Abstract
While it is accepted that protein flexibility plays a role in protein folding, catalysis, and molecular recognition, few techniques are capable of the rigorous measurement of protein motions required to quantify flexibility. Three-pulse photon echo shift spectroscopy can be used to measure the time scale of protein motions, and we have used this technique, along with steady-state spectroscopy and binding and structural data, to examine the immunological evolution of protein flexibility in an anti-fluorescein antibody. Two light chain somatic mutations increase affinity for fluorescein by 12-fold but also significantly affect flexibility. Specifically, a rigidification of the protein is seen in each of three observable motions; two slower motions undergo decreased amplitudes of displacement, by 3- and 20-fold, respectively, in response to an applied force, and the distribution associated with the amplitude of a faster motion is narrowed upon somatic mutation. The somatic mutations appear to rigidify the antibody-fluorescein complex by more strongly anchoring fluorescein to the protein and by more tightly packing the complex. The data demonstrate that in addition to affinity, antibody dynamics are systematically manipulated during affinity maturation, and they imply that the evolution of protein flexibility may be a central component of the immune response. The results also reflect the type of protein rigidification that may be important for other biological interactions, such as protein-protein, protein-ligand or protein-drug, and enzyme-substrate recognition.
Models of molecular recognition, based on conformational selection (1-3), induced-fit (4-6), or lock-and-key-type mechanisms (5, 7), are central to describing virtually all protein-protein and protein-ligand interactions. However, these models have been difficult to test, because they differ only in the flexibility of the protein, and protein flexibility has been difficult to quantify (8). Thus, studies of molecular recognition have not focused directly on flexibility, but instead have searched for manifestations of it, such as changes in on or off rates, varying binding entropies, and structural rearrangements. Nowhere is molecular recognition more important than in the immune system, where a finite number of receptors [antibodies (Ab) and T cell receptors] must bind a virtually infinite range of foreign molecules and peptides (9-16).
While it is apparent that during the initial stages of an immune response, there must be Ab present that have a broad range of specificities, Ab may also be isolated that are highly specific. These highly specific Ab are typically isolated in the later stages of an immune response and are usually highly mutated compared with their corresponding germ-line gene sequences (17). It has thus been argued that during affinity maturation, somatic mutations act to rigidify the Ab, leading to more specific epitope recognition (9, 13, 18-24). In this model, the binding sites of germ-line Ab (Ab before somatic mutation) are flexible, able to adopt many different conformations, and bind a range of epitopes with a conformational selection or induced fit-like mechanism. Although these flexible germ-line Ab are also expected to recognize self epitopes (18), they are not present at concentrations sufficient to be problematic (19). A rapid change in epitope concentration or presentation upon introduction of a foreign substance (25, 26) then induces affinity maturation of the flexible germ-line Ab into a more rigid mature Ab. The rigidity of the mature combining site results in selective recognition of the target epitope with more of a lock-and-key-like mechanism.
Previously, evidence for Ab flexibility has been sought from its structural, kinetic, or thermodynamic manifestations. For example, antigen (Ag)-induced conformational changes in combining sites have been extensively examined by x-ray crystallography (5, 27, 28). While both large and small structural rearrangements have been observed, several studies have shown that somatic mutations preorganize the combining site for Ag binding, as was found for the anti-p-nitrophenyl phosphonate Ab, 48G7 (21, 22). Recent structural and kinetic studies have provided evidence for a conformational selection model of antigen binding in the mature Ab SPE7, in which an isomerization appears to interconvert deep and shallow combining sites before Ag binding (20). Kinetic and thermodynamic studies have also provided evidence for a more negative entropy for Ag association with germ-line Ab, relative to mature Ab (29, 30). Although these results are consistent with the model that affinity maturation transforms flexible receptors into more rigid receptors, the studies do not actually measure flexibility. Testing the model requires knowledge of how the actual protein flexibility changes as a function of affinity maturation.
The flexibility of any material, including a protein, may be quantified by measuring the response to an applied force. For example, a flexible protein will respond to an applied force with low-frequency large-amplitude motions, whereas a more rigid protein will respond to the same force with higher-frequency smaller-amplitude motions. To quantify protein flexibility, both the amplitude and time scale of the motions must be known. We recently developed a technique to apply a force to an Ab combining site and characterize the energy and time scale of the protein response (31, 32). Photoexcitation of a chromophoric Ag bound to an Ab induces an electronic and structural reorganization of the Ag, resulting in an Ab-Ag complex that is out of equilibrium. In this manner, a step-function force induces protein motions within the binding site until equilibrium with the excited Ag is established. A complete description of the induced protein motions, including the amplitudes and time scales, is contained in the spectral density, ρ(ω), which describes the amplitude of the motion as a function of frequency, ω. Thus, ρ(ω) provides a quantitative description of flexibility. ρ(ω) may be determined by using three-pulse photon echo peak shift (3PEPS) spectroscopy, where the experimentally observable decay of the photon echo peak shift reflects the time scales and amplitudes of protein motions (31, 33-35).
Materials and Methods
Ab were cloned and expressed as Fab fragments in Escherichia coli (36). After isolation from the cell lysates by protein G affinity chromatography, Fab fragments were further purified with cation-exchange chromatography on a Mono S 5/5 column (Amersham Pharmacia). The binding properties (Kd and koff) of the germ-line Fab, affinity-matured Fab, and all of the mutants with the hapten were measured by surface plasmon resonance on a Biacore 3000 biosensor by following published methods (37). Briefly, BSA was conjugated with fluorescein (Fl) and was immobilized on a research grade CM5 sensor chip (Biacore). The dissociation constant, Kd, was measured under equilibrium conditions and the dissociation rate constant, koff, was measured under kinetic conditions. The association rate constant, kon, was calculated with the equation kon = koff/Kd. The value determined for the dissociation constant of Ab 4-4-20 is somewhat larger than that reported previously (38), which was determined by using a solution-phase Fl quenching assay. Thus, the differences may correspond to the difference between 4-4-20 binding Fl in solution versus at a surface. The germ-line precursor of the Ab was determined by sequencing and comparing the 5′ untranslated region (5′ UTR) of the hybridoma DNA with BALB/c genomic sequences (www.ensembl.org) (21). Primers were designed on the basis of the 5′ UTR of germ-line genes with the highest homology to the 4-4-20 light ( ) and heavy (
) and heavy (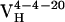 ) chains.
) chains.
The ultrafast laser source used in these measurements was described previously (31, 32). The 3PEPS experiment is analogous to a stimulated spin echo in NMR, and it may be understood by considering the evolution of the system during the three time periods established by the three pulses. During a first coherence period, initiated by the first pulse, the electronic coherence within the ensemble of absorbing molecules dephases (for time τ). During the population period initiated by the second pulse (for time T) no dephasing is possible, but environmental fluctuations continue. Finally, a second coherence period (of duration t) is initiated by the third pulse. During this period, the ensemble may rephase and emit an echo (at t = τ) if protein and chromophore vibrations have not uncorrelated the two coherence period environments. The decay of the experimentally observable peak shift as a function of T closely follows the time correlation function of the protein dynamics, and ρ(ω) may be constructed from the data by using the response function formalism, which assumes the electronic transition is linearly coupled to a harmonic bath (33).
The response function formalism for calculating spectroscopic properties of a molecular system from a model of the nuclear dynamics has been described extensively in the literature (33), thus only a brief summary will be given here. It provides a mathematical formalism to describe the response of matter to a time-dependent light field and may be used to calculate the third-order nonlinear polarization function, which is sufficient to model the 3PEPS, transient grating, and transient absorption signals (34). Furthermore, this formalism provides a means of calculating the steady-state absorption and emission spectra from the spectral density. The system is modeled with a chromophore electronic transition (a ground state, S0, and an excited state, S1) linearly coupled to a bath of harmonic oscillators that represent the Ag and protein vibrations. These oscillators cause the electronic energy gap to fluctuate. In this model fluctuations are probed by following the decay of coherence created within the two-level system by light-matter interactions. The spectral density, ρ(ω), characterizes the frequency distribution of vibrations coupled to the S0-to-S1 electronic transition and contains contributions from both protein fluctuations and chromophore vibrations. The relative weight of each contribution is scaled by its reorganization energy, λ, and its coupling strength, 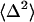 :
:
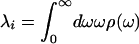 |
1 |
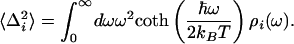 |
2 |
Spectral dynamics of the system are contained within the line-broadening function, g(t), which may be calculated from ρ(ω) by the expression
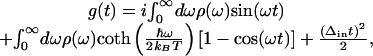 |
3 |
in which Δin is the inhomogeneous broadening, kB is the Boltzmann constant, and T is the absolute temperature. Signals for the various time-resolved experiments such as transient absorption, transient grating, or 3PEPS, as well as steady-state absorption spectra, may be calculated from g(t) by using standard procedures (33).
gaussian 98 was used to calculate the ground electronic state geometry and normal modes of vibration (HF/6-31G*) as well as the excited state geometry (CIS/6-31G*) as described previously (32). Displacements, Δj, were found by projecting the changes in bond lengths onto the normal mode vectors (39). The reorganization energy for each vibration j, λj, was determined from Δj and the normal mode frequency, 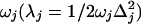 . The calculated frequencies were scaled by 0.9. The frequencies and reorganization energies were then used to calculate ρFl(ω) as described previously (32).
. The calculated frequencies were scaled by 0.9. The frequencies and reorganization energies were then used to calculate ρFl(ω) as described previously (32).
Results
Sequence Maturation of Ab 4-4-20. To examine the flexibility of the 4-4-20-Ag complex as a function of affinity maturation, we identified the germ-line precursor of the Ab by sequencing and comparing the 5′-untranslated region (5′ UTR) of the hybridoma DNA with BALB/c genomic sequences (www.ensembl.org) (21). Sequencing ≈500 nucleotides of the hybridoma 5′ UTR of the 4-4-20 light ( ) and heavy (
) and heavy ( ) chains was sufficient to uniquely identify the germ-line genes that encoded the Ab from which 4-4-20 evolved. The germ-line Ab light chain variable region (
) chains was sufficient to uniquely identify the germ-line genes that encoded the Ab from which 4-4-20 evolved. The germ-line Ab light chain variable region (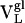 ) results from fusion of the Vκ1A and Jκ1 genes in a frame that yields a Trp-96 junction residue. The germ-line Ab heavy chain variable region (
) results from fusion of the Vκ1A and Jκ1 genes in a frame that yields a Trp-96 junction residue. The germ-line Ab heavy chain variable region (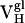 ) results from fusion of a JH606 [VHIII(C)] gene with JH4 and either the DSP2.6 or DSP2.7 alleles. The DNA encoding
) results from fusion of a JH606 [VHIII(C)] gene with JH4 and either the DSP2.6 or DSP2.7 alleles. The DNA encoding 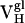 appears to include 7 nucleotides of P/N sequence, which are presumably added randomly during gene rearrangement. Sequence comparison (Fig. 1) shows that
appears to include 7 nucleotides of P/N sequence, which are presumably added randomly during gene rearrangement. Sequence comparison (Fig. 1) shows that 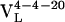 resulted from 2 somatic mutations and
resulted from 2 somatic mutations and 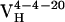 from 10 somatic mutations. We focused our efforts on characterizing the affinity maturation of the 4-4-20 light chain. Thus, the four Ab,
from 10 somatic mutations. We focused our efforts on characterizing the affinity maturation of the 4-4-20 light chain. Thus, the four Ab, 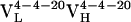 (i.e., Ab 4-4-20),
(i.e., Ab 4-4-20), 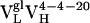 ,
, 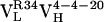 , and
, and 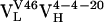 , were cloned and expressed as Fab fragments in E. coli (36).
, were cloned and expressed as Fab fragments in E. coli (36).
Fig. 1.
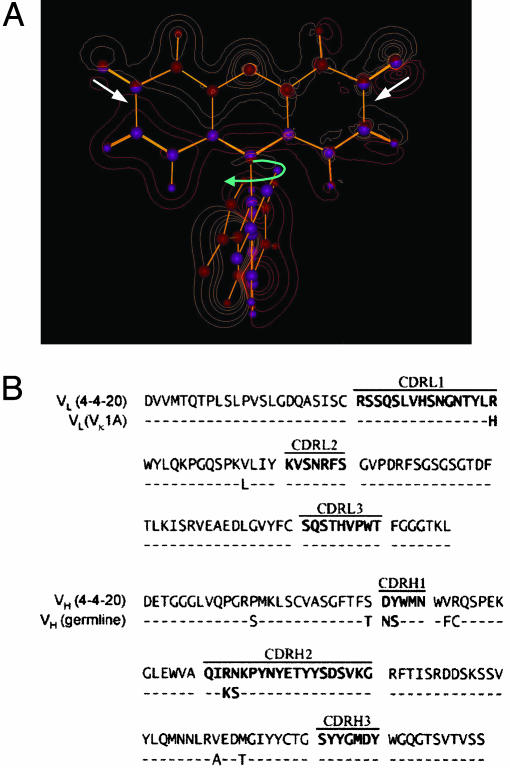
(A) Contour plot of Fl showing structural and electron density differences between ground and excited states. The red contours indicate higher electron density in the ground state, and the gold contours indicate higher electron density in the excited state. Purple and red spheres indicate atomic positions in the ground and excited states, respectively. The most significant electronic and structural reorganizations are highlighted with white (electronic) and blue (structural) arrows. (B) Sequences of 4-4-20 and germ-line precursors. Germ-line residues that are identical to 4-4-20 are indicated with a dash. The complementarity-determining regions (CDRs) are in boldface.
Thermodynamic and Kinetic Maturation of Ab 4-4-20. The dissociation constant (Kd) for each Ag-Fl complex was determined by surface plasmon resonance (Table 1). Ab 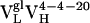 bound Fl with a dissociation constant of 2.6 μM whereas the intermediately evolved receptors,
bound Fl with a dissociation constant of 2.6 μM whereas the intermediately evolved receptors, 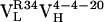 and
and 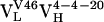 , bound with dissociation constants of 400 nM and 4.2 μM, respectively. The Kd for the mature
, bound with dissociation constants of 400 nM and 4.2 μM, respectively. The Kd for the mature 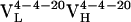 complex with Fl was 220 nM. These affinity constants imply that affinity maturation occurred first by mutation of His-34 to Arg (with an 6.6-fold increase in affinity), followed by Leu-46 to Val (with a 1.8-fold increase in affinity), because the Leu46Val mutation alone does not increase the Kd compared with the light chain germ-line Ab.
complex with Fl was 220 nM. These affinity constants imply that affinity maturation occurred first by mutation of His-34 to Arg (with an 6.6-fold increase in affinity), followed by Leu-46 to Val (with a 1.8-fold increase in affinity), because the Leu46Val mutation alone does not increase the Kd compared with the light chain germ-line Ab.
Table 1. Thermodynamic and kinetic data for VL somatic mutants.
| Ab | Kd, nM | kon, M−1·s−1 | koff, s−1 |
|---|---|---|---|
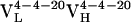 |
220 | 3.1 × 104 | 6.8 × 10−3 |
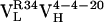 |
400 | 7.5 × 104 | 2.9 × 10−2 |
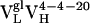 |
2,640 | 1.6 × 104 | 4.3 × 10−2 |
Ag-binding kinetics were also characterized as a function of affinity maturation. Using surface plasmon resonance, we determined the rate constants for Ag association (kon) and dissociation (koff) (Table 1). Affinity maturation is seen to have a consistent effect on the koff, with the dissociation rates slowing by 1.5-fold for the first somatic mutation (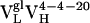 to
to 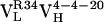 ) and 4.3-fold for the second (
) and 4.3-fold for the second (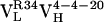 to
to 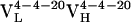 ). Affinity maturation had a less consistent effect on the association rates, increasing by 4.7-fold with the first somatic mutation, and then decreasing by 2.4-fold with the second. Thus, the affinity maturation of the 4-4-20 Ab results predominantly from decreasing off rates for Ab-Ag dissociation.
). Affinity maturation had a less consistent effect on the association rates, increasing by 4.7-fold with the first somatic mutation, and then decreasing by 2.4-fold with the second. Thus, the affinity maturation of the 4-4-20 Ab results predominantly from decreasing off rates for Ab-Ag dissociation.
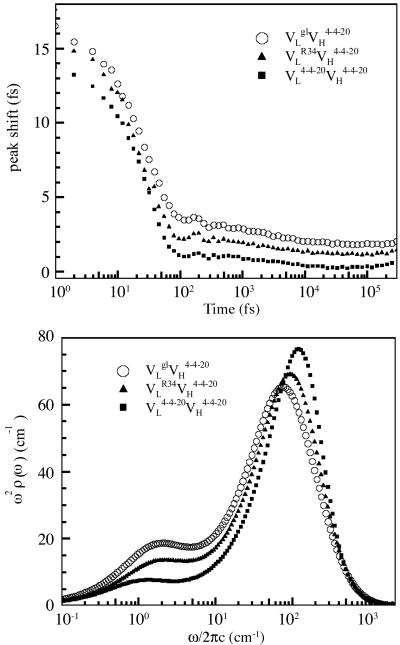
Dynamic Maturation of Ab 4-4-20. The 4-4-20 Fab fragment was characterized by 3PEPS spectroscopy with results that were indistinguishable from those previously reported for the full IgG protein (32). The effect of affinity maturation on Ab flexibility was quantified by performing 3PEPS spectroscopy measurements on each expressed somatic variant (Fig. 2). The data reveal that all three Ab respond to Ag excitation with motions on three discrete time scales: an ultrafast motion (<40 fs), as well as 3-to 5-ps and >3-ns time scale motions. Furthermore, the data show a clear trend: the proportion of low-frequency motion decreases consistently and significantly with affinity maturation. To quantify this trend, the spectral density for each Ab was determined from the 3PEPS data by using the response function formalism (33). Peaks in the spectral density, ρ(ω), correspond to a distribution of motions centered on ω; the amplitude of each peak is proportional to the magnitude of the spatial displacement squared and the coupling of the motion to the electronic transition. The complete spectral density, ρ(ω), is a sum of Ag and Ab spectral densities, ρAg(ω) and ρAb(ω), respectively. ρAg(ω) was determined with quantum chemical methods [this same ρAg(ω) was used in ref. 19]. ρAb(ω) was modeled with three components. A Brownian oscillator
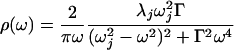 |
4 |
Fig. 2.
(Upper) 3PEPS data for 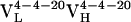 (light chain germ line, ○),
(light chain germ line, ○), 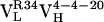 (somatic intermediate, ▴), and
(somatic intermediate, ▴), and 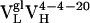 (affinity-matured 4-4-20, ▪). (Lower) Model Ab spectral densities. The functional forms are given in the text, and the parameters are collected in Table 2.
(affinity-matured 4-4-20, ▪). (Lower) Model Ab spectral densities. The functional forms are given in the text, and the parameters are collected in Table 2.
was used for the fastest dynamics. In this expression, λj is the reorganization energy (which corresponds to the amplitude of the motion), ωj is the frequency of the oscillator, and Γ is the damping constant. The intermediate time scale dynamics were modeled as an exponential relaxation (an overdamped oscillator) in the time domain, which gives a Lorentzian function in ρ(ω):
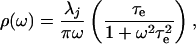 |
5 |
where τe is the exponential decay time. The slowest component (>3 ns) was modeled as static inhomogeneous broadening, Δin. The parameters used to construct ρAb(ω) are shown in Table 2. To facilitate comparison with the amplitudes of the faster motions, the values of inhomogeneous broadening are converted to reorganization energies in Table 2 (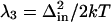 ). Comparison of the damping constants for the ultrafastest motion and the reorganization energies for the pico- and nanosecond motion reveal a significant restriction in protein motion upon affinity maturation.
). Comparison of the damping constants for the ultrafastest motion and the reorganization energies for the pico- and nanosecond motion reveal a significant restriction in protein motion upon affinity maturation.
Table 2. Parameters for VL somatic mutants ρAb(ω).
| Parameter | 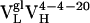 |
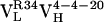 |
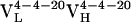 |
|---|---|---|---|
| γ1, cm−1 | 180 | 180 | 180 |
| ω1, cm−1 | 200 | 200 | 200 |
| Γ1, cm−1 | 380 | 480 | 580 |
| γ2, cm−1 | 20 | 35 | 50 |
| τ2, fs | 5,000 | 3,200 | 3,200 |
| γ3, cm−1 | 6 | 56 | 121 |
Discussion
Adaptive immunity requires both the recognition of and the response to foreign substances. Recognition is mediated by the naïve repertoire of germ-line Ab, which must have a sufficiently diverse range of specificities to recognize, with at least modest affinity, virtually anything. This range of specificities would appear to require each individual germ-line Ab to be polyspecific, or cross-reactive, and thus able to bind a range of Ag (13, 23, 24, 29). The ability of a given receptor to bind multiple ligands requires that the binding sites possess sufficient flexibility to adopt the multiple conformations suited to the different ligands. However, the immune response requires that clonally expanded Ab be highly specific to preclude self-recognition; thus these receptors must be sufficiently rigid to prevent population of the conformations suitable for binding self antigens. This apparent contradiction may be resolved if flexible induced-fit like receptors may be evolved by affinity maturation (during clonal expansion) into rigid lock-and-key-like receptors (21, 22, 29, 37). In this model, affinity maturation ensures that any clonally expanded Ab is also rigidified and made specific for its target Ag. Testing this hypothesis requires directly characterizing Ab flexibility as a function of affinity maturation.
Previously, a combination of steady-state and 3PEPS spectroscopy has been used to characterize the flexibility of Ab complexes with the rigid chromophore 8-methoxypyrene-1,3,6-trisulfonate (MPTS) (31), as well as the more flexible chromophore Fl (32). The approach is based on characterizing the energy and time scale of protein motions within the Ab combining site that are induced in response to an excitation-induced change of the Ag electronic structure. For the Ab 6C8-MPTS complex, the protein responded to Ag excitation with three discrete motions, which were suggested to result from heavy chain loop motions. Similarly, for three Ab-Fl complexes (Ab 34F10, 40G4, and 4-4-20), each Ab again responded to Ag excitation on three distinct time scales. The time scales were significantly different from those observed in 6C8, and a comparison of the ρ(ω) for the anti-Fl Ab showed significant differences in the amplitudes of the induced motions. 34F10 is the most flexible, followed by 40G4, then 4-4-20. The structural origins of the observed dynamics were analyzed for the most rigid Ab, 4-4-20. Ab initio calculations and the crystal structure of the complex were used to predict the regions of the protein that were subject to the step-function force. The largest photoinduced changes in the Ag were predicted to include migration of electron density away from the xanthenone carbonyl oxygen atoms into the carbon ring system. This charge migration weakens intermolecular hydrogen bonding and results in the well known correlation of absorption and emission spectral shifts with solvent hydrogen-bond-donating strength (40, 41). The photoinduced charge migration also weakens the intermolecular hydrogen bonds in the 4-4-20 combining site and induces protein side chain reorganization, which was suggested to be a significant component of the observed dynamics (see below).
In this work, the immunological evolution of the 4-4-20 light chain has been characterized, in terms of sequence, affinity, binding kinetics, and most importantly, protein flexibility. Two somatic mutations evolved the 4-4-20 light chain, first His-34 was mutated to Arg, and then Leu-46 was mutated to Val. These mutations were selected based on 6.6- and 1.8-fold decreases in Kd, which are typical of somatic mutations characterized in other systems (17). Increases in affinity during affinity maturation have previously been ascribed to both increases in the rates of association and a decrease in the rate of dissociation. Affinity maturation-dependent increases in on rates have been interpreted in terms of kinetic selection for rapid Ag association (29, 42), whereas decreases in off rates have been interpreted in terms of rigidifying mutations that stabilize favorable binding site configurations from which the Ag associates and dissociates more slowly than it would from a more flexible combining site (30, 43). During the evolution of the 4-4-20 light chain, the 12-fold increase in affinity results largely from a nearly 7-fold decrease in koff. To determine whether these changes are associated with a rigidification of the combining site the flexibility of each Ab was characterized.
A comparison of the ρAb(ω) for each Ab (Fig. 2) shows that affinity maturation affects all of the observed motions. The reorganization energy and frequency of the fastest motion does not change; however, the damping constant decreases by 1.5-fold with maturation (Table 2), indicating a narrowing of the distribution of motions. It is difficult to determine whether the changes result from changes in the damping constant alone (as is assumed here) or whether changes in amplitudes and frequencies also contribute. In the latter case, the frequencies are seen to shift higher, thus, assuming that the changes result from changes in either the damping constant or amplitudes and frequencies leads to the same conclusion: these fast motions in the combing site become more restricted with affinity maturation. The intermediate time scale motions undergo changes in both frequency and amplitude (a 2.5-fold decrease) with maturation. Finally, the amplitude of the longest time scale motions decreases nearly 20-fold upon maturation. Although these changes in amplitude are measured in unitless displacements they are certainly significant. For example, whatever the magnitude of protein deformations (i.e., angstroms, tens of angstroms, etc.) on the picosecond and nanosecond time scales, they are decreased by 2.5- and 20-fold as a result of affinity maturation. The flexibility data unambiguously demonstrate that the Ab is evolved from a flexible germ-line Ab into a significantly more rigid mature Ab. In total, the sequence, thermodynamic, kinetic, and flexibility data imply that two point mutations, His-34 to Arg and Leu-36 to Val, significantly rigidify the Ab-Ag complex and localize the combining site to a configuration appropriate for binding.
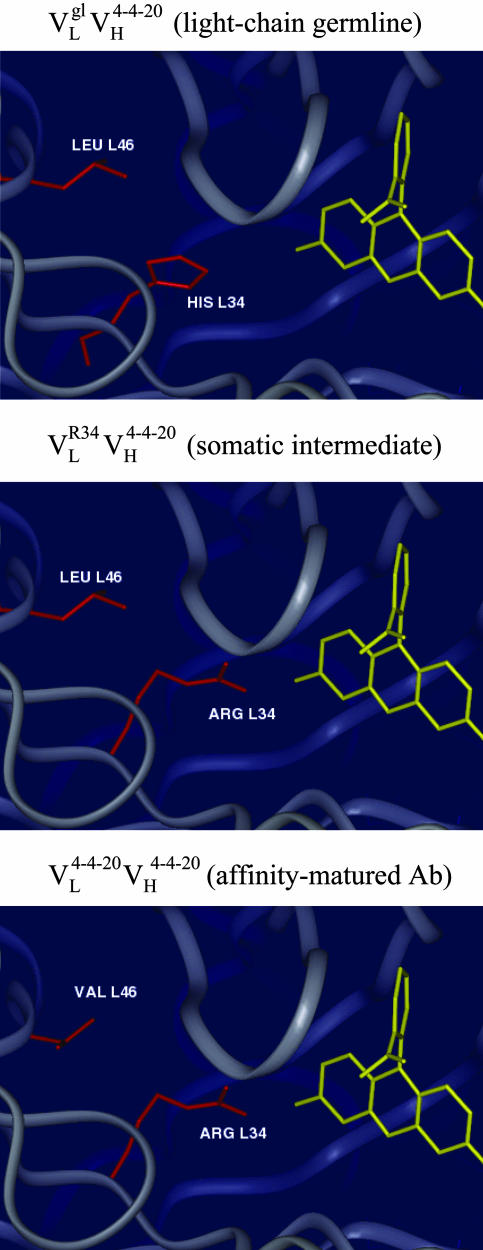
A structural model of the affinity maturation-dependent rigidification may be based on the crystal structure of 4-4-20 and models of the 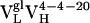 and
and 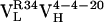 Ab (Fig. 3). Models of the germ-line and somatic intermediate were generated from the structure of the mature Ab (44) by side-chain replacement and optimization (insight ii, Acelerys, San Diego). Interestingly, both of the light chain somatic mutations occur in the region of the combining site that is involved in hydrogen bonding to a xanthenone group of Fl. As mentioned above, excitation of Fl is expected to weaken this hydrogen bond and induce recoiling motion of the residue 34 side chain. In the germ-line Ab, this residue is a histidine (Fig. 3 Top), which provides a relatively weak hydrogen bond that responds to Ag excitation with relatively large-amplitude motions. The first somatic mutation, His34Arg, introduces a stronger hydrogen bond (Fig. 3 Middle), which is also expected to increase the coupling of the side-chain motion to the electronic transition. Thus, the decrease in reorganization energy observed for the two slower motions indicates that somatic mutation results in a decrease in spatial amplitude of the induced motion. The second somatic mutation, Leu46Val (within a protein already containing the His34Arg mutation), is not expected to significantly alter the coupling to the electronic transition. Thus, the second somatic mutation leads to a further reduction in the spatial amplitude of the Arg-34 side-chain motion, by optimizing packing interactions (Fig. 3 Bottom). In this manner two point mutations significantly rigidified the Ab-Ag complex.
Ab (Fig. 3). Models of the germ-line and somatic intermediate were generated from the structure of the mature Ab (44) by side-chain replacement and optimization (insight ii, Acelerys, San Diego). Interestingly, both of the light chain somatic mutations occur in the region of the combining site that is involved in hydrogen bonding to a xanthenone group of Fl. As mentioned above, excitation of Fl is expected to weaken this hydrogen bond and induce recoiling motion of the residue 34 side chain. In the germ-line Ab, this residue is a histidine (Fig. 3 Top), which provides a relatively weak hydrogen bond that responds to Ag excitation with relatively large-amplitude motions. The first somatic mutation, His34Arg, introduces a stronger hydrogen bond (Fig. 3 Middle), which is also expected to increase the coupling of the side-chain motion to the electronic transition. Thus, the decrease in reorganization energy observed for the two slower motions indicates that somatic mutation results in a decrease in spatial amplitude of the induced motion. The second somatic mutation, Leu46Val (within a protein already containing the His34Arg mutation), is not expected to significantly alter the coupling to the electronic transition. Thus, the second somatic mutation leads to a further reduction in the spatial amplitude of the Arg-34 side-chain motion, by optimizing packing interactions (Fig. 3 Bottom). In this manner two point mutations significantly rigidified the Ab-Ag complex.
Fig. 3.
(Top) Model of 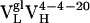 . (Middle) Model of
. (Middle) Model of 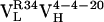 . (Bottom) Crystal structure of mature Ab 4-4-20 (
. (Bottom) Crystal structure of mature Ab 4-4-20 (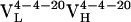 ) (44).
) (44).
These results demonstrate that the immune system is capable of systematically manipulating protein dynamics within a given combining site by means of specific somatic mutations during affinity maturation. This controlled rigidification may play a central role in adaptive immunity by allowing for the rapid evolution of highly specific rigid lock-and-key-like receptors from a finite set of polyspecific flexible induced-fit-like receptors. With the characterization of the 4-4-20 heavy chain somatic mutations, as well as additional Ab-Ag complexes, these results may be prove to be general. In fact, the germ-line gene repertoire may have been selected during evolution, in part, based on their encoding flexible combining sites, and somatic mutations may be selected during affinity maturation, in part, based on their rigidification of these combining sites. This possibility is particularly interesting from a biophysical perspective, where germ-line Ab and their affinity maturation might provide model systems to characterize the general determinants of protein flexibility and how it may be manipulated. In addition to characterizing Ab-Ag complexes, it seems likely that suitable systems might be developed for the 3PEPS characterization of protein flexibility in other biophysical processes where it might play an important role, for example, in protein folding (45), catalysis (46), and drug design (47, 48).
Acknowledgments
We thank Drs. Delmar Larsen and Jeffrey Reimers for help with calculations. Funding was provided by the Skaggs Institute for Chemical Biology.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ag, antigen or antigens; 3PEPS, three-pulse photon echo shift; Fl, fluorescein.
References
- 1.Foote, J. & Milstein, C. (1994) Proc. Natl. Acad. Sci. USA 91, 10370-10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma, B., Shatsky, M., Wolfson, H. J. & Nussinov, R. (2002) Protein Sci. 11, 184-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosshard, H. R. (2001) News Physiol. Sci. 16, 171-173. [DOI] [PubMed] [Google Scholar]
- 4.Koshland, D. E. (1958) Proc. Natl. Acad. Sci. USA 44, 98-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sunderberg, E. J. (2003) Adv. Protein Chem. 61, 119-160. [Google Scholar]
- 6.Berzofsky, J. A. (1985) Science 229, 932-940. [DOI] [PubMed] [Google Scholar]
- 7.Pauling, L. (1946) Chem. Eng. News 24, 1375-1377. [Google Scholar]
- 8.Hagen, S. J., Hofrichter, J. & Eaton, W. A. (1997) J. Phys. Chem. B 101, 2352-2365. [Google Scholar]
- 9.Guigou, V., Guilbert, B., Moinier, D., Tonnelle, C., Boubli, L., Avrameas, S., Fougereau, M. & Fumoux, F. (1991) J. Immunol. 146, 1368-1374. [PubMed] [Google Scholar]
- 10.Garcia, K. C., Degano, M., Pease, L. R., Huang, M., Peterson, P. A., Teyton, L. & Wilson, I. A. (1998) Science 279, 1166-1172. [DOI] [PubMed] [Google Scholar]
- 11.Boniface, J. J., Reich, Z., Lyons, D. S. & Davis, M. M. (1999) Proc. Natl. Acad. Sci. USA 96, 11446-11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilcox, B. E., Gao, G. F., Wyer, J. R., Ladbury, J. E., Bell, J. I., Jakobsen, B. K. & van der Merwe, P. A. (1999) Immunity 10, 357-365. [DOI] [PubMed] [Google Scholar]
- 13.Mason, D. (1998) Immunol. Today 19, 395-404. [DOI] [PubMed] [Google Scholar]
- 14.Ysern, X., Li, H. & Mariuzza, R. A. (1998) Nat. Struct. Biol. 5, 412-414. [DOI] [PubMed] [Google Scholar]
- 15.Hare, B. J., Wyss, D. F., Osburne, M. S., Kern, P. S., Reinherz, E. L. & Wagner, G. (1999) Nat. Struct. Biol. 6, 574-581. [DOI] [PubMed] [Google Scholar]
- 16.Holler, P. D., Chlewicki, L. K. & Kranz, D. M. (2003) Nature Immunol. 4, 55-62. [DOI] [PubMed] [Google Scholar]
- 17.Steele, E. J. (1990) Somatic Hypermutation in V-Regions (CRC, Boca Raton, FL).
- 18.Comtesse, N., Heckel, D., Maldener, E., Glass, B. & Meese, E. (2000) Clin. Exp. Immunol. 121, 430-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Souroujon, M., White-Scharf, M. E., Andre-Schwartz, J., Gefter, M. L. & Schwartz, R. S. (1988) J. Immunol. 140, 4173-4179. [PubMed] [Google Scholar]
- 20.James, L. C., Roversi, P. & Tawfik, D. S. (2003) Science 299, 1362-1367. [DOI] [PubMed] [Google Scholar]
- 21.Patten, P. A., Gray, N. S., Yang, P. L., Marks, C. B., Wedemayer, G. J., Boniface, J. J., Stevens, R. C. & Schultz, P. G. (1996) Science 271, 1086-1091. [DOI] [PubMed] [Google Scholar]
- 22.Wedemayer, G. J., Patten, P. A., Wang, L. H., Schultz, P. G. & Stevens, R. C. (1997) Science 276, 1665-1669. [DOI] [PubMed] [Google Scholar]
- 23.Joyce, G. F. (1997) Science 276, 1658-1659. [DOI] [PubMed] [Google Scholar]
- 24.Hodgkin, P. D. (1998) Immunologist 6, 223-226. [Google Scholar]
- 25.Jegerlehner, A., Storni, T., Lipowsky, G., Schmid, M., Pumpens, P. & Bachmann, M. F. (2002) Eur. J. Immunol. 32, 3305-3314. [DOI] [PubMed] [Google Scholar]
- 26.Baumgarth, N. (2000) Immunol. Rev. 176, 171-180. [DOI] [PubMed] [Google Scholar]
- 27.Rini, J. M., Schulze-Gahmen, U. & Wilson, I. A. (1992) Science 255, 959-965. [DOI] [PubMed] [Google Scholar]
- 28.Nezlin, R. (1998) The Immunoglobulins: Structure and Function (Academic, New York).
- 29.Manivel, V., Sahoo, N. C., Salunke, D. M. & Rao, K. V. S. (2000) Immunity 13, 611-620. [DOI] [PubMed] [Google Scholar]
- 30.Sagawa, T., Oda, M., Ishimura, M., Furukawa, K. & Azuma, T. (2003) Mol. Immunol. 39, 801-808. [DOI] [PubMed] [Google Scholar]
- 31.Jimenez, R., Case, D. A. & Romesberg, F. E. (2002) J. Phys. Chem. B 106, 1090-1103. [Google Scholar]
- 32.Jimenez, R., Salazar, G., Baldridge, K. K. & Romesberg, F. E. (2003) Proc. Natl. Acad. Sci. USA 100, 92-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukamel, S. (1995) Principles of Nonlinear Optical Spectroscopy (Oxford Univ. Press, New York).
- 34.Fleming, G. R. & Cho, M. (1996) Annu. Rev. Phys. Chem. 47, 109-134. [Google Scholar]
- 35.de Boeij, W. P., Pshenichnikov, M. S. & Wiersma, D. A. (1996) J. Phys. Chem. 100, 11806-11823. [Google Scholar]
- 36.Ulrich, H. D., Patten, P. A., Yang, P. L., Romesberg, F. E. & Schultz, P. G. (1995) Proc. Natl. Acad. Sci. USA 92, 11907-11911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin, J., Mundorff, E. C., Yang, P. L., Wendt, K. U., Hanway, D., Stevens, R. C. & Schultz, P. G. (2001) Biochemistry 40, 10764-10773. [DOI] [PubMed] [Google Scholar]
- 38.Kranz, D. M., Herron, J. N. & Voss, E. W. (1982) J. Biol. Chem. 257, 6987-6995. [PubMed] [Google Scholar]
- 39.Reimers, J. R. (2001) J. Chem. Phys. 115, 9103-9109. [Google Scholar]
- 40.Klonis, N., Clayton, A. H. A., Voss, E. W., Jr., & Sawyer, W. H. (1998) Photochem. Photobiol. 67, 500-510. [PubMed] [Google Scholar]
- 41.Reichardt, C. (1990) Solvents and Solvent Effects in Organic Chemistry (VCH, Weinheim, Germany).
- 42.Foote, J. & Milstein, C. (1991) Nature 352, 530-532. [DOI] [PubMed] [Google Scholar]
- 43.Liu, J., Liu, L. & Jemmerson, R. (2000) Mol. Immunol. 37, 95-105. [DOI] [PubMed] [Google Scholar]
- 44.Whitlow, M., Howard, A. J., Wood, J. F., Voss, E. W., Jr., & Hardman, K. D. (1995) Protein Eng. 8, 749-761. [DOI] [PubMed] [Google Scholar]
- 45.Kuhn, L. A. & Thorpe, M. F. (2001) in Protein Flexibility and Folding, ed. Kuhn, L. A. (Elsevier, New York).
- 46.Welch, G. R. (1986) The Fluctuating Enzyme (Wiley, New York).
- 47.Davis, A. M. & Teague, S. J. (1999) Angew. Chem. Int. Ed. 38, 736-749. [DOI] [PubMed] [Google Scholar]
- 48.Carlson, H. A. (2002) Curr. Opin. Chem. Biol. 6, 447-452. [DOI] [PubMed] [Google Scholar]