-negative organisms are mainly responsible for neonatal septicemia. The probable reasons being, newborns most probably acquire these Gram-negative rods from the vaginal and fecal flora of the mother and the environment where the delivery occurs.[1] Importance of both vertical transmission from the mother and postnatal acquisition of infection from the environment has been suggested in the literatures for pathogenesis of neonatal sepsis.
Blood culture positivity in our study was 31.57%, while that in Mehmood et al,[1] (2002) study was 4.76%, Shaw Ck et al,[3] (2007) study was 54.64%, Bhattacharjee et al,[12] (2008) study was 48%, Dias et al,[13] (2010) study was 32%. A preponderance of male infants is apparent in almost all studies of sepsis in newborns.[8] In our study also, culture positivity was more in male infants. In present study, early onset septicemia was observed in 95% cases, while it was 49.6% in Shaw Ck et al,[3] (2007) study, 55.3% in studies by Vinodkumar et al,[14] (2008), 73% in AH Movahedian et al,[15] (2006), 64.7% in Aletayeb et al,[16] (2011). Low birth weight [17] and prematurity were the common risk factors associated with neonatal sepsis.
Present study showed ampicillin resistance in 75% isolates. Increased resistance was also noticed against amikacin and gentamycin, which are commonly used for empirical therapy. According to our antibiogram of our hospital, higher generation cephalosporins and β-lactamase inhibitor combinations, carbapenems were effective against Gram-negative bugs. The drug treatment given according to the protocol showed marked improvement in the outcome of patient. In case of Gram-positive isolates, penicillin resistant was noted in 80% cases, which is a primary drug against Gram-positive organisms. Resistance to macrolides is also increasing. Linezolid and vancomycin were sensitive in all isolates (100%). Initial empiric coverage was with ceftriaxone and amikacin which was changed with combination with β-lactamase inhibitors. Thirty percent of cases in our study were negative for CRP but shows blood culture positive. Although C-reactive protein is a very good septic screen marker, other screening markers also should be looked for.
Studies from different geographical area at different time were compared to see the changing antibiotic susceptibility trend [Figures 3 and 4]. So we can conclude that the resistance to the drugs is increasing year by year.
Figure 3.
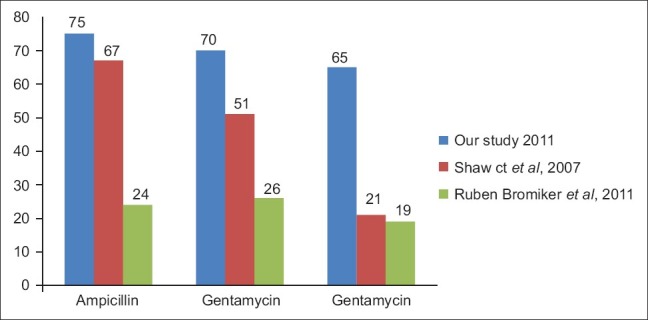
Comparison of % resistance in Gram-negative organism in different studies
Figure 4.
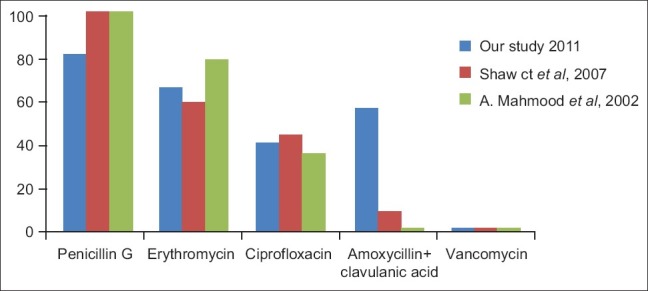
Comparison of % resistance in Gram-positive organism in different studies
CONCLUSIONS
Blood culture is the gold standard in diagnosis and treatment of neonatal septicemia. Multiple antibiotic resistances among neonatal sepsis are currently one of the greatest challenges to the effective management of infections. Slow race in development of newer drugs and rapidity in resistance development are major areas of concern. Wise use of antibiotics should be done or there will be no drugs available in the future for treatment. Longitudinal surveillance should be carried out at regular interval to describe the varied pathogen causing neonatal sepsis as well as their changing antibiotic susceptibility pattern. Each and every hospital must have its own local antibiogram mentioning empirical therapy options.
Present study advises higher generation cephalosporins with β-lactamase inhibitor combination for empirical treatment. Vancomycin should be added to cover Gram-positive ones. The change in antibiotic regime has improved the management of patients. Decrease in the CONS isolates can be done by proper skin treatment before collection of blood, reducing the duration of central and peripheral blood lines. Possible strategies to reduce the load of neonatal sepsis are hand hygiene practice, barrier nursing, promotion of clean deliveries, exclusive breast feeding, rationalization of admissions to and discharge from neonatal units.
ACKNOWLEDGEMENT
This work was supported by dean of Govt. Medical College, Surat, Medical Superintendent and Department of Pediatrics of New Civil Hospital, Surat. We are greatful to them for having graciously provided the opportunity to look for the neonatal sepsis data of our hospital so that if needed we can plan better policies to curtail the mortality and morbidity in NICU.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Mahmood A, Karamat KA, Butt T. Neonatal Sepsis: High Antibiotic Resistance of the Bacterial Pathogens in a Neonatal Intensive Care Unit in Karachi. J Pak Med Assoc. 2002;52:348–50. [PubMed] [Google Scholar]
- 2.Shahian M, Pishva N, Kalani M. Bacterial Etiology and Antibiotic Sensitivity Patterns of Early-Late Onset Neonatal Sepsis among Newborns in Shiraz, Iran 2004-2007. IJMS. 2010;3:293–98. [Google Scholar]
- 3.Shaw CK, Shaw P, Thapaliala A. Neonatal sepsis bacterial isolates and antibiotic susceptibility patterns at a NICU in a tertiary care hospital in western Nepal: A retrospective analysis. Kathmandu Univ Med J. 2007;5:153–160. [PubMed] [Google Scholar]
- 4.Bromiker R, Arad I, Peleg O, Premiger A, Engelhard D. Neonatal Bacteremia: Patterns of Antibiotic Resistance: Infect Control. Hosp Epidemiolo. 2001;22:767–70. doi: 10.1086/501860. [DOI] [PubMed] [Google Scholar]
- 5.Motara F, Ballot DE, Perovic O. Epidemiology of neonatal sepsis at Johannesburg Hospital. South Afr Epidemiol Infect. 2005;20:90–3. [Google Scholar]
- 6.Viswanathan R, Singh AK, Mukherjee S, Mukherjee R, Das P, Basu Aetiology and Antimicrobial Resistance of Neonatal Sepsis at a Tertiary Care Centre in Eastern India: A 3 Year Study. Indian J Pediatr. 2011;78:409–12. doi: 10.1007/s12098-010-0272-1. [DOI] [PubMed] [Google Scholar]
- 7.Shrestha S, Adhikari N, Rai BK, Shreepaili A. Antibiotic Resistance Pattern of Bacterial Isolates in Neonatal Care Unit. J Nepal Med Assoc. 2010;50:277–81. [PubMed] [Google Scholar]
- 8.Ghotaslou R, Ghorashi Z, Nahaei MR. Klebsiella pneuminiae in neonatal sepsis: A 3-year study in pediatric hospital of Tabriz, Iran. JPN J Infect Dis. 2007;60:126–8. [PubMed] [Google Scholar]
- 9.CDC, Trends in Perinatal Group B Streptococcal Disease - United States, 2000-2006. MMWR. 2009;58:109–12. [PubMed] [Google Scholar]
- 10.Waseem R, Khan M, Tahira S, Qureshi AW. Neonatal sepsis. Prof Med J. 2005;12:451–6. [Google Scholar]
- 11.Nidal S. Younis. Neonatal sepsis in Jordan: Bacterial isolates and susceptibility pattern. Rawal Med J. 2011;36(3):169–72. [Google Scholar]
- 12.Bhattacharjee A, Sen MR, Prakash P, Gaur A, Anupurba S. Increased prevalence of extended spectrum β-Lactamase producers in neonatal septicaemic cases at tertiary referral hospital. Indian J Med Microbiol. 2008;26:356–60. doi: 10.4103/0255-0857.43578. [DOI] [PubMed] [Google Scholar]
- 13.Dias E, Vighneshwaran P. The bacterial profile of neonatal septicaemia in a rural hospital in south India. J Clin Diagn Res. 2010;4:3327–30. [Google Scholar]
- 14.Vinodkumar CS, Neelagund YF, Kalsurmath S, Banapurmath S, Kalappannavar NK, Basavarajappa KG. Perinatal risk factors and microbial profile of neonatal septicemia: A multicentred study. J Obstet Gynecol India. 2008;58:32–4. [Google Scholar]
- 15.Movahedian AH, Moniri R, Mosayebi Z. Bacterial Culture of Neonatal Sepsis. Iranian J Publ Health. 2006;35:84–90. [Google Scholar]
- 16.Aletayeb S, Khosravi A, Dehdashtian M, Kompani F, Mortazavi S, Aramesh M. Identification of bacterial agents and antimicrobial susceptibility of neonatal sepsis: A 54-month study in a tertiary hospital. Afr J Microbiol Res. 2011;5:528–31. [Google Scholar]
- 17.Lehner R, Leitich H, Jirecek S, Weninger M, Kaider A. Retrospective analysis of early-onset neonatal sepsis in very low birth-weight infants. Eur J Clin Microbiol Infect Dis. 2001;20:830–32. doi: 10.1007/s100960100622. [DOI] [PubMed] [Google Scholar]


