Abstract
Numerous diseases of wildlife have recently emerged due to trade and travel. However, the impact of disease on wild animal populations has been notoriously difficult to detect and demonstrate, due to problems of attribution and the rapid disappearance of bodies after death. Determining the magnitude of avian mortality from West Nile virus (WNV) is emblematic of these challenges. Although correlational analyses may show population declines coincident with the arrival of the virus, strong inference of WNV as a cause of mortality or a population decline requires additional evidence. We show how integrating field data on mosquito feeding patterns, avian abundance, and seroprevalence can be used to predict relative mortality from vector-borne pathogens. We illustrate the method with a case study on WNV in three species of small songbirds, tufted titmouse (Baeolophus bicolor), Carolina wrens (Thryothorus ludovicianus), and northern cardinals (Cardinalis cardinalis). We then determined mortality, infectiousness, and behavioral response of wrens and titmouse following infection with WNV in laboratory experiments and compared them to a previous study on WNV mortality in cardinals. In agreement with predictions, we found titmouse had the highest mortality from WNV infection, with 100% of eleven birds perishing within seven days after infection. Mortality in wrens was significantly lower at 27% (3/11), but still substantial. Viremia profiles indicated that both species were highly infectious for WNV and could play roles in WNV amplification. These findings suggest that WNV may be killing many small-bodied birds, despite the absence of large numbers of dead birds testing positive for WNV. More broadly, they illustrate a framework for predicting relative mortality in hosts from vector-borne disease.
Keywords: Disease, population, birds, epidemiology, West Nile virus, conservation
Introduction
The impact of disease on wild animal populations has been notoriously difficult to detect and demonstrate, due to problems of attribution and the rapid disappearance of bodies after death (McCallum 2005; McCallum & Dobson 1995). The clearest examples of disease-caused impacts on wildlife populations come from epidemics in large abundant animals such as anthrax and Rinderpest in African mammals (Holdo et al. 2009), experimental or purposeful viral introductions such as myxomatosis and Australian rabbits (Ratcliffe et al. 1952), and experimental studies that remove pathogens from hosts through treatment (Hudson et al. 1998). For many other diseases and populations, impacts are inferred from long term monitoring and observations of sudden declines, and in rare cases scientists have been able to observe a wave of mortality as a pathogen arrives (Hochachka & Dhondt 2000; Kilpatrick et al. 2010; Langwig et al. 2012; Lips et al. 2006; Vredenburg et al. 2010). However, in many cases mortality due to disease is difficult to detect and even striking patterns, such as distributional limits coincident with disease boundaries, required experimental infection studies to confirm impacts of disease (e.g., avian malaria and Hawaiian birds; (Van Riper et al. 1986; Warner 1968)).
A recent introduction of a pathogen to North America, West Nile virus (WNV; Flaviviridae; Flavivirus) in 1999, was also accompanied by waves of mortality in wild birds, with large numbers of dead American crows and Blue jays testing positive for WNV in the northeast USA (Bernard et al. 2001; Nemeth et al. 2007). A decade later, transmission still occurs annually in many bird communities throughout North and South America (Kilpatrick 2011). Several retrospective analyses have shown population declines in birds coincident with the arrival of WNV as it spread south and west from New York, with impacts being largest on corvids (Hochachka et al. 2004; LaDeau et al. 2007; Wheeler et al. 2009). Evidence of WNV-caused mortality in corvids was also provided by experimental infection in laboratory studies (Komar et al. 2003; Reisen et al. 2005). However, evidence of WNV mortality in smaller passerines has been far sparser, with relatively few WNV-infected dead birds collected. The extent to which this is due to poor detectability (Ward et al. 2006) or lack of an mortality is not clear.
Two families of small passerines that may suffer population level impacts from WNV are Paridae (chickadees and titmouse) and Troglodytidae (wrens). Multiple studies have observed declines in one or more species in the family Paridae and Troglodytidae coincident with the arrival of WNV (Bonter & Hochachka 2003; LaDeau et al. 2007), and several other studies have demonstrated feeding on parids and wrens by WNV mosquito vectors (Hamer et al. 2009; Hassan et al. 2003; Kilpatrick et al. 2006a). However, these data are only suggestive and supportive evidence in the form of WNV-infected dead chickadees, titmouse or wrens is mostly lacking.
The gold standard to determine whether a species suffers mortality from a pathogen, part of Koch's postulates (Koch 1893), is through experimental infection. There are far too many species of birds in North America to do this for all taxa, and these studies cannot determine whether in fact birds are exposed to a pathogen in nature. For effective conservation planning, there is clearly a need for a framework to determine assess whether WNV and other vector-borne diseases cause mortality in small avian hosts (and other small wildlife species that are difficult to detect).
Here we describe how one can use field data on the transmission ecology of a vector-borne disease – specifically the feeding patterns of WNV mosquito vectors, avian abundance, and the WNV antibody prevalence of wild-caught birds – to generate hypotheses about differences in mortality from WNV infection between hosts. We illustrate this method with a study on three species of small songbirds, tufted titmouse (Baeolophus bicolor), Carolina wrens (Thryothorus ludovicianus), and northern cardinals (Cardinalis cardinalis). We generate and tested hypotheses about the relative mortality of three species and measuring morbidity and mortality following experimental infection with WNV. Experimental infection studies also provide data on infectiousness for WNV that can be integrated with the aforementioned data on mosquito preferences to determine the role of different species in WNV transmission (Kilpatrick 2011; Kilpatrick et al. 2006a).
Methods
Framework for predicting relative host mortality from a vector-borne pathogen
This framework generates a prediction about the relative mortality from infection with a vector-borne pathogen between two or more species.
The seroprevalence, S, or fraction of a population with antibodies against a pathogen at a point in time is equal to the fraction of the population exposed, e, multiplied by the probability of survival (1-m, where m is the probability of mortality given infection), divided by the total population size after exposure, (e(1-m)+1-e):
| (1) |
The fraction of the population exposed, e, will increase asymptotically with the average number of infective bites, I, each host receives (Smith et al. 2005):
| (2) |
where k is parameter controlling the degree to which mosquitoes feed more on some individuals of a species than others (Dye & Hasibeder 1986). Previous work suggests that in some populations k is approximately 0.25 (Smith et al. 2005). Simulations suggest that using k = 0.25 produces qualitatively correct predictions about which species suffers higher mortality as long as k is not too small (i.e. as long as bites aren't extremely concentrated on just a few individuals).
The number of bites that a population that is exposed to will increase with the host utilization index (sometimes termed mosquito preference, forage ratio, or host selection index) of vectors, U, on that host population, where the utilization index is the fraction of bloodmeals, b, from that host population divided by the relative abundance of that host, a (i.e. the fraction of all hosts made up by that host):
| (3) |
Thus, if data on host utilization, U, and seroprevalence, S, is available for two or more species at the same site(s), they can be used to predict which species has a higher mortality probability, m, given infection. First, it is necessary to invert equation (2) and derive an approximate value of infective bites, I, using the measured seroprevalence, S:
| (4) |
where k = 0.25. We then computed the ratio(s) of predicted mortality for each of the two or more species (i = 1, 2, …):
| (5) |
A ratio greater than one indicates that species 1 suffers higher mortality once infected than species 2. It is worth noting that the ratio derived cannot be used to estimate the actual mortality in a species due to the approximations made in eq. 4, but it does indicate the relative difference in mortality (i.e., a larger ratio indicates a larger difference in mortality, all else being equal).
Sites
We determined mosquito feeding patterns, avian abundance, and WNV antibody prevalence in ~1 km diameter areas at three urban sites (Foggy Bottom, DC, Baltimore, MD, and the National Mall, DC), two residential sites (Takoma Park, MD and Bethesda, MD) and two park sites surrounded by residential development (Rock Creek Park Meadowside Nature Center in Rockville, MD, and Fort Dupont Park in southeast DC) (Kilpatrick et al. 2006a; Kilpatrick et al. 2006b) from 2004 through 2008. Evidence of WNV transmission (infected mosquitoes or antibody-positive resident (non-migratory) hatch year birds) was present at all sites except Rock Creek Park in 2005 (Kilpatrick et al unpub. data).
Mosquito feeding patterns
We trapped mosquitoes at each site with at least 8 CDC light traps, 4 CDC gravid traps and by aspirating the surfaces of vegetation with a large backpack mounted aspirator. Sites were trapped for two nights approximately every 2–3 weeks from May through September each year. Mosquitoes were identified to species and all partially or fully engorged mosquitoes were stored in a freezer at −80 C for subsequent host identification. We used PCR to molecularly identify engorged Culex mosquitoes to distinguish between Cx. pipiens, Cx. restuans, and Cx. salinarius (Crabtree et al. 1995). We only used data from Cx. pipiens or Cx. restuans to estimate feeding utilizations, because these two species have similar feeding patterns, whereas Cx. salinarius feeds on a very different set of hosts (Apperson et al. 2002; Apperson et al. 2004). We identified the vertebrate source of each blood meal by PCR amplification of the cytochrome b gene and nucleotide sequencing of the amplified product (Kilpatrick et al. 2006a). We compared the sequence to known sequences in Genbank using the blastn search tool. As described above, we calculated a mosquito utilization index for titmouse and wrens by dividing the fraction of bloodmeals at each site identified as titmouse or wren by the relative abundance (i.e. fraction of the avian community) of the same species. Abundances were estimated from 4–6 six minute unlimited distance point counts conducted at dawn monthly from May-September and analyzed with program Distance (Thomas et al. 2004). A mosquito utilization index, U, of one indicates that a species is fed on in proportion to their abundance, a value less than one indicates underutilization, and a value greater than one indicates overutilization. In addition to data for wrens and titmouse, we show values of the mosquito utilization index, U, (and seroprevalence, S) for northern cardinal, a common species of bird that shows high WNV seroprevalence, and suffered moderate (22%; 2/9 birds) mortality in the laboratory following experimental infection with WNV (Komar et al. 2005).
Avian Serology
We captured birds in 20 to 40 6–18m long mist nets operated from dawn until early afternoon for 2–3 days at each site approximately monthly from mid-July to early October. Birds were extracted and taken to a banding station where they were aged, sexed, banded with an aluminum USFWS band, weighed, and a 0.1ml blood sample was taken by brachial venipuncture. Blood was tested for flavivirus antibodies using an enzyme-linked immunosorbent assay (ELISA; (Ebel et al. 2002)). We confirmed a random sample of 18% of flavivirus antibody-positive samples (185 of 1026) by a plaque reduction neutralization test (Calisher et al. 1989; Ebel et al. 2002; Wong et al. 2004). None of these samples indicated exposure to St. Louis Encephalitis virus, so we interpreted all flavivirus positive samples as indicating prior exposure to WNV and survival in estimating WNV seroprevalence, S.
Experimental Infection
We captured eleven hatch-year Carolina wrens and twelve tufted titmouse (six hatch-year and six after-hatch year birds) from Montgomery and Anne Arundel Counties in Maryland during the last week of August, 2009. Birds were transported from the National Zoo to the New York State Department of Health where they were held for two weeks for acclimation. On the 15th day, a 0.05ml blood sample was taken by brachial venipuncture to determine whether any birds had flavivirus antibodies by ELISA. All birds tested negative.
Birds in captivity were given water ad libitum, and fed mealworms, waxworms, and a vitamin supplemented “meat mash” consisting of beef, wheat germ, whole grain cereal, boiled egg, carrot, bonemeal, and powdered milk. Titmouse were also fed sunflower seeds.
We initially separated the birds into treatment (infection: 9 wrens, 8 titmouse) and control (mock infection: 2 wrens and 4 titmouse) groups. Treatment birds were infected by subcutaneous injection in the cervical region with 104 PFU of WNV (strain 03–1956 in the WN02 clade (Davis et al. 2005)) in animal diluent, PBS w/ 1% fetal bovine serum. All birds were bled every other day with half the birds bled on day 1 post-infection (PI) and the other half on day 2 so that half the birds were sampled on each day PI, 1–6. All control birds survived until two weeks after mock infection, and by which time all birds infected in this experiment had recovered or perished.
Normally all surviving (control and treatment) birds would have been sacrificed on day 14 post infection. However, in order to maximize sample sizes for survival and viremia profiles, we held the control birds for 14 additional days after the initial infection study was completed (28 days after the start of the first experiment). We then infected these previous “control” birds and bled these birds as described above and measured survival and viremia. Since they served as their own controls in terms of examining the effect of bleeding and handling during the first experiment, we treated all birds similarly in analyses of WNV viremia and mortality from WNV infection described below. Fourteen days after the second infection, all remaining birds were bled for evidence of WNV antibodies and were euthanized by an overdose of pentobarbitol.
WNV viremia was measured by plaque assay on Vero cells (Payne et al. 2006) with a limit of detection of 101.7 PFU/ml, and average daily viremias were calculated after log-transformation. We calculated the host competence for each species by estimating the average infectiousness of each birds on days 1–6 PI using a viremia-infectiousness relationship for Cx. pipiens (% of mosquitoes infectious (transmitting) = 0.1349 × Log10(viremia) – 0.6235; (Kilpatrick et al. 2007)), and multiplied this average by the number of days birds were viremic. Kaplan-Meier survival curves of the two species following infection were compared with a log-rank test on right-censored data using the date of sacrifice for birds surviving infection as the censor date.
We examined the persistence of live virus in bird tissues by harvesting approximately 0.2 g portions of brain, heart, kidney, spleen, lung, and skin (inoculation site) from all birds surviving infection (eight wrens and zero titmice). Tissues were homogenized in BA-1 diluent and were co-cultured as previously described (Tesh et al. 2005). Briefly, samples were homogenized in 2x antibiotics/fungicide and then 0.1mL of the homogenate was inoculated in duplicate on Vero cell monolayers. Cultures were observed for 7 days for cytopathic effect (CPE). If CPE was not present, then cultures were passed to fresh monolayers. Samples were considered negative for persistent infectious virus if after three successive passages CPE was not evident. If CPE was observed, then cultures were confirmed by RT-PCR. Infectious viral loads were not calculated since the assay used (co-culture) is not quantitative.
To determine whether birds showing illness or clinical signs would be evident in the field and to what extent infected birds might be at greater susceptibility to predation we observed the behavior of birds using instantaneous sampling (Altmann 1974) with five 10 second sampling periods each day for each bird spaced over an 30 minute period in the afternoon (1200–1600). We ranked the behaviors on a eight point scale from dead to highly active (Table S1).
Results
Tufted titmouse were present at four of seven sites where they made up 2.8% (±1 SE 2.9%) of the avian community, and we identified Culex pipens or Cx. restuans bloodmeals from them at three sites (Table1). Carolina wrens were present at all 7 sites where they made up 2.7% (±1%) of the avian community and we identified bloodmeals from wrens at 5 of 7 sites. Carolina wrens were fed on by mosquitoes significantly more than expected given their availability at two sites, less than expected at one site and fed on in proportion to their abundance at two sites whereas Culex mosquitoes fed on titmouse slightly more frequently than expected from their abundance at all three sites (Table 1). Northern cardinals were present at all seven sites, made up 8.5% (±5.2%) of the avian community, and were fed on at all seven sites. Feeding on cardinals varied from being fed on half to twice as frequently as expected based on their abundance (Table 1). The antibody prevalence of hatch-year birds caught in mid-July to early October was 17.5% for wrens and 34.8% cardinals, whereas only 3 of the 176 titmouse (1.7%) tested positive for WNV antibodies (Table 1). We used these estimates of seroprevalence, S, to estimate the number of infectious bites, I, (eqn. 4) and combined these with mosquito utilization values to generate multiple predictions (eqn. 5) about the relative mortality of wrens, cardinals and titmouse (Table 1; rightmost three columns): mortality was predicted to be lowest in cardinals (22% or 2/9 birds died in a previous experimental infection; (Komar et al. 2005)), slightly (but not significantly) higher in wrens, and significantly higher in titmouse.
Table 1.
Host utilization (feeding preferences from equation 3), U, West Nile virus seroprevalence, (fraction of hatch-year birds with antibodies, sampled in mid-July to early October, 2004–2008), S, estimated infective bites (using equation 4), I, and calculated mortality ratios (using equation 5). Values in parentheses are the standard error of the estimate. Ranges in parentheses for Mortality ratios are 95% confidence intervals.
| Feeding preferences | Seroprevalence | Infective bites | Mortality ratios | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | C. wren | T. titmouse | N. cardinal | C. wren | T. titmouse | N. cardinal | C. wren | T. titmouse | N. cardinal | Wren/Cardinal | Titmouse/wren | Titmouse/Cardinal |
| Foggy Bottom, DC | 0.93 (0.06) | 2.96 (0.10) | 0.25 (0.15) | |||||||||
| Fort Dupont Park, DC | 0.20 (0.06) | 1.28 (0.14) | 0.74 (0.10) | 0.25 (0.04) | 0.03 (0.03) | 0.48 (0.05) | 0.52 (0.09) | 0.03 (0.03) | 3.28 (0.35) | 1.74 (0.5– 6.1) | 100.40 (20.1 - ) | 174.32 (36.4 - ) |
| Bethesda, MD | 1.77 (0.17) | 0.21 (0.06) | 0.12 (0.06) | 0.00* | 0.23 (0.04) | 0.17 (0.09) | 0.00 | 0.46 (0.07) | >181.8* >(5.2- ) | |||
| Takoma Park, MD | 1.47 (0.17) | 0.19 (0.08) | 0.00 | 0.56 (0.04) | 0.34 (0.14) | 0.00 | 6.53 (3.5) | |||||
| National Mall, DC | 8.90 (0.17) | 1.76 (0.08) | ||||||||||
| Rock Creek Park, MD | 0.87 (0.20) | 1.52 (0.25) | 0.51 (0.14) | 0.12 (0.05) | 0.03 (0.02) | 0.15 (0.03) | 0.16 (0.07) | 0.03 (0.02) | 0.23 (0.04) | 2.41 (0.6 – 15.8) | 8.57 (1.28 - ) | 20.67 (5.2 - ) |
| Baltimore, MD | 7.08 (0.20) | 2.15 (0.12) | ||||||||||
Seroprevalence for this species at this site was zero (0.49), so the mortality ratio is .
We estimated the mortality ratio assuming one of the 49 titmouse had been seropositive and expressed the results as a minimum estimate.
We then performed an experimental infection study in the laboratory. All control birds survived the handling and bleeding regiment during the first experiment. Over the course of the two experiments (see Methods) we experimentally infected eleven wrens and twelve titmouse with WNV (Figure 1a). Three of the eleven (27%) wrens died following WNV infection on days 7, 7, and 8 post-infection (PI) and all twelve of the tufted titmouse died following infection (Figure 2; three birds on day five, seven birds on day six, one bird on day seven, and one an additional bird on day two whose death may have been related to handling). Survival was significantly higher in wrens than titmouse (Log-rank test: = 19.8; df = 1, p < 0.001). It is worth noting that although none of the control birds died from the bleeding regiment, it is still possible that the mortality of birds infected with WNV could have been slightly inflated by being bled every other day.
Figure 1.
Survival curves (bottom) and average daily viremia profiles (bottom) and for eleven Carolina wrens and twelve tufted titmouse following experimental infection with WNV.
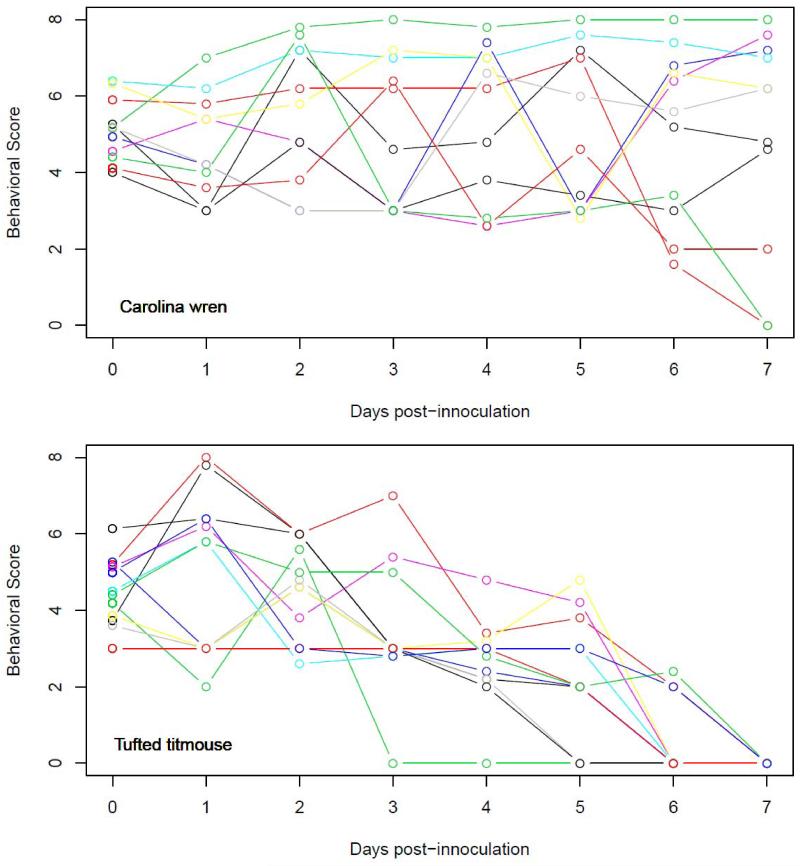
Figure 2.
Behavioral scores for Carolina wrens (top) and tufted titmouse (bottom) before and during the viremic period. Note that analyses only use the non-zero values. See table S1 for explanation of behavioral codes.
The behavior of wrens and titmouse following infection also differed significantly (Fig. 2). Titmouse showed a highly significant decline in behavioral score becoming less active and alert with increasing days since infection (mixed effects model with species interacting with days since infection interaction as fixed effects and bird as a random effect: species effect: titmouse coefficient −0.29±SE = 0.39, t = −0.735, p >0.5; titmouseby-days since infection coef. −0.43±0.10, t = −4.460, p < 0.001). The behavior of the wrens who succumbed to infection were not significantly different from those that survived (mixed effects model with days since infection interacting with succumbed to WNV infection as fixed effects and bird as a random effect: days since infection-by-succumbed coef. 0.13±0.14, t = 0.95, p > 0.3). Only two of the three birds that died showed behaviors associated with sickness and these behaviors were only observed the day before death.
The average WNV viremia (concentration of virus in the blood) of wrens was significantly lower than titmouse, and peaked on day 2 at 107.8 PFU/ml, whereas titmouse viremia peaked on day 4 and remained high through day 6 (Figure 1b; mixed effects model with bird as random effect using the lme4 package in R (v2.15): titmouse species coefficient 2.58± 0.49; p <0.001). The competence (Komar et al. 2003) of titmouse (3.15 or an average infectiousness of 52.4% across six days) was higher than that recorded for any of the other fifty species that have been studied (Kilpatrick et al. 2007), partly due to a six day long viremic period (other highly infectious species like crows and jays died mostly on days 4 and 5 PI; (Komar et al. 2003; Reisen et al. 2005)).
As in previous studies (Nemeth et al. 2009; Reisen et al. 2006; Wheeler et al. 2012), infectious virus or viral RNA was detected several weeks post-infection. We isolated virus from at least one tissue from all wrens surviving infection at 29 days post infection (and viral RNA even more frequently), with the kidneys and spleens being frequently infected (Table S2,S3).
Discussion
Analysis of population trends following the arrival of WNV suggested that tufted titmouse, chickadees, and house wrens were significantly impacted by disease, with the largest drop in mid-Atlantic populations following the intense 2003 WNV epidemic (LaDeau et al. 2007). However, the inference from that study and others (Bonter & Hochachka 2003; Wheeler et al. 2009) that trends in these species, as well as several other small songbirds, were due to WNV was indirect. Similarly, although songbirds have tested positive for WNV in some dead bird surveillance for WNV (Bernard et al. 2001; Nemeth et al. 2007), the relative numbers are often small and thus give little hard evidence for WNV impact. Here we have more rigorously tested the hypothesis that titmouse and wrens are perishing in the field from WNV infection at equal or higher rates than a previously studied species, Northern cardinals. We found strong support for the hypothesis in that titmouse were highly susceptible to mortality from WNV infection, with all birds perishing within seven days after infection. Nearly thirty percent of wrens, which were predicted to suffer lower, but significant mortality, also died following infection. These results which agree well with predictions based on serology and feeding preferences (Table 1) highlight the utility of our framework to predict the relative WNV mortality of different species in the absence of experimental infection studies. With the growing number of studies that estimate mosquito utilization index values (Hamer et al. 2009; Hassan et al. 2003; Kent et al. 2009; Kilpatrick et al. 2006a; Thiemann et al. 2011), and dozens of studies on WNV seroprevalence it is now possible to make predictions about relative susceptibility to mortality from WNV for many species of birds that have not been studied but may be dying from this disease.
These results on wren and titmouse mortality suggest that WNV may be killing many smaller bodied birds that aren't identified in large numbers in WNV dead bird collections. Our behavioral studies suggest that infected titmouse may exhibit sick behavior before death and this might be apparent through citizen science projects like Feederwatch (Hochachka & Dhondt 2000), whereas other species like Carolina wrens show relatively little change in behavior over most of the viremic period, even if they eventually die from the disease. Actions to reduce WNV impacts on these and other species include reducing WNV transmission by reducing mosquito larval habitat of WNV vectors (e.g., Culex pipiens, Cx. restuans, Cx. tarsalis; (Kilpatrick et al. 2005)) by removing man-made containers such as tires, clogged gutters, etc. Disease reduction through habitat modification to reduce vector density should be considered an important part of habitat restoration for birds susceptible to WNV and other mosquito-borne pathogens.
Our results also provide valuable information about the host competence of these two species (and other species in the previously unstudied families Paridae and Troglodytidae, because competence is phylogenetically conserved to some extent; (Kilpatrick et al. 2006a)). These data have proven to be an integral part of determining the contribution of hosts to transmission (Kilpatrick 2011) as well as predicting spatial and temporal patterns of WNV transmission (Hamer et al. 2011; Kilpatrick et al. 2006a). We found that both wrens and titmouse were highly infectious for WNV mosquito vectors, and are fed on by these mosquitoes (Fig. 1; (Hamer et al. 2008)). However, due to their low relative abundance and only moderate feeding utilizations, they are likely to play only minor roles in WNV amplification compared to species such as American robins (Hamer et al. 2009; Kent et al. 2009; Kilpatrick 2011; Kilpatrick et al. 2006a).
Our host competence data also informs our broader understanding of host-parasite relationships. For example, the top fifteen most infectious hosts for WNV (Kilpatrick et al. 2007) now spans twelve families (including four non-passerine families), which demonstrates the ability of WNV to efficiently infect a broad range of hosts, and challenges assertions that only passerines or corvids are highly competent for WNV.
More broadly, our findings demonstrate the value of both field and experimental evidence for understanding disease susceptibility in species conservation. As more pathogens are spread among continents and infect new hosts, there is an urgent need to predict and mitigate the impacts of emerging diseases on wildlife populations.
Supplementary Material
Acknowledgements
We thank Will Janousek for assistance capturing and transporting birds, the Smithsonian's Neighborhood Nestwatch program and residents, the staff of Rock Creek Park (Meadowside Nature Center), Fort Dupont Park, the Smithsonian National Museum of Natural History, the National Gallery of Art and the Hirshhorn museum for permission to use their property. We are indebted to Montgomery County Parks and the Patuxent National Wildlife Refuge for permission to use birds from these lands for this study. Funding was provided by NSF grant EF-0914866, NIH grant 1R01AI090159-01, and NIAID contract #NO1-AI-25490. Birds were collected under USFWS permit MB177891-1. Experimental infection studies were approved by the Wadsworth Center, New York State Department of Health, animal use protocol 09-412.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altmann J. Observation study of be havior - sampling methods. Behaviour. 1974;49:227–267. doi: 10.1163/156853974x00534. [DOI] [PubMed] [Google Scholar]
- Apperson CS, Harrison BA, Unnasch TR, Hassan HK, Irby WS, Savage HM, Aspen SE, Watson DW, Rueda LM, Engber BR, Nasci RS. Host-feeding habits of Culex and other mosquitoes (Diptera: Culicidae) in the Borough of Queens in New York City, with characters and techniques for identification of Culex mosquitoes. Journal of Medical Entomology. 2002;39:777–785. doi: 10.1603/0022-2585-39.5.777. [DOI] [PubMed] [Google Scholar]
- Apperson CS, Hassan HK, Harrison BA, Savage HM, Aspen SE, Farajollahi A, Crans W, Daniels TJ, Falco RC, Benedict M, Anderson M, McMillen L, Unnasch TR. Host feeding patterns of established and potential mosquito vectors of West Nile virus in the eastern United States. Vector-Borne and Zoonotic Diseases. 2004;4:71–82. doi: 10.1089/153036604773083013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard KA, Maffei JG, Jones SA, Kauffman EB, Ebel GD, Dupuis AP, Ngo KA, Nicholas DC, Young DM, Shi PY, Kulasekera VL, Eidson M, White DJ, Stone WB, Kramer LD. West Nile virus infection in birds and mosquitoes, New York State, 2000. Emerging Infectious Diseases. 2001;7:679–685. doi: 10.3201/eid0704.010415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonter D, Hochachka WM. Declines of chickadees and corvids: Possible impacts of West Nile virus. American Birds, 103rd Christmas Bird Count. 2003:22–25. [Google Scholar]
- Calisher CH, Karabatsos N, Dalrymple JM, Shope RE, Porterfield JS, Westaway EG, Brandt WE. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. Journal of General Virology. 1989;70:37–43. doi: 10.1099/0022-1317-70-1-37. [DOI] [PubMed] [Google Scholar]
- Crabtree MB, Savage HM, Miller BR. Development of a species-diagnostic polymerase chain reaction assay for the identification of Culex vectors of St. Louis encephalitis virus based on interspecies sequence variation in ribosomal DNA spacers. Am J Trop Med Hyg. 1995;53:105–109. [PubMed] [Google Scholar]
- Davis CT, Ebel GD, Lanciotti RS, Brault AC, Guzman H, Siirin M, Lambert A, Parsons RE, Beasley DWC, Novak RJ, Elizondo-Quiroga D, Green EN, Young DS, Stark LM, Drebot MA, Artsob H, Tesh RB, Kramer LD, Barrett ADT. Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: Evidence for the emergence of a dominant genotype. Virology. 2005;342:252–265. doi: 10.1016/j.virol.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Dye C, Hasibeder G. Population dynamics of mosquito-borne disease - effects of flies which bite some people more frequently than others. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1986;80:69–77. doi: 10.1016/0035-9203(86)90199-9. [DOI] [PubMed] [Google Scholar]
- Ebel GD, Dupuis Alan P, II, Nicholas D, Young D, Maffei J, Kramer LD. Detection by enzyme-linked immunosorbent assay of antibodies to West Nile virus in birds. Emerging Infectious Diseases. 2002;8:979–982. doi: 10.3201/eid0809.020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer GL, Chaves LF, Anderson TK, Kitron UD, Brawn JD, Ruiz MO, Loss SR, Walker ED, Goldberg TL. Fine-scale variation in vector host use and force of infection drive localized patterns of West Nile virus transmission. Plos One. 2011;6 doi: 10.1371/journal.pone.0023767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer GL, Kitron UD, Brawn JD, Loss SR, Ruiz MO, Goldberg TL, Walker ED. Culex pipiens (Diptera: Culicidae): a bridge vector of West Nile virus to humans. Journal of Medical Entomology. 2008;45:125–128. doi: 10.1603/0022-2585(2008)45[125:cpdcab]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Hamer GL, Kitron UD, Goldberg TL, Brawn JD, Loss SR, Ruiz MO, Hayes DB, Walker ED. Host selection by Culex pipiens mosquitoes and West Nile virus amplification. American Journal of Tropical Medicine and Hygiene. 2009;80:268–278. [PubMed] [Google Scholar]
- Hassan HK, Cupp EW, Hill GE, Katholi CR, Klingler K, Unnasch TR. Avian host preference by vectors of eastern equine encephalomyelitis virus. American Journal of Tropical Medicine and Hygiene. 2003;69:641–647. [PubMed] [Google Scholar]
- Hochachka WM, Dhondt AA. Density-dependent decline of host abundance resulting from a new infectious disease. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5303–5306. doi: 10.1073/pnas.080551197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochachka WM, Dhondt AA, McGowan KJ, Kramer LD. Impact of West Nile virus on american crows in the northeastern United States, and its relevance to existing monitoring programs. Ecohealth. 2004;1:60–68. [Google Scholar]
- Holdo RM, Sinclair ARE, Dobson AP, Metzger KL, Bolker BM, Ritchie ME, Holt RD. A Disease-Mediated Trophic Cascade in the Serengeti and its Implications for Ecosystem C. Plos Biology. 2009;7 doi: 10.1371/journal.pbio.1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson PJ, Dobson AP, Newborn D. Prevention of population cycles by parasite removal. Science. 1998;282:2256–2258. doi: 10.1126/science.282.5397.2256. [DOI] [PubMed] [Google Scholar]
- Kent R, Juliusson L, Weissmann M, Evans S, Komar N. Seasonal blood feeding behavior of Culex tarsalis (Diptera: Culicidae) in Weld County, Colorado, 2007. Journal of Medical Entomology. 2009;46:380–390. doi: 10.1603/033.046.0226. [DOI] [PubMed] [Google Scholar]
- Kilpatrick AM. Globalization, land use, and the invasion of West Nile virus. Science. 2011;334:323–327. doi: 10.1126/science.1201010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick AM, Briggs CJ, Daszak P. The ecology and impact of chytridiomycosis, an emerging disease of amphibians. Trends in Ecology & Evolution. 2010;25:109–118. doi: 10.1016/j.tree.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Kilpatrick AM, Daszak P, Jones MJ, Marra PP, Kramer LD. Host heterogeneity dominates West Nile virus transmission. Proceedings of the Royal Society B-Biological Sciences. 2006a;273:2327–2333. doi: 10.1098/rspb.2006.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick AM, Kramer LD, Campbell S, Alleyne EO, Dobson AP, Daszak P. West Nile virus risk assessment and the bridge vector paradigm. Emerging Infectious Diseases. 2005;11:425–429. doi: 10.3201/eid1103.040364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick AM, Kramer LD, Jones MJ, Marra PP, Daszak P. West Nile virus epidemics in North America are driven by shifts in mosquito feeding behavior. PLoS Biology. 2006b;4:606–610. doi: 10.1371/journal.pbio.0040082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick AM, LaDeau SL, Marra PP. Ecology of West Nile virus transmission and its impact on birds in the western hemisphere. Auk. 2007;124:1121–1136. [Google Scholar]
- Koch R. Über den augenblicklichen stand der bakteriologischen choleradiagnose. Zeitschrift für Hygiene und Infectionskrankheiten. 1893;14:319–333. [Google Scholar]
- Komar N, Langevin S, Hinten S, Nemeth N, Edwards E, Hettler D, Davis B, Bowen R, Bunning M. Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerging Infectious Diseases. 2003;9:311–322. doi: 10.3201/eid0903.020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar N, Panella NA, Langevin SA, Brault AC, Amador M, Edwards E, Owens J. Avian hosts for West Nile virus in St. Tammany Parrish, Louisiana, 2002. Am J Trop Med Hyg. 2005;73:1031–1037. [PubMed] [Google Scholar]
- LaDeau SL, Kilpatrick AM, Marra PP. West Nile virus emergence and large-scale declines of North American bird populations. Nature. 2007;447:710–713. doi: 10.1038/nature05829. [DOI] [PubMed] [Google Scholar]
- Langwig KE, Frick WF, Bried JT, Hicks AC, Kunz TH, Kilpatrick AM. Sociality, density-dependence and microclimates determine the persistence of populations suffering from a novel fungal disease, white-nose syndrome. Ecology Letters. 2012;15:1050–1057. doi: 10.1111/j.1461-0248.2012.01829.x. [DOI] [PubMed] [Google Scholar]
- Lips KR, Brem F, Brenes R, Reeve JD, Alford RA, Voyles J, Carey C, Livo L, Pessier AP, Collins JP. Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3165–3170. doi: 10.1073/pnas.0506889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum H. Inconclusiveness of chytridiomycosis as the agent in widespread frog declines. Conservation Biology. 2005;19:1421–1430. [Google Scholar]
- McCallum H, Dobson A. Detecting disease and parasite threats to endangered species and ecosystems. Trends in Ecology & Evolution. 1995;10:190–194. doi: 10.1016/s0169-5347(00)89050-3. [DOI] [PubMed] [Google Scholar]
- Nemeth N, Young G, Ndaluka C, Bielefeldt-Ohmann H, Komar N, Bowen R. Persistent West Nile virus infection in the house sparrow (Passer domesticus) Archives of Virology. 2009;154:783–789. doi: 10.1007/s00705-009-0369-x. [DOI] [PubMed] [Google Scholar]
- Nemeth NM, Beckett S, Edwards E, Klenk K, Komar N. Avian mortality surveillance for West Nile virus in Colorado. American Journal of Tropical Medicine and Hygiene. 2007;76:431–437. [PubMed] [Google Scholar]
- Payne AF, Binduga-Gajewska I, Kauffman EB, Kramer LD. Quantitation of flaviviruses by fluorescent focus assay. Journal of Virological Methods. 2006;134:183–189. doi: 10.1016/j.jviromet.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Ratcliffe FN, Myers K, Fennessy BV, Calaby JH. Myxomatosis in Australia - a step towards the biological control of the rabbit. Nature. 1952;170:7–11. doi: 10.1038/170007a0. [DOI] [PubMed] [Google Scholar]
- Reisen WK, Fang Y, Lothrop HD, Martinez VM, Wilson J, O'Connor P, Carney R, Cahoon-Young B, Shafii M, Brault AC. Overwintering of West Nile virus in southern California. Journal of Medical Entomology. 2006;43:344–355. doi: 10.1603/0022-2585(2006)043[0344:oownvi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Reisen WK, Fang Y, Martinez VM. Avian host and mosquito (Diptera : Culicidae) vector competence determine the efficiency of West Nile and St. Louis encephalitis virus transmission. Journal of Medical Entomology. 2005;42:367–375. doi: 10.1093/jmedent/42.3.367. [DOI] [PubMed] [Google Scholar]
- Smith DL, Dushoff J, Snow RW, Hay SI. The entomological inoculation rate and Plasmodium falciparum infection in African children. Nature. 2005;438:492–495. doi: 10.1038/nature04024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesh RB, Siirin M, Guzman H, da Rosa A, Wu XY, Duan T, Lei H, Nunes MR, Xiao SY. Persistent West Nile virus infection in the golden hamster: Studies on its mechanism and possible implications for other flavivirus infections. Journal of Infectious Diseases. 2005;192:287–295. doi: 10.1086/431153. [DOI] [PubMed] [Google Scholar]
- Thiemann TC, Wheeler SS, Barker CM, Reisen WK. Mosquito host selection varies seasonally with host availability and mosquito density. PLoS Neglected Tropical Diseases. 2011;5 doi: 10.1371/journal.pntd.0001452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L, Laake JL, Strindberg S, Marques FFC, Buckland ST, Borchers DL, Anderson DR, Burnham KP, Hedley SL, Pollard JH, Bishop JRB. Distance 4.1. Release 2. Research Unit for Wildlife Population Assessment. University of St. Andrews; UK: 2004. [Google Scholar]
- Van Riper C, III, Van Riper SG, Goff ML, Laird M. The epizootiology and ecological significance of malaria in Hawaiian (USA) land birds. Ecological Monographs. 1986;56:327–344. [Google Scholar]
- Vredenburg VT, Knapp RA, Tunstall TS, Briggs CJ. Dynamics of an emerging disease drive large-scale amphibian population extinctions. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9689–9694. doi: 10.1073/pnas.0914111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MR, Stallknecht DE, Willis J, Conroy MJ, Davidson WR. Wild bird mortality and West Nile virus surveillance: Biases associated with detection, reporting, and carcass persistence. Journal of Wildlife Diseases. 2006;42:92–106. doi: 10.7589/0090-3558-42.1.92. [DOI] [PubMed] [Google Scholar]
- Warner RE. The role of introduced diseases in the extinction of the endemic Hawaiian avifauna. Condor. 1968;70:101–120. [Google Scholar]
- Wheeler SS, Barker CM, Fang Y, Armijos MV, Carroll BD, Husted S, Johnson WO, Reisen WK. Differential impact of West Nile virus on California birds. Condor. 2009;111:1–20. doi: 10.1525/cond.2009.080013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler SS, Langevin SA, Brault AC, Woods L, Carroll BD, Reisen WK. Detection of persistent West Nile virus RNA in experimentally and naturally infected avian hosts. American Journal of Tropical Medicine and Hygiene. 2012;87:559–564. doi: 10.4269/ajtmh.2012.11-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SJ, Demarest VL, Boyle RH, Wang T, Ledizet M, Kar K, Kramer LD, Fikrig E, Koski RA. Detection of human anti-flavivirus antibodies with a West Nile virus recombinant antigen microsphere immunoassay. Journal of Clinical Microbiology. 2004;42:65–72. doi: 10.1128/JCM.42.1.65-72.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.