Abstract
In the present study we aimed to determine the prevalence of C9ORF72 GGGGCC hexanucleotide expansion in our cohort of 53 FTLD patients and 174 neurologically normal controls. We identified the hexanucleotide repeat, in the pathogenic range, in 4 (2 bv-FTD and 2 FTD-ALS) out of 53 patients and one neurologically normal control. Interestingly, two of the C9ORF72 expansion carriers also carried two novel missense mutations in GRN (Y294C) and in PSEN-2 (I146V). Further, one of the C9ORF72 expansion carriers, for whom pathology was available, showed amyloid plaques and tangles in addition to TDP-43 pathology. In summary, our findings suggest that the hexanucleotide expansion is probably associated with ALS, FTD or FTD-ALS and occasional comorbid conditions such as Alzheimer’s disease. These findings are novel and need to be cautiously interpreted and most importantly replicated in larger numbers of samples.
Keywords: FTLD, bv-FTD, FTD-ALS, C9ORF72, GRN, PSEN-2, Alzheimer’s disease
Introduction
Frontotemporal lobar degeneration (FTLD, OMIM #600274) is the second most common cause of presenile dementia. It is characterized by progressive degeneration of the frontal and anterior temporal lobes of the brain leading to wide range of clinical symptoms including changes in personality and behavior, language impairment, and cognitive dysfunction (reviewed in Ferrari et al. 2011). Approximately 15% of FTLD patients develop symptoms of motor neuron dysfunction (Lomen-Hoerth et al., 2002). The co-occurrence of FTLD and MND in familial cases suggests that FTLD represents a spectrum of neurological disorders with complicated clinical, pathological and genetic etiology. The most common form of FTLD pathology is FTLD-TDP with TAR DNA-binding protein-43 (TDP-43) immunoreactive neurons, which is also deposited in neurons of ALS patients. This pathological finding links FTLD and ALS (Neumnann et al., 2006; Arai et al., 2006).
To date, mutations in the progranulin gene (GRN) are considered the most prevalent and encompass phenotypes from bvFTD to CBS. The common pathology in GRN mutation carriers is TDP-43 positive inclusions. Mutations in the TDP-43 gene however, are usually associated with the ALS phenotype (Corrado et al. 2009) and rarely found in FTLD (Borroni et al. 2009) or CBS (Huey et al. 2011). The causal link between the mutations in GRN gene and TDP-43 pathology is yet to be established. Mutations in the other FTLD candidate genes, MAPT, VCP, CHMP2B, PSEN1, and PSEN2 explain only very small number of FTLD patients and there is no report of ALS or MND among those mutation carriers.
Linkage studies of cases with ALS, FTD, FTD-ALS with type 2 TDP-43 pathology had suggested a locus on chromosome 9p (Boxer et al., 2011; Morita et al., 2006; Vance et al., 2006) although it was not clear if the GWA and the linkage studies identified the same locus on 9p. Mok et al. in 2011 suggested a common founder for FTLD and ALS on chromosome 9, based on the original Finnish association study reported by Laaksovirta et al in 2010. The identification of hexanucleotide GGGGCC repeat expansion in the non-coding region of C9ORF72 in families linked to 9p21 (DeJesus-Hernandez et al., 2011; Renton et al., 2011) suggested this expansion as a possible cause of FTLD and ALS.
In the present study we screened 53 patients and 174 neurologically normal controls for the expansion of the GGGGCC hexanucleotide. Our cohort encompasses a wide range of the FTLD spectrum which may prove useful in establishing a phenotype/genotype correlation within the FTLD spectrum. In the present study, we identified 4 out of 53 patients and one neurologically normal control who had donated a sample to the Coriell institute who carried the hexanucleotide repeat in the apparent pathogenic range.
Patients and methods
Patients
The study population comprised a sub-group of patients which have been previously described (Huey et al. 2011), consisting of 27 probable bv-FTD, 9 possible bv-FTD, 6 PPA-PNFA, 2 PPA-semantic, 4 FTD-ALS, 4 AD, and 1 MSA diagnosed using appropriate criteria (Neary et al., 1998; Raskovcky et al., Brain 2011; McKhann et al., 1984; Gilman et al., 2008). A full neurological and neuropsychological evaluation was performed for all patients. Table 1 presents a brief family history of all patients.
Table 1.
Samples IDs with corresponding diagnosis, number of hexanucleotide expansion and a brief family history of neurological disorders for each patient. Diagnoses are described following Rascovsky et al., 2011.
| Patient ID |
Diagnosis | Hexanucleotide Repeats # |
Family history of neurological disorders |
|---|---|---|---|
| 158 | Probable bv- FTD |
> 50 | Father - AD; Brother - MS |
| 198 | Probable bv- FTD |
> 45 | Brother - TIA; Brother - alcoholism; Daughter - migraines; Paternal aunt - dementia |
| 211 | FTD-ALS | > 50 | Mother - ALS; Brother - alcoholism; Father - dementia; Brother/Son - depression; Daughter - migraines, depression; Son - anoxic at birth |
| 223 | FTD-ALS | > 50 | Mother - dementia; Father - dementia ; Sister - FTD ; Brother - alcoholism ; Son - depression, drug use |
| 81 | Probable bv- FTD |
1 | Father - died of dementia at 77 |
| 83 | AD | 7 | Paternal grandfather - dementia |
| 85 | FTD-PPA | 7 | Father -vasc. dementia died at 68 |
| 88 | FTD-PPA | 14 | Sister - PD |
| 89 | Probable bv- FTD |
8 | Mother - LOAD; Paternal grandmother - dementia died at 89 |
| 90 | Probable bv- FTD |
4 | Father - CVA; Nephew - OCD |
| 95 | SD | 4 | Maternal aunt - dementia |
| 96 | Probable bv- FTD |
~ 25 | Father - AD onset 58; Maternal uncle - schizophrenia |
| 97 | FTD-PPA | 3 | Son - schizophrenia |
| 98 | Possible FTLD |
15 | Sister CVA. O/w no +FH |
| 99 | Probable bv- FTD |
10 | Father - PD died at 85; Mother dementia, brain tumor; 3 aunts - dementia; Maternal grandmother - dementia |
| 101 | Probable bv- FTD |
3 | Son - schizophrenia, drug abuse; Daughter - schizophrenia; Sister &Brrother - drug abuse |
| 103 | SD | 5 | Maternal grandmother - CVA; Maternal aunt - CVA |
| 107 | Probable bv- FTD |
1 | Paternal uncle - unknown mental illness |
| 111 | Possible FTLD |
2 | Sister - Korsakoff syndrome; Mother - OCD, depression; Sister -alcoholism; Twin brother - alcoholism |
| 114 | FTD-PPA | 2 | Father - AD, PD died at 63; 2 brothers - schizophrenia |
| 115 | Probable bv- FTD |
6 | Father - dementia onset at 72 paternal grandmother Parkinson’s and memory problem died in her 70”s |
| 118 | Probable bv- FTD |
6 | Father - AD; Mother - AD, depression both died in their early 90’s |
| 123 | Probable bv- FTD |
8 | Brother - learning disabilities, alcoholism; Sister - eating disorder, depression |
| 124 | Probable bv- FTD |
7 | Father - PD; aunt AD died at 79 |
| 127 | MSA | 1 | Mother - AD; Brother - CVA; Brother - brain cancer |
| 128 | Probable bv- FTD |
3 | Father - OCD; Mother - Mental disorder NOS |
| 134 | FTD-MND | 1 | |
| 136 | Probable bv- FTD |
2 | Mother - dementia at 87; maternal uncle dementia |
| 139 | FTD-PPA | 10 | Mother - depression, alcoholism |
| 143 | Probable bv- FTD |
17 | Daughter - OCD, depression |
| 144 | Probable bv- FTD |
5 | |
| 163 | FTD-MND | 9 | Paternal uncle - dementia, alcoholism |
| 176 | FTD-PPA | 2 | Mother - depression; Daughter - ADD |
| 177 | AD | 3 | Mother - LOAD; Father - alcoholism; Son - alcoholism |
| 183 | Possible FTLD |
2 | Father PTSD & depression; mother DLB, mutism; mat grandfather PD & stroke; maternal uncle DLB; 2 paternal aunts with MND |
| 194 | Probable bv- FTD |
3 | Sister - dementia; Father - stroke; Mother - depression ; Son - depression/anxiety |
| 195 | Probable bv- FTD |
6 | Mother - action tremor; Paternal grandmother - dementia; maternal great grandmother dementia |
| 199 | Possible FTLD |
2 | Father - dementia ; Daughter - seizures ; Maternal grandmother - dementia |
| 200 | Probable bv- FTD |
6 | Daughter - ADD, anxiety; Son - substance abuse |
| 201 | Possible FTLD |
4 | Father- dementia in his 60’s, stroke; Sister - dementia; Brother - stroke; Brother - epilespy, stroke; Son - epilepsy, mental illness, grandfather dementia in his 60’s |
| 203 | Probable bv- FTD |
3 | Paternal grandmother - late onset dementia at 91; Mother - MS; maternal grandmother Parkinson’s onset at 78, maternal great aunt and uncle late onset AD |
| 205 | Possible FTLD |
10 | 2 Sisters - anxiety; Sister - PSP; Daughter - eating disorder; Daughter - anxiety, depression; Daughter - schizophrenia; Son/2 Daughters - depression |
| 207 | Possible FTLD |
7 | Siblings tremor, anxiety, alcoholism, substance abuse, mother died at 83 had dementia and anxiety; father died at 79 had Parkinson’s and anxiety. |
| 209 | Probable bv- FTD |
7 | Father - stroke ; Brother - born w/hole in skull; Daughter - seizure disorder |
| 210 | Possible FTLD |
7 | Maternal grandfather - dementia |
| 212 | Probable bv- FTD |
5 | |
| 215 | Possible FTLD |
2 | Mother - AD ; Father - dementia ; Brother - depression ; Son - arthrogyrosis |
| 216 | Possible FTLD |
9 | Sister - mild stroke, depression |
| 217 | Probable bv- FTD |
2 | father died at age 82 no dementia but paternal grandfather died of dementia at 81 |
| 218 | Probable bv- FTD |
~ 25 | Mother - dementia; Son - ALS; Maternal aunt - AD |
| 219 | Probable bv- FTD |
2 | Daughter - anx, dep, neurocardiogenic syndrome ; Brothers - depression ; Sister - anx, dep, ADHD ; Father - dementia ; Mother - stroke |
| 221 | Probable bv- FTD |
10 | Father - vascular dementia at 78 |
| 222 | Possible FTLD |
2 | Father - AD, PD, heart disease; Brother - alcohol abuse |
Genetic screening
Blood samples from index patients were collected in accordance with the local Institutional Review Board guidelines and informed written consent form was obtained. The DNA of all patients was screened for MAPT, PGRN, FUS, TDP43, PSEN1, PSEN2, APP and CHMP2B. In order to screen for hexanucleotide expansions in the C9ORF72 gene, we performed the experiments as described in Renton et al. (2011). Although this method is not able to determine the exact number of repeats, it can detect repeat numbers of maximum ~ 60. This method however, can discriminate the normal repeat range detected in the normal population (0-20) from the higher mutated range (≥30). Renton et al. (2011) detected a pathogenic repeat number of ~250 repeats in a carrier whose fluorescence in situ hybridization (FISH) analysis verified the repeat expansion. DeJesus-Hernandez et al. (2011) used a different method to amplify the repeat region of C9ORF72 and for the verification of repeat size they subsequently performed Southern blot analysis. They estimated the number of repeats between 700-1600. As the goal of our study was solely to determine if our cohort has repeat numbers above the range of normal controls, we screened the DNA of 53 patients and 174 neurologically normal controls from Coriell Institute (NDPT 098 and 099). The mutated alleles were considered at the length of 30 repeats and above (Renton et al., 2011). The DNA of these patients had been genotyped on the illumina infinium platform as part of the ongoing international FTD-GWAS consortium project. The genotypes of 42 SNPs which were used to build the chromosome 9p21 haplotype in Finnish population (Laaksovirta et al., 2010) were derived from the GWAS data for the 53 patients to determine if the carriers of the expansion also harbor the same ancestral haplotype as the Finnish carriers of hexanucleotide expansion.
Results
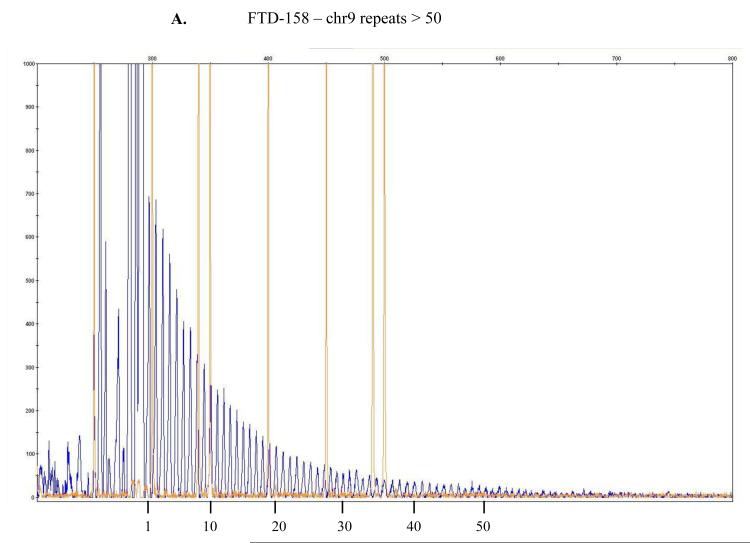
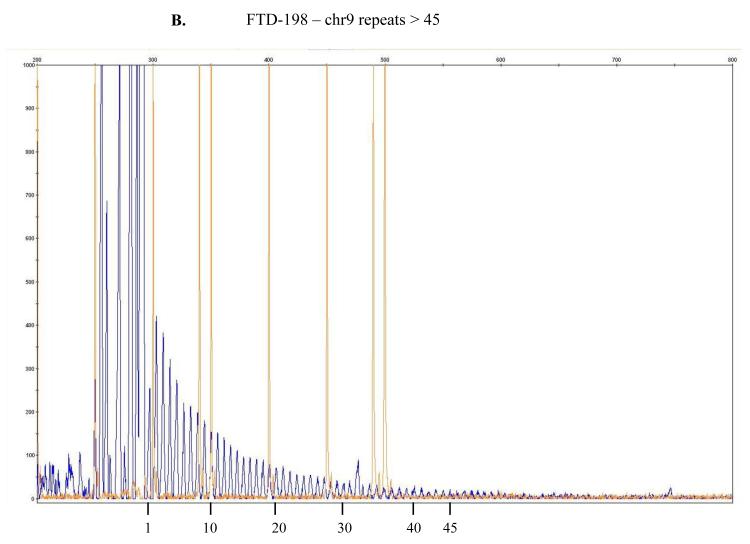
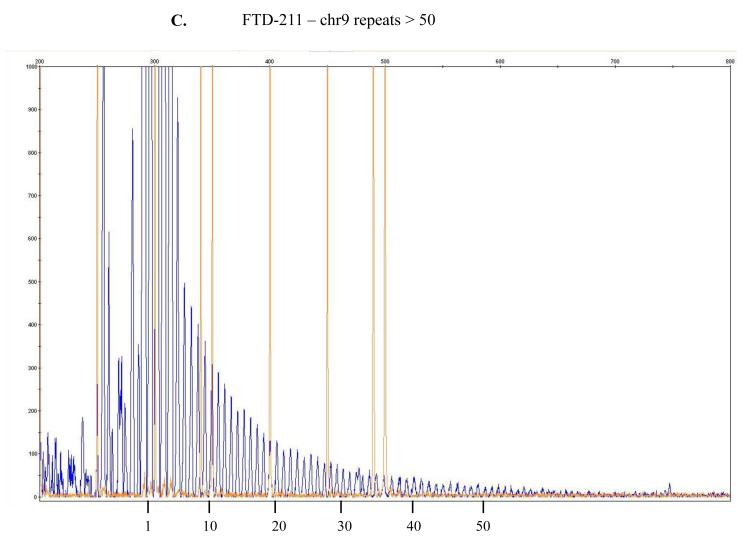
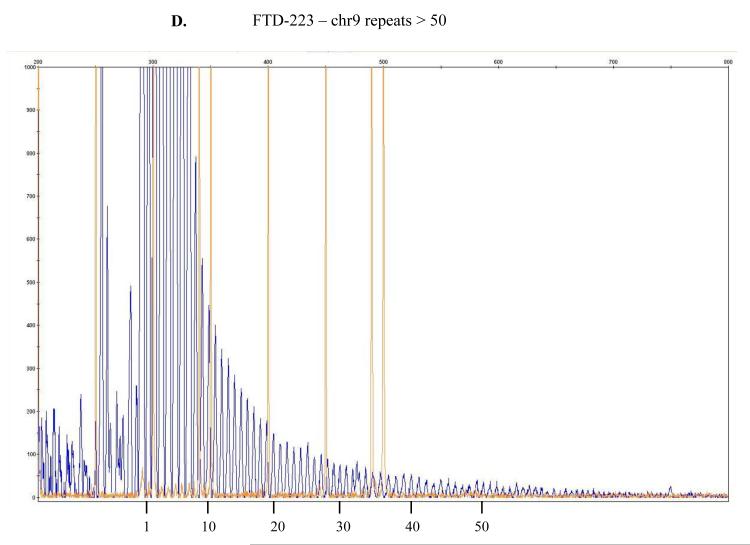
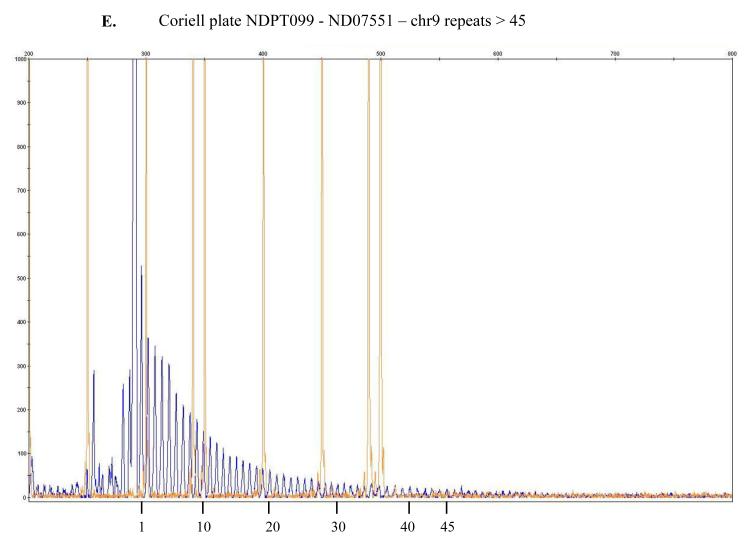
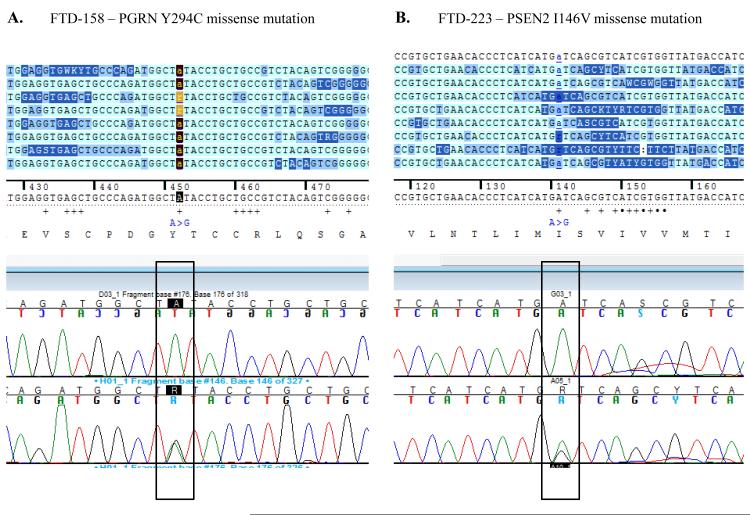
In our cohort we identified 4 out of 53 patients (FTD158, FTD198, FTD211 and FTD223) and one out of 174 neurologically normal controls (Coriell plate NDPT099 - ND07551) with repeat expansion in the pathogenic range (≥30) (Figure 1). Among the carriers of the hexanucleotide expansion, one patient (case 1 FTD158) carries a novel GRN mutation Y294C (Figure 2A) and another patient (case 4 FTD223) carries a novel mutation in PSEN2 I146V (Figure 2B). None of these mutations have been reported previously and there is no functional data available regarding pathogenicity of these variants. Due to lack of informative family members, we could not examine the segregation of these mutations with the disease. The in silico analysis by PolyPhen-2 software (PolyPhen-2: http://genetics.bwh.harvard.edu/pph2/) predicted both variants to be probably damaging.
Figure 1.
The repeat-primed PCR reactions were run on ABI3730 DNA Analyzer .The results were analyzed & visualized using Gene Mapper software. Fluorescence intensities are shown in the vertical axis: the size marker is identified by orange vertical lines and the hexanucleotide repeats that extend beyond the 300 bp marker are marked in blue. The range of > 30 repeats is considered the threshold suggesting presence of expansion even though a direct and precise relatedness between this method and the actual repeat size has not been established yet being, as such, simply predictive of probable repeat expansion. In here, counts of approximately > 45 or > 50 repeats are depicted for, respectively, samples FTD-158 (A), FTD-198 (B), FTD-211 (C), FTD-223 (D) and for Coriell neurologically normal control ND07551 (E).
Figure 2.
Electropherograms of the two novel missense mutations found in PGRN (A) and in PSEN2 (B) in two of the hexanucleotide repeat carriers, respectively individuals FTD-158 (A) and FTD-223 (B). Both missense mutations have not been previously reported. After in silico analysis using Polyphen 2 software, both mutations have been predicted to be probably damaging.
We extracted the genotypes of 42 SNPs of the original Finnish haplotype by Laaksovirta et al 2010 to determine if our patients harbor the original risk haplotype on chr9p21 for FTD-ALS (rs3849942, rs1330921, rs10121765, rs1110264, rs1110155, rs2150336, rs2225389, rs1161680, rs2120718, rs2589054, rs10812596, rs1058326, rs944404, rs765709, rs1316679, rs4406503, rs10511817, rs725804, rs10511816, rs1444533, rs1822723, rs4879515, rs895023, rs868856, rs7046653, rs2440622, rs1977661, rs903603, rs10812610, rs2814707, rs12349820, rs10122902, rs10757665, rs1565948, rs774359, rs2282241, rs1948522, rs1982915, rs2453556, rs702231, rs696826, rs2477518). Our genotyping (Supplementary Table 1) revealed that the complete risk haplotype does not exist in any of our patients. However, Renton et al 2011 reported that SNP rs3849942 is a surrogate marker for the risk haplotype in the populations that they studied. Our 4 patients with expansions carry the risk allele A (rs3849942) and 20 out of 49 of the rest of patients without expansion carry that risk allele as well. Of note, in our patients series there were two individuals diagnosed with FTD-MND, samples FTD134 and FTD163 (Table 1). Patient FTD134 carries neither the expansion nor the risk allele A (rs3849942), while FTD163 does not carry the expansion but the risk allele.
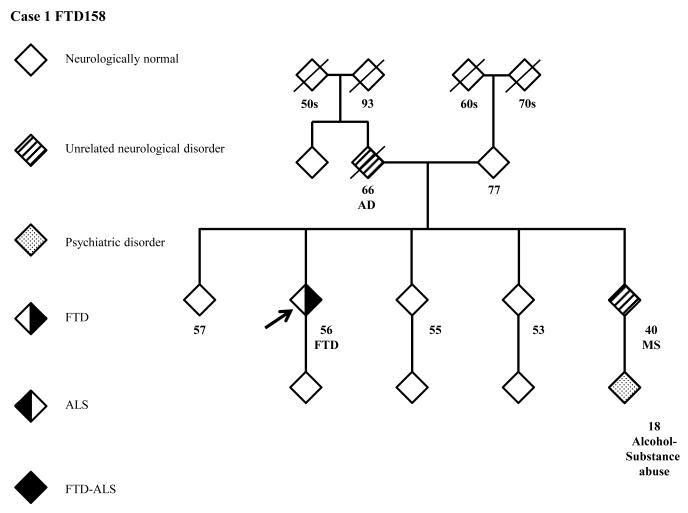
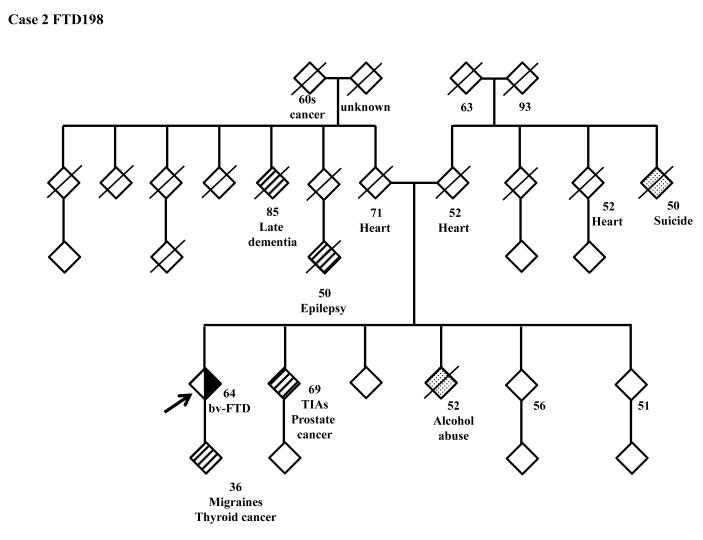
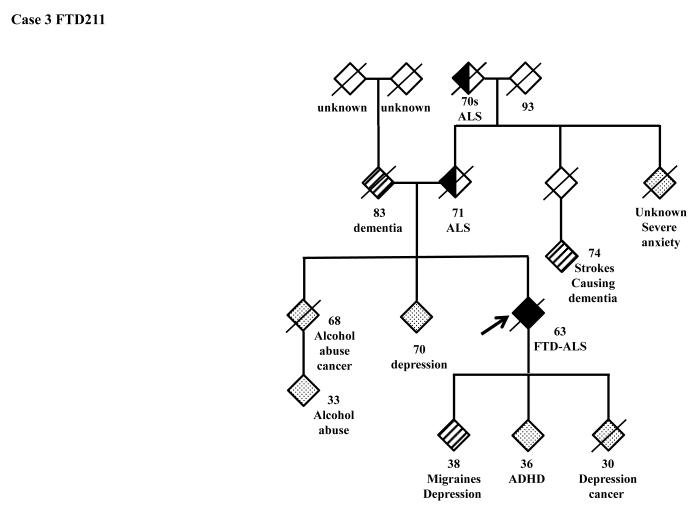
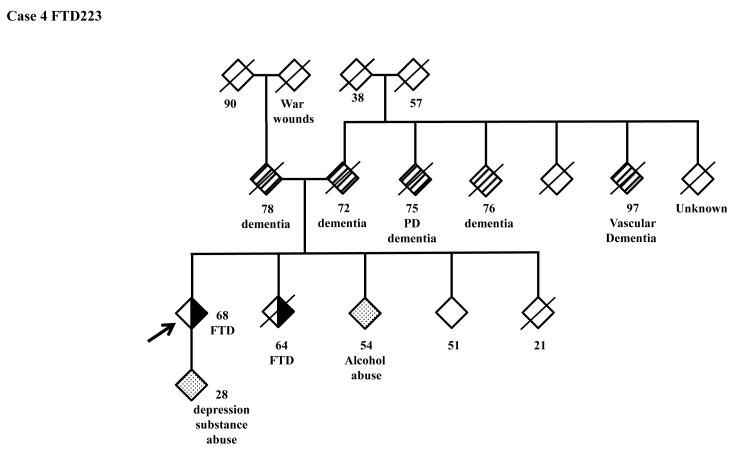
The family history of the patients who carry the hexanucleotide repeat expansion can be seen in more detail in the pedigrees (Figure 3). A full clinical description of these four patients can be found in the supplementary clinical data.
Figure 3.
Pedigrees of the patients carrying the C9ORF72 hexanucleotide expansion. 3A: FTD158, 3B: FTD198, 3C: FTD211, 3D: FTD223.
FTD: Frontotemporal Dementia; ALS Amyotrophic Lateral Scelerosis; AD: Alzheimer’s Disease; MS: Multiple Sclerosis; TIA: Transient Ischemic Attack; ADHD: Attention Deficiency Hyperactivity Disorder; PD: Parkinson’s Disease.
Discussion
Our cohort of FTLD patients was recruited from around the country and was not selected on the basis of family history, known familial mutations, or presence of MND. This sample is likely representative of the general U.S. FTLD population. In the quest to identify the genetic cause of FTLD in our cohort, we screened 53 of our patients for the newly identified locus on chromosome 9p21 (DeJesus et al., 2011; Renton et al., 2011). Patients were referred to NIH primarily with symptoms of frontotemporal dementia and some had developed accompanying neuropsychiatric or motor neuron symptoms as well (Table 1). We identified hexanucleotide expansion of over 30 repeats in 4 patients of which two have a diagnosis of FTD-ALS and two of bv-FTD. As our mutation finding did not reach the statistical significance of the original publications by DeJesus-Hernandez et al. and Renton et al., we aimed to verify the existence of the risk haplotype in our population (Supplementary Table 1). However, neither the risk alleles of the 42 SNPs of the original Finnish risk haplotype (Laaksovirta et al., 2010), nor the phenotypes of the mutation carriers showed a strong correlation to the hexanucleotide expansion compared to those who did not carry the mutation. Among our 53 patients, no one carries the exact same Finnish risk haplotype. Considering the fact that our patients are all recruited in the U.S. and have diverse ethnic background this outcome is not unusual. The haplotype can be explained as a possible genetic background for the carriers of the expansion and not necessarily associated with the hexanucleotide expansion. The phenotype of our cohort was also diverse. As reported previously (DeJesus-Hernandez et al., 2011 Renton et al., 2011; Gliselnick et al., 2011; Troakes et al., 2011), the expansion is detected mainly in FTD-MND cases. As evident in Tables 1 and 2 we had overall 4 patients with FTD-ALS or FTD-MND (of which only two, FTD211 and FTD223, carried the expansion) and 2 bv-FTD cases with family history of ALS (FTD183 and FTD218) who did not carry the hexanucleotide expansion. Additionally, we identified the expansion of more than 45 in one neurologically normal control (see Supplementary clinical data). These findings prompt us to have a more critical view to understand whether the C9ORF72 hexanucleotide repeat expansion is the pathogenic cause of the disease or merely a risk factor contributing to the susceptibility to the disease. As we have previously shown (Huey et al., 2011) in cases such as FUS gene, the variants which were originally reported as pathogenic in ALS cases were later found also in neurologically normal controls. Furthermore, we replicated an interesting finding by Murray et al (Table 2): the C9ORF72 expansion carriers may be more likely to present with memory and psychiatric symptoms rather than typical FTLD symptoms. One of our C9ORF72 expansion positive patients presented with paranoia as his predominant symptom and two others demonstrated significantly impaired memory. The one patient whose autopsy report is available (FTD211) showed amyloid plaques and tangles in addition to TDP-43 pathology (Dr. Ghetti personal communication). Murray et al. also describes one expansion case with both TDP-43 and AD pathology. This suggests that the hexanucleotide expansion may also coexist with forms of dementia other than ALS, FTLD, or FTD-ALS. Although this is only a single case it raises the possibility that some cases of clinical and pathologic TDP-43 positive AD may be associated with C9ORF72 expansions. This association is proven best through screening of pathologically confirmed AD cases which would link C9ORF72 expansions to AD as a cause of the disease, if confirmed.
Table 2.
A review of the findings from the current study and other studies on subjects found to carry the C9ORF72 expansion.
| NIH / NINDS C9ORF72 expansion negative |
NIH / NINDS C9ORF72 expansion positive |
DeJesus- Hernandez et al |
Renton et al |
Murray et al | Gijselnick et al |
Stewart et al | Pearson et al |
|
|---|---|---|---|---|---|---|---|---|
| Sample | FTD spectrum patients not enriched for FH or MND |
FTD spectrum patients not enriched for FH or MND |
374 clinical FTLD, out of total of 696 clinical and pathological patients) |
75 Finnish FTLD cases |
20 expansion+ pathological cases from a brain bank |
305 FTLD cases, 137 ALS, 23 with FTD-ALS |
231 ALS patients |
9 expansion+ cases of ALS-FTD in one family |
|
Expansion
prevalence |
49 | 4 8% overall, 3% of sporadic and 15% of familial were expansion + |
3% sporadic and 11.7% familial were expansion+ |
22 (29.3%) expansion+ |
20 |
Familial: 86% of FTD- ALS 47% of ALS 16% of FTLD Sporadic: 6% of FTD- ALS 5% of ALS 4% of FTLD |
3.6% of sporadic, 27.4% of familal |
N.A. |
|
Clinical
Diagnoses |
4 FTD-ALS 27 probable bv-FTD 9 possible FTLD 6 PPA-PNFA 2 PPA- semantic 4 AD 1 MSA |
2 FTD-ALS 2 bv-FTD |
For +: 25 bv-FTD 1 language- variant 7 of the 26 also had ALS |
For +: 8 PPA- PNFA 16 bv-FTD 8 of the 22 had personal or FH of ALS |
8 FTLD 6 ALS 1 FTD-ALS 4 AD 1 Other |
Selected for cases of FTLD, ALS, and FTD-ALS |
Expansion + cases more frequent bulbar onset and FTD-ALS |
5 presented with ALS, 1 with FTD- ALS, and 3 with bv-FTD. 2 developed psychosis, 3 visuo-spatial dysfunction, 4 |
|
Mean age of
onset |
57 | 58 | For +: 56.2 | Overall 58.4 | 61.75 | FTLD: 55.3 in carriers, 63.2 years in non- carriers ALS: 54.5 in carriers, 60.4 years in non- carriers |
For +: 58.2, for -: 57.4 |
42.7 |
|
+FH of a first-
degree relative with dementia or ALS |
15/44 (34%) | % (75%) | For +: 37/52 (71%) |
Overall 27 (36%) |
12/20 (60%) | FTD-ALS: 30% ALS: 10% FTLD: 25% |
For +: 73% for -: 15% |
N.A. |
|
Evidence of
anticipation |
N/A | Probands earlier age of onset than parents |
Not apparent | Not observed |
-- | Trend towards younger age of onset between generations |
-- | -- |
| FDG-PET | -- | 1 typical for FTLD 3 c/w FTLD or AD |
-- | -- | -- | -- | -- | -- |
| Pathology | -- | 1 FTLD-TDP type B with AD pathology |
11 – all FTLD- TDP type B |
-- | FTLD-TDP type B. One case had AD pathology |
FTLD-TDP type B, more ubiquitin than TDP-43+ staining |
ALS with TDP-ir inclusions and FTLD- TDP type B, more ubiquitin than TDP-43+ staining |
FTLD-TDP type B |
Lastly, it is notable that we found two missense mutations, Y294C in GRN and I146V in PSEN2, in two of our hexanucleotide expansion carriers (FTD158 and FTD223) which had not been reported previously. There are several possible interpretations of this finding: 1. These missense mutations are benign and the disease is caused by the C9ORF72 expansion. However, it seems unlikely that this is the case because we found the expansion in a neurologically normal control and also because, although these two missense mutations are novel and have not been previously reported, in silico analysis by Polyphen 2 predicts highest level of pathogenicity for these two variants. Moreover, while GRN missense mutations have not been considered as a common cause of FTLD, PSEN2 missense mutations have been indisputably shown to be pathogenic. 2. The GRN and PSEN2 missense mutations are the primary pathogenic causes of the disease in these two FTD patients and the C9ORF72 expansion contributes to the disease as a risk factor, increasing the susceptibility to the disease. 3. These are two complex cases where two comorbid conditions are caused by two separate lines of genetic mutations, namely the missense mutations on one hand and C9ORF72 expansion on the other hand. If this is the case, the GRN mutation would most probably lead to TDP-43 pathology and the PSEN-2 mutation to the amyloid plaques. These two patients will probably have an additional pathology due to C9ORF72 expansion which is yet to be identified.
To conclude, in this study we report the presence of hexanucleotide repeats in two bv-FTD cases, two FTD-ALS cases (one of which showed, at autopsy, FTLD-TDP type B and AD pathology), and one neurologically normal control from Coriell cell repository. Our findings suggest that the hexanucleotide expansion is probably associated with ALS, FTD or FTD-ALS and occasional comorbid conditions such as Alzheimer’s disease as evident in our pathologically confirmed case 3 (FTD211). These findings are novel and need to be cautiously interpreted and most importantly replicated in larger numbers of samples.
Supplementary Material
Supplementary Table 1 The genotypes of 42 SNPs in the original Finnish risk haplotype on chr9p21 (Laaksovirta et al, 2010) were extracted from Illumina Infinium genotyping platform 660K for 53 patients (first column on the left of the Table on each page). The risk alleles for each SNP are identified by an orange background.
Acknowledgement
PM molecular genetics work is funded by the office of the Dean of the School of Medicine, department of Internal Medicine, at Texas Tech Health Sciences Center. JH work on FTD GWAS is supported partly by a grant from Alzheimer’s Research UK. This research was supported in part by the Intramural Research Programs of the National Institute on Aging and the National Institute of Neurological Disorders and Stroke. EDH’s work is supported by NIH / NINDS grant 5R00NS060766. JG work is supported by the NINDS Intramural Research Program. DG work was supported by NIH grant PHS P30 AG 10133. We would like to thank Mike Hubank and Kerra Pearce at the Genomic core facility at The Institute of Child Health (ICH), University College of London (UCL) for assisting RF in performing illumina genotyping experiments. We would like to Thank Cynthia Crews, Anne Leopold, and Karen DeTucci for the study coordination.
Footnotes
Disclosure statement
All authors disclose no actual or potential conflicts of interest. All human study protocols were approved by the ethics committee of local institutions. An informed consent was obtained from the persons with the power of attorney for the patient.
References
- 1.Arai T, Hasegawa M, Akiyama H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–11. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 2.Borroni B, Bonvicini C, Alberici A, Buratti E, Agosti C, Archetti S, Papetti A, Stuani C, Di Luca M, Gennarelli M, Padovani A. Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum Mutat. 2009 Nov;30(11):E974–83. doi: 10.1002/humu.21100. [DOI] [PubMed] [Google Scholar]
- 3.Boxer AL, Mackenzie IR, Boeve BF, Baker M, Seeley WW, Crook R, Feldman H, Hsiung GY, Rutherford N, Laluz V, Whitwell J, Foti D, McDade E, Molano J, Karydas A, Wojtas A, Goldman J, Mirsky J, Sengdy P, Dearmond S, Miller BL, Rademakers R. Clinical, neuroimaging and neuropathological features of a new chromosome 9p-linked FTD-ALS family. J Neurol Neurosurg Psychiatry. 2011 Feb;82(2):196–203. doi: 10.1136/jnnp.2009.204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corrado L, Ratti A, Gellera C, Buratti E, Castellotti B, Carlomagno Y, Ticozzi N, Mazzini L, Testa L, Taroni F, Baralle FE, Silani V, D’Alfonso S. High frequency of TARDBP gene mutations in Italian patients with amyotrophic lateral sclerosis. Hum Mutat. 2009 Apr;30(4):688–94. doi: 10.1002/humu.20950. [DOI] [PubMed] [Google Scholar]
- 5.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011 Oct 20;72(2):245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrari R, Kapogiannis D, Huey ED, Momeni P. FTD and ALS: a tale of two diseases. Curr Alzheimer Res. 2011 May 1;8(3):273–94. doi: 10.2174/156720511795563700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, Janssens J, Bettens K, Van Cauwenberghe C, Pereson S, Engelborghs S, Sieben A, De Jonghe P, Vandenberghe R, Santens P, De Bleecker J, Maes G, Bäumer V, Dillen L, Joris G, Cuijt I, Corsmit E, Elinck E, Van Dongen J, Vermeulen S, Van den Broeck M, Vaerenberg C, Mattheijssens M, Peeters K, Robberecht W, Cras P, Martin JJ, De Deyn PP, Cruts M, Van Broeckhoven C. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012 Jan;11(1):54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 8.Gilman S, Wenning GK, Low PA, Brooks DJ, Mathias CJ, Trojanowski JQ, Wood NW, Colosimo C, Dürr A, Fowler CJ, Kaufmann H, Klockgether T, Lees A, Poewe W, Quinn N, Revesz T, Robertson D, Sandroni P, Seppi K, Vidailhet M. Second consensus statement on the diagnosis of multiple system atrophy. Neurology. 2008;71:670–676. doi: 10.1212/01.wnl.0000324625.00404.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huey ED, Ferrari R, Moreno JH, Jensen C, Morris CM, Potocnik F, Kalaria RN, Tierney M, Wassermann EM, Hardy J, Grafman J, Momeni P. FUS and TDP43 genetic variability in FTD and CBS. Neurobiol Aging. 2011 Sep 23; doi: 10.1016/j.neurobiolaging.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laaksovirta H, Peuralinna T, Schymick JC, Scholz SW, Lai SL, Myllykangas L, Sulkava R, Jansson L, Hernandez DG, Gibbs JR, Nalls MA, Heckerman D, Tienari PJ, Traynor BJ. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol. 2010 Oct;9(10):978–85. doi: 10.1016/S1474-4422(10)70184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002 Oct 8;59(7):1077–9. doi: 10.1212/wnl.59.7.1077. [DOI] [PubMed] [Google Scholar]
- 12.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984 Jul;34(7):939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 13.Mok K, Traynor BJ, Schymick J, Tienari PJ, Laaksovirta H, Peuralinna T, Myllykangas L, Chiò A, Shatunov A, Boeve BF, Boxer AL, DeJesus-Hernandez M, Mackenzie IR, Waite A, Williams N, Morris HR, Simón-Sánchez J, van Swieten JC, Heutink P, Restagno G, Mora G, Morrison KE, Shaw PJ, Rollinson PS, Al-Chalabi A, Rademakers R, Pickering-Brown S, Orrell RW, Nalls MA, Hardy J. The chromosome 9 ALS and FTD locus is probably derived from a single founder. Neurobiol Aging. 2012 Jan;33(1):209.e3–8. doi: 10.1016/j.neurobiolaging.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita M, Al-Chalabi A, Andersen PM, Hosler B, Sapp P, Englund E, Mitchell JE, Habgood JJ, de Belleroche J, Xi J, Jongjaroenprasert W, Horvitz HR, Gunnarsson LG, Brown RH., Jr A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology. 2006 Mar 28;66(6):839–44. doi: 10.1212/01.wnl.0000200048.53766.b4. [DOI] [PubMed] [Google Scholar]
- 15.Murray ME, DeJesus-Hernandez M, Rutherford NJ, Baker M, Duara R, Graff-Radford NR, Wszolek ZK, Ferman TJ, Josephs KA, Boylan KB, Rademakers R, Dickson DW. Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol. 2011 Dec;122(6):673–90. doi: 10.1007/s00401-011-0907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, Freedman M, Kertesz A, Robert PH, Albert M, Boone K, Miller BL, Cummings J, Benson DF. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 17.Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Neuhaus J, van Swieten JC, Seelaar H, Dopper EG, Onyike CU, Hillis AE, Josephs KA, Boeve BF, Kertesz A, Seeley WW, Rankin KP, Johnson JK, Gorno-Tempini ML, Rosen H, Prioleau-Latham CE, Lee A, Kipps CM, Lillo P, Piguet O, Rohrer JD, Rossor MN, Warren JD, Fox NC, Galasko D, Salmon DP, Black SE, Mesulam M, Weintraub S, Dickerson BC, Diehl-Schmid J, Pasquier F, Deramecourt V, Lebert F, Pijnenburg Y, Chow TW, Manes F, Grafman J, Cappa SF, Freedman M, Grossman M, Miller BL. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011 Sep;134(Pt 9):2456–77. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M, ITALSGEN Consortium. Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011 Oct 20;72(2):257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Troakes C, Maekawa S, Wijesekera L, Rogelj B, Siklós L, Bell C, Smith B, Newhouse S, Vance C, Johnson L, Hortobágyi T, Shatunov A, Al-Chalabi A, Leigh N, Shaw CE, King A, Al-Sarraj S. An MND/ALS phenotype associated with C9orf72 repeat expansion: Abundant p62-positive, TDP-43-negative inclusions in cerebral cortex, hippocampus and cerebellum but without associated cognitive decline. Neuropathology. 2011 Dec 19; doi: 10.1111/j.1440-1789.2011.01286.x. [DOI] [PubMed] [Google Scholar]
- 20.Vance C, Al-Chalabi A, Ruddy D, Smith BN, Hu X, Sreedharan J, Siddique T, Schelhaas HJ, Kusters B, Troost D, Baas F, de Jong V, Shaw CE. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2-21.3. Brain. 2006 Apr;129(Pt 4):868–76. doi: 10.1093/brain/awl030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 The genotypes of 42 SNPs in the original Finnish risk haplotype on chr9p21 (Laaksovirta et al, 2010) were extracted from Illumina Infinium genotyping platform 660K for 53 patients (first column on the left of the Table on each page). The risk alleles for each SNP are identified by an orange background.