Abstract
Unmet needs in prostate cancer drug development and patient management are the ability to monitor treatment benefit and to identify the target of interest in a tumor at the time treatment is being considered. This review focuses on establishing analytical valid biomarkers for specific contexts of use in patients with castration-resistant prostate cancer (CRPC), emphasizing a biomarker currently in clinical use, circulating tumor cells (CTC). The Oncology Biomarker Qualification Initiative provides a road map for these investigations, which, if followed, will facilitate the incorporation of these types of assays into clinical decision-making.
CTC enumeration at baseline and post-treatment is prognostic of survival, with no threshold effect, and the shedding of cells into the circulation represents an intrinsic property of the tumor, distinct from extent of disease. The clinical utility of monitoring CTC changes with treatment as an efficacy-response surrogate biomarker of survival is currently being tested in large phase III trials with the novel antiandrogen therapies abiraterone acetate and MDV3100.
Molecular biomarkers can be characterized in CTC as potential predictive biomarkers of tumor sensitivity to a therapeutic modality. Additionally, we discuss novel technologies to enrich and characterize CTC from more patients, and the potential clinical uses of CTC's in determining prognosis and monitoring treatment effects, and as a source of tissue to identify predictive markers of drug sensitivity to guide treatment selection. Prospective studies, designed around the biomarker itself and the specific clinical context for which it is applied, are needed to further assess the role of these and novel markers in clinical practice.
Introduction
Prostate cancer demonstrates distinctive difficulties in drug development and patient management. This is because the standard imaging modalities used to assess disease in bone, the most common site of spread, have not been standardized and because the directional change in prostate-specific antigen (PSA) levels, the most frequently altered biomarker in the disease, may not reflect the status of disease accurately (1, 2). Up to 20% of men with castration-resistant prostate cancer (CRPC) who eventually respond to a systemic cytotoxic therapy proven to prolong life have an initial PSA rise before the decline (3, 4); the decline may not occur for up to 12 weeks (5) or not occur at all with immunomodulatory agents proven to prolong life and postulated to slow the disease trajectory through effects on the tumor microenvironment (eg, sipuleucel-T) (6). It is therefore not surprising that the association between a given post-therapy change in PSA and survival is modest, and thus appropriately is not accepted by regulatory agencies for drug approvals (7, 8). Finding more reliable indicators of disease status is a critical unmet need. While it has been particularly encouraging to see several drugs with diverse mechanisms show a survival benefit in phase III trials (9), it is also clear that these agents are active in subsets of patients, and that there remain distinct cohorts that are resistant to therapy de novo. This, coupled with the recognition that the molecular determinants of a tumor that contribute to growth may change as the disease progresses, presents the additional unmet need to profile an individual patient's tumor at the time treatment is considered (10). It is our hypothesis that specific blood-based CTC assays that are obtained in the context of routine clinical practice can fulfill these unmet needs.
Before the role of any assay in medical decision-making can be determined, it is essential that the assay itself meet specific and rigorous performance requirements. This review is focused on the process of establishing the analytical validity of CTC biomarkers for specific contexts of use in patients with CRPC, looking first at what is currently available, and next at other assays that are at different points in the development process. For a discussion of bone turnover, urinary, and tissue-based biomarkers, the reader is referred to key reviews (11-16).
A Disease Framework
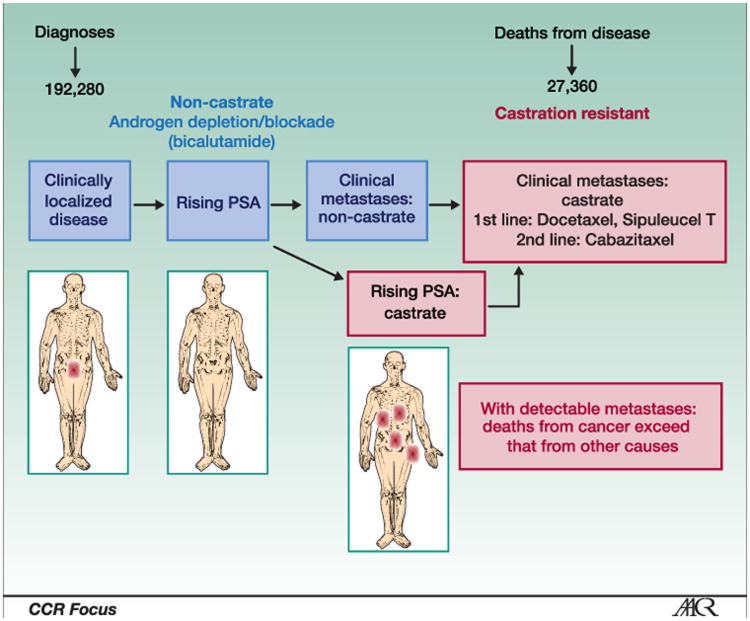
Prostate cancer is a heterogeneous disease that can span decades in some or be lethal in a relatively short time in others. The clinical states model provides a framework to reduce heterogeneity and define therapeutic objectives for an individual patient or group of patients, and to guide drug development (17, 18). This model is the basis of the Prostate Cancer Working Group 2 (PCWG2) guidelines (Figure 1) (8). Importantly, the framework incorporates established standards of care and enables clinical trial questions to focus on the unmet needs for specific patient groups at different points in the disease continuum. The PCWG2 guidelines also recommend reporting outcomes based on changes in each disease manifestation (eg, PSA, soft tissue, and bone) individually. That recommendation enables each parameter to be studied alone or in combination with other measures in relation to a clinical outcome.
Figure 1.
A clinical states framework for clinical practice, clinical research, and biomarker development in prostate cancer, which is the basis of the National Consensus Criteria for clinical trials in this disease. Adapted from Scher and Heller with permission from Elsevier (17).
Contexts of Use for Biomarkers
The road map for biomarker development is outlined in the Oncology Biomarker Qualification Initiative (OBQI), a collaboration between the U.S. Food and Drug Administration (FDA), Centers for Medicare & Medicaid Services (CMS), and National Cancer Institute (NCI), described in the FDA Critical Path Initiative (19). It requires a longitudinal process that starts with developing a robust assay, which is analytically validated across multiple systems and laboratories, and separately, prospective trials developed for the specific biomarker question(s) for the context of use in which the biomarker is being evaluated. A draft guidance was recently issued by the FDA (20). The specific contexts of use where qualified biomarkers would influence medical decisions include:
Detection, use of the biomarker to establish a diagnosis;
Prognosis, measuring the probability of a specific clinical outcome, such as recurrence, progression, or survival;
Prediction, identifying the chance of response to a specific therapy;
Response-indicator biomarkers demonstrate a pharmacologic or physiologic response from the treatment (eg, a decline in PSA), which does not necessarily mean that the patient has benefitted from a treatment;
Efficacy-response biomarkers are surrogates of how a patient feels or functions or how long he survives, extrapolating the clinical benefit;
Treatment resistance biomarkers define biologic determinants of failure or progression, such as second site mutations.
Analytical validation establishes the performance characteristics of an assay and the range of conditions under which an assay gives reproducible and accurate data. In this process, it requires rigorous performance testing to meet Clinical Laboratory Improvement Amendments (CLIA) regulatory requirements. Validation requires rigorous performance that can be considered in three steps: 1) pre-analytical assessment of specimen selection, handling, processing, and storage parameters; 2) validation of analytical characteristics to meet CLIA regulatory requirements, establishing the inter- and intra- assay precision, linearity, analyte recovery, and standardization and developing a comprehensive quality control program; 3) post-analytical parameters require data management and storage.
Clinical qualification, distinct from the analytical validation, is the evidentiary process of linking a biomarker with biological processes or clinical endpoints in the context of an intended use to inform a medical decision as described in the FDA Critical Path (19, 21).
As there are more biomarkers than questions, and given the effort and cost to conduct these investigations, it is essential to develop metrics to determine which biomarkers that appear promising warrant prospective testing in large-scale phase III trials to establish qualification.
Enumerating Circulating Tumor Cells
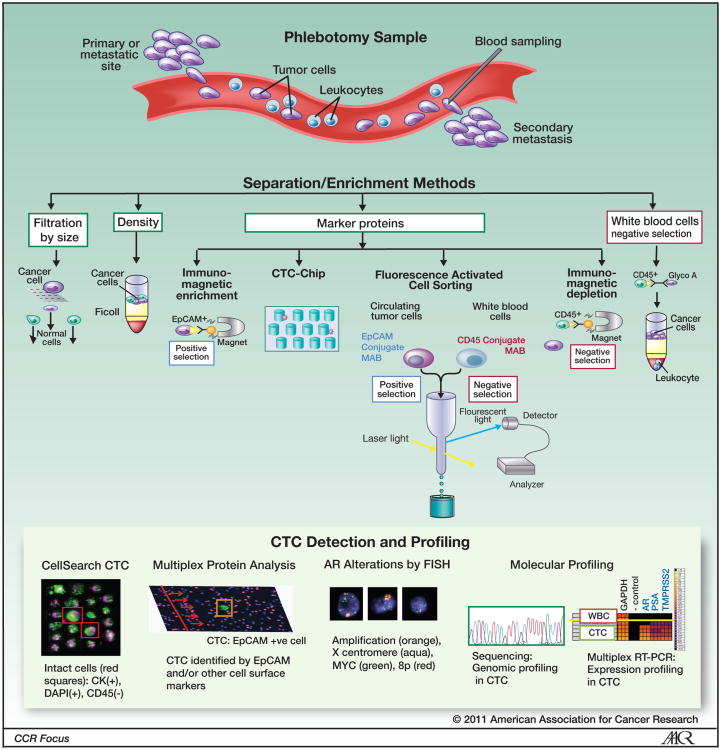
Circulating tumor cells (CTC) are rare events in the peripheral blood of patients with a variety of metastatic carcinomas, and are currently estimated to account for 1 cell in a billion nucleated cells. In contrast to invasive procedures like biopsies, a blood test for CTC is safe and can be performed frequently (Figure 2).
Figure 2.
Circulating tumor cells in patients with prostate cancer. Sampled by phlebotomy at the time when treatment is being considered, CTC have the potential to provide tumor material for molecular profiling for biomarkers informative of tumor sensitivity to the targeted therapy being considered. FISH images reproduced from Leversha et al (15).
Numerous CTC isolation and capture techniques have been reported, but only one method, CellSearch® (Veridex LLC, Raritan, NJ), is analytically valid (22) and FDA cleared for use. The assay enriches CTC based on antibodies to epithelial cell adhesion molecule (EpCAM) conjugated to magnetic beads, which are further classified as CTC based on morphologic limits and rigorous criteria for staining for cytokeratin (CK-6, 8, 18), displays a nucleus (DAPI), and excludes white blood cell (CD-45 staining). The automatically selected images are then reviewed by an operator who makes the final identification. The results are reported as the number of cells meeting the definition per 7.5 mL of blood (Figure 2). CTC isolated by this method from patients with CRPC have been demonstrated to exhibit features of prostate cancer, expressing PSA and alpha-methylacyl-CoA racemase (AMACR), and prostate-specific genomic abnormalities such as androgen receptor (AR) gene copy number amplifications, phosphatase and tensin homolog (PTEN) deletions, and TMPRSS:ETV fusion products (23, 24). It is perhaps not sufficiently appreciated in reports using the CellSearch technology that the proportion of cells visible in the chamber meeting the strict criteria defining a CTC represents only 1-10% of the cells actually present (Figure 2). Cellular fragments, anucleated cells, and necrotic cells are not counted using the FDA cleared definition. A recent report suggested that the sensitivity of the test can be increased if EpCAM+CK+CD45- events are counted and that they are prognostic for survival.(25) This finding, however, will require independent analytical validation and clinical qualification before it can be incorporated into clinical practice.
Based on a series of prospective trials enrolling patients with progressive breast, colorectal, and prostate cancer, the CellSearch assay received FDA clearance as an aid to the monitoring disease status that is to be used in conjunction with other modalities, in patients with metastatic breast (26-29), colorectal (30, 31), and prostate cancer (32, 33). An important finding for prostate cancer was that only a modest association between the number of cells isolated and overall disease burden (based on the level of PSA and burden of disease in bone) was noted, showing that the number of cells isolated reflected an intrinsic property of an individual patient's tumor (32-34). As such, the test provides unique information. Overall, more cells are isolated from more patients with bone and visceral metastases, compared to patients with lymph node disease alone, consistent with the known routes of seeding by hematogenous versus lymphatic spread, respectively (32, 34).
Studying patients with progressive metastatic breast, colon, or prostate cancer who were about to start a new line of chemotherapy showed that CTC number is prognostic pre-therapy as well as post-therapy, using discrete disease-specific cut-off values (≥5 CTC/7.5 ml of blood vs. ≤4 CTC) to define unfavorable and favorable groups (27, 30, 32, 33). In patients with CRPC, post-treatment CTC number was a stronger prognostic factor for survival than a 50% decline in PSA (receiver operating characteristic [ROC] AUC 0.87 vs. 0.62) (32, 33).
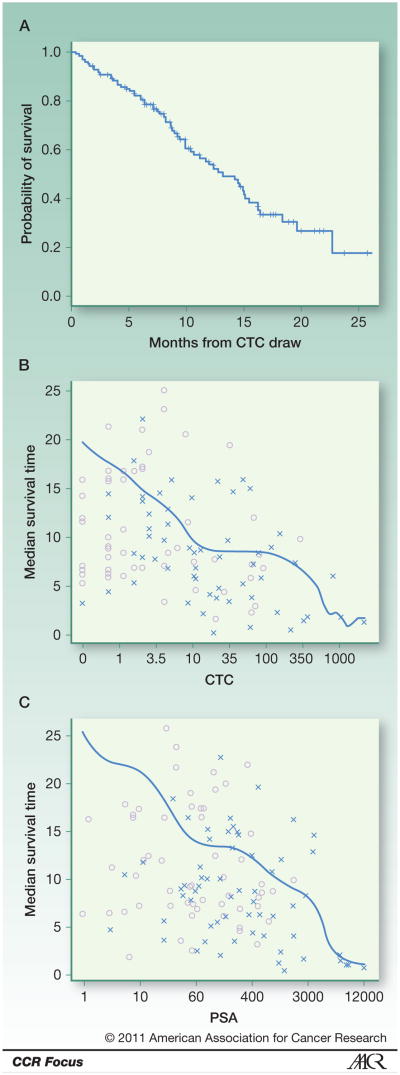
In a separate cohort of patients treated at Memorial Sloan-Kettering Cancer Center (MSKCC), CTC number considered as a continuous variable was also shown to be an independent prognostic factor for survival with no threshold effect (Figure 3) (34). This argues against the use of discrete CTC cutoff values in decision making, a finding confirmed in a reanalysis of patients included in the IMMC38 prostate cancer study (trial registration ID: NCT00133900) upon which the prostate cancer clearance was based (33). For this analysis, baseline pre-treatment variables associated with high risk of death were high lactate dehydrogenase (LDH) concentration (HR 6.44, 95% confidence interval [CI] 4.24-9.79), high CTC count (HR 1.58, 95% CI 1.41-1.77), high PSA levels (HR 1.26, 95% CI 1.10-1.45), low albumin (HR 0.10, 95% CI 0.03-0.39), and low hemoglobin (HR 0.72, 95% CI 0.64-0.81). In the post-treatment analysis at 4 weeks, 8 weeks, and 12 weeks, the changes in CTC number from baseline were strongly associated with risk, whereas changes in PSA were weakly or not associated. The most prognostic factors for survival were baseline LDH levels and the fold change in CTC count from baseline to the follow-up time of measurement (concordance probability estimate 0.72-0.75) (33). The finding was consistent at baseline, 4, 8, and 12 weeks post-therapy, which suggests that CTC number measured at 4 or 8 weeks following treatment provides important prognostic information and can potentially be used as an indicator of a favorable or unfavorable outcome with therapy. In these analyses, the CTC biomarker (baseline or fold change from baseline) was more prognostic than PSA, significantly improving on prior models used to predict the overall survival probability among patients with metastatic CRPC (35).
Figure 3.
CTC number as a biomarker prognostic of overall survival in patients with CRPC. The estimated median survival time decreased continuously with increasing CTC number, such as Kaplan-Meier estimate of survival calculated from time of CTC draw (A), estimated median survival time based on baseline CTC counts (B) and PSA (C). The symbol (x) represents death times and (o) represents the last follow-up times. Reproduced from Danila et al (34).
Data from phase II investigations, while often encouraging, do not establish surrogacy. Now needed are large-scale phase III trials in which a survival benefit is demonstrated and with the CTC biomarker question embedded. Such a stepwise approach was studied in the context of the clinical development in CRPC of abiraterone acetate, a17α-hydroxilase and 17,20-lyase inhibitor that inhibits androgen biosynthesis in the testis, adrenal gland, and prostate tumor (9). Based on activity in a phase I study (trial registration ID: NCT00473512) (36), the CTC enumeration biomarker was incorporated in two independent phase II trials at MSKCC (trial registration ID: NCT00485303) and the Royal Marsden Hospital (trial registration ID: NCT00474383). Patients with CRPC progressing after docetaxel-based chemotherapy were treated with abiraterone acetate with baseline CTC frequencies of ≥5 cells/7.5 mL of 70% (95% CI 54-81) and 79% (95% CI 63-90) of patients, and a similar post-therapy conversion rate from unfavorable to favorable counts of 34% (95% CI 20-53) and 41% (95% CI 25-59) in the two trials, respectively (37, 38). Similar post-therapy CTC conversion rate of 49% (95% CI 35-63) were found in a phase I/II trial (trial registration ID: NCT00510718) of patients with CRPC treated with MDV3100, a novel AR antagonist specifically developed by investigators at MSKCC for activity in prostate cancer cells with overexpressed AR (39). These findings speak to the analytical validity of the assay and formed the basis for exploring CTC enumeration as a potential efficacy-response (surrogate) biomarker of survival for AR-targeted therapies in patients with CRPC progressing post-docetaxel treatment.
Through a longstanding relationship with the FDA, CTC enumeration has entered the first formal collaborative effort to prospectively establish it as an efficacy-response biomarker. As discussed with the Biomarker Qualification Review Team (“A Voluntary Data Submission to Support the Qualification of Circulating Tumor Cells (CTCs) as an Efficacy-response Biomarker in Castration Resistant Prostate Cancer (CRPC),” December 2009), the statistical analysis plan toward qualification of CTC number as an efficacy-response surrogate for overall survival (OS) requires the biomarker to fully capture the net effect of treatment on OS (40), in multiple prospective trials (41). The data analysis of the first randomized phase III trial of patients with post-chemotherapy treated CRPC randomized to receive abiraterone acetate vs. placebo (trial registration ID: NCT00638690), which incorporated a CTC biomarker question, was discussed in a face to face meeting at Center for Drug Evaluation and Research (CDER). The trial, presented at the 2010 European Society of Medical Oncology meeting, showed prolonged survival (HR 0.65, 95% CI 0.54-0.77), time to PSA progression (HR 0.58, 95% CI 0.46-0.73), and radiographic progression-free survival (HR 0.67, 95% CI 0.58-0.78) (42). A second randomized phase III trial treating patients with progressive CRPC post-docetaxel with MDV3100 (trial registration ID: NCT00974311), and including an embedded CTC biomarker as an efficacy-response question, has completed accrual. A similar statistical analysis is planned to explore the relationship between CTC numbers and clinical outcome, according to PCWG2 criteria.
The successful completion of this qualification process will demonstrate the clinical utility of monitoring CTC changes with treatment as an intermediate endpoint for detecting survival benefit from AR-targeted therapies. To extend the context of use, additional surrogacy analyses are embedded in phase III randomized survival-based trials which have the CTC biomarker question embedded, such as TAK-700 (trial registration ID: NCT01193257), and immunotherapy-based therapies such as ipilimumab (trial registration ID: NCT01057810). In addition to an efficacy-response biomarker qualification effort, changes in CTC have been proposed as a pharmacodynamic readout of immune-based therapies, such as the recently FDA-approved vaccine therapy with sipuleucel-T for patients with CRPC (43).
Molecular profiling of CTC
In addition to providing prognostic information and a potential indicator of efficacy, CTC have the potential to provide a snapshot of the molecular makeup of an individual patient's tumor to profile for determinants that predict for sensitivity or resistance to treatment. The molecular determinants that contribute to tumor growth can change during the course of disease. Consequently, to effectively deliver the appropriate targeted approach for an individual patient, it is essential to profile the tumor at the time of treatment decision. Doing so is currently limited by the lack of reliable assays for the biomarkers being studied, and the difficulty in obtaining representative tumor samples in a routine clinical practice setting (44). CTC isolated from the peripheral blood, or disseminated tumor cells from the bone marrow (45, 46) of patients with prostate cancer at any stage are of particular interest because they have the potential to provide tumor material representative of a molecular snapshot of the disease.
Though early work suggested that TMPRSS2-ERG rearrangements are sufficient to initiate prostate neoplasia (24, 47), as are linked ETS rearrangements for more aggressive disease (47), the reported clinical significance of TMPRSS2-ETS rearrangements has been inconsistent. In preclinical animal models, aberrant expression of ERG alone was insufficient to initiate transformation by itself (48), but it was sufficient when concomitant with PI3-kinase activation or PTEN loss (49). Additionally, copy number increase of chromosome 21, with and without rearrangement for TMPRSS2, was associated with high Gleason grade and advanced stage, reflecting generalized aneuploidy (48, 50). Unlike KRAS mutations predicting primary resistance to epidermal growth factor receptor (EGFR) target therapy, there are no known markers predictive of primary resistance in prostate cancer. AR amplification, although infrequent in primary/diagnostic tumor specimens, is detected in upwards of 50% of castration-resistant lesions (10, 51). AR genomic amplification and copy number gain have been documented in CTC from patients with CRPC with frequencies similar to that reported for late-stage tumors (24, 52). Our initial exploratory analysis indicated that AR amplification and copy gain occurring under the selective pressure of androgen deprivation therapy may represent a marker of sensitivity to a second-generation AR antagonist (53). Additionally, cells that have initiated true amplification events also appear to develop the increased chromosomal instability, as demonstrated by additional MYC, TMPRSS-ETV, and PTEN abnormalities (24, 52). Similarly, copy number alterations have been strongly associated with more aggressive tumors, as described in the ongoing comprehensive analysis in the Prostate Oncogenomics Project at MSKCC (54). The prognostic value of HER2/neu, EGFR and IGF-1R expression in CTC is being currently studied in the context of clinical trials with novel therapies targeting these markers, as a potential tumor-sensitivity predictive biomarker, as well as a pharmacodynamic measurement for defining the optimal dose beyond the maximum tolerated dose (23, 52, 55, 56). Although promising, these results still require a rigorous validation across independent cohorts of patients in the appropriate context of proposed use, as currently embedded in larger biomarker efficacy trials.
Moving beyond CellSearch
According to the FDA/OBQI Critical Path Initiative, each new assay needs to be analytically validated and clinically qualified in the intended context of use where the biomarker is tested for the power of association with a specific clinical outcome. Most studies with CellSearch in prostate cancer have evaluated patients with CRPC. CRPC includes patients who have progressed on one or more hormonal therapies, those who are about to start a cytotoxic drug, and those who have progressed in multiple therapies (Figure 1). Table 1 shows the frequency of favorable and unfavorable counts for these patient groups, highlighting a need to develop assays that detect cells in more patients and at a higher frequency (32-34). Based on this, other technologies are being developed to increase CTC detection rates and to obtain more highly enriched samples, such as filtration (38, 57), microfluidics (58-60) and fluorescence activated cell sorting (FACS) (61) (Table 1). Through optimization of the flow in the capturing device, recent CTC-chips have improved detection of CTC in early stages of prostate cancer, including in the context of localized disease (58, 59).
Table 1.
CTC enrichment and characterization techniques.
| Method | Technology | Comments |
|---|---|---|
| Magnetic separation based on Epithelial cell adhesion molecule (EpCAM)-Ab coupled ferrofluid | CellSearch system | CTC are characterized based on rigorous morphological criteria, based on staining for cytokeratin (CK-6, 8, 18), nucleus (DAPI), and excluding white blood cell (CD-45 staining). The only FDA-cleared CTC technology as an aid in monitoring of patients with metastatic breast (26-29), colorectal (30, 31), and prostate cancer (32, 33). |
| MagSweeper | Exploratory enrichment method based on ferro-magnetic conjugated antibodies (62). | |
| Antibody coupled microposts | EpCAM-based separation | Microposts and herring-bone CTC-chips allow sensitive and selective detection of CTC for enumeration and further molecular profiling at genomic, transcriptional and translational levels. (58, 59, 63). |
| Prostate-specific membrane antigen (PSMA)-based separation | Isolation of CTC based on PSMA expression allows enrichment of EpCAM-negative tumor cells (60). Staggered obstacles optimize cell-size dependent flow to maximize enrichment of CTC. | |
| Negative selection | RosetteSep-Applied Imaging Rare Event (RARE) detection method | Depletes erythrocytes and CD-45 positive white blood cells, agnostically negatively enriching tumor cells (64). |
| RT-PCR | Prostate specific gene expression profiling | PSA mRNA detection in peripheral blood of patients with prostate cancer (65-67). |
| Size-based isolation | Micro-pores filtration | Isolation based on micro-filtration separates CTC based on size (38, 57, 68) |
| Imaging based-methods | EPISPOT assay | Immunospot-based method of detecting CTC based on secretion of proteins (PSA from prostate CTC) (69). |
| Fluorescence activated cell sorting | Multimarker cell analysis and isolation for further molecular profiling. (61, 70, 71) | |
| Scan-microscopy | High-speed scanning for detection of CTC by fluorescence microscopy (72, 73). |
The results obtained are typically compared to CellSearch as the standard technology, and all of these approaches detect cells in patients with unfavorable counts, with the recognition that the “definition” of a CTC is different for these assays. Further analytical validation will be required before they can be considered for clearance or qualification in any disease context (Figure 2). Little appreciated, however, is that different CTC assay technologies are not measuring exactly the same biomarker, and thus each new assay is in essence proposing a new definition of a CTC, as there is currently no “standard.”
Our group developed and validated a FACS-based method which isolates and captures EpCAM+, CD45− cells from a mononucleated layer separated through Ficoll-Hypaque gradient from blood collected from patients with CRPC. DAPI is used as a vital stain to exclude permeable and apoptotic cells. The cells isolated by FACS were shown by multiplex RT-PCR to express prostate-specific mRNAs (PSA, AR, TMPRSS2), indicating that these EpCAM+ events are bona fide CTCs. In parallel samples collected from 124 patients with metastatic CRPC, this FACS-based method isolated an average 100-fold more putative CTC than does CellSearch, and more CTC could be isolated in a larger cohort of patients: >50 events/sample in 58% vs. 10% of patients, or >10 events/sample in 88% vs.32%, by FACS vs CellSearch, respectively (Table 2). Of particular interest is that these approaches have the potential to enable in-depth molecular profiling in CTC in chemotherapy-naïve patients where isolating CTC with higher purity will facilitate the study of gene signature patterns prognostic or predictive of sensitivity to a targeted drug, or define the mechanism of acquired resistance that occurs under the selective pressure of a targeted therapy. Additionally, these sensitive technologies may allow clinical testing for the risk of relapse with distant metastasis based on shedding of CTC at time of primary treatment of the prostate cancer, and guide the decision on adjuvant therapy based on post-treatment CTC enumeration.
Table 2.
Flow cytometric CTC enrichment captures more events. In a cohort of 124 patients with CRPC, FACS sorting and analysis of EpCAM+ve, CD45–ve, DAPI–ve events increased the detection rate and the absolute number of events compared to CellSearch enrichment.
| FACS event range | Total N=124 | Chemotherapy exposure | ||
|---|---|---|---|---|
| Pre N=33 | 1st Line N=27 | 2nd Line N=64 | ||
| 0 – 4 | 8 (6%) | 6 (18%) | 2 (7%) | 0 (0%) |
| ≥ 5 - 9 | 7 (6%) | 2 (6%) | 1 (4%) | 4 (6%) |
| ≥ 10 | 109 (88%) | 25 (75%) | 24 (89%) | 60 (94%) |
| > 50 | 72 (58%) | 11 (33%) | 19 (70%) | 42 (66%) |
| CellSerach Events “Meets CTC Definition” | Total N=124 | Chemotherapy exposure | ||
|---|---|---|---|---|
| Pre N=33 | 1st Line N=27 | 2nd Line N=64 | ||
| 0 – 4 | 74 (60%) | 27 (82%) | 13 (49%) | 34 (54%) |
| ≥ 5 - 9 | 10 (8%) | 3 (9%) | 2 (8%) | 5 (7%) |
| ≥ 10 | 40 (32%) | 3 (9%) | 12 (45%) | 25 (39%) |
| > 50 | 13 (10%) | 1 (3%) | 7 (26%) | 5 (8%) |
Future Directions
The approval of new therapies for patients with prostate cancer based on overall survival requires large cohorts of patients with lengthy follow-up. Qualified biomarkers for discrete contexts of use have the potential to shorten the drug development process. To do so requires validated assays, and, separately, the design and conduct of prospective clinical trials that generate evidence toward the qualification. More significant power to discriminate between low and high risk of a specific outcome could be obtained by combining multiple biomarkers, such as LDH, albumin, and PSA with baseline CTC (33, 34), which will ultimately result in a “basket” biomarker meant to influence medical decision-making.
Since it is not clear which biomarker will be most informative for a medical decision to address a specific context of use, there must be a process for prioritizing biomarker development. A biomarker should be tested in phase I/II efficacy trials to measure the robustness of its association with predicted clinical outcome in the selected context of use, before beginning full development of the biomarker in large, costly phase III trials.
Acknowledgments
Financial support: Supported by: The Sidney Kimmel Center for Prostate and Urologic Cancers. Supported in part by NCI SPORE in Prostate Cancer (P50 CA92629); Prostate Cancer Foundation; Department of Defense Prostate Cancer Research Program Physician Research Award W81XWH-09-1-0307; Department of Defense – Prostate Cancer Clinical Trials Consortium.
References
- 1.Scher HI, Morris MJ, Kelly WK, Schwartz LH, Heller G. Prostate cancer clinical trial end points: “RECIST”ing a step backwards. Clin Cancer Res. 2005;11:5223–32. doi: 10.1158/1078-0432.CCR-05-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newling DWW. Issues with the use of prostate-specific antigen as a surrogate end point in hormone-resistant prostate cancer. Eur Urol Suppl. 2009;8:13–9. [Google Scholar]
- 3.Berthold DR, Pond GR, Roessner M, de Wit R, Eisenberger M, Tannock AI. Treatment of hormone-refractory prostate cancer with docetaxel or mitoxantrone: relationships between prostate-specific antigen, pain, and quality of life response and survival in the TAX-327 study. Clin Cancer Res. 2008;14:2763–7. doi: 10.1158/1078-0432.CCR-07-0944. [DOI] [PubMed] [Google Scholar]
- 4.Madan RA, Pal SK, Sartor O, Dahut WL. Overcoming chemotherapy resistance in prostate cancer. Clin Cancer Res. 2011;17 doi: 10.1158/1078-0432.CCR-10-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleming MT, Morris MJ, Heller G, Scher HI. Post-therapy changes in PSA as an outcome measure in prostate cancer clinical trials. Nature Clin Pract Oncol. 2006;3:658–67. doi: 10.1038/ncponc0664. [DOI] [PubMed] [Google Scholar]
- 6.Gulley JL, Drake CG. Immunotherapy for prostate cancer: recent advances, lessons learned, and areas for further research. Clin Cancer Res. 2011;17 doi: 10.1158/1078-0432.CCR-10-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bubley GJ, Carducci M, Dahut W, Dawson N, Daliani D, Eisenberger M, et al. Eligibility and response guidelines for phase II clinical trials in androgen-independent prostate cancer: recommendations from the Prostate-Specific Antigen Working Group. J Clin Oncol. 1999;17:3461–7. doi: 10.1200/JCO.1999.17.11.3461. [DOI] [PubMed] [Google Scholar]
- 8.Scher HI, Halabi S, Tannock I, Morris M, Sternberg C, Carducci MA, et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: Recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol. 2008;26:1148–59. doi: 10.1200/JCO.2007.12.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massard C. Targeting continued androgen receptor signaling in prostate cancer. Clin Cancer Res. 2011;17 doi: 10.1158/1078-0432.CCR-10-2815. Clin Cancer Res 2011;17. [DOI] [PubMed] [Google Scholar]
- 10.Holzbeierlein J, Lal P, LaTulippe E, Smith A, Satagopan J, Zhang L, et al. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am J Pathol. 2004;164:217–27. doi: 10.1016/S0002-9440(10)63112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Detchokul S, Frauman AG. Recent developments in prostate cancer biomarker research: therapeutic implications. Br J Clin Pharmacol. 2011;71:157–74. doi: 10.1111/j.1365-2125.2010.03766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang SY, Adelstein J, Kassis AI. Putative molecular signatures for the imaging of prostate cancer. Expert Rev Mol Diagn. 2010;10:65–74. doi: 10.1586/erm.09.73. [DOI] [PubMed] [Google Scholar]
- 13.LeBeau AM, Kostova M, Craik CS, Denmeade SR. Prostate-specific antigen: an overlooked candidate for the targeted treatment and selective imaging of prostate cancer. Biol Chem. 2010;391:333–43. doi: 10.1515/BC.2010.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rioja J, Rodriguez-Fraile M, Lima-Favaretto R, Rincon-Mayans A, Penuelas-Sanchez I, Zudaire-Bergera JJ, et al. Role of positron emission tomography in urological oncology. BJU Int. 2010;106:1578–93. doi: 10.1111/j.1464-410X.2010.09510.x. [DOI] [PubMed] [Google Scholar]
- 15.Messiou C, Cook G, deSouza NM. Imaging metastatic bone disease from carcinoma of the prostate. Br J Cancer. 2009;101:1225–32. doi: 10.1038/sj.bjc.6605334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ploussard G, de la Taille A. Urine biomarkers in prostate cancer. Nat Rev Urol. 2010;7:101–9. doi: 10.1038/nrurol.2009.261. [DOI] [PubMed] [Google Scholar]
- 17.Scher HI, Heller G. Clinical states in prostate cancer: towards a dynamic model of disease progression. Urology. 2000;55:323–7. doi: 10.1016/s0090-4295(99)00471-9. [DOI] [PubMed] [Google Scholar]
- 18.Scher HI, Sawyers C. Biology of progressive castration resistant prostate cancer: directed therapies targeting the androgen receptor signaling axis. J Clin Oncol. 2005;23:8253–61. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 19.FDA/NCI/CMS Oncology Biomarker Qualification Initiative. Memorandum of understanding between the FDA, NCI, and CMS for the FDA/NCI/CMS Oncology Biomarker Qualification Initiative. [Accessed March 16, 2011];Document MOU 225-06-8001. 2006 Jan; Last updated April 30,2009 http://www.fda.gov/AboutFDA/PartnershipsCollaborations/MemorandaofUnderstandingMOUs/DomesticMOUs/ucm115681.htm.
- 20.Center for Drug Evaluation and Research (CDER), Food and Drug Adminstration (FDA) [Accessed March 26 2011];Draft guidance for industry: qualification process for drug development tools. 2010 Oct; http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM230597.pdf.
- 21.Altar CA. The Biomarkers Consortium: on the critical path of drug discovery. Clin Pharmacol Ther. 2008;83:361–4. doi: 10.1038/sj.clpt.6100471. [DOI] [PubMed] [Google Scholar]
- 22.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 23.Shaffer DR, Leversha MA, Danila DC, Lin O, Gonzalez-Espinoza R, Gu B, et al. Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:2023–9. doi: 10.1158/1078-0432.CCR-06-2701. [DOI] [PubMed] [Google Scholar]
- 24.Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A'Hern R, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–8. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 25.Coumans FA, Doggen CJ, Attard G, de Bono JS, Terstappen LW. All circulating EpCAM+CK+CD45- objects predict overall survival in castration-resistant prostate cancer. Ann Oncol. 2010;21:1851–7. doi: 10.1093/annonc/mdq030. [DOI] [PubMed] [Google Scholar]
- 26.Budd GT, Cristofanilli M, Ellis MJ, Stopeck A, Borden E, Miller MC, et al. Circulating tumor cells versus imaging--predicting overall survival in metastatic breast cancer. Clin Cancer Res. 2006;12:6403–9. doi: 10.1158/1078-0432.CCR-05-1769. [DOI] [PubMed] [Google Scholar]
- 27.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 28.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005;23:1420–30. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 29.Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12:4218–24. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 30.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–21. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 31.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann Oncol. 2009;20:1223–9. doi: 10.1093/annonc/mdn786. [DOI] [PubMed] [Google Scholar]
- 32.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–9. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 33.Scher HI, Jia X, de Bono JS, Fleisher M, Pienta KJ, Raghavan D, et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233–9. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053–8. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 35.Halabi S, Small EJ, Kantoff PW, Kattan MW, Kaplan EB, Dawson NA, et al. Prognostic model for predicting survival in men with hormone-refractory metastatic prostate cancer. J Clin Oncol. 2003;21:1232–7. doi: 10.1200/JCO.2003.06.100. [DOI] [PubMed] [Google Scholar]
- 36.Attard G, Reid AH, A'Hern R, Parker C, Oommen NB, Folkerd E, et al. Selective inhibition of CYP17 with abiraterone acetate is highly active in the treatment of castration-resistant prostate cancer. J Clin Oncol. 2009;27:3742–8. doi: 10.1200/JCO.2008.20.0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danila DC, Morris MJ, de Bono JS, Ryan CJ, Denmeade SR, Smith MR, et al. Phase II multicenter study of abiraterone acetate plus prednisone therapy in patients with docetaxel-treated castration-resistant prostate cancer. J Clin Oncol. 2010;28:1496–501. doi: 10.1200/JCO.2009.25.9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reid AH, Attard G, Danila DC, Oommen NB, Olmos D, Fong PC, et al. Significant and sustained antitumor activity in post-docetaxel, castration-resistant prostate cancer with the CYP17 inhibitor abiraterone acetate. J Clin Oncol. 2010;28:1489–95. doi: 10.1200/JCO.2009.24.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scher HI, Beer TM, Higano CS, Anand A, Taplin ME, Efstathiou E, et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1-2 study. Lancet. 2010;375:1437–46. doi: 10.1016/S0140-6736(10)60172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prentice RL. Surrogate endpoints in clinical trials: definition and operational criteria. Stat Med. 1989;8:431–40. doi: 10.1002/sim.4780080407. [DOI] [PubMed] [Google Scholar]
- 41.Collette S, Collette L, Budiharto T, Horiot JC, Poortmans PM, Struikmans H, et al. Predictors of the risk of fibrosis at 10 years after breast conserving therapy for early breast cancer: a study based on the EORTC Trial 22881-10882 ‘boost versus no boost’. Eur J Cancer. 2008;44:2587–99. doi: 10.1016/j.ejca.2008.07.032. [DOI] [PubMed] [Google Scholar]
- 42.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone acetate and survival of patients with metastatic prostate cancer. N Engl J Med. 2011 in press. [Google Scholar]
- 43.Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363:411–22. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 44.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–4. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 45.Kollermann J, Weikert S, Schostak M, Kempkensteffen C, Kleinschmidt K, Rau T, et al. Prognostic significance of disseminated tumor cells in the bone marrow of prostate cancer patients treated with neoadjuvant hormone treatment. J Clin Oncol. 2008;26:4928–33. doi: 10.1200/JCO.2007.15.0441. [DOI] [PubMed] [Google Scholar]
- 46.Holcomb IN, Grove DI, Kinnunen M, Friedman CL, Gallaher IS, Morgan TM, et al. Genomic alterations indicate tumor origin and varied metastatic potential of disseminated cells from prostate cancer patients. Cancer Res. 2008;68:5599–608. doi: 10.1158/0008-5472.CAN-08-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 48.Carver BS, Tran J, Chen Z, Carracedo-Perez A, Alimonti A, Nardella C, et al. ETS rearrangements and prostate cancer initiation. Nature. 2009:457–E1. doi: 10.1038/nature07738. discussion E2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gopalan A, Leversha MA, Satagopan JM, Zhou Q, Al-Ahmadie HA, Fine SW, et al. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–6. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scher HI, Kelly WK. The flutamide withdrawal syndrome: its impact on clinical trials in hormone-refractory prostatic cancer. J Clin Oncol. 1993;11:1566–72. doi: 10.1200/JCO.1993.11.8.1566. [DOI] [PubMed] [Google Scholar]
- 52.Leversha MA, Han J, Asgari Z, Danila DC, Lin O, Gonzalez-Espinoza R, et al. Fluorescence in situ hybridization analysis of circulating tumor cells in metastatic prostate cancer. Clin Cancer Res. 2009;15:2091–7. doi: 10.1158/1078-0432.CCR-08-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danila DC, de Bono J, Ryan CJ, Denmeade S, Smith M, Taplin M, et al. Phase II multicenter study of abiraterone acetate (AA) plus prednisone therapy in docetaxel-treated castration resistant prostate cancer (CRPC) patients (pts): Impact of prior ketoconazol (keto) J Clin Oncol. 2009;25 doi: 10.1200/JCO.2009.25.9259. Abstract 5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Bono JS, Attard G, Adjei A, Pollak MN, Fong PC, Haluska P, et al. Potential applications for circulating tumor cells expressing the insulin-like growth factor-I receptor. Clin Cancer Res. 2007;13:3611–6. doi: 10.1158/1078-0432.CCR-07-0268. [DOI] [PubMed] [Google Scholar]
- 56.Karp DD, Pollak MN, Cohen RB, Eisenberg PD, Haluska P, Yin D, et al. Safety, pharmacokinetics, and pharmacodynamics of the insulin-like growth factor type 1 receptor inhibitor figitumumab (CP-751,871) in combination with paclitaxel and carboplatin. J Thorac Oncol. 2009;4:1397–403. doi: 10.1097/JTO.0b013e3181ba2f1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ, et al. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A. 2007;1162:154–61. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 58.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–9. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392–7. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, et al. Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab Chip. 2010;10:27–9. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Racila E, Euhus D, Weiss A, Rao C, McConell J, Terstappen L, et al. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci U S A. 1998;95:4589–94. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci U S A. 2009;106:3970–5. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Helzer KT, Barnes HE, Day L, Harvey J, Billings PR, Forsyth A. Circulating tumor cells are transcriptionally similar to the primary tumor in a murine prostate model. Cancer Res. 2009;69:7860–6. doi: 10.1158/0008-5472.CAN-09-0801. [DOI] [PubMed] [Google Scholar]
- 64.Naume B, Borgen E, Tossvik S, Pavlak N, Oates D, Nesland JM. Detection of isolated tumor cells in peripheral blood and in BM: evaluation of a new enrichment method. Cytotherapy. 2004;6:244–52. doi: 10.1080/14653240410006086. [DOI] [PubMed] [Google Scholar]
- 65.Helo P, Cronin AM, Danila DC, Wenske S, Gonzalez-Espinoza R, Anand A, et al. Circulating prostate tumor cells detected by reverse transcription-PCR in men with localized or castration-refractory prostate cancer: concordance with CellSearch assay and association with bone metastases and with survival. Clin Chem. 2009;55:765–73. doi: 10.1373/clinchem.2008.117952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghossein RA, Carusone L, Bhattacharya S. Molecular detection of micrometastases and circulating tumor cells in melanoma prostatic and breast carcinomas. Vivo. 2000;14:237–50. [PubMed] [Google Scholar]
- 67.Ghossein RA, Scher HI, Gerald WL, Kelly WK, Curley T, Amsterdam A, et al. Detection of circulating tumor cells in patients with localized and metastatic prostatic carcinoma: clinical implications. J Clin Oncol. 1995;13:1195–200. doi: 10.1200/JCO.1995.13.5.1195. [DOI] [PubMed] [Google Scholar]
- 68.Hofman VJ, Ilie MI, Bonnetaud C, Selva E, Long E, Molina T, et al. Cytopathologic detection of circulating tumor cells using the isolation by size of epithelial tumor cell method: promises and pitfalls. Am J Clin Pathol. 2011;135:146–56. doi: 10.1309/AJCP9X8OZBEIQVVI. [DOI] [PubMed] [Google Scholar]
- 69.Alix-Panabieres C, Rebillard X, Brouillet JP, Barbotte E, Iborra F, Segui B, et al. Detection of circulating prostate-specific antigen-secreting cells in prostate cancer patients. Clin Chem. 2005;51:1538–41. doi: 10.1373/clinchem.2005.049445. [DOI] [PubMed] [Google Scholar]
- 70.Scholtens TM, Schreuder F, Ligthart ST, Swennenhuis JF, Tibbe AG, Greve J, et al. CellTracks TDI. An image cytometer for cell characterization. Cytometry A. 2011 doi: 10.1002/cyto.a.21024. Epub Feb 18. [DOI] [PubMed] [Google Scholar]
- 71.Bocsi J, Varga VS, Molnar B, Sipos F, Tulassay Z, Tarnok A. Scanning fluorescent microscopy analysis is applicable for absolute and relative cell frequency determinations. Cytometry A. 2004;61:1–8. doi: 10.1002/cyto.a.20061. [DOI] [PubMed] [Google Scholar]
- 72.Hsieh HB, Marrinucci D, Bethel K, Curry DN, Humphrey M, Krivacic RT, et al. High speed detection of circulating tumor cells. Biosens Bioelectron. 2006;21:1893–9. doi: 10.1016/j.bios.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 73.Ntouroupi TG, Ashraf SQ, McGregor SB, Turney BW, Seppo A, Kim Y, et al. Detection of circulating tumour cells in peripheral blood with an automated scanning fluorescence microscope. Br J Cancer. 2008;99:789–95. doi: 10.1038/sj.bjc.6604545. [DOI] [PMC free article] [PubMed] [Google Scholar]