Abstract
Alternative pre-mRNA splicing yields functionally distinct splice variants in regulating normal cell differentiation as well as cancer development. The putative tumor suppressor gene GT198 (PSMC3IP), encoding a protein also known as TBPIP and Hop2, has been shown to regulate steroid hormone receptor–mediated transcription and to stimulate homologous recombination in DNA repair. Here, we have identified 6 distinct GT198 splice variant transcripts generated by alternative promoter usage or alternative splicing. Various splice variant transcripts preserve a common open reading frame, which encodes the DNA binding domain of GT198. The splice variants act as dominant negatives to counteract wild-type GT198 activity in transcription and to abolish Rad51 foci formation during radiation-induced DNA damage. In fallopian tube cancer, we have identified 44 point mutations in GT198 clustered in 2 mutation hotspot sequences. The mutation hotspots coincide with the regulatory sequences responsible for alternative splicing, strongly supporting that imbalanced alternative splicing is a selected consequence in cancer. In addition, splice variant–associated cytoplasmic expression is found in tumors carrying germline or somatic GT198 mutations. An altered alternative splicing pattern with increased variants is also present in lymphoblastoid cells derived from familial breast cancer patients carrying GT198 germline mutations. Furthermore, GT198 and its variant are reciprocally expressed during mouse stem cell differentiation. The constitutive expression of the GT198 variant but not the wild type induces tumor growth in nude mice. Our results collectively suggest that mutations in the GT198 gene deregulate alternative splicing. Defective alternative splicing promotes antagonizing variants and in turn induces a loss of the wild type in tumorigenesis. The study highlights the role of alternative splicing in tumor suppressor gene inactivation.
Keywords: alternative splicing, GT198, tumor suppressor gene, somatic mutation, DNA repair
Introduction
Alternative splicing control is an integral step in pre-mRNA transcription, and more than 90% of multiexon genes in the human genome are alternatively spliced.1 Various choices of alternative splicing include exon skipping, intron retention, alternative 5′ or 3′ splice site usage, and multiple promoters.2,3 Alternative splicing is regulated by enhancing or silencing regulatory sequence elements located at introns and exons.3 Alternative splicing choice is also controlled by promoter activity since transcription and alternative splicing are coupled.4 Splice variants often yield proteins functionally distinct from the wild type in regulating normal differentiation and development.5 In contrast, defective alternative splicing is frequently associated with disease and cancer.6,7 For example, the breast and ovarian cancer gene BRCA1 regulates homologous DNA recombination.8,9 The protein domain encoded by its large exon 11 is responsible for Rad51-mediated DNA repair.10,11 The splice variant BRCA1Δ11b, lacking a large sequence in exon 11, is abnormally overexpressed in breast cancer, resulting in a potential suppression of wild-type BRCA1.12 Consistently, germline mutations in BRCA1 exon 11 promote the expression of BRCA1Δ11b.13 Additional germline mutations in BRCA1 have also been identified, causing aberrant splicing14,15 or altering alternative splicing.16-18 Since a single pre-mRNA can be processed to generate multiple functional isoforms, it is essential to analyze the interrelationship between wild-type and splice variants when characterizing gene functions in cancer or disease.
Genetic mutations including silent or missense mutations in exons and splicing mutations in introns have often been observed in genetic studies but were previously less characterized than truncating mutations.19 This is in part because they minimally alter protein sequences, and their effects on alternative splicing are underestimated. One of the examples is the TP53 gene encoding tumor suppressor p53. The p53 transcript is alternatively spliced to generate 12 distinct isoforms, all of which preserve a functional DNA binding domain (DBD).20 Isoform imbalance in p53 has been previously shown in various tumors.20,21 The reported silent mutations spread throughout the gene, and multiple silent or missense mutations are found in one tumor.22,23 The hypermutation pattern strongly suggests that at least a significant proportion of TP53 mutations are associated with alternative splicing deregulation,24 although individual cases remain to be further confirmed.
The human GT198 gene (gene symbol PSMC3IP, encoding a protein also known as TBPIP or Hop2) is originally reported in refined BRCA1 locus mapping at the chromosome 17q21 locus,25 which has been linked to breast and ovarian cancer predisposition. The GT198 protein was later characterized as a nuclear receptor co-regulator participating in estrogen, androgen, glucocorticoid, and progesterone receptor–mediated gene regulation.26,27 GT198 has also been shown to regulate DNA recombination in DNA repair28 and to stimulate Rad51-mediated DNA strand exchange.29-31 GT198 possesses DNA recombinase activity, an activity present in Rad51 homologs.32 A germline deletion in GT198 has been identified in a diseased family of ovarian dysgenesis, supporting its critical role in ovarian development.33 In an accompanying study of familial breast and ovarian cancer (Peng et al., in this issue), we have identified GT198 germline and somatic mutations including splicing mutations. Although GT198 splice variant mRNA are evident in the National Center for Biotechnology Information (NCBI) databases, their regulation has not been described. We sought to characterize GT198 alternative splicing to understand its role in cancer.
In this report, we have identified multiple GT198 splice variant transcripts. All variant transcripts contain premature stop codons, causing wild-type truncation and resulting in the expression of a common variant protein. The splice variants act as dominant negatives to counteract GT198 in transcription and in DNA repair. In human primary fallopian tube tumors, we have identified a total of 44 GT198 point mutations clustered in 2 mutation hotspots. The specific hotspot sequences provide clear evidence that alternative splicing regulation is targeted by mutations. We have further confirmed that altered splicing patterns are present in the lymphoblastoid cells of breast cancer patients carrying germline mutations. The cytoplasmic GT198, a variant-specific phenotype, is also found in tumors carrying somatic mutations. To our knowledge, this is the first report of alternative splicing regulation of GT198. Our results suggest that GT198 is functionally inhibited by its splice variants and imply that mutation-induced wild-type loss in cancer is largely due to defective alternative splicing.
Results
Identification of splice variant transcripts in GT198
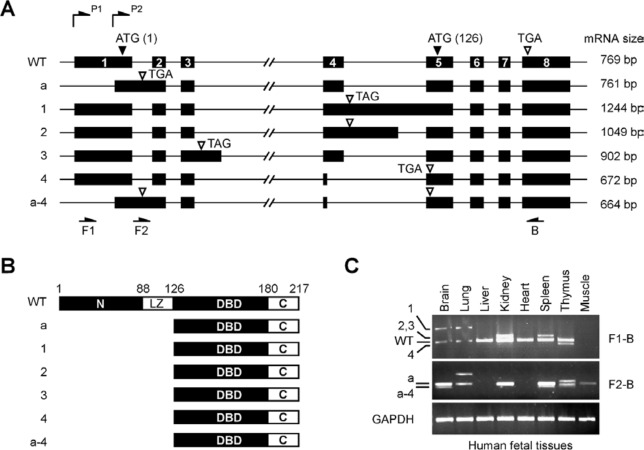
The human GT198 gene has 8 exons spanning approximately 5.5 kb (Fig. 1A). GT198 encodes a 217–amino acid protein containing an N-terminus, a leucine zipper domain required for dimerization, a DBD, and a C-terminus (Fig. 1B).26,31 Using RT-PCR and 5′ RACE analyses, we have identified 6 distinct alternative splicing events occurring strictly at GT-AG consensus sites in human fetal mRNA (Fig. 1C). Fetal mRNA contain more variety of alternative splicing events since GT198 expression is embryonically regulated.26 Still, there may be additional less abundant splice variants present but not yet cloned or characterized. The identified variant cDNA sequences have been deposited to NCBI databases. Interestingly, all identified splice variant transcripts contain premature stop codons (open arrows in Fig. 1A), causing wild-type truncation but preserving an undisrupted open reading frame at the 3′ half of the gene. This open reading frame permits the expression of the DBD and C-terminus by utilizing an internal start codon ATG in exon 5. Thus, even though alternative splicing events differ, they all encode the DBD (Fig. 1B). Four of 6 identified variants are alternatively spliced at the exon 4–intron 4 junction through partial exon exclusion or intron inclusion, suggesting that this splicing junction is frequently regulated. In addition, dual transcription initiation sites driven by 2 promoters (P1 and P2) were found in GT198 through 5′ RACE analysis (Fig. 1A), resulting in longer (expressing GT198 and variants 1-4) and shorter 5′ ends of transcripts (expressing variants a and a-4). The latter have very short 5′ untranslated regions (UTRs) at 10 base pairs and retain intron 1, causing a frameshift, and thereby encode the DBD. The differential usage of 2 promoters appears to link to the choice of intron 1 inclusion (Fig. 1A). The functional effects of these variants need to be analyzed since their mRNA levels are often comparable to or exceed the wild type (Fig. 1C). Taken together, our data suggest that GT198 pre-mRNA is extensively alternative spliced to produce various tissue-specific variants.
Figure 1.
Alternative splicing events in the human GT198 gene. (A) Schematic diagrams of the human GT198 gene and identified alternative splicing events (not to scale). Introns are lines, and numbered exons are bars. Two transcription initiation sites are labeled as P1 and P2. Translation start codons including an internal start codon at amino acid 126 (ATG) are indicated by filled arrows. Translation stop codons including premature stop codons in splice variants (TGA and TAG) are indicated by open arrows. Identified GT198 alternative splicing variants are designated at the left. The sizes of deposited sequences, absent of 3′ UTR, are labeled at the right. (B) Schematic diagrams of protein domains encoded by GT198 and its splice variants. Numbers indicate amino acids. (C) RT-PCR analysis of GT198 mRNA in human fetal tissues. Splice variants labeled on the left correspond to those in A. The primer positions and sizes of PCR products are indicated in A.
GT198 splice variants antagonize wild-type activities
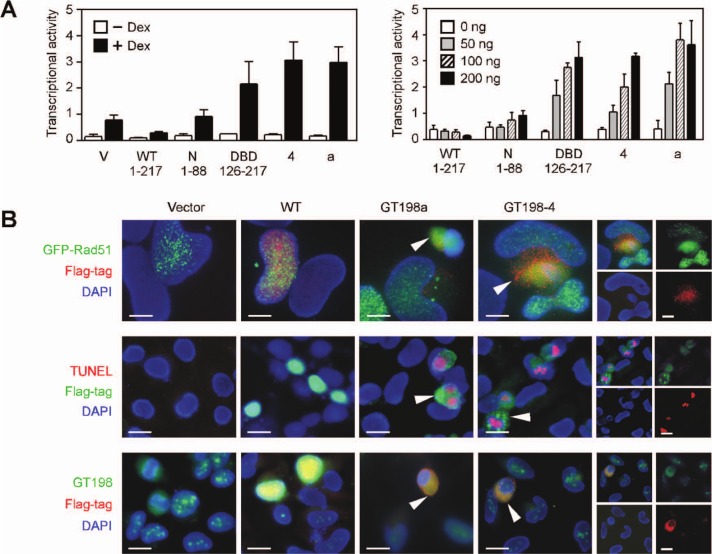
To understand the function of GT198 splice variants, we analyzed the variant activities in transcription and in DNA repair in which their wild type was previously characterized. We found that GT198 variants act as dominant negatives. Variants GT198a and GT198-4, encoded by their full-length cDNA, were chosen to be tested because GT198a represents intron 1–containing variants and GT198-4 is alternatively spliced at the exon 4–intron 4 splice junction. In a transcriptional assay that we previously characterized,26 GT198 regulates mouse mammary tumor virus (MMTV) promoter activity mediated by a glucocorticoid receptor. In HeLa cells, wild-type GT198 suppressed MMTV–luciferase reporter activity, while splice variants GT198a and GT198-4 stimulated transcription (Fig. 2A). The variant stimulation was robust and dose dependent. As controls, the DBD-containing fragment but not the N-terminus also significantly stimulated transcription, suggesting a similar action of the variants and the DBD. These data show that splice variants or their encoded DBD antagonize wild-type activity in transcription.
Figure 2.
GT198 splice variants antagonize wild-type activities. (A) Transient transfection in HeLa cells using GT198 expression plasmids (200 ng) together with the MMTV–luciferase reporter (100 ng), MAP kinase (20 ng), and glucocorticoid receptor (10 ng) that bind to the MMTV promoter. Cells were induced by ligand dexamethasone (100 nM) overnight. Transcriptional activity is measured by luciferase light units shown as mean ± SEM of triplicate transfections (n = 3). A similar transfection is shown on the right using different amounts of GT198 plasmids in the presence of ligand dexamethasone (100 nM). (B) In top panels, fluorescent double staining of HeLa cells transfected with Flag-tagged GT198 plasmids (red) and GFP-Rad51 (green). Rad51 foci formation was induced by γ-irradiation at 8 Gy. The middle panels show TUNEL assays for cell death with DNA fragmentation (red) induced by GT198 plasmids (green). The bottom panels show the cytoplasmic translocation of GT198 variants. Individual color for GT198-4 is shown in the right panels. Sections are counterstained with DAPI. Arrows indicate variant-transfected cells. Scale bars, 10 µm.
In addition to transcription, GT198 is also known to stimulate DNA recombination during DNA repair.29-31 Rad51 recombinase is involved in DNA recombination and forms nuclear foci when DNA damage is induced by γ-irradiation.34,35 Foci formation serves as a readout to test proteins that influence Rad51-mediated DNA repair. GT198 was previously shown to facilitate Rad51-mediated DNA recombination.31 When tested in γ-irradiated HeLa cells expressing GFP-Rad51, GT198 splice variants but not the wild type abolished Rad51 foci formation, resulting in diffuse Rad51 expression (Fig. 2B, top panels). Since the endogenous GT198 protein in HeLa cells is abundant (Fig. 2B, vector control in lower panels), a wild-type stimulatory effect may not be apparent, but a variant inhibitory effect on foci formation was significant. Cell death was also detected by TUNEL assay in cells expressing variants but not the wild type (Fig. 2B, middle panels), explaining their apparent disintegrated nuclei. The data also suggested that GT198 variant activity is able to trigger apoptosis. When protein expression level was examined by Western blots, wild-type GT198 was highly expressed, whereas variants GT198a and GT198-4 produced scant variant proteins corresponding to the DBD (126-217) detected by immunoprecipitation (Suppl. Fig. S1). The variant protein was also detectable by fluorescent staining (Fig. 2B). The low level of variant proteins was possibly due to the apoptosis of transfected cells or due to nonsense-mediated decay in transcripts carrying premature stop codons, nonetheless suggesting that variants must have potent activities in stimulating transcription or inhibiting Rad51 foci formation.
In addition, GT198 variants were expressed in the cytoplasm, while its wild type was in the nucleus (Fig. 2B).26 Although the reason for altered subcellular localization in GT198 variants has not been clear, a similar cytoplasmic localization is well observed in splice variant or mutant forms of tumor suppressors such as p53,20,36,37 Rb,38 or BRCA1.12 The ability to stimulate transcription, induce apoptosis, and impair Rad51 foci formation minimally suggests that GT198 variants are functionally effective. Since variants encode a fragment of the wild-type protein, these data also imply that wild-type activity may not be exposed until regulated so that the variants appear to gain functions due to constitutive activity. In summary, our results suggest that GT198 splice variants counteract with the wild type.
GT198 DBD is autoinhibited by its C-terminus
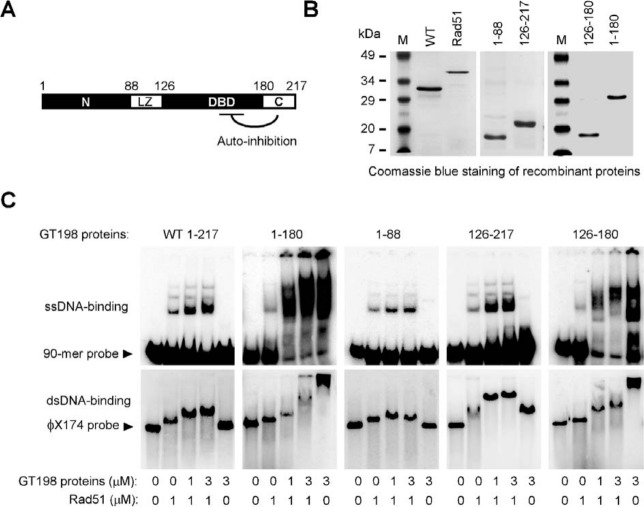
To understand the mechanism of dominant-negative variants, we further characterized GT198 protein domains in DNA binding assays. We purified recombinant GT198 protein fragments (Fig. 3A and 3B) and tested in Rad51-mediated single- or double-stranded DNA binding. Rad51 bound DNA by itself, and wild-type GT198 facilitated DNA binding in the presence of Rad51 (Fig. 3C), which was consistent with previous reports.29,32 The DBD-containing protein encoded by splice variants (126-217), but not the N-terminus (1-88), showed comparable DNA-binding activities than the wild type (1-217). This result potentially explains the dominant-negative effect in the DBD observed above. We also found that the C-terminal deletion (1-180) showed very robust DNA-binding activity even in the absence of Rad51, and the DBD alone with C-terminal deletion (126-180) showed similar strong binding to both single- or double-stranded DNA (Fig. 3C). These data indicate the presence of autoinhibitory regulation in the GT198 molecule in which the C-terminus inhibits the DNA-binding activities of the DBD. The C-terminal deletion, or potential Rad51 interaction, releases this inhibition and renders GT198 active in DNA binding. Consistently, a GT198 germline deletion mutation causing familial ovarian dysgenesis is located within the C-terminus at amino acid Glu201,33 supporting a regulatory role of the C-terminus. The N-terminus would have physiological activities but itself may not act as a dominant negative due to the lack of DNA binding activity to compete. The DBD is likely protected by its surrounding protein sequences especially in a dimer form in the wild type26; the splice variants encoding a less protected DBD incapable of dimerization are more constitutively active in DNA binding so that a desired GT198 activity in cells can be controlled through alternative splicing. Accordingly, uncontrolled constitutive DNA-binding activity would be induced by a truncated mutant protein similar to the DBD (126-217) caused by a germline mutation c.310C>T that we have identified in the accompanying study, (Peng et al., in this issue) thereby suppressing the wild-type function.
Figure 3.
GT198 DBD is autoinhibited by its C-terminus. (A) GT198 protein domains are shown with numbers indicating amino acids. (B) Coomassie blue staining of purified recombinant human Rad51 and His-tagged GT198 proteins in SDS-PAGE. (C) GT198 proteins were analyzed for their single- and double-stranded DNA binding in the presence or absence of human Rad51 in vitro. Single-stranded 90-mer oligonucleotide (0.5 µM nt) and double-stranded φX174 linear DNA (RF I, 5 µM nt) were 32P-labeled as probes and were incubated with Rad51 and GT198 proteins at indicated concentrations. ssDNA binding was analyzed in 6% polyacrylamide gel and dsDNA binding in 0.8% agarose gel. Arrows indicate free probes.
GT198 mutation hotspots in alternative splicing regulatory sequences
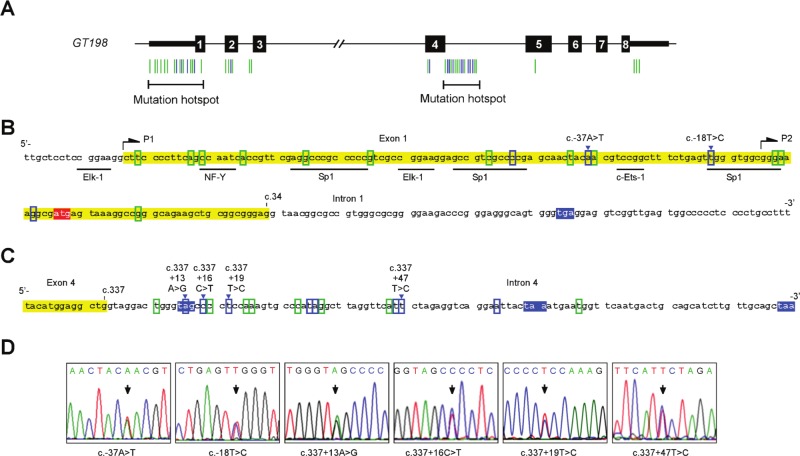
The alternative splicing regulatory sequences in GT198 became more apparent when we identified prevalent GT198 point mutations in fallopian tube cancers (Fig. 4). Initially, during the analysis of germline mutations in GT198 in cancer families, we unexpectedly found GT198 somatic mutations in an ovarian cancer patient (Peng et al., in this issue). We then systematically searched GT198 mutations in tumors. In a set of 29 cases of early stages of fallopian tube cancer, we have identified 44 distinct GT198 point mutations in 25 cases (Fig. 4A and Suppl. Table S1). Some mutations are reoccurring, and multiple different mutations can be found in 1 case (Suppl. Fig. S2). In contrast, mutation was not detected in a set of 12 cases of high-grade serous fallopian tube carcinomas at advanced stages, implying that GT198 mutations are possibly associated with early lesions of the tumor. Most identified mutations are potentially somatic in origin because germline mutations are rare. In the absence of blood samples to verify, it does not exclude the possibility of the presence of germline mutations. The entire gene was analyzed by Sanger sequencing except for its large intron 3, which contains Alu repeats. Within the coding sequences, the identified mutations are missense or silent mutations. When searched against the single-nucleotide polymorphism (SNP) databases, most mutations are not SNPs except that 3 are present at very low frequencies (Suppl. Table S1). Interestingly, the identified mutations are not randomly distributed in the gene but are mostly clustered in 2 mutation hotspots: in the 5′ UTR (Fig. 4B) and in the exon 4–intron 4 splice junction (Fig. 4C). The 2 hotspots coincide with the regulatory sequences responsible for the alternative splicing events that we identified above. Potentially, the differential usage of P1 and P2 promoters by 5′ UTR, which contains predicted transcription factor binding sites (Fig. 4B), determines the choice of intron 1 inclusion. The splicing of intron 1 may also be controlled by the flanking exon 2, which contains several mutations. Noticeably, no mutation was found within intron 1, implicating the absence of the regulatory element. In the second mutation hotspot, high-density mutations including reoccurring ones are located near the splice junction in intron 4. Some of these splicing mutations are identical to what we have identified in a familial cancer patient and altered intron 4 splicing when tested in the accompanying study (Peng et al., in this issue).. Therefore, this hotspot strongly indicates the presence of an intronic splicing regulatory element in the mutated sequence in intron 4, which may be responsible for frequent alternative splicing events surrounding this splice junction (Fig. 1A).
Figure 4.
GT198 mutation hotspots in alternative splicing regulatory sequences. (A) Schematic diagram of the human GT198 gene (not to scale). Introns are shown as lines, numbered exons as bars, and coding regions as thick bars. Identified mutations are indicated by vertical colored lines below the diagram and listed in Supplementary Table S1. (B, C) Nucleotide sequences of the 2 mutation hotspots at the 5′ UTR (B) and the exon 4–intron 4 splice junction (C). GT198 mutations are denoted with recurrent mutations in blue and nonrecurrent mutations in green. Premature stop codons are highlighted in blue, start codon in red, and exons in yellow. Mutation numbering is based on the cDNA reference sequence. (D) Sequence traces of representative recurrent mutations. Additional sequence traces are shown in Supplementary Figure S2. Arrows indicate allelic changes.
Given that mutations are consequences of functional selection, the 2 mutation hotspots inferred the 2 major alternative splicing regulatory sequences in the GT198 gene. Conversely, GT198 alternative splicing would affect function since its defects were selected in tumors. Because alternative splicing can be more disrupted by multiple mutations, it explains the accumulation of multiple somatic mutations in a single tumor. Consistent with this explanation, identified GT198 mutations are missense, silent, and intronic splicing mutations, which will affect alternative splicing more than the protein sequence. Potentially, alternative splicing is one of the major regulatory mechanisms to induce functional defects in GT198 in view of all identified mutations (Fig. 4A). In conclusion, our results demonstrate that GT198 gene activity is critically controlled by alternative splicing.
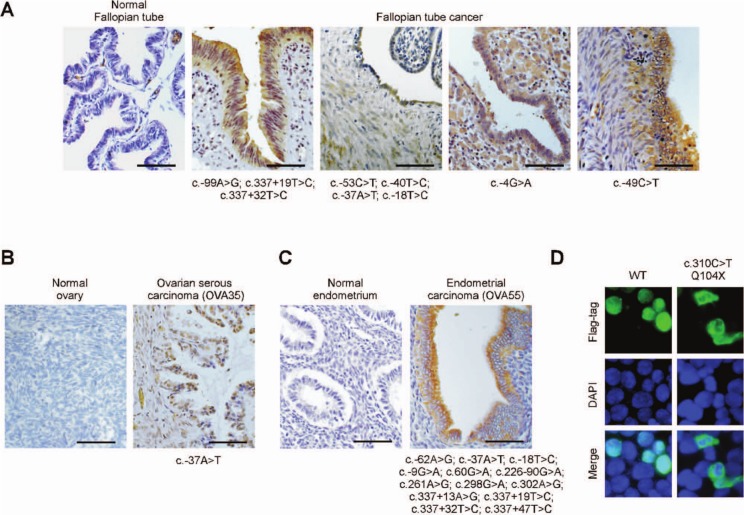
Cytoplasmic expression of GT198 in tumors carrying somatic mutations
Wild-type GT198 is predominantly a nuclear protein,26 and its variant is expressed in the cytoplasm (Fig. 2B). We utilized cytoplasmic GT198 as a variant-associated phenotype to examine tumor tissues carrying GT198 mutations. In fallopian tube cancers carrying mutations, including those carrying a single 5′ UTR mutation, cytoplasmic GT198 was found in tumor cells as well as in tumor-associated stromal cells (Fig. 5A). Almost all mutation-positive fallopian tube cancers were found to express various levels of cytoplasmic GT198 (not shown). In an ovarian cancer derived from a familial cancer patient, cytoplasmic GT198 was detected, although nuclear GT198 was also present (Fig. 5B). In endometrial cancer carrying multiple mutations, strong cytoplasmic GT198 was found in the epithelium (Fig. 5C). When the truncating germline mutation c.310C>T was tested in cells, the truncated protein was also expressed in the cytoplasm (Fig. 5D). These results consistently indicate that the variant phenotype with cytoplasmic expression is associated with GT198 mutations.
Figure 5.
Cytoplasmic expression of the GT198 protein in tumors carrying GT198 mutations. Immunohistochemical staining of GT198 in fallopian tube cancers (A), in ovarian serous papillary carcinoma of patient OVA35 (B), and in endometrial cancer of patient OVA55 (C). Patients OVA35 and OVA55 are sisters and were identified in ovarian cancer family OVA #1. Normal corresponding tissues are controls. Identified mutations are shown below each tumor sample. Polyclonal anti-GT198 antibody (1:200) was affinity purified. Sections are counterstained with hematoxylin. Scale bars, 50 µm. (D) Cytoplasmic expression of GT198 in a truncating mutant. A GT198 cDNA carrying the truncating mutation c.310C>T and encoding a C-terminal Flag-tagged mutant protein was expressed in 293 cells using the wild type as a control. Cells were fluorescent stained by anti-Flag antibody (green) and counterstained with DAPI (blue).
Altered alternative splicing patterns in lymphoblastoid cells carrying GT198 germline mutations
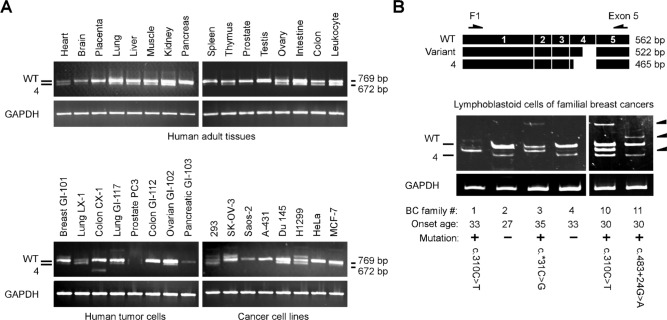
We also analyzed mRNA expression to examine if GT198 mutations alter splice variant expression. We first characterized the normal mRNA expression of GT198 splice variants. Using a 5′ UTR primer, we found that wild-type GT198 is present in all human adult normal tissues and that GT198-4 is a predominant variant (Fig. 6A). GT198 alternative splicing patterns appear to be tissue specific. In contrast, more variety of splice variants is present in cancer cell lines analyzed under the same condition, implicating potential altered alternative splicing in cancer (Fig. 6A). We then analyzed lymphoblastoid cells derived from familial breast cancer patients (Peng et al., in this issue). Similar to the expression pattern in normal blood leukocytes (Fig. 6A), the wild type and GT198-4 are 2 predominant transcripts in lymphoblastoid cells derived from mutation-negative cancer patients in families BC #2 and BC #4 (Fig. 6B). In contrast, several upregulated splice variants (indicated by arrows in Fig. 6B) are present in cells carrying GT198 germline mutations. One detected variant with a fragment at 522 bp between the wild type and GT198-4 was cloned. Sequencing analysis of this variant showed partial exon 4 exclusion of 40 nt, causing a frameshift deletion. This variant was upregulated in cells derived from BC #1, BC #3, and BC #10 (Fig. 6B). Loss of the GT198-4 variant was present in cells from BC #1 and BC #3. Loss of the wild type was present in cells from BC #1. Extra uncharacterized variants were also present in mutant cells. It is unclear why the same c.310C>T mutation resulted in different splice variants; however, the alternative splicing regulation might also be influenced by the copy number changes in BC #10. Our data nonetheless suggest that GT198 mutations are associated with altered alternative splicing, albeit causing complex splicing patterns.
Figure 6.
GT198 germline mutations induce altered alternative splicing. (A) RT-PCR analysis using primers F1 and B for wild-type GT198 in human adult tissues and in cancer cell lines (MTC panels, Clontech). GT198-4 is a predominant variant detected in human normal tissues. GAPDH is the control. PCR products were resolved in 0.8% agarose gel with their sizes labeled at the right. (B) RT-PCR analysis in B-lymphoblastoid cells derived from the blood of breast cancer patients from selected breast cancer families. The onset ages of patients and identified germline mutations are indicated below. The patients without the GT198 mutation served as negative controls. Three arrows at the right indicate abnormally increased GT198 variants. Schematic diagrams above show the primer locations and one of the variants (522 bp) with partial exon 4 exclusion. PCR products were resolved in 6% polyacrylamide gel to increase resolution.
The GT198 activity induced by variant imbalance was further tested in 293 cells using a designed siRNA in exon 4. The designed siRNA has potential significance since its target sequence overlaps with the c.244A>G mutation and the binding site of microRNA miR-493-3p (Suppl. Fig. S3A). The results showed that siRNA inhibition in exon 4 suppressed wild-type but not GT198-4 mRNA expression and caused an increase in transcriptional activity (Suppl. Fig. S3B and S3C). The data confirm that total GT198 activity is contributed by a delicate balance between the wild type and variant. This balance is potentially targeted by genetic mutations.
Splice variant of GT198 induces tumor growth in nude mice.
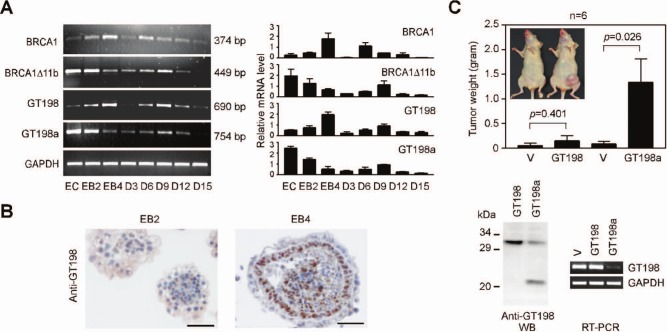
We further obtained evidence supporting the antagonizing splice variant of GT198 by analyzing its expression pattern in a physiological condition. A stem cell differentiation system was preferred because alternative splicing switches in various genes are often observed.39-41 During embryoid body formation,40 we found that wild-type GT198 mRNA was upregulated, whereas variant GT198a mRNA was downregulated (Fig. 7A). GT198a was found as a predominant variant in mouse P19 stem cells as well as in human BG01 stem cells (not shown). A similar reciprocal mRNA expression was also detected between mouse BRCA1 and its splice variant BRCA1Δ11b (Fig. 7A), consistent with the functional importance of alternative splicing in BRCA1.24 In addition, immunohistochemical staining of GT198 confirmed a switch from cytoplasmic to nuclear expression in embryoid bodies (from EB2 to EB4). Wild-type GT198 was increased in a specific ectoderm layer (Fig. 7B). These results clearly suggest that the GT198 variant is downregulated during stem cell differentiation. Then, aberrant variant upregulation might disrupt stem cells if wild-type GT198 activity is required in the process. To test this idea, P19 stem cells stably expressing GT198a were injected into the mammary gland in nude mice. GT198a stable cells, with reduced wild-type mRNA or protein expression, promoted tumor growth in nude mice (Fig. 7C). As a control, wild-type GT198 stable cells did not significantly promote tumors. Our data suggest that GT198 is involved in stem cell differentiation, and variant-induced wild-type loss in differentiating stem cells is potentially tumorigenic. This observation implies a possible role for mutation-induced wild-type GT198 inactivation in human cancers.
Figure 7.
GT198 variant promotes tumor growth in nude mice. (A) Reciprocal expression of endogenous GT198 mRNA and its splice variant during stem cell differentiation. RT-PCR analysis of GT198 and variant GT198a, BRCA1 and its splice variant BRCA1Δ11b, during mouse P19 stem cell differentiation. EC = undifferentiated embryonal carcinoma; EB = embryoid body; D = days of further differentiation. Quantification by real-time PCR is shown on the right. (B) Immunostaining of GT198 in mouse embryoid bodies at stages EB2 and EB4. Nuclear GT198 is upregulated in the ectoderm layer in EB4. Sections are counterstained with hematoxylin. Scale bars, 50 µm. (C) Stably selected mouse P19 stem cells expressing GT198 or GT198a were injected into mammary glands of nude mice (n = 6, Student t test). Representative tumors are shown in the insert. In lower panels, Western blot and RT-PCR analyses of GT198 showing that the variant GT198a inhibits GT198 expression. Endogenous GT198 mRNA was analyzed using a 5′ UTR primer to avoid detecting transfected cDNA in stable cells.
Discussion
Our study underscores the importance of alternative splicing imbalance in cancer genes. Alternative splicing is coupled with transcription and is controlled by multiple elements including promoters, UTRs, and exonic and intronic regulatory sequences.4 A coordinated regulation of these elements in BRCA1 has been demonstrated during estrogen-stimulated transcription.42 Consistently, BRCA1 is known to be alternatively spliced with many splicing mutations identified. Human BRCA1 transcripts,43 as well as human p53 transcripts,44 are also driven by 2 promoters with dual transcription initiation sites. Although a detailed mechanism remains to be further studied, the differential usage of promoters or 5′ UTR sequences has often been shown to link to alternative splicing decisions.4,45-48 In GT198, the identified splice variants invariably truncate the wild type through premature stop codons and serve as dominant negatives. The choice of intron 1 inclusion or exclusion is likely regulated in the 5′ UTR, which explains its section as one of the mutation hotspots. Frequent GT198 mutations in the 5′ UTR or in the splice junction site, and silent or missense mutations in the coding regions, are consistent with the alternative splicing regulation in these sequences and suggest that defective alternative splicing is a selected consequence in cancer. Similarly, missense mutations in many other breast cancer genes have also been observed.49-51 More than half of the TP53 mutations are missense mutations,52 and p53 is alternative spliced with multiple splice variants.20 It is possible that defective alternative splicing, in addition to amino acid change, is one of the direct consequences of missense mutations in cancer genes.
An on-off switch during stem cell differentiation between GT198 and the variant GT198a, and between BRCA1 and its variant BRCA1Δ11b, suggests that alternative splicing control is physiologically relevant. In fact, mutation-induced GT198 splice variants are similar to normal splice variants in specific human embryonic tissues (Fig. 1), where wild-type GT198 may need to be suppressed during normal development. Thus, GT198 mutations could pose problems only in specific tissues or cell types requiring wild-type activity, such as in the ovary in which a wild-type protein was previously detected.26 Since splicing forms are competitively expressed and often functionally counteract, the mutation-induced upregulation of variants is phenotypically equivalent to its wild-type deficiency, that is, loss of a tumor suppressor.
GT198 shows intriguing characteristics as a potential tumor suppressor. The human GT198 gene is located at the chromosome 17q21 locus previously linked to breast and ovarian cancer predisposition. Germline mutations in GT198 are present at a low frequency in breast and ovarian cancer families (Peng et al., in this issue). Somatic mutations in GT198 are prevalent in fallopian tube cancer tissues. Various mutations would result in the upregulation of dominant-negative splice variants, causing a loss of the wild type. Similar to the TP53 gene, GT198 has prevalent mutations, is extensively alternative spliced, and encodes a DBD preserved in alternatively spliced variants.20 These analogical characteristics imply that alternative splicing may be a common and shared mechanism in human cancer genes.
Materials and Methods
Cloning of GT198 splice variants and RT-PCR
Endogenous human GT198 and its splice variant mRNA were analyzed by RT-PCR followed by sequencing analysis using first-strand cDNA from human fetal and adult tissues and cancer cell lines (MTC panels, Clontech, Mountain View, CA). Full-length GT198-1 to -4 cDNA containing a longer 5′ end similar to that of the wild type were obtained by 5′ RACE (Invitrogen, Carlsbad, CA). Subsequent 5′ RACE using the intron 1 primer identified shorter 5′ end transcripts 10 base pairs upstream of the start codon in GT198a and GT198a-4 variants. Mouse GT198a was isolated from mouse P19 stem cells with the retention of intron 1. Expression pattern analysis of P19 stem cells relied on specific primers at 5′ for wild-type GT198 and at intron 1 for variant GT198a. For RT-PCR, total RNA was isolated using Trizol reagent (Invitrogen), treated with DNase I, reverse-transcribed to cDNA using SuperScript III (Invitrogen), and normalized for their concentrations before use. GAPDH was a control. Human and mouse RT-PCR primers are as follows: human wt, cttccccttcagccaatcac (F1), gtgggcccctcaggggtctg (B); human intron 1 specific, cagtgggtgaggaggtcggttga (F2); human exon 5, ggtagctgctttaatgttcttcaa; mouse wt, agctctcgagtttggtggcg, tcaggggtcggggagca aaac; mouse intron 1 specific, ccgggaggggaataggagagg; mouse BRCA1 wt, gaagtggatgggggttttagtt, gcttgctcatttggctccatta; mouse BRCA1Δ11b, cagtaactaagccaggtgattg, tgacatgtttggttccagatct (detect BRCA1Δ11b but not BRCA1 due to the large size of exon 11 in BRCA1); GAPDH, accacagtccatgccatcac, tccaccaccctgttgctgta.
GT198 splice variant accessions
Sequence data have been deposited at NCBI databases under the following accessions: hGT198, FJ952179; hGT198-1, FJ952180; hGT198-2, FJ952181; hGT198-3, FJ952182; hGT198-4, FJ952183; hGT198a, GQ851964; hGT198a-4, GQ851965; mGT198, FJ937966; and mGT198a, FJ937967.
Luciferase assay
HeLa cells were transfected in triplicate in 24-well plates using a MMTV–luciferase reporter (100 ng), glucocorticoid receptor (10 ng), MAP kinase (20 ng), and various amounts of plasmids containing cDNA of GT198, variants GT198a and GT198-4 (without 5′ UTR), the DBD (126-217), and N-terminus (1-88). For siRNA transfection, 293 cells were similarly transfected with a MMTV–luciferase reporter (100 ng), glucocorticoid receptor (10 ng), and increasing amounts of siRNA tgaaggucagcaucacucac (0, 25, 50, and 100 nM). Cells were incubated with ligand dexamethasone (100 nM) to induce the expression of the MMTV–luciferase reporter for 16 hours before harvest. Relative luciferase activities were measured by a Dynex Luminometer ( Dynex Technologies, Chantilly, VA, USA). Data are shown as mean ± SEM of triplicate transfections (n = 3).
Rad51 foci formation assay and immunofluorescence
HeLa cells were co-transfected with GFP-Rad51 (green) and Flag-tagged expression plasmids of GT198 or variants (red). Cells were γ-irradiated at 8 Gy and recovered for 4 hours before methanol-fixed for immunofluorescence. Immunofluorescent double staining was carried out using affinity-purified rabbit anti-GT198 (1:200) and mouse anti-Flag (M2, 1:1,000) (Sigma, St. Louis, MO) antibodies and Cy3- and FITC-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA). For GT198 expression plasmid vectors, the cDNA were inserted into pcDNA3 (Invitrogen) under the control of a cytomegalovirus (CMV) promoter. GT198a and GT198-4 were C-terminal Flag-tagged. The GFP-Rad51 plasmid contained full-length Rad51 in a pEGFP-C3 vector (Clontech). For TUNEL assay, immunofluorescent antibody binding was performed prior to the TUNEL assay using an In Situ Cell Death Detection Kit (Roche, Basel, Switzerland). Sections were counterstained with DAPI.
DNA binding assays
Human Rad51 was purified through spermidine precipitation followed by chromatography using Affi-Gel Blue (Bio-Rad, Hercules, CA), Heparin (Bio-Rad), and Mono-Q columns (GE Healthcare, Little Chalfont, UK).53 N-terminal His-GT198 proteins were expressed in Escherichia coli BL21(DE3) LysS and purified through Ni-NTA-agarose (Qiagen, Hilden, Germany) and Mono-Q columns. For ssDNA binding, 32P-labeled 90-mer (0.5 µM nt) was incubated with Rad51 and His-GT198 proteins at indicated concentrations at 37°C for 30 minutes in a reaction buffer (40 mM HEPES, pH 7.0, 50 mM NaCl, 10% glycerol, 1 mM ATP, 1 mM MgCl2, 1 mM DTT). The 90-mer oligonucleotide is aattctcattttacttaccggacgctattagcagtggcagattgtactgagagtgcaccatatgcggtgtgaaataccgcacagatgcgt. For dsDNA binding, a 32P-labeled φX174 linear dsDNA probe (RF I, 5 µM nt) was incubated at 25°C for 30 minutes with proteins in the above binding buffer. ssDNA binding was analyzed in 6% polyacrylamide gel and dsDNA binding in 0.8% agarose gel. The results were visualized using PhosphorImager (GE healthcare, Piscataway, NJ, USA).
Stem cell differentiation
Undifferentiated mouse P19 stem cells were induced for differentiation by 500 nM all–trans retinoic acid to form embryoid bodies and further differentiate in the absence of retinoic acid for 15 days.39 Total RNA was isolated for RT-PCR analysis using mouse GT198 or mouse BRCA1 primers. For immunohistochemistry, mouse embryonic stem cells were grown on γ-irradiated feeder fibroblasts and were differentiated into embryoid bodies by serum deprivation. Anti-GT198 was affinity-purified as previously described.26
Mutation analysis
Informed consent from the individuals was obtained following institutional guidelines. Fallopian tube cancer tissues were obtained from Georgia Regents University, Dana-Farber Cancer Institute, US Biomax, and the Cooperative Human Tissue Network. Two groups of tumor samples were analyzed for somatic mutations. The first group contained premalignant lesions or early-stage tumors derived from the fallopian tube. These included 14 fallopian tube carcinomas at early stages, 10 premalignant lesions of the fallopian tube derived from female genital cancer patients, and 5 premalignant lesions in the fallopian tube adjacent to fallopian tube cancer. A total of 44 distinct GT198 mutations were identified in 25 of the 29 samples. The second group contained 12 high-grade serous carcinomas, which were at the advanced stages of fallopian tube cancer. This group was found absent of the GT198 mutation. Selected positive samples were histopathologically verified and analyzed by immunohistochemistry using anti-GT198. Corresponding blood DNA was unavailable for each tumor sample. Most identified mutations are potentially somatic in origin because germline mutations are rare. However, the possibility for the presence of germline mutations also exists. Genomic DNA was isolated using the DNeasy Tissue Kit (Qiagen). The entire gene (c.-131G to c.*316C) was Sanger sequenced excluding its large intron 3 (c.225+160A to c.226-96G). Nucleotide numbering is based on the cDNA reference sequence. The identified 44 mutations were searched against the SNP databases including dbSNP, HapMap, Exome Variant Server, and USCS genome browser. Only 3 mutations were present at low frequencies (Suppl. Table S1). To confirm weak sequencing signals, amplified PCR products containing mutant copies were cloned into the pGEM-T Easy TA cloning vector (Promega, Fitchburg, WI). Multiple clones were sequenced individually to confirm the presence of mutations on each allele.
Prediction of transcription factor binding site
Transcription factor binding sites in the 5′ UTR of GT198 were predicted using Match 1.0 at TRANSFAC (www.gene-regulation.com).
Western blot analysis
Polyclonal anti-GT198 antibody was generated by immunizing rabbits (Covance, Princeton, NJ).26 Western blot analysis was performed using whole cell lysates of transfected 293 cells or stably selected P19 cells. Immunoprecipitation was carried out using anti-Flag M2 agarose beads (Sigma) and incubated with 1:10 diluted 293 cell extracts. The precipitates were washed and subjected to Western blot analysis using anti-GT198 antibody. The blots were probed with anti-GT198 at a dilution of 1:200 and detected with the ECL system (Amersham Pharmacia Biotech UK Ltd., Little Chalfont, UK).
Tumor formation in nude mice
The animal work was performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of Georgia Regents University/Medical College of Georgia under approved protocols. Mouse P19 stem cells were stably transfected with GT198 or GT198a cDNA driven by a CMV promoter. Stable cells (105) were injected subcutaneously into female nude mouse mammary glands. In each group of 6, the right side was injected with vector-transfected control P19 cells, and the left side was injected with either GT198- or GT198a-transfected stable cells. Mice were examined for tumor weight 48 days after injection. Results are shown as mean ± SEM (n = 6). Statistical analysis was performed by a Student t test.
Acknowledgments
The authors thank Drs. William W. Chin, Nelly Auersperg, Ronny I. Drapkin, Quinten Waisfisz, Teresa A. Coleman, Shuang Huang, Dorothy Tuan, Ying Meng, and Wendy W. Kuhne for their help. They also thank Kimerly K. Smith, Doris B. Cawley, and Kimya Howard for technical assistance.
Footnotes
Supplementary material for this article is available on the Genes & Cancer website at http://ganc.sagepub.com/supplemental.
Declaration of Conflicting Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Lan Ko, the corresponding author, is the inventor of patent applications concerning potential diagnosis and drug development using the GT198 gene. No financial agreement and licensing has been involved.
Funding: The author(s) received the following financial support for the research, authorship, and/or publication of this article. This work is supported in part by the Georgia Cancer Coalition Distinguished Cancer Scholar Award (to L.K.) and by the National Institutes of Health (CA062130 and CA132640 to N.F.M.).
References
- 1. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413-5 [DOI] [PubMed] [Google Scholar]
- 2. Xing Y, Lee C. Alternative splicing and RNA selection pressure: evolutionary consequences for eukaryotic genomes. Nat Rev Genet. 2006;7:499-509 [DOI] [PubMed] [Google Scholar]
- 3. Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386-98 [DOI] [PubMed] [Google Scholar]
- 4. Kornblihtt AR. Promoter usage and alternative splicing. Curr Opin Cell Biol. 2005;17:262-8 [DOI] [PubMed] [Google Scholar]
- 5. Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12: 715-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre- mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342-57 [DOI] [PubMed] [Google Scholar]
- 7. Venables JP. Unbalanced alternative splicing and its significance in cancer. Bioessays. 2006;28:378-86 [DOI] [PubMed] [Google Scholar]
- 8. Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864-74 [DOI] [PubMed] [Google Scholar]
- 9. Welcsh PL, Owens KN, King MC. Insights into the functions of BRCA1 and BRCA2. Trends Genet. 2000;16:69-74 [DOI] [PubMed] [Google Scholar]
- 10. Chen JJ, Silver D, Cantor S, Livingston DM, Scully R. BRCA1, BRCA2, and Rad51 operate in a common DNA damage response pathway. Cancer Res. 1999;59:1752s-6s [PubMed] [Google Scholar]
- 11. Huber LJ, Yang TW, Sarkisian CJ, et al. Impaired DNA damage response in cells expressing an exon 11-deleted murine Brca1 variant that localizes to nuclear foci. Mol Cell Biol. 2001;21:4005-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wilson CA, Payton MN, Elliott GS, et al. Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-delta11b. Oncogene. 1997;14:1-16 [DOI] [PubMed] [Google Scholar]
- 13. Anczukow O, Buisson M, Salles MJ, et al. Unclassified variants identified in BRCA1 exon 11: consequences on splicing. Genes Chromosomes Cancer. 2008;47:418-26 [DOI] [PubMed] [Google Scholar]
- 14. Brose MS, Volpe P, Paul K, et al. Characterization of two novel BRCA1 germ-line mutations involving splice donor sites. Genet Test. 2004;8:133-8 [DOI] [PubMed] [Google Scholar]
- 15. Zhang L, Chen L, Bacares R, et al. BRCA1 R71K missense mutation contributes to cancer predisposition by increasing alternative transcript levels. Breast Cancer Res Treat. 2011;130:1051-6 [DOI] [PubMed] [Google Scholar]
- 16. Orban TI, Olah E. Emerging roles of BRCA1 alternative splicing. Mol Pathol. 2003;56:191-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet. 2001;27:55-8 [DOI] [PubMed] [Google Scholar]
- 18. Claes K, Vandesompele J, Poppe B, et al. Pathological splice mutations outside the invariant AG/GT splice sites of BRCA1 exon 5 increase alternative transcript levels in the 5’ end of the BRCA1 gene. Oncogene. 2002;21:4171-5 [DOI] [PubMed] [Google Scholar]
- 19. Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285-98 [DOI] [PubMed] [Google Scholar]
- 20. Khoury MP, Bourdon JC. p53 isoforms: an intracellular microprocessor? Genes Cancer. 2011;2:453-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hofstetter G, Berger A, Fiegl H, et al. Alternative splicing of p53 and p73: the novel p53 splice variant p53delta is an independent prognostic marker in ovarian cancer. Oncogene. 2010;29:1997-2004 [DOI] [PubMed] [Google Scholar]
- 22. Strauss BS. Silent and multiple mutations in p53 and the question of the hypermutability of tumors. Carcinogenesis. 1997;18:1445-52 [DOI] [PubMed] [Google Scholar]
- 23. Kanjilal S, Strom SS, Clayman GL, et al. p53 mutations in nonmelanoma skin cancer of the head and neck: molecular evidence for field cancerization. Cancer Res. 1995;55:3604-9 [PubMed] [Google Scholar]
- 24. Okumura N, Yoshida H, Kitagishi Y, Nishimura Y, Matsuda S. Alternative splicings on p53, BRCA1 and PTEN genes involved in breast cancer. Biochem Biophys Res Commun. 2011;413:395-9 [DOI] [PubMed] [Google Scholar]
- 25. Rommens JM, Durocher F, McArthur J, et al. Generation of a transcription map at the HSD17B locus centromeric to BRCA1 at 17q21. Genomics. 1995;28:530-42 [DOI] [PubMed] [Google Scholar]
- 26. Ko L, Cardona GR, Henrion-Caude A, Chin WW. Identification and characterization of a tissue-specific coactivator, GT198, that interacts with the DNA-binding domains of nuclear receptors. Mol Cell Biol. 2002;22:357-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Satoh T, Ishizuka T, Tomaru T, et al. Tat-binding protein-1 (TBP-1), an ATPase of 19S regulatory particles of the 26S proteasome, enhances androgen receptor function in cooperation with TBP-1-interacting protein/Hop2. Endocrinology. 2009;150:3283-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Petukhova GV, Romanienko PJ, Camerini-Otero RD. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev Cell. 2003;5:927-36 [DOI] [PubMed] [Google Scholar]
- 29. Enomoto R, Kinebuchi T, Sato M, et al. Stimulation of DNA strand exchange by the human TBPIP/Hop2-Mnd1 complex. J Biol Chem. 2006;281:5575-81 [DOI] [PubMed] [Google Scholar]
- 30. Pezza RJ, Voloshin ON, Vanevski F, Camerini-Otero RD. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 2007;21:1758-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Enomoto R, Kinebuchi T, Sato M, et al. Positive role of the mammalian TBPIP/HOP2 protein in DMC1-mediated homologous pairing. J Biol Chem. 2004;279:35263-72 [DOI] [PubMed] [Google Scholar]
- 32. Petukhova GV, Pezza RJ, Vanevski F, et al. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat Struct Mol Biol. 2005;12:449-53 [DOI] [PubMed] [Google Scholar]
- 33. Zangen D, Kaufman Y, Zeligson S, et al. XX ovarian dysgenesis is caused by a PSMC3IP/HOP2 mutation that abolishes coactivation of estrogen-driven transcription. Am J Hum Genet. 2011;89:572-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115-23 [DOI] [PubMed] [Google Scholar]
- 35. Pellegrini L, Yu DS, Lo T, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287-93 [DOI] [PubMed] [Google Scholar]
- 36. Ghosh A, Stewart D, Matlashewski G. Regulation of human p53 activity and cell localization by alternative splicing. Mol Cell Biol. 2004;24:7987-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pickard A, Wong PP, McCance DJ. Acetylation of Rb by PCAF is required for nuclear localization and keratinocyte differentiation. J Cell Sci. 2010;123:3718-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang Z, Sui Y, Xiong S, et al. Switched alternative splicing of oncogene CoAA during embryonal carcinoma stem cell differentiation. Nucleic Acids Res. 2007;35:1919-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brooks YS, Wang G, Yang Z, et al. Functional pre- mRNA trans-splicing of coactivator CoAA and corepressor RBM4 during stem/progenitor cell differentiation. J Biol Chem. 2009;284:18033-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gabut M, Samavarchi-Tehrani P, Wang X, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132-46 [DOI] [PubMed] [Google Scholar]
- 42. Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci U S A. 2008;105: 5160-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chodosh LA. Expression of BRCA1 and BRCA2 in normal and neoplastic cells. J Mammary Gland Biol Neoplasia. 1998;3:389-402 [DOI] [PubMed] [Google Scholar]
- 44. Khoury MP, Bourdon JC. The isoforms of the p53 protein. Cold Spring Harb Perspect Biol. 2010;2:a000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burch JB, Davis DL. Alternative promoter usage and splicing options result in the differential expression of mRNAs encoding four isoforms of chicken VBP, a member of the PAR subfamily of bZIP transcription factors. Nucleic Acids Res. 1994;22:4733-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anderson CL, Zundel MA, Werner R. Variable promoter usage and alternative splicing in five mouse connexin genes. Genomics. 2005;85:238-44 [DOI] [PubMed] [Google Scholar]
- 47. Pamula J, Krzesniak M, Zientek H, et al. Functional impact of sequence alterations found in BRCA1 promoter/5’UTR region in breast/ovarian cancer families from Upper Silesia, Poland. Hered Cancer Clin Pract. 2006;4:20-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hitchins MP, Rapkins RW, Kwok CT, et al. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5’UTR. Cancer Cell. 2011;20:200-13 [DOI] [PubMed] [Google Scholar]
- 49. Le Calvez-Kelm F, Lesueur F, Damiola F, et al. Rare, evolutionarily unlikely missense substitutions in CHEK2 contribute to breast cancer susceptibility: results from a breast cancer family registry case-control mutation-screening study. Breast Cancer Res. 2011;13:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tavtigian SV, Oefner PJ, Babikyan D, et al. Rare, evolutionarily unlikely missense substitutions in ATM confer increased risk of breast cancer. Am J Hum Genet. 2009;85:427-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Park DJ, Lesueur F, Nguyen-Dumont T, et al. Rare mutations in XRCC2 increase the risk of breast cancer. Am J Hum Genet. 2012;90:734-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2:466-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tombline G, Fishel R. Biochemical characterization of the human RAD51 protein, I: ATP hydrolysis. J Biol Chem. 2002;277:14417-25 [DOI] [PubMed] [Google Scholar]