Abstract
Genetic, biochemical, and structural data support an essential role for the ribosomal RNA in all steps of the translation process. Although in vivo genetic selection techniques have been used to identify mutations in the rRNAs that result in various miscoding phenotypes and resistance to known ribosome-targeted antibiotics, these are limited because the resulting mutant ribosomes must be only marginally disabled if they are able to support growth of the cell. Furthermore, in vivo, it is not possible to control the environment in precise ways that might allow for the isolation of certain types of rRNA variants. To overcome these limitations, we have developed an in vitro selection system for the isolation of functionally competent ribosomal particles from populations containing variant rRNAs. Here, we describe this system and present an example of its application to the selection of antibiotic resistance mutations. From a pool of 4,096 23S rRNA variants, a double mutant (A2058U/A2062G) was isolated after iteration of the selection process. This mutant was highly resistant to clindamycin in in vitro translation reactions and yet was not viable in Escherichia coli. These data establish that this system has the potential to identify mutations in the rRNA not readily accessed by comparable in vivo systems, thus allowing for more exhaustive ribosomal genetic screens.
Protein synthesis is carried out by the ribosome, a large ribonucleoprotein complex composed of three RNA molecules (rRNAs) and >50 proteins. Genetic, biochemical, and structural data point to the RNA component of the ribosome being centrally involved in all steps of protein synthesis, consistent with its ancient origins during an evolutionary period that is thought to have been nucleic acid based, the “RNA world.” In light of this, the isolation of rRNA mutants with biochemically assignable defects, enhanced functions, or altered capabilities is a potentially powerful tool for dissecting the molecular mechanics of the ribosome. Indeed, a variety of genetic screens and selections have been used to identify mutations in the components of the ribosome that result in resistance to different classes of antibiotics and in a variety of miscoding phenotypes (e.g., nonsense and missense suppression and frameshifting) (1-3). Other screens for temperature and cold-sensitive mutations have identified critical elements within the rRNA (4-7). Second-site suppressor analyses have revealed functionally relevant interactions between different regions of the ribosome or with other components of the translation machinery (8-10). Now, with the availability of high-resolution structures for both the large and small subunits of the ribosome (11-14), many of these previously identified genetic interactions can be partly understood.
However, most classic genetic approaches have a number of limitations. The most evident one is the inability to identify mutations that are not viable in vivo and yet may well be informative (i.e., have interesting, tractable phenotypes in vitro). This quandary exists because of the central role of protein synthesis for cell viability. An in vivo strategy to overcome the general problem of dominant lethality was pioneered by de Boer (15) and further developed by Cunningham and colleagues (16). In this system the mutant ribosome pool carries a modified Shine-Dalgarno (SD) region in the 16S rRNA so that variant ribosomes in principle only translate a reporter messenger RNA (mRNA) with complementarity to the altered SD. Although this in vivo-based system has successfully been used to identify functional variants within the rRNA, it remains limited to studying mutations in 16S rRNA because they must be physically coupled to the altered SD. Also, there is no current evidence that dominant lethal rRNA variants are viable in this system where leaky translation of a variety of mRNAs by these mutant ribosomes could easily cause dominant growth defects (15).
In vitro genetic approaches have been previously used for analysis and isolation of known and novel functional RNAs (17). This strategy is most accessible when the functional moiety of interest is RNA or DNA because rare isolated variants can be amplified by reverse transcription (RT) and/or PCR. Iterative selection cycles allow for successive enrichment of the functionally competent variants, and their molecular identities are revealed by sequencing. Recent innovative approaches have extended in vitro nucleic acid selections to the in vitro evolution of proteins by linking the protein product of translation to the mRNA encoding it (18-21). Such in vitro selection approaches allow for fine-tuning of the selective pressures in ways not permitted by the cellular environment and thus for the isolation of mutants that might not emerge from an in vivo-based selection.
In this work, we describe a system that combines strategies from both RNA and protein in vitro evolution and that has the capability to identify functional mutations within any of the rRNAs. The overall scheme involves the assembly of a mutant ribosome pool in vivo, thus avoiding the inefficiency of in vitro reconstitution (22-26). The pool of variant ribosomes isolated from Escherichia coli is then subjected to selective pressure in vitro, where the definition of “functional” can be exquisitely manipulated. As a test of our system, we generated a pool of 4,096 (46) 23S rRNA ribosome variants and isolated ribosomes that are resistant to the antibiotic clindamycin (clin). Parallel isolation of clin-resistant mutants in vivo allowed for direct comparison between these two populations. We show that this in vitro selection system allows for the isolation of ribosomal variants that are competent for protein synthesis under the desired in vitro conditions and yet are lethal when expressed in vivo.
Materials and Methods
Aminoacylation, N-Acetylation, and Biotinylation of tRNAPhe. E. coli Phe-tRNAPhe (Sigma) and N-Ac-Phe-tRNAPhe were prepared as described (27). Biotin-Phe-tRNAPhe was made by incubating Phe-tRNAPhe with 1 mg/ml Sulfo-NHS-LC-Biotin (Pierce) in Na-phosphate buffer (pH 7) overnight at 16°C. Biotinylated aminoacyl-tRNA was precipitated twice with NaOAc (pH 5) and EtOH and stored in 20 mM KOAc (pH 5.5).
Cloning and Transcription of tRNASerGAA. tRNASerGAA was amplified from MRE600 genomic DNA with primers specific for the GGA anticodon isoacceptor, carrying a mismatch that introduces the anticodon mutation, and EcoRI and BamHI restriction sites that allowed for cloning into pNEB193 (New England Biolabs). The construct was verified by sequencing. A BstNI site at the 3′ end allows for run-off transcription by T7 RNA polymerase (28, 29).
Poly(U)-Directed in Vitro Translation. In vitro translation reactions were carried out according to ref. 30 with some modifications. Ribosomes were purified as subunits or tight couples (31). Poly(U) mRNA was from Pharmacia. Elongation was carried out in the presence of tRNASerGAA-free serine ([14C]serine was used for quantitation) and free phenylalanine (1:10 ratio with serine) to prevent inhibition by deacylated tRNAPhe. For quantitative assays, translation was initiated with N-Ac-Phe-tRNAPhe and the reaction stopped with 0.5 M KOH. For streptavidin selection, translation was initiated with biotin-Phe-tRNAPhe, and at given times the reaction was diluted to 0.5 ml in 50 mM Tris·HCl (pH 7.5)/150 mM NH4Cl/50 mM MgCl2 and allowed to bind to 40 μl of prewashed streptavidin matrix (SA-MagnaBeads, Promega) for 30 min at RT in a rotary shaker. Beads were washed four times (0.5 ml of buffer each) and then vigorously shaken for 30 min in 0.3 ml of 0.3 M NaOAc (pH 5)/0.25% SDS/12.5 mM EDTA to cause ribosome disassembly. The rRNA was phenol extracted, EtOH precipitated, and resuspended in water for RT-PCR or analysis on 4% urea-polyacrylamide gels.
Construction and Expression of Mutant rRNA Library. The mutant ribosome library was constructed in plasmid pLK45, carrying the rrnB operon under control of λPL promoter (32) but lacking the mutation A2058G. A XhoI site (1999-2004) and a SpeI site (2077-2081) were introduced in the 23S gene by site-directed mutagenesis (QuikChange, Stratagene), as were two substitutions conferring priming specificity for plasmid-derived rRNA (pLK-XSp). All mutations were phenotypically silent. A cassette spanning positions 1999-2018, flanked by the restriction sites, and containing random sequence at 2057-2062 was generated by filling in two overlapping oligonucleotides with Taq polymerase. The cassette was digested and ligated into pLK-XSp, and the resulting library was transformed into DH10 (Invitrogen) carrying a plasmid with a temperature-sensitive allele of the λ repressor cI. Mutant pools were expressed by growing the cells at 30°C to saturation, diluting the cultures (1/50), and further growing for 2 h at 42°C. This scheme typically yields populations with 20% plasmid-derived ribosomes (33).
Reverse Transcription and PCR. RNA recovered from the SA beads was annealed to a primer (2185-2221, CGTTAGAACATCAAAC) for RT in 50 mM K-Hepes (pH 7.0)/100 mM KCl/4 mM primer by heating to 95°C for 3 min and slow-cooling to 25°C. RT with SuperScript II (Invitrogen) was at 42°C for 20 min, and the cDNA was then diluted 1/20 into a PCR with a 5′ primer specific for the plasmid-derived cDNA (2000-2011, TCGAGACTCGGC) and the 3′ primer used for RT. With this scheme, all rRNA extracted from the streptavidin support is reverse transcribed, but only plasmid-derived cDNA is amplified by PCR, reducing background from chromosome-encoded rRNA. In addition, only plasmid-derived cassettes will contain both restriction sites, and thus ligation of the product into pLK-XSp is specific for the targeted variant population.
Affinity Purification of Tagged Mutant Ribosomes. Specific mutations were introduced by site-directed mutagenesis (QuikChange) into a pLK-derived vector that contains an MS2 aptamer inserted at position 2797 in 23S rRNA (p278-MS2). This allows for affinity purification of plasmid-derived ribosomes by using a GST-MS2 binding protein fusion and a glutathione FPLC column (E. Youngman, J. Brunelle, A. Kochaniak, and R.G., unpublished work). Mutant 70S ribosomes of >95% purity were eluted with glutathione and concentrated over an Amicon 100 concentrator to a 5-10 μM final concentration.
In Vivo Selection for clinR Mutants. Strain SQZ10Δ7 (S. Quan and C. Squires, unpublished data) with all seven chromosomal operons deleted and living on a plasmid carrying a copy of rrnC and the counterselectable marker, sacB (prrnC-sacB, kanr) (34), was transformed with the 2057-2062 randomized library in pLK45. The number of transformants obtained under nonselective conditions covered the complexity of the library ≈15 times (6 × 104 transformants). Transformants were selected first on 350 μg/ml clin and then on 5% sucrose/350 μg/ml clin.
Results
Design of the System. The first task was to establish a system to generate diverse pools of mutant ribosomes. Ribosomes are large complex assemblies of various RNAs and proteins, and although individual components can be produced and assembled in vitro, particles reconstituted from in vitro transcribed rRNA are only marginally functional (24, 26). As an alternative, we chose to assemble mutant ribosomes in E. coli despite the limitation in pool complexity resulting from the need to transform a mutant rDNA library into cells (106 to 109). Additionally, because many interesting mutations will have dominant effects on cell growth, we based our mutagenesis on a system that allows for inducible expression of the variant ribosome population in the background of a wild-type ribosome pool (32). Growth under repressive conditions followed by a short induction minimizes the likelihood of underrepresentation of deleterious mutants in the total population.
The mutant library was constructed by replacing a cassette of wild-type rRNA sequence with a mutagenized cassette, using phenotypically silent restriction sites inserted into a plasmid-borne copy of the rrnB operon. The mutagenized cassette is flanked by other phenotypically silent changes that allow pool specific primers to be used during the RT-PCR amplification step (35), thus preventing amplification of the wild-type ribosomes that survive the selective step. This feature is important because mutant pools are coexpressed with the host chromosome-encoded wild-type ribosomes. In a final step, the amplified cassette is ligated back into the rDNA carrying plasmid, allowing the selectively enriched set of variants to be reproduced as intact ribosomes in vivo for another cycle of selection.
Selective steps can be designed for a wide variety of purposes. Here, we present a strategy for the isolation of functional ribosomes from a diverse, largely inactive population based on the ribosome display technology for the in vitro evolution of proteins (20). In this system, synthesis by the ribosome of a peptide that emerges from the exit channel is used as an affinity tag to isolate the peptide-ribosome-mRNA complex (Fig. 1A). The system was modified to depend on the ribosome's ability to translate a poly uridine [poly(U)] mRNA into a polyphenylalanine [poly(Phe)] peptide that has been initiated with biotinylated phenylalanine. Biotinylated chains elongated beyond a length of ≈35 aa emerge from the ribosome's exit channel and can bind to streptavidin, allowing functional ribosomes to be isolated as peptide-ribosome-mRNA complexes.
Fig. 1.
(A) Selection scheme. A pool of mutant ribosomes is used in an in vitro translation reaction where biotin-Phe-tRNAPhe is used as the “initiator” tRNA. Only variants active under the chosen reaction conditions will extend a polypeptide beyond the length of the ribosome's exit channel. Binding to a streptavidin affinity support allows for the isolation of functional ribosomes as peptide-ribosome-mRNA fusions. (B) Denaturing PAGE analysis of rRNA recovered after in vitro selection. Translation of a poly(U) mRNA by wild-type (MRE600) ribosomes was initiated with either biotin-Phe-tRNAPhe or N-Ac-Phe-tRNAPhe by using as substrates Phe-tRNAPhe only (lanes 1-3) or both Phe-tRNAPhe and Ser-tRNASerGAA in a 1:10 ratio (lanes 4-6). Dependence of the reaction on poly(U) mRNA was tested (lanes 2 and 5). The positions of 16S and 23S rRNAs are shown.
Optimization of the Functional Ribosome Isolation Step. The parameters of the selection system were initially tested by asking how efficiently ribosomes could be isolated on the streptavidin matrix after a typical in vitro translation reaction. Wild-type ribosomes (MRE600) were incubated at 37°C for 20 min with poly(U) as mRNA and N-biotinylated Phe-tRNAPhe to “initiate” translation, and then combined with an elongation mix containing tRNAPhe and an S100 extract that supplies phenylalanyl-tRNA synthetase (PheRS) and elongation factors EF-Tu, EF-Ts, and EF-G. In addition, free phenylalanine, ATP, and GTP, and the components of an energy regenerating system were supplied. After incubation of the translation reaction, peptide-ribosome-mRNA complexes were bound to the streptavidin affinity matrix and extensively rinsed before the RNA from the adhering complexes was isolated by phenol extraction. The efficiency of the system was assessed by analysis of the RNA content of the eluted population on a denaturing polyacrylamide gel. Of the 20% of the ribosomes that are active in poly(Phe) synthesis (data not shown), only 5-10% are recovered on the streptavidin matrix (“pulled-down”) (i.e., a total of 1-2% of the ribosomal input) (Fig. 1B, lane 3). In addition, although ribosome pull-down was dependent on the presence of poly(U) mRNA, it was less dependent on the addition of the biotinylated initiator tRNA (Fig. 1B, lane 1). Both marginal pull-down efficiency and lack of dependence on biotin might be attributable to the hydrophobicity of poly(Phe), which might result in difficulty in passing the peptide through the exit channel and might also decrease the accessibility of the biotin moiety (36).
Problems associated with poly(Phe) were minimized by using a modified tRNASer that carries a mutation in its anticodon (GGA to GAA), allowing it to read poly(U) mRNA rather than its cognate codon (37). tRNASerGAA was generated in vitro by using T7 RNA polymerase and can be efficiently aminoacylated in vitro (data not shown), consistent with the observation that seryl-tRNA synthetase (SerRS) does not depend on the anticodon region for tRNA discrimination (38). In principle, by using this modified tRNA we avoid problems associated with phenylalanine hydrophobicity but still maintain the simplicity and robustness of poly(U)-directed in vitro translation. Indeed, by initiating with biotinylated Phe-tRNAPhe, and supplying a 10-fold excess of tRNASerGAA in addition to both free serine and phenylalanine, the performance of the system is substantially improved. Both the overall efficiency of pull-down and its dependence on the biotin moiety are increased by the incorporation of serine into the polypeptide chains (Fig. 1B, lanes 4-6). About 50% of the 10-20% of ribosomes that are active in this modified in vitro translation system were recovered from the streptavidin matrix (i.e., a total of 5-10% of the ribosomal input).
Selective Enrichment of Clindamycin-Resistant Ribosomes. We next asked whether the system could be used to enrich a diverse population of ribosomes in those that are functional under a given set of reaction conditions. The isolation of ribosomal variants resistant to the lincosamide antibiotic clindamycin was chosen as the initial test. We chose this antibiotic because there are known resistance mutations (39), because there is structural data indicating which regions of the rRNA are involved in direct interactions with this compound (40) and, importantly, because in our in vitro system clin effectively inhibits poly(U) translation. Lincosamide antibiotics bind to the central loop region of domain V in 23S RNA within the peptidyl transferase center and so we chose to randomize six contiguous nucleotides (2057-2062) in this region (Fig. 2A).
Fig. 2.
(A) 3D rendition of the binding pocket for clin in the central loop of domain V of 23S rRNA (40). Clindamycin is shown in pink, the six nucleotides randomized for the selection are shown in blue (numbered 2057-2062), and the two positions where mutations were selected for in vitro are shown in darker blue. The figure was made in ribbons (49). The 2D structure of clin is shown at bottom right. (B) Cassette mutagenesis strategy. Details of the sequence changes made in pLK-45 to construct a DNA cassette to be removed by restriction digestion and replaced with one containing randomized sequence.
Randomization was accomplished by cassette mutagenesis in plasmid pLK-XSp containing a copy of the rrnB operon under the control of λ PL promoter and a few modifications with respect to the original pLK45 (41): two phenotypically silent, unique restriction sites flanking the region of interest created an 80-bp cassette that can be replaced with randomized sequence, and two additional single nucleotide substitutions conferred priming specificity and thus selective amplification of plasmid-derived rRNA (Fig. 2B). The randomized cassette was generated by filling in two overlapping oligonucleotides covering the region followed by digestion with the appropriate restriction enzymes. Ligation of this cassette into pLK-XSp resulted in a library containing 46 (4,096) 23S rRNA variants that was transformed into E. coli (DH10) carrying a plasmid with a temperature-sensitive cI repressor allele, allowing for inducible expression of the variant rRNAs. Growth under nonpermissive conditions (30°C) followed by a short induction period (42°C) results in reasonably high expression levels (20-30%) from the plasmid-borne 23S rRNA gene, even in cases where the introduced mutations cause lethality or slow growth (42). The nucleotide degeneracy in the region of interest of the starting rRNA variant pool (pool 0) before selection was confirmed by sequencing (data not shown).
Ribosomes from pool 0 were passaged through the in vitro selection procedure in the presence of a high concentration of clin (500 μM), at which the in vitro translation of wild-type ribosomes is reduced to background levels. After selection, functional variants from pool 0 were isolated on the streptavidin matrix as described above. The rRNA contained in the selected peptide-ribosome-mRNA complexes was isolated by phenol extraction and the mutagenized cassettes were amplified by RT-PCR and cloned back into pLK-XSp. The resulting library was transformed into E. coli to allow for the in vivo synthesis of the functionally enriched ribosome pool (pool 1). This process was iterated six successive times. After each round of selection, the level of clin-resistant, in vitro translation activity of the pool was examined (Fig. 3). After round 6, there was substantial enrichment in clin-resistant activity, and the mutagenized cassettes from 31 independent clones were sequenced (Table 1). The first observation is the absence of wild-type variants in the selected population, confirming that the selection effectively eliminates nonfunctional ribosomes. The second striking observation is that >50% of the selected variants carry the double substitution A2058U/A2062G, whereas a small fraction carry single substitutions at A2058 or A2059, positions where mutations had been found by using classic approaches (39).
Fig. 3.
Clindamycin-resistant translation of successive ribosome pools. Enriched ribosome pools were assayed for translation activity by using a poly(U) mRNA and both Phe-tRNAPhe and [14C]Ser-tRNASerGAA as substrates in the presence of 500 μM clin, by following trichloroacetic acid-precipitable counts.
Table 1. In vitro selection.
| Sequence | Times represented |
|---|---|
| GUAAGG | 17 |
| GUUAGG | 2 |
| GUAAUG | 1 |
| GUGAGC | 2 |
| GUAAGA | 1 |
| GUCAGC | 1 |
| GAAACG | 1 |
| GAGAGA | 1 |
| GAGAGC | 2 |
| GAGAUC | 1 |
| GAGAUG | 1 |
| GAUACA | 1 |
Number of times each different sequence appears in 31 independent clones isolated from in vitro selection for clin-resistant ribosomes. Underlined letters indicate deviations from wild type (GAAAGA).
Characterization of Isolated rRNA Mutants. To dissect the relative contributions from each of the two single substitutions (A2058U and A2062G) to the resistance of the predominant selected variant, we isolated pure populations of single- and double-mutant ribosomes for in vitro analysis. The mutations were reconstructed in a 23S RNA backbone, lacking both restriction sites and the silent priming site but carrying an MS2 aptamer sequence that is exposed on the back of the 50S subunit, allowing for affinity purification of mutant ribosomes by binding to a GST-MS2 coat protein fusion immobilized on a glutathione FPLC column (E. Youngman, J. Brunelle, A. Kochaniak, and R.G., unpublished work). In addition, we constructed the A2058G mutant that had previously been isolated in vivo (39), and the A2058G/A2062G double mutant that we did not find in our isolated pool but would address whether the A2062G substitution requires a specific mutation at A2058.
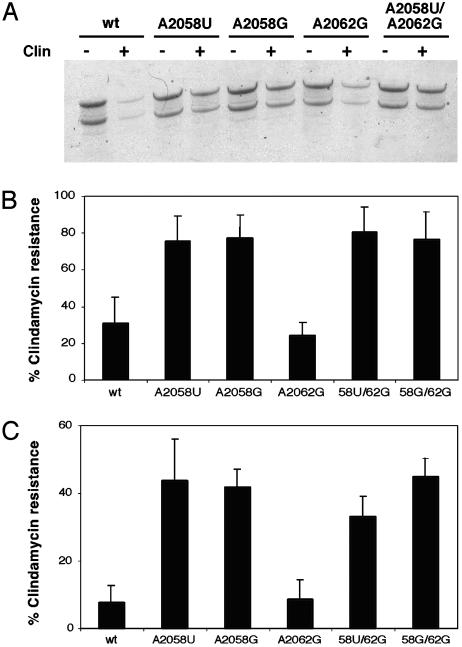
The clin-resistant translation activity of each of the purified mutant ribosomes was evaluated in three independent assays. First, the relative efficiency of streptavidin pull-down after translation in the absence or presence of clin was monitored (Fig. 4A). This experiment mimics the actual selection and directly tests whether these variants display the phenotype for which they were selected. These data indicate that the double-mutant A2058U/A2062G ribosomes and both single-mutant variants at position 2058 (U and G) are more efficiently pulled-down than either the wild-type or A2062G mutant ribosomes in the presence of clin. In the absence of clin, all of the ribosomes perform equivalently.
Fig. 4.
Analysis of the clin-resistant activity of pure populations of mutant ribosomes. (A) Denaturing PAGE analysis of the streptavidin isolated rRNA after in vitro translation in the absence or presence of clin (500 μM) with the different mutant ribosome populations. (B) Poly(U) translation followed by incorporation of [14C]serine into trichloroacetic acid-precipitable chains. Percent resistance is obtained from the ratio of activities in the presence and absence of clin. (C) Clindamycin-resistant translation of an mRNA containing an 88-aa ORF. Translation was initiated with f[35S]Met-tRNAfMet, and translation products were resolved on 4-12% denaturing PAGE. Percent resistance is obtained as in B.
Because of the qualitative nature of this assay, we also measured poly(U) translation in the absence or presence of clin by following incorporation of [14C]serine. Here, overall translation efficiency is monitored independent of whether the peptide chain succeeds in escaping the ribosomal exit channel. Again, this experiment shows that both double mutants are at least as resistant to clin as the ribosomes carrying a single substitution at A2058 (either U or G) (Fig. 4B). Finally, we assessed the activity of the mutants in an in vitro translation assay by using a natural mRNA encoding an 88-aa protein. As in the previous assays, wild-type and A2062G mutant ribosomes are sensitive to clin, whereas the other single- and double-mutant ribosomes are resistant to the antibiotic (Fig. 4C). Although we reasoned that substantial differences in clin-resistant activity might be detectable in these quantitative assays, we have been unable to identify conditions where these are evident and thus simply explain why the A2058U/A2062G double mutant overtook the in vitro selection.
In Vivo Isolation of Clindamycin-Resistant rRNA Mutants. To allow fair comparison between variants isolated in in vitro and in vivo systems, we asked which mutants in the same initial pool of 4096 variants emerged as clin resistant in an in vivo selection. To do this, the same rRNA library with six random positions was transformed into E. coli strain SQZ10Δ7 that has all seven rrn operons deleted, and a plasmid, prrnC-sacB, carrying the rrnC operon under control of its own strong promoter (34, 43). This plasmid also carries the counterselectable marker sacB to select for loss of the plasmid carrying the wild-type rrn operon. This system thus allows for the isolation of rRNA mutants that both confer resistance to clin and are sufficient to support growth in the absence of wild-type ribosomes. By growing the transformed SQZ10Δ7 strain on LB agar plates with 350 μg/ml clin and then replica plating to plates with both clin (350 μg/ml) and 5% sucrose, resistant colonies were isolated at a frequency of ≈1/2,000. Sequences from 31 individual clones are shown in Table 2. Although a variety of mutants were isolated multiple times, the double mutant that dominated the in vitro selection (A2058U/A2062G) was not isolated by the in vivo selection procedure. The in vivo isolates are dominated by the single mutants at positions A2058 and A2059.
Table 2. In vivo selection.
| Sequence | Times represented |
|---|---|
| GUAAGA | 8 |
| GGAAGA | 5 |
| GAUAGA | 8 |
| GAGAGA | 2 |
| GUCAGA | 4 |
| UGAAGA | 2 |
| AGAAGA | 1 |
| CGAAGA | 1 |
Number of times each different sequence appears in 31 independent clones isolated from in vivo selection for clin-resistant ribosomes. Underlined letters indicate deviations from wild type (GAAAGA).
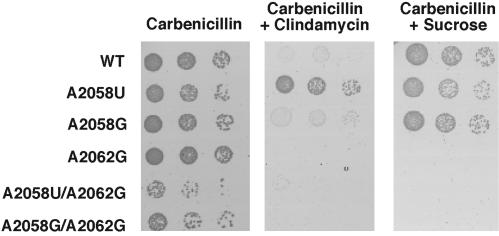
We next directly asked why the double mutant (A2058U/A2062G) selected by the in vitro approach was not isolated in the parallel in vivo selection. When this double mutant was transformed into SQZ10Δ7 and selected on clin, or on sucrose in the absence of clin, we observed strong growth defects resulting from these mutations (Fig. 5). Indeed, the substitution of only A2062 with either G or C resulted in substantial growth defects (Fig. 5 and data not shown). In contrast, single substitutions at position 2058 display normal growth and some resistance to clin. Thus, despite the robust behavior of ribosomes carrying mutations at position 2062 in our in vitro translation assays, these ribosomes are not able to support normal cellular growth.
Fig. 5.
Phenotypes of the different rRNA mutant ribosomes in strain SQZΔ7. Plasmid pLK-XSp (carbr) carrying the rrnB operon bearing the different 23S mutations was transformed into strain SQZΔ7 carrying the plasmid prrnC-sacB (kanr). (Left) Growth on carb (100 μg/ml). (Center) Growth on carb (100 μg/ml) and clin (350 μg/ml). (Right) Growth on carb (100 μg/ml) and sucrose (5%) to select for loss of the prrnC-sacB plasmid.
Discussion
We have developed a system for in vitro selection of functional ribosomal variants from diverse populations of largely nonfunctional particles. As an initial test of our system, we isolated ribosomal variants resistant to the lincosamide antibiotic clindamycin. The rRNA library used in the selection covered six positions in the antibiotic binding pocket (2057-2062 in E. coli). The winning variant isolated after six rounds of selection carries two mutations, A2058U and A2062G, and exhibits high-level resistance to clin in each of three independent translation assays. The A-to-U transversion at position 2058 has previously been studied as it confers resistance to macrolides and lincosamides both in vivo and in vitro (44). However, in our isolated population, the substitution at 2058 is almost always accompanied by the second substitution, an A-to-G transition at position 2062.
A question that immediately falls out of these results is why the double-mutant (A2058U/A2062G) but not the single-mutant (A2058U) ribosomes are isolated. Three different biochemical assays argue that there are at best modest differences in activity between the double- and single-mutant ribosome variants; and, although a small difference between the double- and single-mutant ribosomes in the poly(U) translation assay might be amplified by the iterative nature of the selection procedure, it still seems surprising to us that the single mutants are found at such low frequency in the final selected population. Although nucleotide A2062 has not previously been implicated in interactions with clin by genetic approaches, recent structural and biochemical data suggest that this nucleotide closely approaches the antibiotic (40, 45). Interestingly, A2062 has been shown to directly interact with the 16-membered ring macrolides by forming a covalent bond to their ethylaldehyde moiety (46). Other studies found that an A2062C substitution in Streptococcus pneumoniae confers resistance to 16-membered macrolides and streptogramins but not 14- or 15-membered macrolides, ketolides, or lincosamides (47). Indeed, macrolides, lincosamides, and streptogramins have overlapping binding sites (MLS family of antibiotics) and thus it is not surprising that a mutation affecting the action of one class of molecules might have effects on the other class. We speculate that there may be more subtle differences in activity between the single- and double-mutant ribosomes that we cannot detect in our in vitro assays but that might explain the statistical bias in the isolated population. Other possible explanations for this bias include selective amplification, a skewed starting oligonucleotide population, or selection during the induction period in E. coli, although sequencing of pool 0 variants and robust expression of clear dominant lethal mutants (42) argue against these possibilities.
The in vivo selection for clin resistance that we performed allowed us to make direct comparisons between the variants isolated by the two different approaches. This selection yielded resistant variants at positions G2057-A2058-A2059 in various combinations, but no variation was observed at positions A2060-G2061-A2062. Consistent with these data, we found that the A2058U/A2062G variant that dominated the in vitro selection did not support life in E. coli and thus could not have been isolated by the in vivo selection. Thus, we have demonstrated that it will be possible with the in vitro selection approach to isolate ribosomal variants not accessible by analogous in vivo screens and selections. The constraints of life are clearly more stringent than the requirements for in vitro protein synthesis.
Although in vivo-based genetic approaches have been applied to the study of the ribosome with considerable success, there are a variety of selective pressures that one might like to impose on the protein synthesis machinery that would be precluded by the need to function in a cellular milieu. We can imagine a number of different types of selections that would fit these criteria. For example, a selection that involves unnatural substrates (that cannot be readily generated in vivo) could lead to the identification of ribosome variants capable of translating DNA or other unnatural polymers or capable of synthesizing altered polypeptide backbones. A selection where the temperature is substantially altered could allow for the identification of potentially mobile regions of the rRNA (4, 5, 48). Finally, a selection where specific translation components are omitted could shed light on the mechanism of the processes these factors promote, such as decoding and translocation. We anticipate that this in vitro selection system will open new paths for dissecting the molecular mechanics of ribosome function.
Acknowledgments
We thank S. Quan and C. Squires for strain SQZΔ7, C. Merryman for reading the manuscript, and J. Williamson for discussions leading to these experiments. This work was supported by the Packard Foundation, the Howard Hughes Medical Institute, and the National Institutes of Health.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: clin, clindamycin; RT, reverse transcription.
References
- 1.Parker, J. (1989) Microbiol. Rev. 53, 273-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Connor, M. & Dahlberg, A. E. (1993) Proc. Natl. Acad. Sci. USA 90, 9214-9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigmund, C. D. & Morgan, E. A. (1982) Proc. Natl. Acad. Sci. USA 79, 5602-5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dammel, C. S. & Noller, H. F. (1993) Genes Dev. 7, 660-670. [DOI] [PubMed] [Google Scholar]
- 5.Triman, K., Becker, E., Dammel, C., Katz, J., Mori, H., Douthwaite, S., Yapijakis, C., Yoast, S. & Noller, H. F. (1989) J. Mol. Biol. 209, 645-653. [DOI] [PubMed] [Google Scholar]
- 6.Sweeney, R., Yao, C. H. & Yao, M. C. (1991) Genetics 127, 327-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lodmell, J. S., Gutell, R. R. & Dahlberg, A. E. (1995) Proc. Natl. Acad. Sci. USA 92, 10555-10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweeney, R. & Yao, M. C. (1998) Genetics 149, 937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lodmell, J. S. & Dahlberg, A. E. (1997) Science 277, 1262-1267. [DOI] [PubMed] [Google Scholar]
- 10.Dammel, C. S. & Noller, H. F. (1995) Genes Dev. 9, 626-637. [DOI] [PubMed] [Google Scholar]
- 11.Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., Bashan, A., Bartels, H., Agmon, I., Franceschi, F. & Yonath, A. (2000) Cell 102, 615-623. [DOI] [PubMed] [Google Scholar]
- 12.Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F. & Yonath, A. (2001) Cell 107, 679-688. [DOI] [PubMed] [Google Scholar]
- 13.Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. (2000) Science 289, 905-920. [DOI] [PubMed] [Google Scholar]
- 14.Wimberly, B. T., Brodersen, D. E., Clemons, W. M., Jr., Morgan-Warren, R. J., Carter, A. P., Vonrhein, C., Hartsch, T. & Ramakrishnan, V. (2000) Nature 407, 327-339. [DOI] [PubMed] [Google Scholar]
- 15.Hui, A. & de Boer, H. A. (1987) Proc. Natl. Acad. Sci. USA 84, 4762-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, K., Varma, S., SantaLucia, J., Jr., & Cunningham, P. R. (1997) J. Mol. Biol. 269, 732-743. [DOI] [PubMed] [Google Scholar]
- 17.Wilson, D. S. & Szostak, J. W. (1999) Annu. Rev. Biochem. 68, 611-647. [DOI] [PubMed] [Google Scholar]
- 18.Roberts, R. W. & Szostak, J. W. (1997) Proc. Natl. Acad. Sci. USA 94, 12297-12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merryman, C., Weinstein, E., Wnuk, S. F. & Bartel, D. P. (2002) Chem. Biol. 9, 741-746. [DOI] [PubMed] [Google Scholar]
- 20.Mattheakis, L. C., Bhatt, R. R. & Dower, W. J. (1994) Proc. Natl. Acad. Sci. USA 91, 9022-9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nemoto, N., Miyamoto-Sato, E., Husimi, Y. & Yanagawa, H. (1997) FEBS Lett. 414, 405-408. [DOI] [PubMed] [Google Scholar]
- 22.Krzyzosiak, W., Denman, R., Nurse, K., Hellmann, W., Boublik, M., Gehrke, C. W., Agris, P. F. & Ofengand, J. (1987) Biochemistry 26, 2353-2364. [DOI] [PubMed] [Google Scholar]
- 23.Nierhaus, K. H. & Dohme, F. (1974) Proc. Natl. Acad. Sci. USA 71, 4713-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Semrad, K. & Green, R. (2002) RNA 8, 401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green, R. & Noller, H. F. (1999) Biochemistry 38, 1772-1779. [DOI] [PubMed] [Google Scholar]
- 26.Green, R. & Noller, H. F. (1996) RNA 2, 1011-1021. [PMC free article] [PubMed] [Google Scholar]
- 27.Moazed, D. & Noller, H. F. (1989) Cell 57, 585-597. [DOI] [PubMed] [Google Scholar]
- 28.Sampson, J. R. & Uhlenbeck, O. C. (1988) Proc. Natl. Acad. Sci. USA 85, 1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milligan, J. F., Groebe, D. R., Witherell, G. W. & Uhlenbeck, O. C. (1987) Nucleic Acids Res. 15, 8783-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rheinberger, H. J., Geigenmuller, U., Wedde, M. & Nierhaus, K. H. (1988) Methods Enzymol. 164, 658-670. [DOI] [PubMed] [Google Scholar]
- 31.Moazed, D. & Noller, H. F. (1986) Cell 47, 985-994. [DOI] [PubMed] [Google Scholar]
- 32.Gourse, R. L., Takebe, Y., Sharrock, R. A. & Nomura, M. (1985) Proc. Natl. Acad. Sci. USA 82, 1069-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sigmund, C. D., Ettayebi, M., Borden, A. & Morgan, E. A. (1988) Methods Enzymol. 164, 673-690. [DOI] [PubMed] [Google Scholar]
- 34.Zaporojets, D., French, S. & Squires, C. (2003) J. Bacteriol. 185, 6921-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powers, T. & Noller, H. F. (1993) Gene 123, 75-80. [DOI] [PubMed] [Google Scholar]
- 36.Odom, O. W., Picking, W. D., Tsalkova, T. & Hardesty, B. (1991) Eur. J. Biochem. 198, 713-722. [DOI] [PubMed] [Google Scholar]
- 37.Picking, W., Picking, W. D. & Hardesty, B. (1991) Biochimie 73, 1101-1107. [DOI] [PubMed] [Google Scholar]
- 38.Normanly, J. & Abelson, J. (1989) Annu. Rev. Biochem. 58, 1029-1049. [DOI] [PubMed] [Google Scholar]
- 39.Vester, B. & Douthwaite, S. (2001) Antimicrob. Agents Chemother. 45, 1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlunzen, F., Zarivach, R., Harms, J., Bashan, A., Tocilj, A., Albrecht, R., Yonath, A. & Franceschi, F. (2001) Nature 413, 814-821. [DOI] [PubMed] [Google Scholar]
- 41.Douthwaite, S., Powers, T., Lee, J. Y. & Noller, H. F. (1989) J. Mol. Biol. 209, 655-665. [DOI] [PubMed] [Google Scholar]
- 42.Powers, T. & Noller, H. F. (1990) Proc. Natl. Acad. Sci. USA 87, 1042-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asai, T., Zaporojets, D., Squires, C. & Squires, C. L. (1999) Proc. Natl. Acad. Sci. USA 96, 1971-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vester, B. & Garrett, R. A. (1987) Biochimie 69, 891-900. [DOI] [PubMed] [Google Scholar]
- 45.Tenson, T., Lovmar, M. & Ehrenberg, M. (2003) J. Mol. Biol. 330, 1005-1014. [DOI] [PubMed] [Google Scholar]
- 46.Hansen, J. L., Ippolito, J. A., Ban, N., Nissen, P., Moore, P. B. & Steitz, T. A. (2002) Mol. Cell 10, 117-128. [DOI] [PubMed] [Google Scholar]
- 47.Depardieu, F. & Courvalin, P. (2001) Antimicrob. Agents Chemother. 45, 319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staley, J. P. & Guthrie, C. (1999) Mol. Cell 3, 55-64. [DOI] [PubMed] [Google Scholar]
- 49.Carson, M. (1997) Methods Enzymol 277, 493-505. [PubMed] [Google Scholar]