SUMMARY
We describe the use of a unique DNA-modification-dependent restriction endonuclease AbaSI coupled with sequencing (Aba-seq) to map high-resolution hydroxymethylome of mouse E14 embryonic stem cells. The specificity of AbaSI enables sensitive detection of 5-hydroxymethylcytosine (5hmC) at low-occupancy regions. Bioinformatic analysis suggests 5hmCs in genic regions closely follow the 5mC distribution. 5hmC is generally depleted in CpG islands and only enriched in a small set of repetitive elements. A regularly spaced and oscillating 5hmC pattern was observed at the binding sites of CTCF. 5hmC is enriched at the poised enhancers with the monomethylated histone H3 lysine 4 (H3K4me1) marks, but not at the active enhancers with the acetylated histone H3 lysine 27 (H3K27Ac) marks. Non-CG hydroxymethylation appears to be prevalent in the mitochondrial genome. We propose that some amounts of transiently stable 5hmCs may indicate a poised epigenetic state or demethylation intermediate, whereas others may suggest a locally accessible chromosomal environment for the TET enzymatic apparatus.
INTRODUCTION
The dynamic epigenomic landscape of the mammalian cell, including chromatin states and DNA modifications, is set forth by past cellular events, and it affects the developmental trajectory of the cell by regulating the genome-wide expression. DNA modifications such as 5-methylcytosine (5mC) are laid onto the genome by various DNA methyltransferases (e.g., DNMT1, DNMT3A, and DNMT3B) and undergo cycles of erasure and re-establishment at both genomic and regional levels. The unique methylomes are largely maintained in differentiated cell types, making them a key feature in understanding the differentiation potential of the cell. Recently, it was shown that 5-hydroxymethylcytosine (5hmC) is an oxidation product of the TET1/2/3 and is enriched in neuronal tissues as well as in embryonic stem cells (ESCs) (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). Successive oxidation of 5hmC by the TET enzymes produces 5-formylcytosine (5fC) and 5-carboxycytosine (5caC), which are found at a lower abundance in the genome (He et al., 2011; Ito et al., 2011). It was then realized that conventional cytosine methylation profiling methods, including bisulfite sequencing, cannot distinguish between 5mC and 5hmC (Huang et al., 2010; Jin et al., 2010). Hence, dissection of the hydroxymethylome from the methylome has become a priority in understanding its role in the genome regulation circuitry.
A number of methods have been developed to investigate the genome-wide distribution of 5hmC and its dynamics. For example, based on the differential sensitivity of MspI and HpaII to modified cytosines, 5hmC/5mC/C levels can be quantitatively estimated at any CCGG sites (Kinney et al., 2011). Several genome-wide studies have used 5hmC-specific antibodies (hMeDIP) or chemical affinity tags to enrich 5hmC-containing DNA for sequencing (Song et al., 2011; Ficz et al., 2011; Pastor et al., 2011; Williams et al., 2011; Wu et al., 2011). Although limited by resolution, these studies yielded many insightful observations. Recently, two methods of base-resolution hydroxymethylome mapping were reported (Booth et al., 2012; Yu et al., 2012). Both methods utilized the differential readout of various modified cytosines after bisulfite conversion and sequencing. 5mC and 5hmC are read as cytosine after bisulfite sequencing, whereas further oxidized products of 5hmCs (5fC or 5caC) are read as thymine. One method (oxoBS-seq) achieved the selective chemical oxidation of 5hmC to 5fC by using potassium perruthenate (KRuO4) (Booth et al., 2012). The other method (Tetassisted bisulfite sequencing, TAB-seq) first blocked the existing 5hmC by glucosylation and then converted all genomic 5mC to 5caC by utilizing the in vitro enzymatic oxidation of TET1 (Yu et al., 2012). Both methods used traditional bisulfite conversion and sequencing to decipher methylome and hydroxymethylome. However, several technical hurdles may still persist. For example, DNA complexity and integrity are further compromised after the treatment which may hamper the amplification steps and the subsequent read mapping. In addition, sequencing depth for each cytosine must be considerably high to detect low-level 5hmCs (Yu et al., 2012).
Alternative methods, ideally with a facile application format, are needed to complement the existing approaches. In previous work we characterized a family of 5hmC-dependent restriction endonucleases (Wang et al., 2011). One member in the family, AbaSI, recognizes 5-glucosylatedmethylcytosine (5gmC) with high specificity when compared to 5mC and C, and cleaves at a narrow range of distances away from the recognized modified cytosine. By mapping the cleaved ends, the exact 5hmC locations could be determined (Wang et al., 2011). Here we report the genome-wide high-resolution hydroxymethylome of the mouse E14 ESCs using AbaSI-coupled sequencing (Aba-seq).
RESULTS
Aba-seq and 5hmC Site Validation
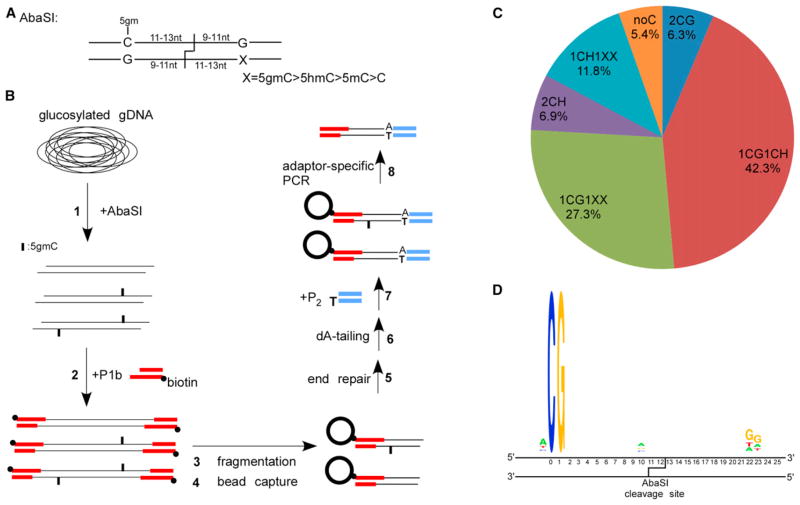
DNA modification-dependent restriction endonuclease AbaSI recognizes 5gmC with high specificity and generates double-stranded breaks at the 3′ side of the binding sites (Wang et al., 2011). AbaSI does not digest 5mC or C under the conditions used (Figure S1A). The distances between the cleavage sites and the modified cytosine are fixed within a narrow range, with the majority 11–13 nucleotides away in the top strand and 9–10 nucleotides away in the bottom strand (Wang et al., 2011). The recognition and cleavage sites of AbaSI are shown in Figure 1A. AbaSI prefers sites with two cytosines positioned symmetrically around the cleavage site: one is 5gmC and the other can be modified or not (Figure 1A). AbaSI cleaves with a lower efficiency when only one 5gmC is present (Wang et al., 2011). Conversion of genomic 5hmC to 5gmC was completed by using the T4 β-glucosyltransferase (Terragni et al., 2012) (Figure 1B). The AbaSI-digested DNA was first ligated to a biotinylated DNA adaptor P1b containing 2-base randomized 3′-overhang (Figure 1B). After fragmentation and end polishing, bead-captured DNA was ligated to the second adaptor P2 (Figure 1B). Sequencing was initiated from the P1b end, and the AbaSI cleavage sites were revealed after mapping the reads to the reference genome. We determined that as low as 50 ng of E14 genomic DNA is sufficient for successful library construction (Figure S1B).
Figure 1. Overview of Aba-seq and Validation of the Identified 5hmC Sites.
(A) Recognition and cleavage sites of AbaSI.
(B) Schematic overview of the Aba-seq method. 1: glucosylated genomic DNA was digested with AbaSI. 2: digested DNA was ligated to a biotinylated adaptor P1b with 2-base 3′-overhang. 3: DNA fragmentation by sonication. 4: capture of the biotinylated DNA onto avidin-coated beads. 5 and 6: on-bead DNA end-repair and dA-tailing. 7: on-bead ligation with adaptor P2 containing one blunt end and one unligatable end. 8: PCR using primers specific to the P1b and P2.
(C) Distribution of different categories of identified AbaSI cleavage sites.
(D) Representative sequence logos of the largest categories 1CG1CH.
Putative 5hmC sites can be deduced by checking the presence of cytosine at expected distances (N13/N11,N12/N10,N11/N9) from either side of the mapped cleavage sites (Figure 1A). Based on the cytosine presence around the cleavage sites, we categorized them into groups (Figure 1C), e.g., 1CG1CH signifies one CG and one CH (H = A/C/T) at either side of the cleavage site respectively; similarly, 1CG1XX signifies one CG on one side and no cytosine on the other side. In groups with cytosines on both sides, we prioritized the 5hmC assignment in the CG context over the CH context. For example, 5hmC is assigned to the cytosine in the CG context in the 1CG1CH group. For over 82% of the cleavage sites, e.g., 1CG1CH, 1CG1XX, 1CH1XX, 5hmC can be assigned to a single cytosine with high confidence. There is assignment ambiguity to the exact cytosine base in categories such as 2CGs or 2CHs and accounts for 13% of all identified cleavage sites. These sites were included in our analysis as one 5hmC site. A representative sequence logo is shown in Figure 1D for the 1CG1CH sites, demonstrating little bias exists besides the recognized cytosines. There are only 5% of the mapped cleavage sites without suitable AbaSI recognition cytosines, in contrast to an expected 25% by chance (Figure S1C). With two technical replicates starting from AbaSI digestion, we obtained 242.0 (library R1) and 228.0 (library R2) million reads, of which 131.8 (54%) and 114.3 (50%) million reads were unambiguously mapped to the reference genome. Within mapped reads, 124.7 and 107.8 million 5hmC-induced reads were called, corresponding to 17.6 and 15.2 million 5hmC sites respectively (see Methods). There is a high degree of reproducibility between the two replicates with 12,229,723 unique 5hmC sites in common, which was used in the following analyses (Figure S1D). The mean and median of the combined sequencing coverage for these common 5hmC sites are 12.3X and 9X respectively (Figure S1E). In addition, within each library, sequencing coverage for the common 5hmC sites is significantly higher than that for the library-specific 5hmC sites (Figure S1E).
We further validated Aba-seq results using MspI and T4 β-glu-cosyltransferase (Kinney et al., 2011). We quantified 5hmC percentage at a number of CCGG sites with different read coverage in the 1CG1CH and 1CG1XX categories. A strong positive correlation between the read coverage and 5hmC percentage was observed (r = 0.73, Figure S1F). Background levels of 5hmC at random, unreported CCGG sites are below the detection limit (0.35% ± 0.56%). Levels of 5hmC for sites with read coverage equal to 1 and 2 are significantly higher than the background (mean = 2.2%, p value < 0.02, one-sided Student’s t test), confirming the presence of 5hmC. The lowest detectable 5hmC percentage in CCGG sites was around 2%, giving an estimate on the sensitivity of Aba-seq. To validate 5hmC in the CH sites, we quantified AbaSI cleavage percentage at these sites by individual quantitative PCR (qPCR), as shown in Table S3. Similarly, all reported CH sites has significantly higher 5hmC readings than randomly chosen CH sites (p value < 1 ×10−9). Overall, our validation confirms Aba-seq resulted a high-quality hydroxymethylome of the E14 cells. In addition, regional 5hmC levels, as reflected by the cumulative read numbers in a genomic region, show strong correlation with the normalized 5hmC site density (Figure S1G). Therefore, we will only show results by using the 5hmC site density in subsequent analysis.
Comparison with the earlier hMeDIP data (Williams et al., 2011) suggests that 95% of the enriched peaks contain at least one Aba-seq 5hmC site. Importantly, the local 5hmC density of those sites outside the hMeDIP peaks is much lower than that of those inside (p value < 1 × 10−6, Figure S1H), which would suggest that the 5hmC antibody used in hMeDIP may favor the densely populated 5hmC regions. A recent study (TAB-seq) also reported base-resolution hydroxymethylome in mouse E14 cells with 2,057,636 5hmCs (Yu et al., 2012), about one sixth of our results in size. A comparison between Aba-seq and TAB-seq resulted in 1,246,351 common 5hmC sites (60.5% of TAB-seq data). Among our validated 5hmCG sites, 12 (~15%) were reported in the TAB-seq data set. A notable difference between the two methods is that TAB-seq may preferentially detect sites with high 5hmC occupancy (Yu et al., 2012). However, comparison in read numbers between Aba-seq-specific sites and the Aba/TAB-seq common sites yielded no significant difference. In addition, we compared our data and TAB-seq data with earlier base-resolution data from the single-molecule sequencing method (Song et al., 2011). Of the 744 hmCG sites reported by that method (Song et al., 2012), 522 (~70.2%) overlap with Aba-seq and 123 (~16.5%) overlap with TAB-seq. Further biological replicates and technical investigations are required to interpret the discrepancies between the different methods.
Genomic View of the Hydroxymethylome in Mouse ESCs
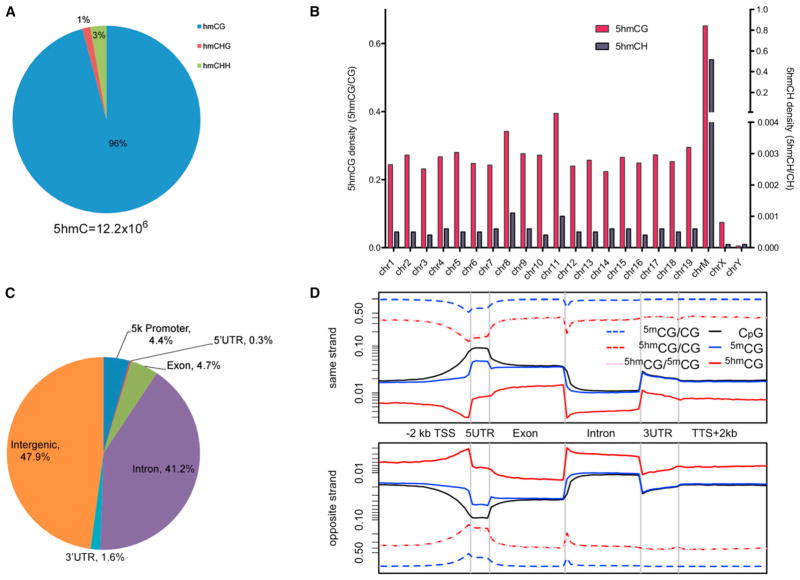
In ESCs, a significant amount (20%–30%) of 5mC exists in CH context, whereas this number reduces to negligible levels in somatic cells (Lister et al., 2009, 2011; Stadler et al., 2011). Based on our data, we estimate that 95.9% of all 5hmCs exists in CG context, whereas 4.1% exist in CH context (1.4% CHG and 2.7% CHH) (Figure 2A). These numbers differ from those reported by TAB-seq, where close to 99% of 5hmCG were observed (Yu et al., 2012). The average read number of the CH group (10.0) is significantly smaller than that of the CG group (12.4, p value < 1 × 10−6, two-sided Student’s t test), suggesting lower 5hmC occupancy at the CH sites. Using 5hmCG sites with significant read coverage by a bionomial distribution (p = 0.5), we estimate that the percentage of the symmetrically hydroxymethylated sites within the 5hmCG sites exceeds 90% (Figure S2A), arguing against strand bias in hydroxylation of the fully methylated CG sites.
Figure 2. Genomic View of the Mouse ESC Hydroxymethylome.
(A) Distribution of 5hmCG and 5hmCH sites in ESCs.
(B) 5hmCG (left y axis) and 5hmCH (right y axis) site densities by each chromosome.
(C) Distribution of 5hmC in different genomic elements.
(D) Strand-specific metaplot of normalized 5hmC and 5mC (Stadler et al., 2011) distribution near coding sequences.
See also Figure S2.
Examination of the 5hmC site density in chromosomes reveals comparable levels across the autosomes, with a slightly increase in chromosome 11 (Figure 2B). Sex chromosomes are depleted in 5hmC, with little 5hmCs in the Y chromosome (Figure 2B). Depletion of 5mC and 5hmC (Szulwach et al., 2011) in X chromosomes has been observed in neuronal tissues and stem cells. Together these results suggest a different epigenetic regulation mechanism between the autosomes and sex chromosomes. In addition, we observed the highest 5hmC site density in the mitochondria genome in both CG and CH contexts (Figure 2B). Considering that mitochondria genomes are present in multiple copies in the cell, it remains inconclusive whether they are enriched in 5hmC. Nevertheless, inside mitochondria DNA, normalized CH 5hmC site density is nearly as high as the normalized CG 5hmC site density, whereas it is less than 1% of the latter for all other chromosomes (Figure 2B). A set of key DNA epigenetic components, including DNMT1, TET enzymes, and their products 5mC and 5hmC, were reported in mitochondria (Shock et al., 2011). Our results further suggest that the prevalent pattern of hydroxymethylation, possibly methylation, in mitochondria genome might exist in an unbiased manner toward CG or CH. Overall, our results suggest a different epigenome in the mitochondria than that in the nucleus.
Most of the identified 5hmC sites reside in intergenic regions, followed by introns (Figure 2C). Compared with a base-resolution methylome map in the mouse ESCs (Stadler et al., 2011) (Figure S2B), 5hmCGs are slightly enriched in the intragenic regions and depleted in the intergenic regions. This is consistent with the previous findings using hMeDIP approaches (Ficz et al., 2011). A gene-centric view of 5hmC as well as 5mC distribution in both DNA strands is shown in Figure 2D. There is no noticeable difference for the 5hmCG distribution between the sense and the antisense strands (Figure 2D). We normalized the 5hmCG site density by the 5mCG density, which accounts for the local substrate concentration effect presented to the TET enzymes. The normalized 5hmCG site density is reduced at the transcription start sites (TSSs) and 5′ UTR regions (Figure 2D), and a similar trend is observed for the 5hmCH site (data not shown). Interestingly, although the absolute 5hmCG and the 5mCG densities are higher in exons than in introns, the normalized 5hmCG site density does not show difference between exons and introns (Figure 2D). Overall, our results suggest that the 5hmC profile closely follows the 5mC distribution in the genic regions (Lister et al., 2009).
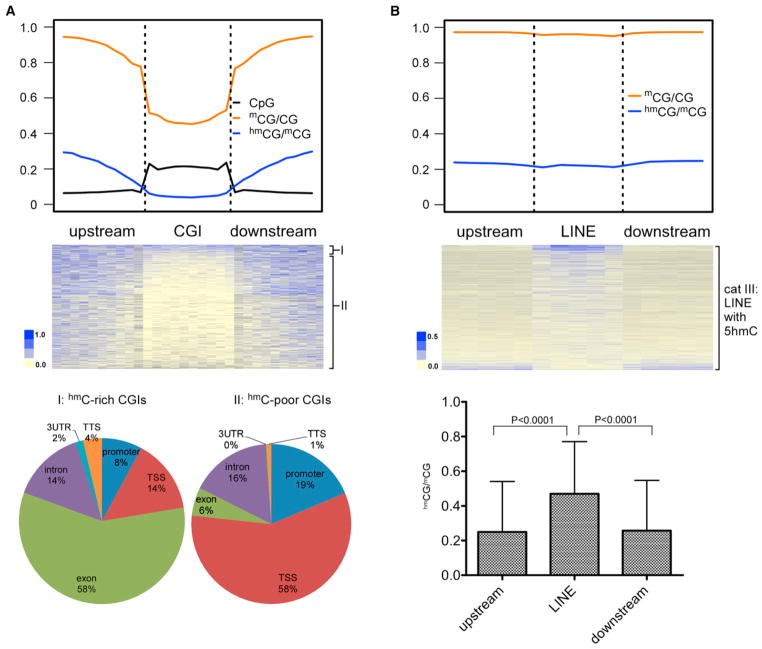
Similar to 5mC, 5hmC is depleted in the CpG islands (CGIs) at the genome scale (Figure 3A). Levels of 5hmC in CGIs, as estimated by the cumulative reads, are inversely correlated with the CpG density (Figure S2C), consistent with an earlier report (Booth et al., 2012). We manually separated all CGIs into two groups based on the overall ranked 5hmC density: group I (top 5%) and group II (lower 95%) (Figure 3A, middle panel). In group I, even with the increased 5hmC density inside CGIs, there is no significant enrichment compared with the flanking regions (data not shown). Most of the group I CGIs appear in exonic regions, whereas most of the group II CGIs appear in the TSS/promoter regions (Figure 3A). The higher 5hmC density in exonic CGIs may be due to their higher 5mC levels compared with the TSS/promoter CGIs (Lister et al., 2009; Deaton and Bird, 2011).
Figure 3. 5hmC Is Depleted in CGIs and Enriched in a Small Set of Repetitive Elements.
(A) Upper panel: metaplot of CpG (black), normalized 5hmCG (red) and 5mCG (Stadler et al., 2011) (blue) distribution around the CGIs. Middle panel: heatmap representation of 5hmC site density in all the genomic CGIs, ranked by the overall 5hmC density in each CGI. We manually separate the upper 5% into group I and the lower 95% into group II. Lower panel: distribution of group I and group II CGIs in different genomic elements.
(B) Upper panel: metaplot of normalized 5hmCG and 5mCG (Stadler et al., 2011) around the LINE elements. Middle panel: heatmap representation of all the LINE elements with 5hmC, ranked by the overall 5hmC content. Lower panel: averaged 5hmC density inside the selected LINE elements is significantly higher than that in their flanking regions.
Error bars represent 1 SD. See also Figure S3.
An observed enrichment of 5hmC in repetitive DNA, such as LINE elements, has been previously reported (Ficz et al., 2011). However, our results show that the genome-averaged 5hmC density inside LINE is at the same level as their flanking regions (Figure 3B). Detailed analysis separated all LINE elements into three categories: (1) those that do not contain CpG sites inside the elements (~25%); (2) those that contain CpG sites but no 5hmCG sites (~42%); and (3) those that contain both CpG sites and 5hmCG sites (32%). Although category II can be regarded as depleted of 5hmCG, there is moderate but significant 5hmCG enrichment in category III (Figure 3B, middle and lower panel). Similar patterns were observed for the SINE and LTR repetitive elements (Figure S3). Overall, our results argue that there is only a small fraction of repetitive elements showing 5hmC enrichment in ESCs.
5hmC at the Protein-DNA Interaction Sites
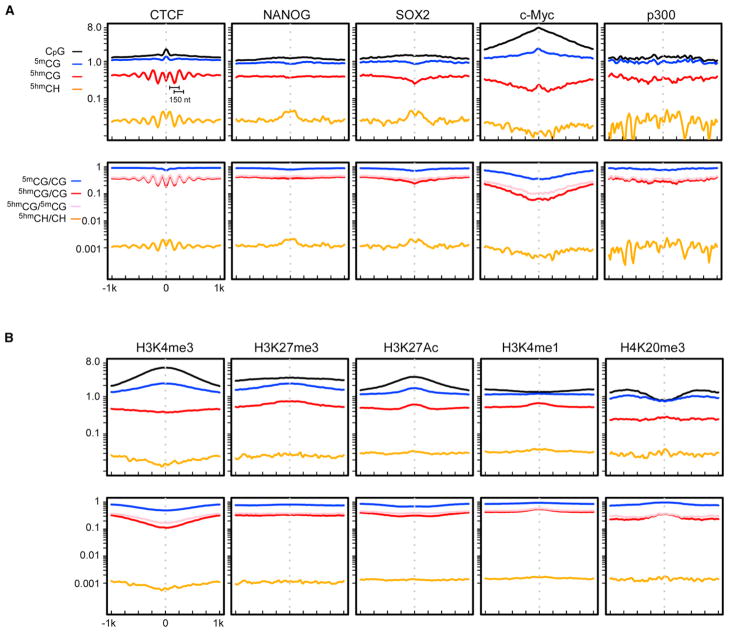
5mC is generally depleted at the cis-elements to facilitate transcription factor (TF) binding (Lister et al., 2009). By correlating with a set of chromatin immunoprecipitation sequencing (ChIP-seq) data, Figure 4A illustrates the representative 5hmC distributions around various protein-DNA interaction sites (Chen et al., 2008).
Figure 4. 5hmC at the Protein-DNA Interacting Sites and Regions with Various Chromatin Modifications.
Both absolute (top panel) and normalized (bottom panel) densities are displayed.
(A) 5hmC distribution around the ChIP-seq peaks of various DNA-interacting factors.
(B) Correlation between 5hmC and various chromatin marks. See also Figures S4 and S5.
The first intriguing observation is a symmetrical, regularly spaced oscillating distribution of 5hmC around the CTCF binding sites in both CG and CH context (Figure 4A). The center of the CTCF binding sites are slightly CG-rich (Figure 4A, CpG line) and depleted in 5mCG (Figure 4A, 5mCG/CG line), consistent with the methylation sensitivity for the CTCF binding (Kurukuti et al., 2006). Accordingly, 5hmCG and 5hmCH are depleted at the center of the CTCF binding sites (Figure 4A). The distance between the neighboring 5hmC peaks on either side of the binding site is about 150 nucleotides (Figure 4A), reminiscent of the unit length of the nucleosome-wound DNA. It was suggested that CTCF binds to the linker DNA between the nucleosomes and the binding event leads to highly organized local nucleosome array on both sides (Fu et al., 2008; Cuddapah et al., 2009). The observed regularity of the 5hmC distribution is in line with such local nucleosomal structure. This further suggests that the local nucleosome environment may play an important role in defining the 5hmC profile, possibly by affecting the 5mC accessibility to the TET enzymes.
A second interesting observation is for the Nanog binding sites, which show noticeable accumulation of 5hmCH at the center, but not 5hmCG (Figure 4A). Similar patterns are also observed for Sox2, Oct4, STAT3, Esrrb, and Tcfcp2I1 (Figures 4A and S4). Nanog/Oct4/Sox2 are known to co-occupy a same set of loci in ESCs and their predicted binding motifs do not contain CG dinucleotides (Chen et al., 2008). To investigate whether non-CG 5-hydroxymethylation affects binding, we then compared the binding affinity of Sox2 to the unmodified, methylated, and hydroxymethylated sites (Figure S5). Our results suggest that cytosine modifications within the binding site do not affect Sox2 binding. In addition, structural modeling reveals that Sox2 binds DNA in the minor groove, whereas the five-position modification of cytosine is located in the major groove and thus not recognized (Figure S5). Further investigation is needed to explain the non-CG accumulation of 5hmC around the binding sites of these transcription factors.
For the other factors we studied, including c-myc, nmyc, Klf4, E2f1, Zfx, and 5hmC, distribution was either slightly depleted or relatively constant around the binding sites (Figures 4A and S4). Binding sites of the histone acetyltransferase p300 showed unique fluctuating 5hmC distributions (Figure 4A).
Correlation between 5hmC and Chromatin States
We next analyzed the 5hmC distribution around the ChIP-seq peaks of various chromatin modification marks (Mikkelsen et al., 2007), as shown in Figure 4B. Both 5mC and 5hmC are depleted in regions marked with H3K4me3, an activation mark. Furthermore, 5hmC is depleted in the RNA polII (Mikkelsen et al., 2007) occupied regions (Figure S4), suggesting 5hmC is generally excluded from active transcriptions. For regions marked with H3K27me3, a repression mark, 5hmC does not show obvious depletion or enrichment. Previous reports have documented 5hmC enrichment in enhancer regions (Stroud et al., 2011; Szulwach et al., 2011). We investigated a few histone marks that are associated with enhancers. For the H3K27Ac marked regions, which may act as active enhancers (Creyghton et al., 2010), 5hmC is slightly depleted as well as 5mC. In contrast, for the H3K4me1 marked regions, which may indicate poised enhancers (Creyghton et al., 2010), both 5mC and 5hmC are enriched toward the center of the regions. Lastly, both 5mC and 5hmC are enriched in the H4K20me3 marked regions, which are associated with specific classes of repetitive elements (Martens et al., 2005).
DISCUSSION
The epigenetic processes of 5hmC regulation remain elusive and controversial after its rediscovery. Base-resolution genome-wide hydroxymethylomes as well as methylomes provide the first steps toward probing their biological relevance. Compared with other bisulfite-conversion based methods, Aba-seq has several unique advantages: (1) simple library construction even with low amount of input DNA (100 ng in our experiment); (2) 5hmC sites with low occupancy can be reliably detected; and (3) data processing is more straightforward. Its potential limitations, on the other hand, largely stem from the enzymatic properties of AbaSI (Wang et al., 2011). For example, target sites are potentially cleaved with varying efficiency, with 5gmC at both sides of the cleavage sites showing the highest efficiency (Wang et al., 2011). However, this appears not a big concern as such sites are only slightly overrepresented in our current data set (Figure S1I). For a small portion of data (~13% in our data set), there is ambiguity in assigning 5hmC to the exact base. Nevertheless, approximately 12.2M putative 5hmC sites are detected in the mouse ESC genome and bioinformatic analysis has provided sufficiently fine details about the E14 hydroxymethylome.
Based on our results, we propose that factors affecting the local 5mC accessibility to the TET enzymes play important roles in the 5hmC deposition. These factors can include chromatin compaction, nucleosome positioning, or TF binding. The most striking example is the regularly oscillating 5hmC profile around the CTCF-binding sites, suggesting 5hmC “writers” may be sensitive to the nucleosomal environment. Similarly, the elevated 5hmC levels in the exonic regions are possibly due to a higher concentration of more accessible 5mC sites. The 5hmC level after normalization is actually comparable for the inter- and intragenic regions, arguing against specific enrichment of 5hmC in either region. Depletion of 5hmCs is often observed when there is depletion of 5mC, as seen in the TSS regions, binding sites of RNA polII, and some TFs. For 5hmCs in other regions—in particular, poised enhancers marked by H3K4me1—the presence of 5hmC may indicate a transition or a poised state between methylation and demethylation. Finally, the unique mitochondrial hydroxymethylome and the locally increased CH hydroxymethylation at TF-binding sites such as NANOG and SOX2 await further biological investigation.
EXPERIMENTAL PROCEDURES
Aba-seq Library Construction
The undifferentiated E14 cells was cultured as previously described (Kinney et al., 2011). E14 cells were harvested and DNA extracted with a QIAGEN Pure-gene Core Kit (catalog no. 1042601) per manufacturer’s specifications. E14 genomic DNA was then treated with QIAGEN RNase A (catalog no. 19101).
Genomic DNA was glucosylated overnight at 37°C by incubating 100 ng of E14 genomic DNA with 200 μM UDP-glucose and 20 units of T4-β-glucosyl-transferase (NEB: M0357S) in NEB buffer #4. Glucosylated genomic DNA was subsequently purified by phenol-chloroform extraction and ethanol precipitation. AbaSI digestion was done by incubating the glucosylated genomic DNA with four units of AbaSI in NEB buffer #4 at room temperature (25°C) for 1 hr in a total reaction volume of 20 μl. The AbaSI digested DNA was then purified by phenol-chloroform extraction and ethanol precipitation. Biotinylated P1 adapters (Table S1) were ligated onto the AbaSI digested DNA by incubating the digested DNA with 300 nM of P1 and 2 μl of T4 DNA quick ligase (NEB: M2200) in the 1 × quick ligation buffer in a total volume of 42 μl at room for 30 min. Ligated DNA was then purified by running samples on a 6% Tris-borate-EDTA (TBE) gel followed by gel excision of the DNA. Gel extraction was done by crushing and soaking the isolated gel fragment (0.3M NaOAC [pH 7.4], 2 mM EDTA) overnight at 37°C followed by ethanol precipitation of the DNA. Covaris s-series sonicator was used to shear the P1-ligated genomic DNA to a range around 300 bp with the following settings: duty, 10; intensity, 4; cycles per burst, 100; six cycles at 60 s. P1-ligated DNA was then captured by mixing 50 μl of fragmented DNA with 12.5 μl of Dynabeads MyOne Streptavidin C1 beads (Life Technologies, catalog no. 650–01) according to the manufacturer’s specifications. End repair was done by using the NEBNext End Repair Module (NEB: E6050S) on the DNA-bound beads for 30 min at 20°C. The DNA-bound beads were then washed two times with a buffer containing 5 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, and 1 M NaCl; followed by wash with H2O. A-tailing of the end-repaired DNA on beads was carried out by using the NEBNext dA-Tailing Module (NEB: E6053S) for 30 min at 37°C. The DNA-bound beads were then washed as the previous step. P2 adapters (Table S1) were ligated by incubating the DNA-bound beads with 250 nM of P2 and 2 μl of T4 DNA quick ligase in the 1× quick ligation buffer at room temperature for 30 min. The DNA-bound beads were then washed as the previous steps. Amplification of the DNA library was carried out by adding the entirety of the DNA-bound beads to a reaction mixture containing 0.2 mM of each dNTP, 1× Phusion HF Buffer, 300 nM forward primer (PCR_I), 300 nM reverse primers (PCR_IIpe) (Table S1), and ten units of Phusion DNA polymerase (NEB: M0531S). The following thermocycler conditions were utilized for amplification: 30 s 98°C, 16 cycles (10 s 98°C, 15 s 70°C, 15 s 72°C), 10 min 72°C, and hold at 4°C. The beads were then removed from the PCR and the library was extracted by gel purification as described above.
Real-Time PCR Validation of Genomic 5hmC Sites
E14 genomic DNA used in this assay was same batch to that of the Aba-seq library construction. Validation of genomic 5hmC at CCGG sites were carried out by using the EpiMark 5hmC and 5mC Analysis Kit according to manufacturer’s specifications (NEB: E3317S). Real-time PCR of digested DNA was done with iQ SYBR Green Supermix (BIO-RAD: 170–8882) per manufacturer’s specifications.
Validation of genomic 5hmC at CH sites were done on AbaSI-digested glucosylated E14 genomic DNA followed by real-time PCR. Real-time PCR of digested DNA was done with iQ SYBR Green Supermix (BIO-RAD: 170–8882).
All real-time PCR primers and the method used to calculate 5hmC percentage are listed in Table S2 for CCGG sites and Table S3 for CH sites.
Processing Aba-Seq Reads and Identification of 5hmC
All sequencing runs (two lanes) were done on the Illumina HiSeq platform with read length of 50 bp. All the raw reads were mapped to the mouse reference (UCSC mm9) using the Bowtie (Langmead et al., 2009) aligner with parameters (-v 2 -m 1). We then interrogated neighboring sequences of the aligned AbaSI cleavage sites in search of putative 5hmC sites at expected distance (−CN11–13 or +N9–11G). Copy number is defined as the total number of reads that resulted from AbaSI digestion of a particular 5hmC site. Sequence logos were constructed using weblogo (Crooks et al., 2004). All scripts were written in PERL and are available upon request.
Comparison to the Methylome in ESCs
CpG methylome of mouse ESC by Bisulfite sequencing (BisSeq) was acquired from NCBI Gene Expression Omnibus (GEO) database GSE30202 (Stadler et al., 2011) and was used as a reference. 99% (11,066,961) of Aba-seq 5hmCG sites overlap with the 5mCG sites in this BisSeq data set. The 11,066,961 overlapped 5hmCG sites were used for the subsequent genomic analyses of 5hmCG in this study.
Genome-wide 5hmC Profile in Different Genomic Regions
All genomic regions and their genomic coordinates were defined based on the mm9 genome annotation tracks downloaded from UCSC Genome Browser (Fujita et al., 2011) in February 2012. Different genomic elements were identified from the RefSeq genes track. Promoter is defined as 5 kb upstream of nonredundant TSS. We define intergenic regions as 5 kb downstream of transcription termination sites (TTS) that do not overlap with any known genic regions.
To generate genome-wide metagene plots we divided 2 kb upstream region of TSS, 5′ UTR, exon, intron, 3′ UTR, and 2 kb region downstream of each nonredundant RefSeq gene into equal-sized bins respectively. For CGIs and repetitive elements, we binned the same-length upstream and downstream flanking regions into same number of intervals. CpG, 5mCG, 5hmCG, and 5hmCH sites were mapped to individual regions using BEDTOOL (Quinlan and Hall, 2010). Site density was calculated as number of sites divided by region length. 5hmCG density was also corrected by background CpG and 5mCG. Average densities were then computed for each bin position and were used for metaplots or global trend plots.
Genomiccoordinates of the CGIs wereacquired from the cpgIslandExt table in UCSC database. Depending on their locations, we classified CGIs into promoter CGI, TSS CGI, exon CGI, intron CGI, 3′ UTR CGI, and TTS CGI (a TSS or TTS region is defined as −500 bp to +500 bp of a TTS or a TTS). Each type of CGIs has at least 50% overlap with a region of that particular genomic feature.
Repetitive elements (LINE, SINE, and LTR) were retrieved from the Repeat-Masker track in UCSC database.
Comparison to Other 5hmC Mapping Data Sets
We compared our Aba-seq data set to three different types of 5hmC mapping data sets: hydroxymethylated DNA immunoprecipitation sequencing (Williams et al., 2011); selective chemical labeling of 5hmC with single-molecule real-time DNA sequencing (SMRT) (Song et al., 2012) and TAB-seq (Yu et al., 2012).
hMeDIP: We downloaded BED files containing genomic coordinates of mapped sequencing reads from the NCBI GEO database (GSE24843). 5hmC-enriched regions were identified by the MACS (Zhang et al., 2008) program using the following criteria: peak p value < 10−5 and fold enrichment over immunoglobulin (Ig) G > 10.
TAB-seq: Genomic coordinates of 2,057,636 reported high-confidence TAB-seq hmC sites were kindly provided by the authors.
SMRT: Genomic locations of 744 selected 5hmC in mouse ESCs from initial shotgun SMRT DNA sequencing were obtained from Table S2 of the published paper (Song et al., 2012).
All the above data sets were converted to BED format. Nonredundant 26-mer fragments flanking the AbaSI cleavage sites were used to compare with the 5hmC-enriched regions derived from hMeDIP data. Comparison between AbaSI-seq, TAB-seq, and SMRT were carried out at base resolution by BEDTOOL.
Correlating 5hmC to ChIP-seq Data Sets
To create 5hmC profile at ChIP-seq peaks, we downloaded data sets from NCBI.
TET1 binding sites: BED files of mapped sequencing reads from a Tet1 ChIP-seq data set were downloaded from the GEO database (GSE24843). TET1 binding regions were identified by the MACS program using the following criteria: peak p value <10−8, fold enrichment over IgG > 10.
Histone modification marks: genome-wide maps of chromatin state (H3K4me3, H3K27me3, H3K36me3, H4K20me3) were downloaded from NCBI GEO database (GSE12241)(Mikkelsen et al., 2007). ChIP-seq data sets of two enhancer histone mark H3K27ac and H3K4me1 were downloaded from NCBI GEO database (GSE24165) (Creyghton et al., 2010). Genomic locations of mapped sequencing reads (mm8) were remapped to mm9 via the LiftOver tool. Genomic intervals enriched with a specific chromatin mark were identified by the MACS program using the following criteria: peak p value < 10−5, fold enrichment over control H3 > 10.
TFBS: Binding sites in mm8 of 13 TFs (Nanog, Oct4, STAT3, Smad1, Sox2, Zfx, c-Myc, n-Myc, Klf4, Esrrb, Tcfcp2l1, E2f1, and CTCF) and two transcription regulators (p300 and Suz12) were downloaded from the GEO database (GSE11431)(Chen et al., 2008). Genomic coordinates were remapped to mm9 by the LiftOver tool.
RNA polymerase II: locations of mapped sequencing reads of chromatin IP against RNA polymerase II were downloaded from NCBI GEO database (GSE12241)(Mikkelsen et al., 2007). Genomic coordinates were remapped to mm9 by the LiftOver tool. RNA polymerase II binding intervals were identified by the MACS program using the following criteria: peak p value < 10−5, fold enrichment over control > 10.)
A region of −1 to +1 kbp around the peak summits or the binding centers was selected and binned into 50 bp sliding windows at 25 bp steps. 5mCG, 5hmCG, 5hmCH, and CpG sites were aligned to individual bins and site density profiles were normalized to the site density per 1 bp per bin. Global trend plots were generated by calculating the average site density at each bin position.
Supplementary Material
Acknowledgments
We thank Dr. Shannon Kinney for providing E14 genomic DNA, Drs. Rich Rob-erts and William Jack for helpful comments, and Drs. Peng Jin and Chuan He for providing us TAB-seq dataset of mouse E14 cells. We thank Cynthia Hen-drickson and HudsonAlpha Institute for Biotechnology for sequencing. This work is supported by NIH SBIR grants (GM096723 to Z.Z. and GM095209 to Y.Z.) and New England Biolabs. Subjects of this paper may be potential future products of New England Biolabs.
Footnotes
ACCESSION NUMBERS
All data have been submitted to the GEO database under the accession number GSE42898.
Supplemental Information includes five figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.01.001.
LICENSING INFORMATION
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39(Database issue):D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcyto-sine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney SM, Chin HG, Vaisvila R, Bitinaite J, Zheng Y, Estève PO, Feng S, Stroud H, Jacobsen SE, Pradhan S. Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. J Biol Chem. 2011;286:24685–24693. doi: 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethyl-cytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci USA. 2011;108:3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Clark TA, Lu XY, Kislyuk A, Dai Q, Turner SW, He C, Korlach J. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nat Methods. 2012;9:75–77. doi: 10.1038/nmeth.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. Integrating 5-hydroxymethylcyto-sine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terragni J, Bitinaite J, Zheng Y, Pradhan S. Biochemical characterization of recombinant β-glucosyltransferase and analysis of global 5-hy-droxymethylcytosine in unique genomes. Biochemistry. 2012;51:1009–1019. doi: 10.1021/bi2014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Guan S, Quimby A, Cohen-Karni D, Pradhan S, Wilson G, Roberts RJ, Zhu Z, Zheng Y. Comparative characterization of the PvuRts1I family of restriction enzymes and their application in mapping genomic 5-hydroxymethylcytosine. Nucleic Acids Res. 2011;39:9294–9305. doi: 10.1093/nar/gkr607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxyme-thylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.