Abstract
Bacterial magnetosomes are intracellular compartments that house highly ordered magnetite crystals. By using Magnetospirillum sp. AMB-1 as a model system, we show that magnetosome vesicles exist in the absence of magnetite, biomineralization of magnetite proceeds simultaneously in multiple vesicles, and biomineralization proceeds from the same location in each vesicle. The magnetosome-associated protein, MamA, is required for the formation of functional magnetosome vesicles and displays a dynamic subcellular localization throughout the growth cycle of magnetotactic bacteria. Together, these results suggest that the magnetosome precisely coordinates magnetite biomineralization and can serve as a model system for the study of organelle biogenesis in noneukaryotic cells.
Membrane-bounded organelles are generally thought to be unique to eukaryotic cells and absent from bacteria and archaea. The magnetosome compartments of magnetotactic bacteria, however, appear to possess many of the defining features of eukaryotic membrane-bounded organelles. Magnetosomes, which are intracellular chains of magnetite or greigite crystals (Fig. 1A), form a magnet that allows magnetotactic bacteria to orient in geomagnetic fields and possibly search for microaerophillic environments more efficiently (1). The unique properties of bacterial magnetites have made them a potential biomarker for geobiologists and an ideal system for studying biomineralization (2, 3).
Fig. 1.
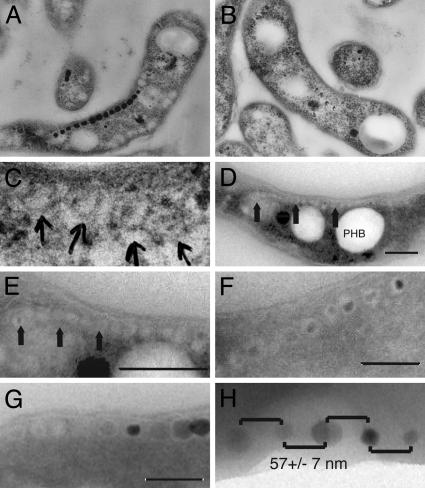
Characterization of magnetosome dynamics by electron microscopy. (A) TEM image of an ultrathin section of Magnetospirillum sp. AMB-1 grown under iron-rich conditions and embedded in epoxy resin. A chain of magnetite crystals can be observed in the cells. (B) AMB-1 cells grown without iron do not contain magnetite chains. (C) Higher magnification of the cell shown in B reveals potential empty magnetosome vesicles. (D) TEM image of cryo-ultrathin section of iron-starved AMB-1 showing a long chain of empty magnetosome vesicles. The three large white inclusions are polyhydroxybutyrate (PHB) granules. A magnetosome chain (arrows) is seen on top of these PHB granules. (E) Higher magnification of magnetosomes in D. Arrows point to magnetosome chains devoid of magnetite. (F) TEM image of a cryo-ultrathin section of AMB-1 cells 7 h after the addition of iron to iron-starved cultures. Small magnetite crystals show the early stages of crystal growth within vesicles having relatively uniform size and shape. It is evident that simultaneous nucleation and growth of magnetite crystals occurs within the full-sized vesicles. (G) TEM image of a cryo-ultrathin section of AMB-1 after 20 h of growth in iron-rich medium. Structures resembling empty magnetosomes are seen at the ends of chains of fully grown magnetite crystals. (H) Regular spacing of magnetite crystals 2 h after the addition of iron. Whole cells of AMB-1 were imaged in TEM without sectioning. The spacing between magnetite crystals is very even, with an average of 57 ± 7 nm. (Scale bars: 200 nm in D-G.)
Despite the detailed characterization of these biogenic magnetite crystals, the cell biology of the bacterial magnetosome has largely been ignored. Previous work has shown that each magnetite crystal within a magnetosome is surrounded by a lipid bilayer that is separate from the cell membrane (4, 5). Additionally, these vesicles contain a unique set of transmembrane and soluble proteins that are thought to direct the biomineralization of magnetite crystals (6-8). These and other results suggest that to build a magnetosome a bacterium must be able to generate vesicles, target the appropriate set of proteins to these vesicles, and control their number and position within a cell. These properties make magnetosomes an attractive system for the study of protein sorting and organelle dynamics in prokaryotes.
If magnetosomes are true membranous organelles that control the biomineralization of magnetite, empty chains of vesicles should be present before magnetite formation. Magnetosome chains devoid of magnetite have not been observed previously mainly because most studies have characterized the magnetosome membrane under iron-rich conditions where magnetite is present (4, 9). Furthermore, in studies where cells were grown without magnetite formation, the presence of empty magnetosomes was not explored (10). In this work, we show that magnetosome vesicles can exist before magnetite biomineralization. Once initiated, magnetite biomineralization proceeds simultaneously in multiple vesicles in a temporally and spatially coordinated fashion. In addition, using genetic and cell biological techniques, we show that the magnetosome-associated protein MamA displays a dynamic growth-dependent subcellular localization and is required for the activation of magnetosome vesicles.
Materials and Methods
Growth Conditions and Medium Composition. For all experiments we used the genetically tractable organism, Magnetospirillum sp. AMB-1 (11) and a modified version of Magnetospirillum growth medium (MSGM) containing per liter: 5 ml of Wolfe's mineral solution (prepared without iron), 0.68 g of potassium phosphate, 0.12 g of sodium nitrate, 0.07 g of sodium acetate, 0.035 g of ascorbic acid, 0.37 g of tartaric acid, 0.37 g of succinic acid, and 0.05 g of sodium thiosulfate. The pH was adjusted to 6.9, and the medium was autoclaved. Most experiments were done by using sealed Hungate tubes containing 10 ml of medium and a 20-ml headspace. The medium was bubbled with nitrogen gas before autoclaving, and the headspace was flushed with nitrogen after autoclaving. Before inoculation with bacteria, 100 μl of Wolfe's vitamin solution, 100 μl of 3 mM ferric malate, and 1-5 ml of air were added to each tube (providing 1-5% oxygen in the headspace). For iron-limited growth, ferric malate was eliminated from the growth medium. Additionally, all glassware was soaked in oxalic acid for 24 h before use. All incubations were at 30°C. For agar plates, 7 g of agar was added per liter of growth medium. Before pouring, 10 ml of vitamin solution and 0.5 ml of 50 mM ferric malate were added per liter of growth medium. Plates were incubated in jars with 2-3% oxygen. In general, colonies appeared 5-7 days after plating. Kanamycin was added at 10-25 μg/ml to plates and 2-10 μg/ml to liquid cultures.
Time-Course Experiments. For the time-course experiments looking at iron uptake and magnetism (Fig. 2B and C) cultures were grown in 10-ml Hungate tubes overnight, and while in log phase, were centrifuged, washed with growth medium, and inoculated into a fresh anaerobic tube containing ferric malate (30-40 μM). For visualization with transmission electron microscopy (TEM), 100-ml cultures were grown overnight without iron to logarithmic phase. They were centrifuged and inoculated into fresh 100-ml anaerobic bottles with ferric malate added to a concentration of 30 μM. At various time points, 20 ml of each culture was centrifuged with 0.1 mol/liter PBS and resuspended in 1 ml of PBS containing 2.5% glutaraldehyde.
Fig. 2.
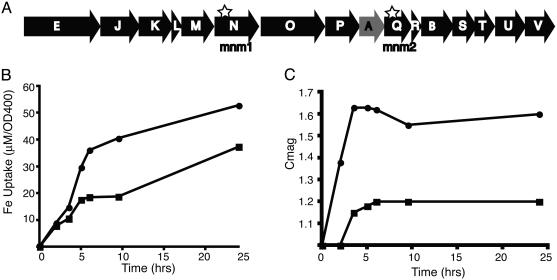
Characterization of ΔmamA. (A) Schematic of the mamABE gene cluster (25). mamA, mamB, and mamE are designated as A, B, and E, respectively. Stars indicate the positions of transposon insertions in mnm1 and mnm2. mamA, deleted in subsequent experiments, is shaded gray. (B) Representative time-course experiment after the addition of iron to iron-starved WT (•) and ΔmamA (▪) strain. ΔmamA cells have taken up less iron than WT cells after 24 h of growth. Iron uptake is normalized to cell number by dividing the μM of iron depleted from the growth medium by the OD at 400 nm. (C) Measurement of response (turning) to a magnetic field for the experiment shown in B. OD400 measurements were made with a magnet parallel or perpendicular to the light source. ΔmamA cells have a clear defect in turning in a magnetic field compared to WT cells.
Conventional Sectioning and TEM. Conventional thin sectioning, staining, and TEM were performed as described (12).
Cryo-Ultramicrotomy. For TEM, whole-cell pellets from the time-course experiments were washed several times with 0.1 mol/liter PBS to remove excess growth medium. The bacteria were fixed by redispersing the pellet in a solution of 2.5% glutaraldehyde in 0.1 mol/liter sodium cacodylate buffer overnight at 4°C. Five microliters of the solution was transferred onto 300-mesh Formvar and carbon-coated Cu grids and allowed to dry at room temperature. Cryofixation and cryo-ultramicrotomy were used to investigate the internal structure of the bacteria, magnetosome membrane, and vesicles. These techniques enable the imaging of biological structures in their near-natural state. Ultrathin cryosections of magnetotactic bacteria were obtained following a modified procedure described by Tokuyasu (13) and Zierold (14). A small pellet of bacteria was fixed by dispersing in a solution of 2.5% glutaraldehyde in 2.3 M sucrose in PBS, a medium that serves as both cryoprotectant and cryoglue. To allow for complete infiltration, the dispersion was incubated for 1 h at room temperature. Fifty microliters of the dispersion was transferred onto a stub, rapidly frozen by plunging into liquid propane (-190°C), and stored in liquid nitrogen (-196°C). The stub was mounted on the specimen holder of an Ultracut-S ultramicrotome equipped with a FCS cryosystem (Reichert-Jung, Vienna). Ultrathin sections (70-100 nm thick) were cut from the frozen pellet at -80°Cto -100°C with a dry glass knife. The sections were transferred onto 300-mesh Formvar and carbon-coated Cu grids by using a gold loop with a droplet of 2.3 M sucrose/PBS. The sucrose was removed by washing the grids a few times with deionized water. The sections were stained with 2% uranyl oxalic acid and cryo-protected from drying by a thin film of methyl cellulose. The specimens were studied with a JEOL JEM-2000FX TEM operating at 80 kV in bright-field mode. Chemical composition was determined by energy dispersive spectroscopy using the PGT Prism System (Princeton Gamma-Tech, Princeton).
Statistics. Random numbers were generated in Microsoft excel. To determine limits for possible range of distances between magnetite particles in neighboring vesicles, we used the average size of the crystals at the 2-h time point to determine the size of the crystals. The boundaries of two neighboring magnetosomes were measured by taking the average distance between the outer edges of two adjacent magnetite crystals after overnight growth. These numbers led to the estimate that for random positioning of crystals at the 2-h time point the average distance could range from 19 to 98.8 nm. Actual distances were measured by using the measure tool in PHOTOSHOP 7.0 (Adobe Systems, Mountain View, CA). All statistics were performed with excel.
Transposon Mutagenesis. We used a hyperactive mariner transposon for the mutagenesis (15). The transposon delivery plasmid, PSC189 (National Center for Biotechnology Information accession no. AY115560) was transformed into the donor bacterial strain Escherichia coli WM3064, [a derivative of B2155 (16) provided by W. Metcalf (University of Illinois, Urbana)], which can grow only in the presence of diaminopimelate (DAP). WM3064 carrying PSC189 was grown overnight in LB ampicillin medium containing 0.3 mM DAP; 0.5 ml of this overnight was washed twice in LB DAP medium, mixed with 10 ml of iron-starved AMB-1 overnight culture concentrated in a final volume of 50 μl, and plated on an MSGM plate for 4-8 h. This mating reaction was harvested and inoculated into 10 ml of MSGM containing 30 μM ferric malate and grown overnight. Small magnets were subsequently placed next to the tubes, and the nonmagnetic cells were removed the next day and plated on MSGM kanamycin plates. Single colonies (which appeared 5-7 days after plating) were grown in 400 μl of MSGM (with ferric malate) in white 96-well plates (Nalg) in sealed jars with 2% oxygen/98% nitrogen atmosphere. After 1-3 days, growth was observed as a pellet of cells at the bottom of each well. To check for nonmagnetic mutants, the 96-well plates were placed on top of a plate containing 24 magnets designed to fit in the middle of each group of four wells on a 96-well plate. Magnetic cells were identified as they were pulled to the sides of the well and nonmagnetic cells remained at the bottom of the well. Southern blots were carried out for four magnetosome mutants (mnm1-4) and two were found to be unique. To identify these we took advantage of the R6K origin present on the mariner transposon. Genomic DNA from mnm1 and mnm2 was digested with StuI, self-ligated, and transformed into E. coli DH5α-λpir. To identify the insertion point, rescued plasmids were sequenced by using the following primers: Mar3 (5′-CTTCTTGACGAGTTCTTCTGAGC-3′) and Mar4 (5′-TAGGGTTGAGTGTTGTTCCAGTT-3′).
Generation of ΔmamA. The mamA sequence was submitted to GenBank (accession no. AY508230). A two-step recombination method was used to generate the ΔmamA strain (17). Approximately 750 bases upstream and downstream of mamA were amplified by using the following primer sets: Mam22KO1 (5′-GGACTAGTTGGCAGCAGGGCCGGTCGCGGCG-3′) and Mam22KO2 (5′-CCCATCCACTAAATTTAAATAATCCGACGGCTTGCTAGACAT-3′) for upstream sequence and Mam22KO3 (5′-TATTTAAATTTAGTGGATGGGGACGAAGGGGCCTCGGTCTGA-3′) and Mam22KO4 (5′-GGACTAGTGTGATGAACGCCAGATAGTTCC-3′) for the downstream sequence. Using fusion PCR of these two fragments, we generated a 1.5-kb fragment that was cloned into pWM91, a plasmid containing the counterselectable sacB gene. Because we found that kanamycin resistance was a more robust selection, we modified this plasmid by cloning a blunt-ended MluI fragment from PSC189 into the ScaI site within the ampicillin resistance gene of pWM91, generating the pAKO plasmid. The fusion PCR product contains the first seven and the last seven codons of mamA, in case regulatory sequences for upstream and downstream genes are contained within them, and a spacer used to generate fusion PCR products. Thus the following peptide is expected to be produced in the place of MamA: MSSKPSDYLNLVDGDEGASV. pAKO was conjugated into AMB-1 by using WM3064 as the donor strain, and colonies were grown and checked for the presence of the deletion construct. To select for recombinants that lost the integrated plasmid, cells were grown for three to four passages in 10 ml of growth medium without kanamycin and then plated on MSGM containing 1% sucrose. These sucrose-resistant colonies were checked for the presence of the deletion marker and the absence of mamA by using PCR.
Iron Uptake. Cultures were grown with 30 μM ferric malate, and at various time points, 0.5-ml aliquots were spun in a filter (Costar) to remove cells and checked for total iron by using the ferrozine assay (18). Iron uptake was normalized as the ratio of the amount of iron depleted from the medium to the absorbance at 400 nm.
Light Scattering. We modified the light scattering assay to check for turning in a magnetic field (19). We used a large bar magnet with a Spec20 spectrophotometer. The bar magnet was placed parallel or perpendicular to the sample holder outside the Spec 20, and maximum and minimum absorbance readings at 400-nm wavelength were recorded. The ratio of the maximum to minimum was designated as Cmag.
Complementation of ΔmamA. We used the pBBR1MCS vectors as the base for our complementing plasmids (20). Using PCR, a plasmid containing the tac promoter (21), mamA without its stop codon and GFP (22), was constructed in pBBR1MCS2, creating pAK20. For complementation, pAK20 and pBBR1MCS2 were conjugated into the ΔmamA strain and grown in the presence of 5 μM kanamycin and 30 μM ferric malate. We found that kanamycin often caused the ferric malate to precipitate. Thus, after overnight growth in kanamycin, 1 ml of each culture was inoculated into fresh iron-containing medium without kanamycin. We found that ΔmamA cells carrying pAK20 had higher Cmag values than cells carrying pBBR1MCS2 (1.43 versus 1.15).
Construction of mamA-GFP. mamA-GFP was amplified from pAK20 and cloned into the XhoI-SpeI sites of pAKO in place of the deletion construct. ORF10, the gene downstream from mamA, was cloned into this plasmid by using a PCR fragment with SpeI-NotI sites. This plasmid (mamAGFPint) was inserted into the AMB-1 chromosome by using the same technique described above for generating deletions.
Fluorescence Microscopy. Microscopy was carried out by using a Zeiss Axioplan microscope and a Hamamatsu (Middlesex, NJ) C474-95 charge-coupled device camera. Images were obtained with the ×100 oil objective. Cell membranes were stained by the addition of FM4-64 (Molecular Probes) to a final concentration of 1 μM. After 15-45 min of staining, cells were washed once in PBS and placed on thin agarose pads on microscope slides. FM4-64 was visualized with a standard Texas red filter. Images were acquired, false-colored, and overlaid in metamorph imaging software (Universal Imaging, Media, PA) and cropped in PHOTOSHOP 7.0.
Results
Imaging of Empty Magnetosome Vesicles and Visualization of Magnetosome Dynamics. To achieve conditions where no magnetite production took place, cells of Magnetospirillum sp. AMB-1 were grown for several passages in growth medium lacking all sources of iron in the components of the growth medium. The absence of added iron did not affect the growth rate or final cell number of AMB-1 presumably because of the presence of trace amounts of iron in glassware or the chemicals used. Under these conditions, the cultures were clearly nonmagnetic as confirmed by sensitivity to magnets and TEM (Fig. 1B). To search for empty magnetosome vesicles, iron-starved cells of AMB-1 were prepared for TEM by either conventional embedding techniques and ultramicrotomy or cryo-ultramicrotomy. When samples were prepared with conventional embedding techniques, no minerals were observed, and on rare occasions structures that resembled chains of empty vesicles of a size similar to magnetite-containing magnetosomes could be seen (Fig. 1 B and C). However, because of low contrast with the rest of the cytoplasm, it was difficult to visualize and characterize these structures further. In contrast, imaging of cryosectioned iron-starved cells by TEM consistently (>10% of the sections) showed long chains of empty vesicles resembling magnetosomes in size and number (Fig. 1 D and E). These structures are likely to be empty magnetosomes for several reasons. First, the size and arrangement of these vesicles in a chain is very similar to magnetosomes. Second, upon induction of magnetite formation by addition of iron to iron-starved cells, small magnetite crystals were seen within similar structures (Fig. 1F). Last, cells with full-sized magnetite crystals contain similar structures at the ends of their chains (Fig. 1G). These results demonstrate that magnetosome vesicles are present before the formation of magnetite by magnetotactic bacteria. Combined with previous work showing that magnetosomes are distinct from the cell membrane (5), they lend support to the model that the magnetosome vesicles are freestanding organelles that direct the biomineralization of magnetite crystals.
The dynamics of magnetite precipitation were monitored by TEM after the addition of iron to iron-starved cultures of AMB-1. Within 2 h, small crystals arranged in chains could be observed in these cells (Fig. 3A). These crystals are magnetite because the cells are responsive to magnetic fields at this time point. In addition, high-resolution TEM revealed that the magnetic crystals within the vesicles are magnetite (data not shown). The size of these crystals increased at each time point, and by 21 h after the addition of iron, most cells contained long chains of full-sized magnetite crystals (Fig. 3 C and E). Other than monitoring the timing for nucleation and growth of magnetite crystals, this time-course experiment revealed two important properties of magnetosomes. First, as observed in the early stages of the experiment, magnetite nucleation and growth proceeds simultaneously within several vesicles of the same chain (Fig. 3 A and C). Note that the small magnetite crystals are present within the preexisting full-sized and relatively uniform vesicles (Fig. 1F). Second, the spacing of crystals within a chain appears to be quite regular even at the early stage of crystal growth. To determine whether this observation is statistically significant, the average distance between the centers of 51 pairs of adjacent magnetite crystals at the 2-h time point was compared to a set of randomly generated numbers covering the possible distances for crystals of similar sizes within two neighboring magnetosomes (Fig. 1H). The average distance between the centers of adjacent crystals at the 2-h time point was 57 ± 7 nm, whereas a randomly generated data set predicted an average distance of 57 ± 21 nm. An F test comparison of the variances revealed a 9.6 × 10-10 probability that these two data sets were the same, suggesting that the spacing of the crystals at the early stages of magnetite growth is very regular. Thus, it is likely that magnetite precipitation begins simultaneously from the same location within preexisting magnetosome vesicles.
Fig. 3.
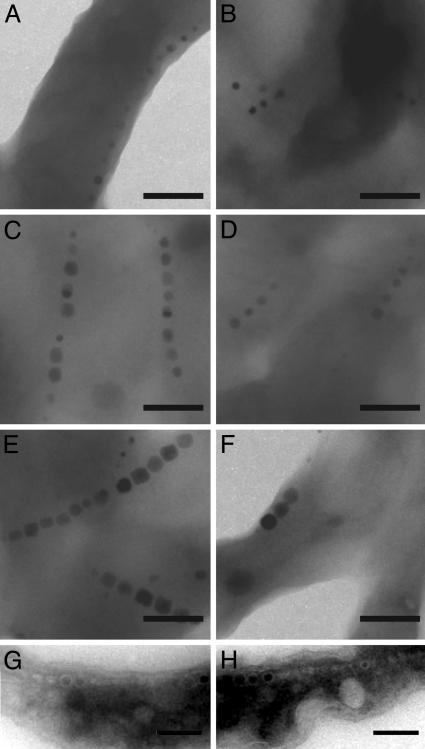
ΔmamA cells have a defect in activating their magnetosome vesicles. (A-F) TEM images of whole cells of WT and ΔmamA after the addition of iron to iron-starved cells. At 2 h both WT (A) and ΔmamA (B) cells have started synthesizing magnetite. The crystals get progressively larger at 3.5 h (C and D) and are full-sized after 21 h (E and F). At all times, however, WT cells have more crystals per chain than ΔmamA cells. (G and H) TEM images of cryo-ultrathin sections of ΔmamA cells grown in the absence (G) or presence (H) of iron, revealing the presence of chains of empty vesicles similar to WT (Fig. 1 E and F). (Scale bars: 200 nm.)
Genetic Identification of Factors Responsible for Magnetosome Functioning. To identify factors involved in magnetite precipitation and magnetosome biogenesis, a genetic screen was performed. Cells were first grown without iron and mutagenized with a hyperactive mariner transposon. Mutagenized cells were then challenged to make magnetite by the addition of iron, WT cells were removed with a magnet, and the cells in the nonmagnetic fraction were plated for single colonies. After screening through ≈700 colonies, two mnms, with severe defects in magnetite formation were chosen for further analysis. Interestingly, both mutants were the result of insertions in the mamABE gene cluster that has been implicated in magnetite formation through biochemical and genome comparison studies (Fig. 2A) (6). Previous screens with AMB-1 have not uncovered insertions in this gene cluster (23, 24). Although the mamABE cluster was part of a large number of genes deleted in a spontaneous nonmagnetic mutant (25), there is little genetic evidence directly implicating these genes in magnetite formation. Because the transposon insertions in mnm1 and mnm2 appear to be polar to downstream genes in the mamABE gene cluster, their severe nonmagnetic phenotypes are likely a result of the loss of several gene products (data not shown). Therefore, to understand the function of individual genes in this cluster we generated nonpolar deletions of mamA to study its phenotype in detail.
Nonpolar Deletion of mamA. MamA (also known as Mam22) contains several tetratricopeptide repeat domains and is predicted to be a soluble protein (8). The ΔmamA strain does not have a growth defect, and our qualitative assays showed that it was still magnetic. However, quantitative measurements of iron uptake showed that ΔmamA cells consistently take up less iron than the WT cells (Fig. 2B). To measure the turning of ΔmamA cells in a magnetic field, we took advantage of the differential light scattering properties of magnetic cells whose long axes are aligned parallel or perpendicular to a light source. Previous work has shown that light scattering approaches a maximum when the alignment is parallel and a minimum when it is perpendicular (19). Cmag, which is the ratio between the maximum and the minimum scattering values, correlates well with the average number of magnetite crystals in a chain (19). Consistently lower Cmag values were measured for ΔmamA cells than WT cells, indicating a lower level of turning in magnetic fields (Fig. 2C). To ensure that the observed phenotypes were caused by nonpolar deletion of mamA, the mutants were complemented with mamA-GFP expressed from the tac promoter. mamA-GFP was used because it functions as well as WT mamA (as shown later in this section), and the stability of the plasmid and expression from the tac promoter could be monitored by microscopy. ΔmamA cells carrying the complementing plasmid had significantly higher Cmag values than those carrying a control plasmid (1.43 versus 1.15), although the mutants were not complemented to WT levels. This is likely because expression of mamA from a heterologous promoter carried on a plasmid.
To understand this defect at the level of magnetosome formation, we analyzed mutant and WT cells with electron microscopy. As expected, WT cells contained 8-12 magnetite crystals per cell. However, ΔmamA cells had shorter chains of only one to five magnetite crystals per cell (Fig. 3). These results could be explained by a delay in initiation of magnetite formation or slower growth of the crystals in cells lacking mamA. To investigate this possibility, a time-course experiment comparing magnetite formation in ΔmamA to WT cells was carried out after the addition of iron to iron-starved cultures. Two hours after the addition of iron, chains of small crystals could be observed in both the WT and ΔmamA cells, indicating that the initiation of magnetite formation is not affected (Fig. 3 A and B). Furthermore, at subsequent time points, magnetite crystals in both WT and ΔmamA cells continued to grow at approximately similar rates (Fig. 3 C-F). However, at all time points the ΔmamA cells contained fewer magnetite crystals per cell than the WT. Therefore, although the timing and kinetics of magnetite formation do not appear to depend on MamA, the formation of the expected number of magnetite crystals in each cell requires its presence. One potential explanation for these results is that ΔmamA cells cannot produce the expected number of magnetosome vesicles. Accordingly, ΔmamA mutants were examined by cryo-ultramicrotomy and electron microscopy for the presence of magnetosome vesicles (Fig. 3 G and H). As can be seen, ΔmamA cells still contained long chains of empty vesicles, indicating that in the absence of MamA, cells can still produce the correct number of magnetosome vesicles but not all these vesicles are functional for the production of magnetite.
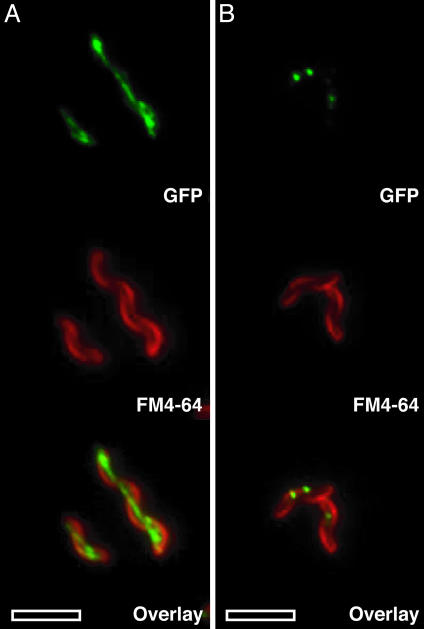
Subcellular Localization Dynamics of MamA-GFP. To understand the dynamics of MamA, we characterized its expression and subcellular localization by using GFP fusions to its C terminus. WT conditions were approximated by replacing the genomic copy of mamA with the mamA-GFP gene fusion. This fusion was fully functional as assessed by Cmag and nearly every cell in the population expressed MamA-GFP under all conditions tested (data not shown). MamA-GFP had a very distinct localization pattern that depended on the stage of growth (Fig. 4). During logarithmic growth, MamA-GFP localized to a thin spotted line extending end to end in nearly all of the cells (Fig. 4A). This localization pattern is reminiscent of the magnetosome chain as visualized by electron microscopy. However, magnetite chains rarely extend end to end within the cell, indicating that MamA might interact with the magnetosome and other parts of the cell. Consistent with this view, MamA-GFP also appeared to localize to the cell membrane occasionally (Fig. 4A). As cells entered stationary phase, MamA-GFP localization became more punctate and at the end of growth all cells contained one to four spots of MamA-GFP (Fig. 4B). This dynamic localization pattern was also seen in cells grown without iron, confirming the existence of magnetosome vesicles before magnetite formation. These results indicate that dynamic MamA localization does not depend on active magnetite formation and might be a useful tool for assessing the presence of magnetosome vesicles in mutants. The dynamic pattern of MamA localization also suggests that various parts of the same magnetosome chain might have different properties throughout the growth cycle of magnetotactic bacteria.
Fig. 4.
MamA displays a dynamic localization during the growth of AMB-1. The top images are the GFP localization, the middle images are the cell membrane stain with FM4-64, and the bottom images are the overlay of the GFP and FM4-64 images. (A) MamA-GFP localizes to a thin line within the cell during the logarithmic growth phase. The localization is from one end of the cell to the other, and at times some membrane localization is also observed. (B) In stationary phase, MamA-GFP displays a punctate pattern with one to four spots within the cell. (Scale bars: 5 μm.)
Discussion
Magnetosomes appear to be true membrane-bounded organelles controlling magnetite formation in a temporally and spatially coordinated fashion. Chains of empty magnetosome vesicles have not been reported previously to our knowledge, and we believe that our success in imaging them is in part because of the use of cryo-ultramicrotomy. This technique does not require multiple fixation and embedding steps that are used in conventional ultrathin sectioning and might be more effective in preserving the integrity of the specimen. The ability to visualize empty magnetosomes has significant implications for the study of biomineralization of magnetite and characterization of magnetosomes as organelles. For example, using this technique, mutants that are defective in the enzymatics of biomineralization can be separated from those that lack the ability to produce magnetosome vesicles. Additionally, this technique might be applied to investigate whether magnetosome vesicles originate from the cell membrane by visualizing vesicle intermediates during budding.
Collectively, our physical characterization of magnetosomes suggests that the organelle is primed for magnetite formation. Our work and previous studies have shown that magnetotactic bacteria can thrive and divide normally under laboratory conditions where no magnetite biomineralization takes place. Thus, it is possible that these organisms can exist in a nonmagnetic state in the environment as well. In this context, the ability to transition quickly from a nonmagnetic to magnetic state may be an extremely important selective advantage. This may explain the observation that magnetite biomineralization was initiated simultaneously within multiple vesicles in a magnetosome chain. Using such a mechanism, cells might achieve magneto-sensitivity before magnetite biomineralization is completed. This idea is supported by our observations that Cmag, which is an indicator of turning in a magnetic field, was nearly maximal at 3.5 h after the addition of iron even though magnetite crystals had not been mineralized to their mature size (Figs. 2C and 3C). However, if magnetite formation was to proceed one crystal at a time, the bacteria might require a longer period to become responsive to magnetic fields.
Similarly, the even spacing of nucleation sites within each vesicle might allow the interactions between partially completed crystals to be uniform within a chain thus creating a response to magnetic fields before completion of biomineralization. The uniform spacing of nucleation sites within a chain might be achieved by localization of nucleation factors or magnetite formation enzymes within each vesicle. This implies the existence of polarity at the level of protein localization in these 50-nm vesicles, providing another layer of complexity to the cell biology of magnetosomes.
The properties of the MamA suggest that magnetosome proteins can display a dynamic localization behavior and that the activity of individual magnetosome vesicles can be regulated. Although we were able to describe the phenotypes associated with the loss of mamA in detail, its exact role remains unclear. It is possible that MamA is part of general magnetosome assembly and maintenance processes such as protein sorting or activation of magnetosome vesicles in response to external cues and in its absence these processes become inefficient.
Another possibility is that MamA may be part of a more specific strategy used by magnetotactic bacteria to regulate the length of their magnetosome chains in response to availability of iron or other environmental conditions. Consistent with this model our TEM analysis shows that ΔmamA cells have shorter magnetosome chains (Fig. 3). In addition, previous work has shown that there could be great variety in the length of magnetosome chains under different conditions (26-28). Further work, such as identification of MamA-interacting proteins, is needed to uncover the exact mechanism of its action.
Recently, other specialized membranous organelles have also been described in prokaryotes (29, 30). The presence of organelles in bacteria has significant evolutionary implications. Although mitochondria and chloroplasts appear to have arisen by symbiosis between eukaryotic and prokaryotic cells, the origins of other membrane-bounded organelles such as the endoplasmic reticulum, the nucleus, and the Golgi complex are unclear (31). Understanding the processes of vesicle biogenesis and protein sorting in magnetosomes at the molecular level might provide insight into the evolutionary origins of other bacterial and eukaryotic organelles.
Acknowledgments
We thank D. Moyles for assistance with electron microscopy; S. Chiang for providing the PSC189 plasmid; M. S. Simon and J. Kirschvink for thoughtful discussions and endless enthusiasm for this project; and J. Mui and Dr. S. K. Sears of the McGill Facility for Electron Microscopy Research for assistance. D.K.N. is supported by grants from the Packard Foundation and the Luce Foundation. A.K. is a Senior Research Fellow of the Beckman Institute and is supported by the Arnold and Mabel Beckman Foundation. H.V. acknowledges financial support from the Natural Sciences and Engineering Research Council of Canada. T.J.B. is funded through grants from the Natural Science and Engineering Research Council of Canada and the U.S. Department of Energy Natural and Accelerated Bioremediation Research program. Electron microscopy was performed at the Natural Sciences and Engineering Research Council Guelph Regional Scanning Transmission Electron Microscopy Facility at the University of Guelph, which is partially funded through a Natural Sciences and Engineering Research Council Major Facilities Access grant (to T.J.B.).
Abbreviations: TEM, transmission electron microscopy; mnm, magnetosome mutant; MSGM, Magnetospirillum growth medium.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY508230).
References
- 1.Schuler, D. (1999) J. Mol. Microbiol. Biotechnol. 1, 79-86. [PubMed] [Google Scholar]
- 2.Kirschvink, J. L. & Hagadorn, J. W. (2000) in Biomineralization: From Biology to Biotechnology and Medical Application, ed. Baeuerlein, E. (Wiley, Weinheim, Germany), pp. 139-147.
- 3.Mann, S., Sparks, N. H. & Board, R. G. (1990) Adv. Microb. Physiol. 31, 125-181. [DOI] [PubMed] [Google Scholar]
- 4.Gorby, Y. A., Beveridge, T. J. & Blakemore, R. P. (1988) J. Bacteriol. 170, 834-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vali, H. & Kirschvink, J. L. (1990) Iron Biominerals (Plenum, New York).
- 6.Grunberg, K., Wawer, C., Tebo, B. M. & Schuler, D. (2001) Appl. Environ. Microbiol. 67, 4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arakaki, A., Webb, J. & Matsunaga, T. (2003) J. Biol. Chem. 278, 8745-8750. [DOI] [PubMed] [Google Scholar]
- 8.Okuda, Y., Denda, K. & Fukumori, Y. (1996) Gene 171, 99-102. [DOI] [PubMed] [Google Scholar]
- 9.Schuler, D. & Baeuerlein, E. (1997) in Transition Metals in Microbial Metabolism, eds. Wikelmann, G. & Carrano, C. J. (Harwood, Amsterdam), pp. 159-185.
- 10.Schuler, D. & Baeuerlein, E. (1998) J. Bacteriol. 180, 159-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsunaga, T., Sakaguchi, T. & Tadokoro, F. (1991) Appl. Microbiol. Biotechnol. 35, 651-655. [Google Scholar]
- 12.Beveridge, T. J., Popkin, T. J. & Cole, R. M. (1994) in Methods for General and Molecular Bacteriology, ed. Krieg, N. R. (Am. Soc. Microbiol., Washington, DC), pp. 42-71.
- 13.Tokuyasu, K. T. (1980) Histochem. J. 12, 381-403. [DOI] [PubMed] [Google Scholar]
- 14.Zierold, K. (1987) in Cryotechniques in Biological Electron Microscopy, ed. Zierold, K. (Springer, Berlin), pp. 132-148.
- 15.Chiang, S. L. & Rubin, E. J. (2002) Gene 296, 179-185. [DOI] [PubMed] [Google Scholar]
- 16.Dehio, C. & Meyer, M. (1997) J. Bacteriol. 179, 538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metcalf, W. W., Jiang, W. H., Daniels, L. L., Kim, S. K., Haldimann, A. & Wanner, B. L. (1996) Plasmid 35, 1-13. [DOI] [PubMed] [Google Scholar]
- 18.Stookey, L. L. (1970) Anal. Chem. 42, 779-781. [Google Scholar]
- 19.Schuler, D., Rainer, U. & Bauerlein, E. (1995) FEMS Microbiol. Lett. 132, 139-145. [Google Scholar]
- 20.Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M. & Peterson, K. M. (1995) Gene 166, 175-176. [DOI] [PubMed] [Google Scholar]
- 21.Deboer, H. A., Comstock, L. J. & Vasser, M. (1983) Proc. Natl. Acad. Sci. USA 80, 21-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cormack, B. P., Valdivia, R. H. & Falkow, S. (1996) Gene 173, 33-38. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura, C., Burgess, J. G., Sode, K. & Matsunaga, T. (1995) J. Biol. Chem. 270, 28392-28396. [DOI] [PubMed] [Google Scholar]
- 24.Wahyudi, A. T., Takeyama, H. & Matsunaga, T. (2001) Appl. Biochem. Biotechnol. 91-93, 147-154. [DOI] [PubMed] [Google Scholar]
- 25.Schubbe, S., Kube, M., Scheffel, A., Wawer, C., Heyen, U., Meyerdierks, A., Madkour, M. H., Mayer, F., Reinhardt, R. & Schuler, D. (2003) J. Bacteriol. 185, 5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blakemore, R. P., Short, K. A., Bazylinski, D. A., Rosenblatt, C. & Frankel, R. B. (1985) Geomicrobiol. J. 4, 53-71. [Google Scholar]
- 27.Guerin, W. F. & Blakemore, R. P. (1992) Appl. Environ. Microbiol. 58, 1102-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heyen, U. & Schuler, D. (2003) Appl. Microbiol. Biotechnol. 61, 536-544. [DOI] [PubMed] [Google Scholar]
- 29.Sinninghe Damste, J. S., Strous, M., Rijpstra, W. I., Hopmans, E. C., Geenevasen, J. A., van Duin, A. C., van Niftrik, L. A. & Jetten, M. S. (2002) Nature 419, 708-712. [DOI] [PubMed] [Google Scholar]
- 30.Seufferheld, M., Vieira, M. C., Ruiz, F. A., Rodrigues, C. O., Moreno, S. N. & Docampo, R. (2003) J. Biol. Chem. 278, 29971-29978. [DOI] [PubMed] [Google Scholar]
- 31.Dyall, S. D. & Johnson, P. J. (2000) Curr. Opin. Microbiol. 3, 404-411. [DOI] [PubMed] [Google Scholar]