SUMMARY
The mammalian target of rapamycin (mTOR)-pathway serves as a key sensor of cellular-energetic state, and functions to maintain tissue homeostasis. Hyperactivation of the mTOR pathway impairs hematopoietic stem cell (HSC) function and is associated with leukemogenesis. However, the roles of the unique mTOR complexes (mTORCs) in hematopoeisis and leukemogenesis have not been adequately elucidated. We deleted the mTORC1 component, Raptor (regulatory-associated protein of mTOR), in mouse HSC and its loss causes a non-lethal phenotype characterized by pancytopenia, splenomegaly, and the accumulation of monocytoid cells. Furthermore, Raptor is required for HSC regeneration, and plays largely non-redundant roles with Rictor (rapamycin-insensitive companion of mTOR), in these processes. Ablation of Raptor also significantly extends survival of mice in models of leukemogenesis evoked by Pten deficiency. These data delineate critical roles for mTORC1 in hematopoietic function and leukemogenesis, and inform clinical strategies based on chronic mTORC1 inhibition.
INTRODUCTION
mTOR is a serine/threonine(Thr)-protein kinase that has been implicated in regulating key processes in hematopoiesis (Warr et al., 2011). The mTOR pathway is activated under conditions of favorable nutrient availability and directly regulates several downstream-metabolic processes, such as mRNA translation, lipid biosynthesis, autophagy, and mitochondrial biogenesis. mTOR exists in two independent multiprotein-containing complexes that have distinct functions in embryonic development and in adult tissue. mTORC1 and mTORC2 are defined by the scaffolding/substrate-guiding proteins, Raptor and Rictor, respectively (Laplante and Sabatini, 2012).
Perturbation of the mTOR pathway in adult HSC of mice, either by introducing loss-of-function alleles of Pten or Tsc1 (both negative regulators of mTOR [Engelman et al., 2006; Laplante and Sabatini, 2012]), or gain-of-function mutations in mTOR activators, RHEB2 or AKT, results in HSC cycling and depletion of long-term (LT) reconstituting activity (Yilmaz et al., 2006; Zhang et al., 2006; Chen et al., 2008; Gan et al., 2008; Campbell et al., 2009; Kharas et al., 2010). Chronic mTOR activation can evokee myeloproliferative neoplasms (MPN), and in some cases acute leukemias, suggesting a differential requirement for mTOR between HSC and leukemia-stem cells (LSC). Furthermore, pharmacological inhibition of mTOR with rapamycin, can restore HSC activity and/or deplete LSC function in these models. These studies, strongly implicate a role for chronic-mTOR activity in HSC and LSC functions, but do not directly address the required roles of the individual mTORCs in these systems. In particular, it is now appreciated that rapamycin is only a partial mTORC1 antagonist (Choo et al., 2008; Hsu et al., 2011; Yu et al., 2011) and can inhibit mTORC2 activity in some cell types (Sarbassov et al., 2006). As ATP-competitive inhibitors of mTOR have recently been demonstrated to have more potent anti-leukemic activity than rapamycin in leukemias (Janes et al., 2010), knowledge of which mTORC to target to enhance therapeutic index would be of value in developing novel therapeutic interventions. To determine the role the mTORCs in hematopoiesis and leukemogenesis we utilized mice containing conditional loss-of-function alleles for Raptor and Rictor. We observe that Raptor is required for HSC regeneration under stress conditions while not absolutely required during homeostasis, and that Raptor and Rictor play largely non-redundant roles in these processes. Of note, loss of Raptor prolongs survival of mice in models of leukemogenesis evoked by Pten deficiency. These data clarify the roles of the mTORCs in benign and malignant hematopoiesis, while indicating potential deleterious responses due to chronic mTORC1 inhibition in the hematopoietic system.
RESULTS
Deletion of Raptor in HSPC Ablates mTORC1 Activity
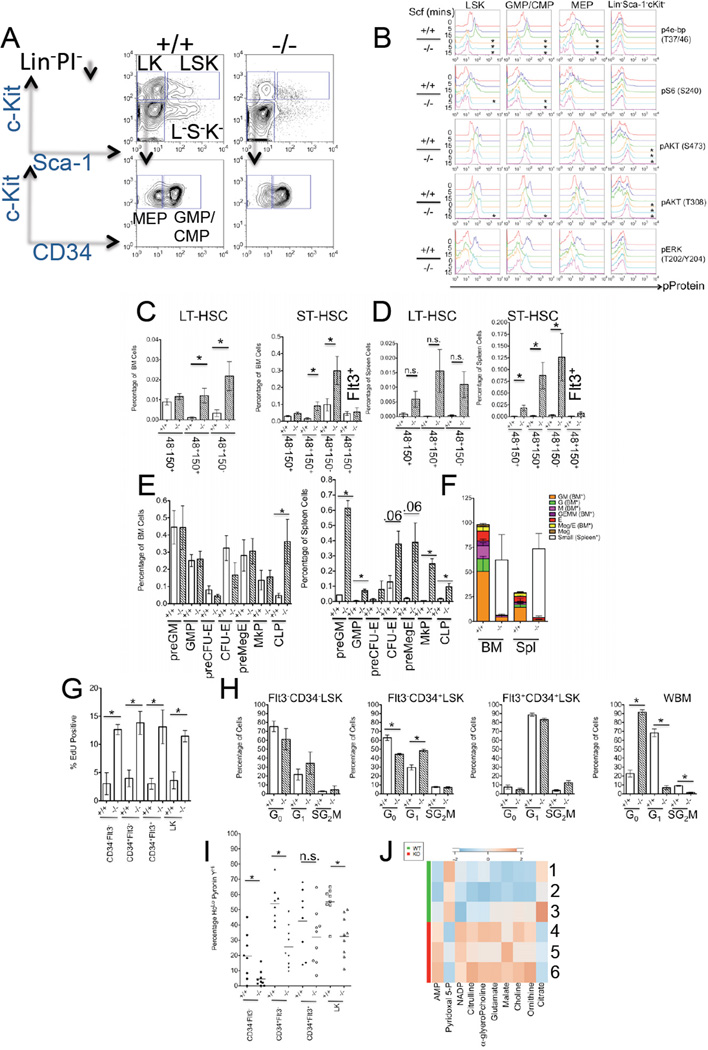
We investigated the effects of mTORC1 loss in hematopoiesis by utilizing mice containing loxP sites flanking (floxed [Fl]) exon 6 of the Raptor gene, that upon Cre recombinase-expression produce null Raptor alleles (Sengupta et al., 2010; Peterson et al., 2011). We conditionally deleted Raptor alleles through use of interferon-inducible transgenic-Mx1Cre mice, that induces recombination upon administration of polyinosinic-polycytidylic acid (pIpC) (Kuhn et al., 1995). Injection of pIpC into 4–8 week old RaptorFl/+, MxCre or RaptorFl/Fl, MxCre mice resulted in efficient deletion of Raptor allele(s) and diminished mRNA and protein expression compared controls (Figures S1A–S1D and data not shown). Deletion of Raptor in HSPC led to decreased basal and Scf-stimulated levels of phospho(p)-4e-bp, as well as Scf-stimulated levels of pS6 (Figures 1A–1B, for quantification see Figure S1E). Since mTORC1 inhibition can evoke increased AKT phosphorylation (due to the negation of a negative-feedback loop) (Carracedo et al., 2008), we also examined pAKT levels in HSPC. Although phosphorylation at Ser473 was normal in Raptor-null HSPC, phosphorylation of AKT at Thr308 was slightly prolonged in Scf-stimulated Raptor-null LSK and GMP/CMP (Figure 1B). Strikingly, relative basal levels of pAKT at either site in the more mature Lin−Sca−1−c-Kit−cells were markedly elevated in Raptor-null cells, indicating cell-type specificity in the mTORC1-PI3K feedback loop in hematopoietic cells (Figure 1B). Levels of pERK were not different between control and Raptor-deleted HSPC, indicating that Raptor deletion does not disrupt all c-Kit signaling (Figure 1B).
Figure 1. Deletion of Raptor Ablates mTORC1 Activity and Leads to HSPC Pertubations.
(A) Representative flow-cytometry plots of sorted Lin− PI− cells post fixation and permeabilization from pIpC-treated RaptorFl/Fl (+/+) and Raptor Fl/Fl , Mx-Cre (−/−) mice (Lin− PI− Sca-1+c-Kit+ [LSK], Lin− PI− Sca-1−c-Kit+[LK], Lin− PI− Sca-1−c-Kit+CD34+ [GMP/CMP], Lin− PI− Sca-1−c-Kit+CD34− [MEP], and Lin− PI− Sca-1−c-Kit[L− S− K−]).
(B) Flow cytometry was performed on sorted Lin−PI− cells from pIpC-treated RaptorFl/Fl (+/+) and Raptor Fl/Fl, Mx-Cre (−/−) mice that were stimulated ex vivo with 20 ng/mL of Scf for the indicated time points in minutes (mins), and then processed for flow cytometry. Representative histograms are shown (% maximal value on the y-axis) and for quantification see Figure S1E (*p<0.05 relative to unstimulated +/+ controls).
(C) Quantification of CD48/CD150/CD34/Flt3/LSK subsets from BM and spleen
(D) of RaptorFl/Fl (+/+) and Raptor Fl/Fl, Mx-Cre (−/−) mice 5–7 months post pIpc. (n=5 +/+, n=6 −/−). For gating see Figure S1N.
(E) Quantification of immunophenotypic HPC subsets from mice described in (C), from BM (left panel) and spleen (right panel). Immunophenotypes are defined as in Pronk et al., 2007.
(F) CFC formation from pIpC-treated RaptorFl/Fl (+/+) and Raptor Fl/Fl, Mx-Cre (−/−) mouse BM and spleen cells. (n=3–4).
(G) Assessment of EdU incorporation in cells from pIpC-treated RaptorFl/Fl (+/+) and Raptor Fl/Fl , Mx-Cre (−/−) mice. (n=3).
(H) Ho 33342/Ki-67 staining from the indicated FACS-sorted LSK populations or whole BM (WBM) and pIpC-treated genotypes. G0 (2N DNA-Ki67Lo), G1 (2N DNA-Ki67Hi), and SG2M (>2N DNA-Ki67Hi) (n=3).
(I) Ho 33342/Pyronin Y in the indicated surface-stained populations of the indicated genotypes (as in [B]) by flow cytometry). HoLo = 2N DNA. (n=8–9)
(J) Heat-map of metabolite measurements from pIpC-treated RaptorFl/Fl (WT [1–3]) and Raptor Fl/Fl, Mx-Cre (KO [4–6]) LSK cells. (p<0.057, citrate p=0.077) (A–J) Data are expressed as mean or mean ± SEM (*p<0.05, n.s. p>0.05).
mTORC1 Regulates Differentiation Along Multiple Hematopoietic Lineages
Raptor null mice were viable surviving at least 20 months post pIpC and containing predominately deleted alleles (data not shown). However, within 1 month of deletion, 100% of mice developed a persistent and rapid pancytopenia, splenomegaly with extramedullary hematopoiesis and disrupted splenic architecture, and decreased thymic mass (Figures S1F–S1G and S1K and data not shown). Despite the pancytopenia, Raptor-deleted BM maintained BM cellularity (Figure S1G and S1K). However, Raptor-deleted BM did show signs of defective hematopoiesis with emergence of monocytoid features (Figures S1G, S1H, and S1I), but without evidence of dysplasia or progression to leukemia in old age (data not shown). Raptor loss leads to an accumulation of Mac-1+Gr-1Mid/Lo monocytes as early as 2 weeks and persisting to at least 5–7 months post deletion (Figure S1H–S1J and S1M). Raptor-deletion resulted in increased levels of B220+IgM−CD43+ proB cells in the BM and decreased levels of B220+IgMloIgDHi mature B cells in spleen (Figures S1I–S1H and S1M and data not shown). Finally, Raptor loss perturbed erythroblast frequencies in the BM and spleen (Figure S1H–S1I). Deletion of Raptor alleles via tamoxifen administration in RaptorFl/Fl mice expressing a UBIQUITIN C promoter-driven tamoxifen-inducible Cre-estrogen receptor (Cre-ER) (Ruzankina et al., 2007) also led to monocyte expansion (Figure S1M and data not shown), suggesting, the effects observed in the Mx-Cre model were not due to interferon responses.
Ablation of Raptor Expands LSK Subsets and Mobilizes HSPC
Raptor loss evoked a rapid (within 2 weeks), sustained (5–7 months), and pIpC-independent increase in the frequency of several LSK subsets (Figures 1C–1D and Figure S1N and S1O). In the LT-HSC-enriched Flt3−CD34−LSK subset (Osawa et al., 1996; Christensen and Weissman, 2001; Yang et al., 2005) Raptor loss increased the frequency of both BM CD48+CD150+-, and CD48+CD150− cells, while CD48−CD150+Flt3−CD34−-LT-HSC (Kiel et al., 2005; Morita et al., 2010) were largely unaffected (Figures 1C–1D). In the short-term HSC (ST-HSC)-enriched Flt3−CD34+LSK subset Raptor loss produced an increase in the frequency of BM CD48+CD150+-, CD48+CD150− cells, as well as an increase in CD48+CD150+-Flt3−CD34+ LSK cells in the spleen (Figures 1C–1D). Raptor deletion led to an expansion of several splenic Lin−c-Kit+-Sca-1−-(LK) subsets (Figure 1E) consistent with increased splenic LSK cells and extramedullary hematopoiesis. While Raptor deletion did not affect BM progenitor frequencies, ex vivo colony-forming activity was severely compromised, forming mostly small irregular colonies in complete-cytokine media, which was also observed with total splenocytes (Figure 1F).
Raptor Loss Affects the Cell-Cycle and Induces Metabolic and Gene-Expression Alterations in HSPC
To investigate the effects of Raptor loss on HSPC cell cycle, 5-ethynyl-2’-deoxyuridine (EdU) incorporation 2 days post injection was assessed in HSPC. LSK subtypes as well as LK cells null for Raptor incorporated higher levels of EdU (Figure 1G). Furthermore, Raptor-deleted ST-HSC were more likely to be in the G1 phase of the cell cycle than controls (Figure 1H). However, Raptor-deleted LT-HSCs did not show steady-state cell cycle differences, and neither did Flt3+CD34+LSK (Figure 1H). Moreover, whole BM cells from Raptor-null mice were in the G0 phase of the cell cycle (Figure 1H). We did not detect differences in either caspase activity or reactive oxygen species levels due to Raptor loss (data not shown). Raptor-deleted LSK subsets, as well as LK cells, did show lower levels of RNA (based on Pyronin Y staining) (Figure 1I). To ascertain other metabolic perturbations caused by Raptor loss in HSPC, we performed metabolite profiling on LSK cells.. This analysis revealed elevated intracellular concentrations of AMP and NADP+ in Raptor null cells, indicative of effects in energy and redox homeostasis, respectively, and increased levels of intermediates involved in lipid metabolism (choline and α-glycerophosphocholine). We also observed diminished levels of pyridoxal 5’-phosphate, a key co-factor in transamination reactions and other metabolites involved in nitrogen metabolism (citrulline, ornithine, and glutamate) (Figure 1J). Finally, we assessed gene-expression alterations induced by Raptor loss in HSPC-enriched Flt3-LSK cells. Gene-set enrichment analysis indicated multiple changes in Raptor-null HSPC, including alterations in known mTOR-regulated pathways or processes such as cholesterol biosynthesis and the hypoxia inducible factor pathway, as well as potentially novel mTORC1 -regulated pathways (ie, depletion of NUP98-HOXA9 target genes) (Table S1). Collectively, under homeostatic conditions mTORC1 is not absolutely required for HSC maintenance, but its loss does lead to HSPC mobilization, and evokes significant cell cycle, metabolic, differentiation, and gene-expression changes.
Raptor is Required for HSC Function Post Transplantation
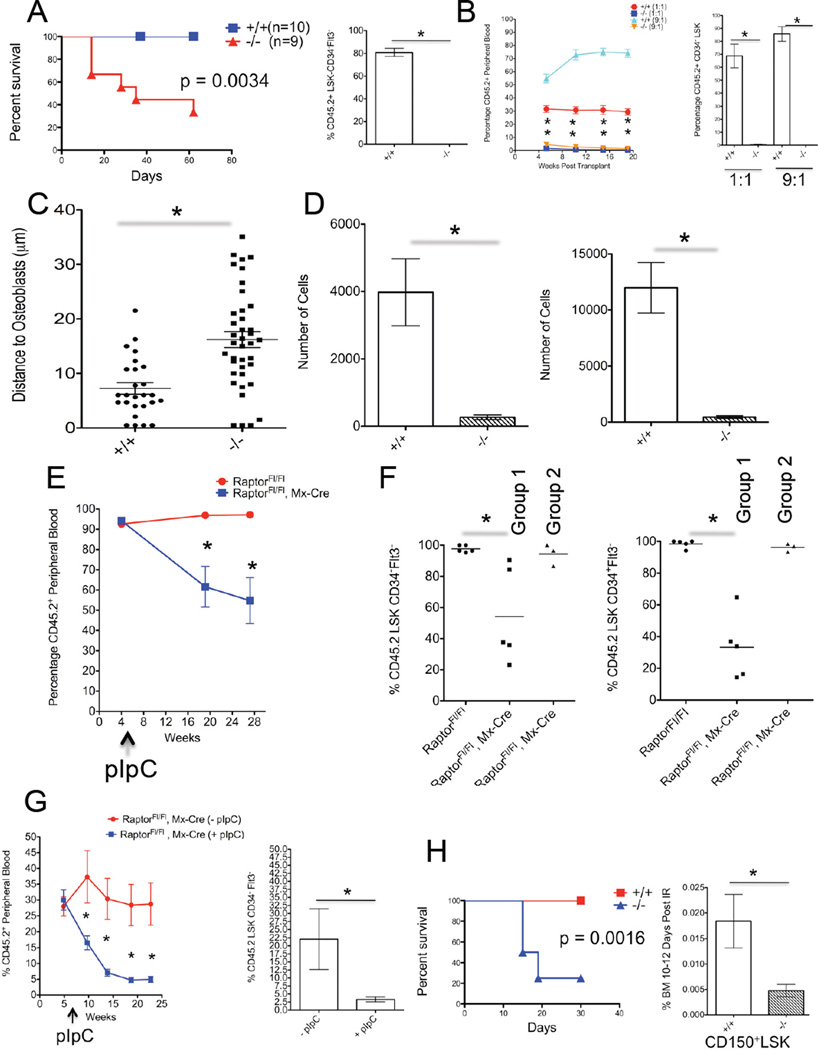
We next assessed the role of Raptor in hematopoietic regeneration by transplanting BM marked by CD45.2+ cells from RaptorFl/Fl or RaptorFl/Fl, MxCre+ mice treated with pIpC (+/+ and −/−, respectively) prior to transplantation into lethally-irradiated congenic CD45.1+ B6.SJL mice, and found that most mice receiving Raptor-null BM did not survive (Figure 2A). We then transplanted cells as above along with whole BM marked as CD45.1+ cells from B6.SJL mice into lethally-irradiated CD45.1+ B6.SJL mice at 1:1 and 9:1 test:competitor ratios. Mice transplanted with −/− showed minimal engraftment beginning as early as 5 weeks post transplantation (Figure 2B). We also transplanted splenocytes at the same manner as above, and again found minimal engraftment from Raptor-null cells (data not shown).
Figure 2. Raptor Is Required for HSC Regeneration in a Cell-Autonomous Manner.
(A) 8 × 105 whole BM cells from pIpC-treated RaptorFl/Fl (+/+) or Raptor Fl/Fl, Mx-Cre (−/−) mice (both CD45.2+) were transplanted into lethally-irradiated CD45.1 mice and survival was monitored. Numbers of recipient mice are indicated (left panel). (p value was derived by Log-rank Test). BM from surviving mice was analyzed for CD45.2 cell contribution to the CD34−Flt3−LSK pool 20 weeks post pIpC (n=3) (right panel).
(B) BM cells from RaptorFl/Fl (+/+) and Raptor Fl/Fl, Mx-Cre (−/−) mice, 6 weeks post pIpC treatment (both CD45.2+), were mixed at the indicated ratios with CD45.1+ BM cells and transplanted into lethally-irradiated CD45.1 recipients. Percentage of CD45.2+ cells in PB is shown over time (n=5) (left panel). 22 weeks post transplantation the percentage of CD45.2+ CD34LSK cells in BM was assessed (n=5) (right panel).
(C) FACS-sorted DiD-labeled LSK-CD48−CD150+ from pIpC-treated RaptorFl/Fl(+/+) and Raptor Fl/Fl, Mx-Cre (−/−) mice were transplanted into lethally-irradiated Col2.3GFP mice and ~24 hrs later live imaging was performed to assess relative distance of HSPC to osteoblasts. Shown are results from 2 independent experiments performed with 5,000–10,000 HSPC.
(D) 200 LT-HSC or ST-HSC from pIpC-treated RaptorFl/Fl (+/+) and Raptor Fl/Fl, Mx-Cre (−/−) mice were used to initiate cultures in serum-free conditions that promote HSC expansion ex vivo. 7–9 days post culture the number of cells was enumerated. Data are from 3 independent experiments performed in duplicate.
(E) 8 × 105 BM cells from non pIpC-treated RaptorFl/Fl or Raptor Fl/Fl, Mx-Cre mice were transplanted into lethally-irradiated CD45.1 recipients. 5 weeks post transplantation mice were injected with 3 doses of pIpC over 5 days. PB chimerism was assessed by measuring the contribution of CD45.2+ cells over the indicated time (n=9–10 recipients).
(F) BM from mice from (E) was analyzed for CD45.2 chimerism in CD34−Flt3-LSK fractions 25 weeks post pIpC. Raptor Fl/Fl, Mx-Cre recipients were separated into two CD45.2+ groups, one with low CD45.2 chimerism that retained deletion (Group1) and ones with high CD45.2 chimerism that escaped deletion (Group 2). (n=4–5). (See Figure S2C).
(G) 2.5 × 105 BM cells from uninduced Raptor Fl/Fl, Mx-Cre were mixed with 2.5 × 105 CD45.1 cells and transplanted into lethally-irradiated CD45.1 recipients. 5.7 weeks post transplantation ½ of the mice were injected with pIpC as in (D) (+pIpC). Contribution of CD45.2 cells to PB was assessed over the indicated time points. (n=6–8) (left panel). Recipient BM was analyzed for CD45.2 cell contribution to the CD34−Flt3-LSK pool 21 weeks post pIpC (n=6–8) (right panel).
(H) Kaplan-Meier (KM) survival curve from pIpC-treated RaptorFl/Fl (+/+), n=10, and Raptor Fl/Fl, Mx-Cre (−/−), n=4, mice that received sub-lethal irradiation. The one surviving −/− mouse had escaped deletion (data not shown) (left panel). (p value was derived by Log-rank Test). pIpC-treated RaptorFl/Fl (+/+) and Raptor Fl/Fl, Mx-Cre (−/−) mice received sub-lethal irradiation and shown is the frequency of BM CD150+-LSK cells 10–12 days post irradiation (IR). (n=4).
(A–H) Data are expressed as mean or mean ± SEM (*p<0.05).
To assess the homing capacity of Raptor null HSPC. LSK-CD48−CD150+ cells were isolated from pIpC-treated RaptorFl/Fl or RaptorFl/Fl, MxCre+ mice, labeled with DiD and transplanted into mice with GFP-expressing osteoblasts (Col2.3-GFP) (Lo Celso et al., 2009). By using live in vivo imaging, we did not observe any obvious defects in homing. Raptor-null HSPC however did localize further from osteoblast cells (Figure 2C). This could reflect a requirement for mTORC1 in integrating niche signals correctly. In fact, Raptor-null LT-HSC or ST-HSC couldn’t initiate and/or sustain ex vivo cultures in serum-free conditions containing niche-secreted factors (Zheng et al., 2011) (Figure 2D). Overexpression of several downstream mTORC1 effectors, as well as potential novel targets (namely Hoxa9), could not restore ex vivo growth in the absence of Raptor.
Raptor is Required Cell-Autonomously for HSC Regeneration
To examine the cell-autonomous phenotypes, we transplanted BM from non-pIpC treated RaptorFl/Fl or RaptorFl/Fl, MxCre+ mice (without helper/competitor) into lethally-irradiated CD45.1+ B6.SJL mice, waited ~5 weeks, then induced deletion with pIpC. Approximately 1 month post pIpC treatment all mice receiving RaptorFl/Fl, MxCre cells developed leukopenia, while mice receiving RaptorFl/Fl did not (Figure S2A). Furthermore, CD45.2+ cells from pIpC-treated mice receiving RaptorFl/Fl, MxCre+ cells were mostly Mac-1+Gr-1Mid/Lo(Figure S2B). After 16 weeks host CD45.1+ cells began to contribute substantially to peripheral chimerism only in pIpC-treated mice receiving RaptorFl/Fl, MxCre+ cells (Figure 2E). We then analyzed the BM and spleens of transplant recipients and observed that mice from pIpC treated RaptorFl/Fl, MxCre+ could be segregated into two groups. One (Group 1) had maintained relatively low percentages of CD45.2+ cells in the BM and spleen, and had retained deleted-Raptor alleles, whereas the other (Group 2) contained a high percentage of CD45.2+, but contained mostly floxed Raptor alleles (ie, had escaped deletion) (Figure S2C). This differential chimerism was also observed in LT-HSC and ST-HSC fractions (Figure 2F), suggesting a cell autonomous role for Raptor in HSC self-renewal. Finally, we transplanted BM cells from RaptorFl/Fl, MxCre+ mice with BM cells from B6.SJL mice at a 1:1 ratio, waited for stable engraftment (~5 weeks), then induced deletion of Raptor alleles in half the mice. Mice receiving pIpC displayed a steady reduction in peripheral chimerism, and Raptor-null grafts were outcompeted (Figure 2G).
To assess the regenerative capacity of Raptor-null HSC in non-transplantation setting, pIpC-treated RaptorFl/Fl (+/+) or RaptorFl/Fl, MxCre+ (−/−) mice were treated with a sub-lethal dose of irradiation. While all +/+ mice survived this treatment and showed signs of BM regeneration over time, most −/− died rapidly, without signs of normal BM regeneration (Figure 2H and data not shown). Furthermore, while +/+ LT-HSC enriched CD150+-LSK cells were reduced by 5.8+/−1.3 fold relative to unirradiated mice 10–12 days post irradiation, there was a 16.9+/−3.7 fold decrease in this population in −/− mice (p<0.05) (Figure 2H). Taken together, Raptor/mTORC1 activity appears to be generally required in regenerative hematopoietic settings.
Neither mTOR complex is Absolutely Required In Hematopoeitic Homeostasis
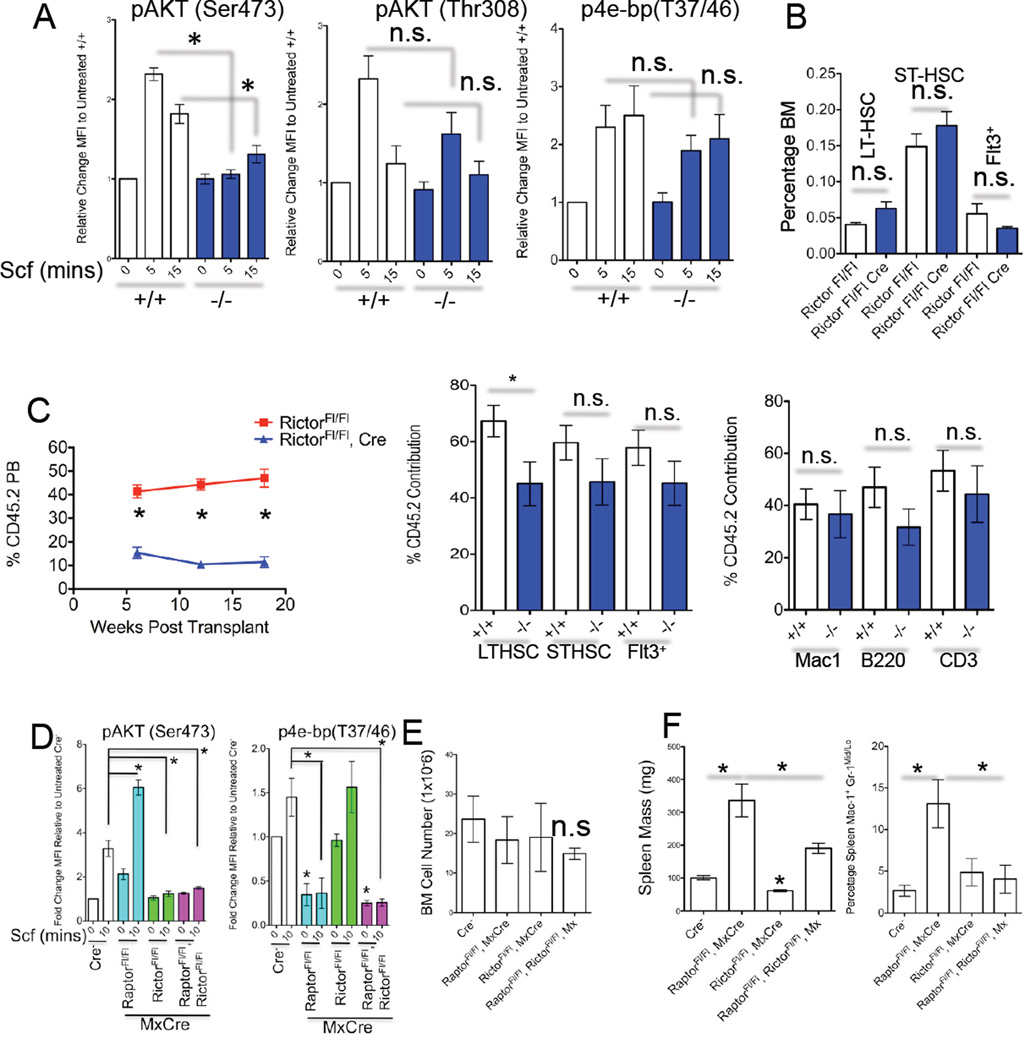
To determine the role of mTorc2 in adult hematopiesis we utilized mice with a floxed Rictor exon 3 allele (Shiota et al., 2006) and generated RictorFl/Fl or RictorFl/Fl, MxCre+ animals. pIpC treatement of RictorFl/Fl, MxCre+ mice ablated mTORC2 activity in HSPC in response to Scf, but did not affect phosphorylation of AKT at Thr308 nor mTORC1 activity (Figure 3A). Deletion of Rictor in the hematopoietic system was tolerated and primarily resulted in diminished peripheral blood (PB) lymphocytes, reduction in spleen mass, and a decrease in splenic B220+IgMLoIgD+ cells (Figure S3A–S3C and data not shown), phenotypes consistent with loss of PI3K/AKT signaling (Fruman et al., 1999; Calamito et al., 2010). Rictor loss did not affect HSPC number by immunophenotype (Figure 3B and data not shown) and did not diminish HSPC chimerism at 30 weeks in a competitive transplant (Figure 3C). However, we did observe that Rictor-null BM contributed to lower levels of PB cells particularly of the lymphoid lineages (Figure 3C and Figure S3D).
Figure 3. Rictor is Required for mTORC2 Activity in HSPC, but Not LT-Hematopoietic Regeneration.
(A) Flow cytometry was performed on sorted LIN−PI− cells from pIpC-treated RictorFl/Fl (+/+) and Rictor Fl/Fl, Mx-Cre (−/−) mice that were treated with Scf for the indicated time (mins). Shown are data from fixed/permeablized LSK-gated events and fold change in MFI for the indicated p-protein is shown relative to untreated +/+ cells. (n=3–4 mice).
(B) Frequency of BM LSK subsets from RictorFl/Fl (+/+) and Rictor Fl/Fl, Mx-Cre (−/−) mice 4 months post pIpC treatment. (n=4–5).
(C) 5 × 105 whole BM cells from pIpC-treated RictorFl/Fl (+/+) and Rictor Fl/Fl, Mx-Cre (−/−) mice, (both CD45.2+), were mixed with the same amount of CD45.1+ BM cells and transplanted into lethally-irradiated CD45.1 recipients. The percentage of CD45.2+ cells in PB is shown over the indicated time (n=6–7 recipients). Panels on the right show the % contribution of CD45.2+ cells to the indicated BM populations 30 weeks post transplantation. Data are from 1 of 2 experiments.
(D) The levels of indicated p-proteins were assessed as in (A) in Scf-treated LSK-gated cells from pIpC-treated mice of the indicated genotypes. (n=3–4 mice per genotype)
(E) BM cell number (n=2–9), spleen mass (F) (n=5–9) and percentage of Mac-1 +Gr-1Mid/Lo splenic cells (F right panel) (n=3–5) are shown from mice of the indicated genotype 1–3 months post pIpC treatment.
(A–F) Data are expressed as mean ± SEM (*p<0.05, n.s. p>0.05).
Finally, we assessed the requirement of both mTORC1 and mTORC2 in hematopoietic homeostasis by generating compound RaptorFl/Fl, RictorFl/Fl, MxCre+. pIpC-treatment of RaptorFl/Fl, RictorFl/Fl, MxCre+ led to reduced phosphorylaton of both mTORC1 and mTORC2 substrates in HSPC (Figure 3D). Compound-deleted mice had phenotypes largely resembling hematopoeitic Raptor-deletion, and did not develop BM failure (Figures 3E and 3F and Figures S3E–S3I and data not shown). (Figure 3E). Lastly, compound mice retained deleted alleles of both Raptor and Rictor up to 10 months post deletion (data not shown). Taken together, mTORC1 and/or mTORC2 play largely non-redundant roles in hematopoiesis.
Raptor Deletion Prolongs Survival in Models of Pten-loss-Evoked Leukemogenesis
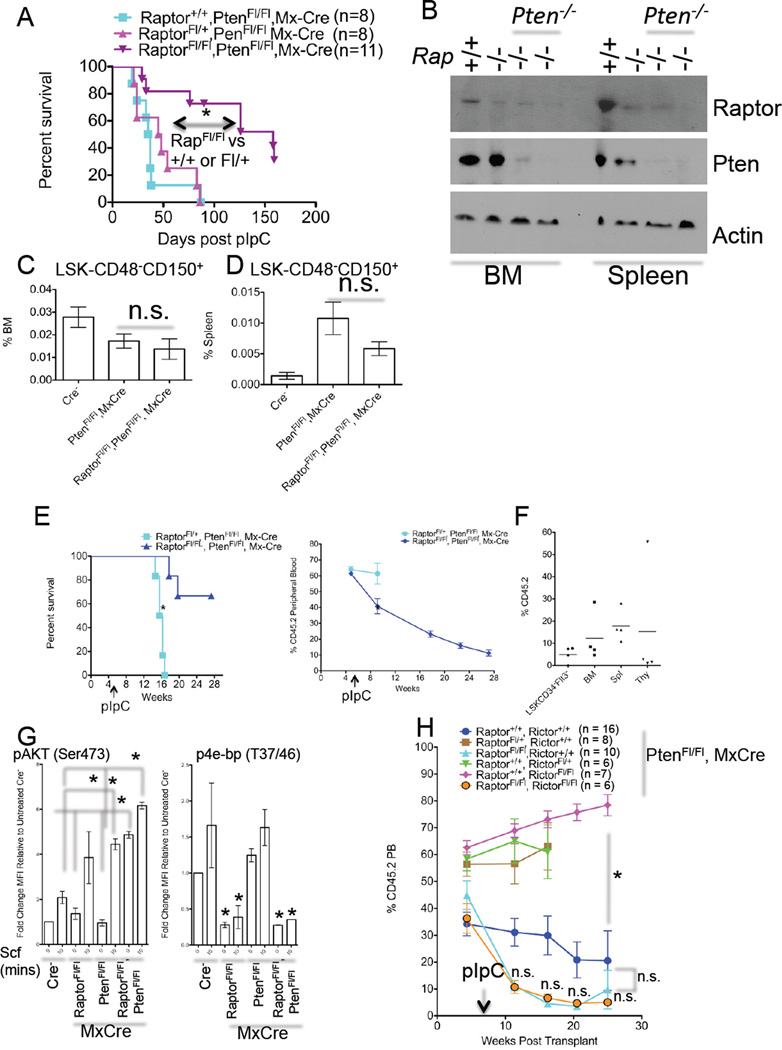
Since hyperactivation of mTOR is observed in many hematological malignancies, (Chapuis et al., 2011), we asked whether complete loss of function of mTORC1 could impact leukemogenesis in vivo. We utilized PtenFl/Fl, MxCre mice that develop symptoms of MPN, can progress to acute leukemia, and whose leukemias are sensitive to rapamycin (Yilmaz et al., 2006; Zhang et al., 2006). We produced PtenFl/Fl mice that were either Raptor+/+, RaptorFl/+, or RaptorFl/Fl (all MxCre positive) and induced deletion. Homozygous, but not heterozygous, Raptor deletion significantly prolonged survival of Pten-null mice, and ameliorated some but not all neoplasia-associated phenotypes (Figures S4B–S4E). However, homozygous Pten-deletion did lead to lethality in the majority of compound mice (Figure 4A). Surviving doubly-deleted mice displayed slightly enlarged spleens and significantly larger livers relative to induced RaptorFl/Fl, MxCre mice (532 +/− 83.8 vs. 318 +/− 37.72 mg for spleen weights, p<0.05 and 2.44 +/− .347 vs. 1.16 +/− .113 g for liver weights, p<0.05, n=4–6). WBC liver infiltrates were apparent in some animals, while some animals displayed large amounts of liver lipid deposits (data not shown). Efficient deletion in most cells for both Raptor and Pten alleles was detected (Figure 4B and Figure S4A). Finally, Raptor loss had no effect on the frequency of Pten-loss evoked changes to BM or splenic LSK-CD48−CD150+ cells (Figure 4C).
Figure 4. Raptor is Required for Efficient Leukemogenesis Evoked by Pten Loss.
(A) KM curve from PtenFl/Fl, MxCre mice of the indicated Raptor genotypes that were treated with pIpC. Number of mice is indicated (*p<0.05 Log-rank Test).
(B) Western blots were performed with BM and spleen extracts from mice of the indicated genotypes ~6 months post pIpC using the indicated antibodies. Shown are samples from 2 individual pIpC-treated RaptorFl/Fl, PtenFl/Fl, MxCre mice.
(C) The frequency of BM and Splenic (D) LSK-CD48−CD150+ cells was assessed from mice of the indicated genotypes 2.5–5 weeks post pIpC treatment (n=6–10).
(E) 1 × 106 BM cells from non pIpC-treated mice of the indicated genotypes (CD45.2) were mixed with 5 × 105 CD45.1 cells and transplanted into lethally-irradiated CD45.1 mice. 5.5 weeks post transplant mice received 3 injections of pIpC over 6 days. Survival (left) and chimerism (right) were then assessed (n=4–6, *p<0.05 Log-rank Test).
(F) Chimerism was assessed from mice described in (E). (n=4)
(G) Assessment of p-protein levels in LSK-gated events from Scf-treated LIN-PI-cells of the indicated gentoypes (n=2–3).
(H) BM cells from mice of the indicated genotypes (all from PtenFl/Fl, MxCre backgrounds) were transplanted and treated as in (D). Number of transplant recipients is indicated.
(A–H). Data are expressed as mean or mean ± SEM. (*p<0.05, n.s. p>0.05).
To investigate if these effects were cell autonomous, we transplanted PtenFl/Fl, MxCre+ cells that were either RaptorFl/+ or RaptorFl/Fl (both CD45.2+) along with CD45.1+ helper/competitor cells at a 2:1 ratio into B6.SJL mice, waited ~5 weeks for engraftment, then treated with pIpC. In this system, most mice receiving PtenFl/Fl, MxCre BM develop hematological malignancies most prevalent being T-acute lymphoblastic leukemia/lymphoma (T-ALL) (Lee et al., 2010). Heterozygous deletion of Raptor in this model led to a median survival of 11 weeks post pIpC, while homozygous deletion of Raptor only lead to lethality in a few of the transplant recipients (Figure 4E). Notably, after 22 weeks homozygous deletion of Raptor lead to reduced chimerism (Figure 4E–4F). Therefore, Raptor is required in a cell-autonomous manner for efficient Pten lossevoked leukemogenesis.
Although diminished, we did observe T-cell lymphoma/leukemia development in a minority of Pten/Raptor double knockouts (that could be transplanted [data not shown]). As mTORC2 is functionally active in Raptordeleted cells, we sought to determine the role of mTORC2 in Pten-loss disease in the absence of Raptor. We observed the highest levels of AKT phosporylation at Ser473 in Raptor/Pten double homozygous-deleted LSK cells (Figure 4G). Hyperactivation of pAKT should predictably result in diminished Foxo activity (Guertin et al., 2006), which has previously been associated with depletion of HSC function (Tothova et al., 2007) as well as impaired leukemogenesis (Sykes et al., 2011). We thus generated RaptorFl/Fl, RictorFl/Fl, PtenFl/Fl, MxCre+ triple-knockout mice and induced deletion [Figure S4F]). However, all triple-knockout mice died relatively rapidly (median survival of 19 days [n=6] without signs of BM failure [data not shown]). We next transplanted non pIpC-treated BM from RaptorFl/Fl, RictorFl/Fl, PtenFl/Fl, MxCre mice along with controls (Figure 4H) as above, and induced deletion 5–7 weeks post transplantation. The additional loss of Rictor (in the Raptor/Pten double knockout cells) did not lead to any differences relative to Raptor-deleted Pten-homozygous chimerism (Figure 4H). Notably, homozygous deletion of Rictor alone did allow for stable LT PB chimerism and prolonged survival (Figure 4H and data not shown).
DISCUSSION
We found that Raptor/mTORC1 plays critical functions in HSPC, hematopoietic differentiation, and leukemogenesis. Previous work has established chronic mTOR activation as depleting HSC function. Herein, we identify mTORC1 to be required for HSC function under regenerative settings. It is likely then that the level of mTORC1 activity needs to be balanced in HSC, whereas either increased or decreased mTORC1 levels result in diminished capacity for HSC function. As mTOR lies downstream of several hematopoietic growth factor/cytokine receptors this could reflect a particular need for this signaling axis in self-renewal under regenerative conditions. In fact, thrombopoietin (Thpo), which partially signals through mTOR, has been shown to be essential for HSC function (Qian et al., 2007). Of note, Thpo deficiency evokes similar decreases in several self-renewal associated HoxA genes which are also downregulated in Raptor-null HSPC (Table S1). We propose that mTORC1 enables proper HSC sensing of both niche factors and cellular energetic state, and loss of function of mTORC1 leads to imprecise integration of these signals and loss of regenerative potential.
While loss of Raptor leads to multiple hematopoietic abnormalities, homozygous-Raptor deletion in hematopoietic cells is tolerated in mice, as is complete ablation of both mTORC1 and mTORC2 activity under homeostatic hematopoietic settings. We have demonstrated that mTORC1 loss via homozygous Raptor ablation is sufficient to extend survival in mouse models of Pten-loss evoked leukemogenesis in a cell-autonomous manner. Previous results have shown that Pten-loss evoked HSC depletion is rapamycin sensitive (Yilmaz et al., 2006). In contrast, our results show that homozygous deletion of Pten and Raptor leads to a gradual loss in reconstituting capacity (Figure 4D). It is possible that rapamycin-sensitive functions of mTORC1 downstream of Pten loss, perhaps the upregulation of the p53/p19Arf/p16Ink4a axis, lead to HSC depletion (Lee et al., 2010), while rapamycin-insensitive functions of mTORC1 are required for HSC regeneration independent of Pten levels. These rapamycin-sensitive functions of mTORC1 are likely downstream of mTORC2 signaling in response to Pten loss. In fact, Magee et al. (2012) demonstrate that loss of Rictor extends survival of pIpC-treated PtenFl/Fl, MxCre mice, while also showing that rapamycin does not affect mTORC2 activity in HSC in vivo. Unlike some Raptor-deleted mice in this setting (MxCre), Rictor-deletion was not compatible with acute leukemia/lymphoma development. Furthermore, like rapamycin treatment and unlike Raptor deletion, Rictor deletion restores normal function to Pten-deleted HSC (Magee et al., 2011). Taken together, both mTORC1 and mTORC2 functions appear to be required downstream of Pten loss in HSC function as well as in leukemogenesis, while mTORC1 functions appear to be important for HSC regeneration regardless of Pten status.
This work demonstrates previously uncharacterized functions of mTORC1 in hematopoiesis, non-redundant hematopoietic functions of mTORCs, and confirms mTORC1 requirements in cancers with hyperactivation of the PI3K pathway. This work also provides mechanistic insight into the role of specific signaling pathways that control hematopoietic regeneration and will inform on therapeutic strategies.
EXPERIMENTAL PROCEDURES
Mouse Experiments
Mouse strains are detailed in Supplemental Experimental Procedures. Deletion of floxed alleles was induced by intraperitoneal (IP) injections of 4–8 week old Mx-1-cre mice with 15 µg/g body weight pIpC three times over 5–6 days, or UBC-cre-ER compound mice with 200 µg/g body weight Tamoxifen (Sigma, St. Louis, MO) once daily for 3 days. For transplantation experiments mice received lethal doses of irradiation (2 doses at 550 rad 3 hours apart) prior to transplantation of BM cells by retroorbital injection on the same day. Complete blood counts were performed on PB samples using a Hemavet (Drew Scientific, Dallas, TX).
Flow Cytometry
Antibodies utilized for flow cytometry are detailed in Supplemental Experimental Procedures. Cells were isolated from BM either by flushing long bones or alternatively crushing bones in a mortar and pestle, RBC lysed with BD Pharmlyse™ lysis buffer (BD), and cells passed through 40 or 70 µm nylon cell strainers (BD). BM cells used in sorting experiments were enriched for Lin− cells by staining with PE-Cy5-conjugated rat anti-mouse antibodies, followed by incubation with sheep anti-rat Dynabeads (Invitrogen, Carlsbad, CA), and cell separation with magnetic columns. Intracellular flow cytometry was performed as described previously (Kalaitzidis and Neel, 2008). EdU incorporation was assessed by IP injection of mice with 60 mg/kg EdU, and incorporation analyzed in FACS-sorted cell populations with the Click-iT™ EdU (Alexa Fluor-647) Assay kit (Invitrogen). For Ki67/DNA staining, FACS-sorted cells were washed once with PBS, resuspended in cold 70% Ethanol and left at −20°C overnight. Cells were pelleted, washed once with 0.5%BSA/PBS, and stained with a PE-Cy7-conjugated antibody to Ki67 (B56 [BD]), and 30 µM Hoechst 33342 (Invitrogen) for 30 minutes at 20°C prior to preparation for flow-cytometric analysis. For DNA/RNA content measurements, BM cells in 10%FBS/IMDM/50µg/mL verapamil were incubated with 30 µM Hoechst (Ho) 33342 for 45 mins at 37°C, cells were washed then stained for surface markers and 3 µg/mL Pyronin Y (Sigma) for 20 min at 37°C. Enumeration of 200 FACS-sorted LT-HSC or ST-HSC grown in 500 µL in wells of 12-well plates in conditions described previously (Zheng et al., 2011), was with Countbright™ Absolute Counting Beads (Invitrogen).
Gene-Expression Analysis
RNA from FACS-sorted cells was isolated with the Arcturus® PicoPure® RNA isolation Kit (Applied Biosystems, Carlsbad, CA). cDNA was synthesized using the High-Capacity cDNA Reverse Transciption Kit (Applied Biosystems). Quantitative PCR was performed using Taqman® probes to murine Gapdh, and Raptor (Mm00712697_m1 spanning exons 5–6, Mm01242616_g1 spanning exons 8–9). qPCR reactions were run on either Applied Biosystems 7300 or 7500 RT PCR Systems. Details of microarray experiments are supplied in Supplemental Experimental Procedures.
Metabolite Profiling
For metabolite profiling BM cells from 4 mice per genotype were pooled and used to FACS sort 100,000–150,000 LSK cells (1 biological replicate), cells were washed with PBS, and resuspended in 200 µL of 100% methanol. Metabolite measurements are from 3 biological replicates per genotype. A detailed description of the methods used to obtain raw data is in the Supplemental Experimental Procedures.
In vivo Imaging
In vivo imaging was performed on 5,000–10,000 Vybrant DiD-dye labeled (Invitrogen) LSK-CD48−CD150+ FACS-sorted cells using Col2.3-GFP recipient mice as described previously (Lo Celso et al., 2009) and detailed in Supplemental Experimental Procedures.
Statistical Analysis
The statistical significance of differences between population means was assessed by two-tailed unpaired Student’s t test, unless otherwise indicated.
Supplementary Material
HIGHLIGHTS.
Homozygous deletion of Raptor reduces mTORC1 activity in HSPC.
Inactivation of mTORC1 leads to expansion of monocytes and pancytopenia.
Raptor/mTORC1 activity is required for HSC regeneration.
Raptor/mTORC1 is required for Pten-loss evoked leukemogenesis.
ACKNOWLEDGMENTS
We wish to thank Rebekka Schneider-Kramann for assistance with pathology, Kristina M. Brumme for assistance with mouse husbandry, and Michael G. Kharas for review of the manuscript. D.K. was supported by NIDDK grant K01DK092300, C.B.C. was partially supported by the Nestle Research Center, D.G.G. is currently a full-time employee of Merck and Co. Inc., D.T.S was supported by NIH grants HL097794, HL097748, HL100402 and DK050234, D.A.G. was supported by NIH grant R00 CA129613 and the PEW Charitable Trust, S.A.A. was supported by grants from the Leukemia and Lymphoma Society, NIH grant CA66996, and Charles H. Hood Foundation (CA105423).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Microarray data have been deposited at the NCBI Gene-Expression Omnibus with accession code GSE32265.
SUPPLEMENTAL INFORMATION
Supplemental Information includes 4 supplemental figures and figure legends, 1 supplemental table, and Supplement Experimental Procedures.
REFERENCES
- Campbell TB, Basu S, Hangoc G, Tao W, Broxmeyer HE. Overexpression of Rheb2 enhances mouse hematopoietic progenitor cell growth while impairing stem cell repopulation. Blood. 2009;114:3392–3401. doi: 10.1182/blood-2008-12-195214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 2008;27:5527–5541. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu R, Ikenoue T, Guan K-L, Zheng P. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J. Exp. Med. 2008;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis N, Tamburini J, Green AS, Willems L, Bardet V, Park S, Lacombe C, Mayeux P, Bouscary D. Perspectives on inhibiting mTOR as a future treatment strategy for hematological malignancies. Leukemia. 2010;23:1686–1699. doi: 10.1038/leu.2010.170. [DOI] [PubMed] [Google Scholar]
- Christensen JL, Weissman IL. Flk-2 is a marker in hematopoeietic stem cell differentiation: A simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. USA. 2001;98:14541–14546. doi: 10.1073/pnas.261562798. Genet. 7, 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Sahin E, Jiang S, Sanchez-Aguilera A, Scott KL, Chin L, Williams DA, Kwiatkowski DJ, DePinho RA. mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc. Natl. Acad. Sci. USA. 2008;105:19384–19389. doi: 10.1073/pnas.0810584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Saitoh M, Kinkel S, Crosby K, Sheen J-H, Mullholland DJ, Magnuson MA, Wu H, Sabatini DM. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell. 2009;15:148–159. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Shubbert S, Chen JY, Valamehr B, Mosessian S, Shi H, Dang NH, Garcia C, Theodoro MF, Varella-Garcia M, et al. Suppression of leukemia development caused by PTEN loss. Proc. Natl. Acad. Sci. USA. 2011;108:1409–1414. doi: 10.1073/pnas.1006937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1 -mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes MR, Limon JJ, So L, Chen J, Lim RJ, Chavez MA, Vu C, Lilly MB, Mallya S, Ong ST, et al. Effective and selective targeting of leukemia cells using a TORC1/2 inhibitor. Nat. Med. 2010;16:205–214. doi: 10.1038/nm.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaitzidis D, Neel BG. Flow-cytometric phosphoprotein analysis reveals agonist and temporal differences in responses of murine hematopoietic stem/progenitor cells. PLoS One. 2008;11:e3776. doi: 10.1371/journal.pone.0003776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhost C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kharas MG, Okabe R, Gani JJ, Gozo M, Khandan T, Paktinat M, Gilliland GG, Gritsman K. Constitutive active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood. 2010;115:1406–1415. doi: 10.1182/blood-2009-06-229443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Nakada D, Yilmaz OH, Tothova Z, Joseph NM, Lim MS, Gilliland DG, Morrison SJ. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell. 2010;7:593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Celso C, Fleming HE, Wu JW, Zhao CX, Lye, Miake-Lye S, Fujisaki J, Cote D, Rowe DW, Lin CP, Scadden DT. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 2009;457:92–96. doi: 10.1038/nature07434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee JA, Ikenoue T, Nakada D, Lee JY, Guan K-L, Morrison SJ. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 2012 doi: 10.1016/j.stem.2012.05.026. X, X-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopietic stem cell compartment. J. Exp. Med. 2010;207:1178–1182. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Hanada K-I, Hamada H, Nakauchi H. Long-term lymphohematoopietic reconstitution by a single CD34-low/negative hematpoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. mTOR Complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Qian H, Buza-Vidas N, Hyland CD, Jensen CT, Antonchuk J, Mansson R, Thoren LA, Ekblom M, Alexander WS, Jacobsen SEW. Critical role of thrombopoietin in maintaining adult quiescent hematopoietic stem cells. Cell Stem Cell. 2007;1:671–684. doi: 10.1016/j.stem.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Ruzankin Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 1997;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen J-H, Hsu PP, Bagley AF, Markhand AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- Shiota C, Woo JT, Shelton KD, Magnuson MA. Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev. Cell. 2006;4:583–589. doi: 10.1016/j.devcel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Sykes SM, Lane SW, Bullinger L, Kalaitzidis D, Yusuf R, Saez B, Ferraro F, Mercier F, Singh H, Brumme KM, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;5:697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiological oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Warr MR, Pietras EM, Passegue′ E. Mechanisms controlling hematopoietic stem cell functions during normal hematopoiesis and hematological malignancies. Wiley Interdiscip. Syst. Biol. Med. 2011;3:681–701. doi: 10.1002/wsbm.145. [DOI] [PubMed] [Google Scholar]
- Yang L, Bryder D, Adolfsson J, Nygren J, Mansson R, Sigvardsson M, Jacobsen SE. Identfication of Lin−Sca1+kit+CD34+Flt3− short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 1995;105:2717–2723. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Ville’n J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. Phophoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedermann LM, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- Zheng J, Umikawa M, Zhang S, Huynh H, Silvany R, Chen BPC, Chen L, Zhang CC. Ex Vivo expanded hematopoietic stem cells overcome the MHC barrier in allogeneic transplantation. Cell Stem Cell. 2011;9:119–130. doi: 10.1016/j.stem.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.