Abstract
The evolution of animal diversity depends on changes in the regulation of a relatively fixed set of protein-coding genes. To understand how these changes might arise, we examined the organization of shared sequence motifs in four coordinately regulated neurogenic enhancers that direct similar patterns of gene expression in the early Drosophila embryo. All four enhancers possess similar arrangements of a subset of putative regulatory elements. These shared features were used to identify a neurogenic enhancer in the distantly related Anopheles genome. We suggest that the constrained organization of metazoan enhancers may be essential for their ability to produce precise patterns of gene expression during development. Organized binding sites should facilitate the identification of regulatory codes that link primary DNA sequence information with predicted patterns of gene activity.
Enhancers are the most prevalent class of regulatory DNAs that determine where and when a given gene is expressed during development. The typical metazoan enhancer is ≈500 bp in length and contains multiple binding sites for two or more sequence-specific transcriptional activators and at least one repressor. The best-characterized enhancers contain densely linked binding sites, at least one site every 40-50 bp across the length of the enhancer. There are two opposite extreme views of enhancer organization. They might serve as unstructured templates that bring different combinations of activators and repressors into close (but random) proximity, or they might be highly structured, so that the integration of disparate activators and repressors depends on a variety of organizational constraints, such as helical phasing between neighboring binding sites. The enhancer that regulates the mammalian IFN-β gene exhibits fixed linkage of neighboring sites (1), but it is possible that this “enhanceosome” is exceptional and reflects its specialized function in mediating rapid response to viral infection. Thus far, there is no evidence that “typical” enhancers, such as those mediating tissue-specific gene expression during development, possess a higher-order structure.
The dorsal-ventral patterning of the early Drosophila embryo (2-5) provides a favorable system for determining whether coregulated enhancers share constrained organizational features. Dorsal-ventral patterning is controlled by a sequence-specific transcription factor, Dorsal, that is distributed in a broad gradient in the early embryo. High levels of the gradient activate target genes required for the differentiation of the mesoderm, whereas intermediate and low levels activate gene expression in ventral and dorsal regions of the neurogenic ectoderm, respectively. Microarray screens have identified at least 30 Dorsal target genes that are regulated by different levels of the gradient (6). The combination of classical gene fusion assays and bioinformatics methods have identified enhancers for 12 of these genes (5-9). Four of the 12 enhancers, from the rhomboid (rho), ventral nervous system defective (vnd), brinker (brk), and vein (vn) genes, are coregulated by intermediate levels of the Dorsal gradient in ventral regions of the neurogenic ectoderm. These enhancers represent the largest collection of coregulated enhancers for any metazoan developmental process. They thereby provide a unique opportunity to determine whether coordinate enhancers contain similar arrangements of regulatory elements.
The four coregulated enhancers were previously shown to share binding sites for Dorsal (GGGWWWWCYS, GGGW4-5CCM), Twist (CACATGT), Suppressor of Hairless [Su(H)] (YGTGDGAA), as well as an unknown regulatory element (the “mystery site,” CTGWCCY). The present study identified specialized forms of the Dorsal (SGGAAANYCSS), Su(H) (CGTGGGAAAWDCSM), and mystery sites (CTGRCCBKSMM) within each enhancer. These specialized motifs exhibit a number of organizational constraints within a 300-bp core domain of each enhancer. First, the specialized Dorsal site maps within 20 bp of an oriented Twist site. Second, the specialized mystery site is located 108-153 bp downstream of the Twist site, and the exact distance exhibits a periodicity of 15 bp among the different enhancers. And third, the specialized Su(H) site is located on the same side of the helix in each enhancer. The core structure seen in the Drosophila neurogenic enhancers is largely retained in the Anopheles vnd enhancer, even though the fly and mosquito enhancers have diverged for >230 million years and lack simple sequence similarity. We suggest that metazoan enhancers possess fixed organizational constraints that are essential for the integration of transcriptional activators and repressors during development.
Materials and Methods
Bioinformatics. Whole-genome scans for sequences matching enhancer models of various types were conducted by using multiple implementations to double-check results. These methods included searching fly and mosquito genomes by using UNIX command-line perl regular expressions for all structured queries. This method became increasingly efficient as various aspects of enhancer organization were revealed. Other utilities included Auilix Biopharma's genegrokker software for mixed probabilistic models involving regular expressions/position-weighted matrices/consensus sequences for the fly genome, flyenhancer (www.flyenhancer.org) for Boolean models with simple consensus sequences for the fly and mosquito genomes (courtesy of www.opengenomics.org), and Target Explorer (http://trantor.bioc.columbia.edu/Target_Explorer) for searches based on position-weighted matrices of fly and mosquito (10). Analysis for shared motifs and sequence comparison for Fig. 3C involved use of the Discriminator and Mirror tools, respectively, in Auilix Biopharma's genegrokker system as reported previously (6). For a complete list of utilities see www.alumni.caltech.edu/~aerives/animal_cisreg.html. Relevant results are shown in Tables 1-3, which are published as supporting information on the PNAS web site.
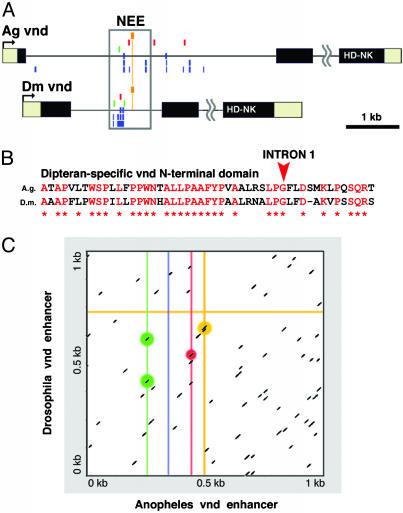
Fig. 3.
The Anopheles vnd locus contains an intronic cluster of NEE motifs. (A) There are only several composite clusters of neurogenic motifs in the Anopheles genome (Table 3). One of these clusters maps within the first intron of the vnd ortholog, as determined by reciprocal blast homology to the homeodomain and an NK-2 specific domain located in the third exon (HD-NK). Motifs described in the text, as well as degenerate Dorsal motifs (lower rows of blue clusters), are only shown for intron 1. Intron 2 is not to scale. (B) We find that the Drosophila and Anopheles vnd loci share only one other region of significant conservation, an N-terminal protein-coding sequence spanning intron 1 in Drosophila. (C) Specific comparison of the 1-kb regions encompassing the cluster of sites for the tested enhancers shows that the two sequences lack any extensive sequence alignment spanning the enhancers. Each line in the graph represents a window of sequence alignment containing at least a perfect 7-bp core alignment. Substituting the vnd enhancer sequence with its reverse-complement gives similar results (data not shown). Thus, the actual points of correspondence between sites for Twist (green), Dorsal-like (blue line), Su(H) (red line), and μ (orange lines) do not rise appreciably above background levels of random matches (nonhighlighted dashes). The Anopheles vnd cluster is no more closely related by primary sequence alignment to the Drosophila vnd NEE than to any of the other Drosophila NEEs (data not shown).
DNA Constructs. DNA fragments encompassing identified clusters were amplified from genomic DNA with the primer pairs listed below. PCR products were cloned either into pGEM T-Easy or directly into -42eve-lacZCasper (e.g., ref. 6). The Anopheles vnd (Ag-vnd) enhancer was amplified as a 908-bp fragment from genomic DNA isolated from the Anopheles gambiae PEST strain by using the primers listed below and subcloned into the pGEM-Teasy vector for sequencing. The Ag-vnd fragment was subsequently cloned as an EcoRI fragment into the EcoRI site of the -42 eve-lacZ CaSpeR vector. Eight independent transformed lines were obtained for this construct. Anopheles vnd neurogenic ectoderm enhancer (NEE) primers were Mosq-vnd-5′-GGG ATT TTG TTT CGC CGC TTC G and Mosq-vnd-3′-CTA CTT CAT GTT GTG TAC TTT GGC C.
In Situ Hybridization. Embryos were hybridized with digoxigenin-labeled antisense RNA probes as described (e.g., ref. 6). An antisense lacZ RNA probe was used to examine the staining patterns in transgenic embryos collected from each of the transformed lines.
Fly Stocks. yw was used for P-element transformations and in situ hybridizations.
Results
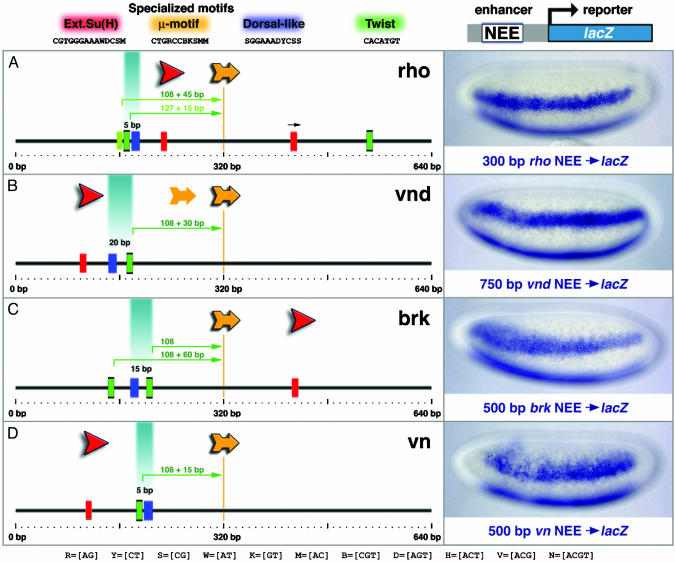
Previous studies identified neurogenic ectoderm enhancers (NEEs) for rho (7), vnd (6), brk (8), and vn (9). All four enhancers direct lateral stripes of gene expression in ventral regions of the neurogenic ectoderm in response to intermediate levels of the Dorsal gradient (Fig. 1). The rho enhancer is located ≈1.7 kb 5′ of the transcription start site (Fig. 2D) and was identified in classical gene fusion assays (7). The minimal rho enhancer is 300 bp in length, although stronger staining is obtained with a larger, ≈600-bp genomic DNA fragment that encompasses the minimal enhancer. The vnd enhancer is located in the first intron of the gene, ≈1 kb downstream of the transcription start site (Fig. 2D). It was identified on the basis of containing a putative cluster of Dorsal-binding sites (6). Full staining is obtained with a 700-bp fragment that encompasses all four putative Dorsal sites, although a normal pattern is directed by a smaller, 350-bp fragment that contains just three of the sites (9). The brk enhancer is 500 bp in length and maps >10 kb 5′ of the transcription start site (Fig. 2D). It was identified in a whole-genome survey of optimal Dorsal-binding sites (8). Finally, the 500-bp vn enhancer was identified in a bioinformatics survey of the Drosophila genome for linked Dorsal, Twist, Su(H), and mystery sites (9). It is located deep within the first intron of the gene, nearly 8 kb downstream of the transcription start site (Fig. 2D). Thus, the four enhancers map in different 5′ and intronic positions, and are associated with unrelated genes encoding transcription factors (Vnd and Brk), a membrane protease (Rho), and epidermal growth factor (EGF) signaling molecule (Vein). Nonetheless, the enhancers direct similar patterns of lacZ reporter gene expression in transgenic embryos (Fig. 1). In all cases, lacZ staining is restricted to ventral regions of the neurogenic ectoderm where there are intermediate levels of the Dorsal gradient.
Fig. 1.
Shared organization of Drosophila enhancers. (Left) Each diagram shows a 640-bp region encompassing the minimal rho (A), vnd (B), brk (C), and vn (D) NEE. Different sequences from these enhancers were attached to a lacZ reporter gene and expressed in transgenic embryos (photomicrographs, Right). Embryos are undergoing cellularization and are oriented with anterior to the left and dorsal up. The staining patterns were determined by in situ hybridization using an antisense RNA probe against lacZ. Red boxes indicate the occurrences of the extended Su(H) site (YGTGRGAAM), whereas the red arrows indicate the orientation of the motif within each enhancer. The specialized μ motif and Dorsal-like sites are shown in yellow and blue, respectively. All CACATGT sequences are shown in green, as well as the related Twist-binding site CATATGT (light green). The distance between linked Twist and Dorsal-like sites is shown directly above each pair. Also indicated are the Twist/μ distances starting from the 3′ position of the palindromic ACATGT core of the Twist site. The centerline at 320 bp bisects the μ core CTGRCC in each enhancer.
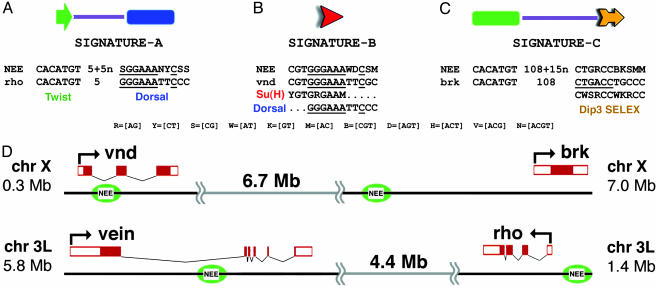
Fig. 2.
Organized sequence motifs in the rho, vnd, brk, and vn NEEs. All four enhancers contain three organizational features: Twist is linked to a Dorsal-like site (A), the extended Su(H) motif shares 3′ sequences related to the Dorsal-binding site and is located on the same strand of the double helix (B), and there is a Twist site located at a characteristic position upstream of μ (C). A consensus sequence for each signature is shown, as well as an example from a specific enhancer. For example, the Dorsal-like motif in the rho NEE is a perfect match to an optimal Dorsal-binding site (GGGW4-5CCC). The extended Su(H) motif in the vnd enhancer matches overlapping Su(H) and Dorsal-binding sites. The extended μ motif is related to the previously identified Dip3 consensus sequence (14). The genetic organization of the neurogenic genes and their enhancers are summarized in D. The vnd and brk genes are linked on the X chromosome, whereas the vein and rho genes are linked on the left arm of chromosome 3 (D).
Identification of Specialized Sequence Motifs. Shared sequence motifs were identified among a set of 640-bp fragments that encompass each of the four minimal enhancers. We consider only those sequences that are present in all four enhancers, and underrepresented in a total of 117 kb of control genomic DNA. The control sequences derive from genetic loci that are expressed along the anterior-posterior axis (rather than the dorsal-ventral axis) of early embryos. The longest sequence motif containing the fewest degenerate positions is a 9-bp sequence, CGTGGGAAA (see Fig. 2 and Table 1), that matches the optimal Su(H) consensus sequence: YGTGRGAAM (11). There is weak conservation of five additional 3′ nucleotides to yield the following extended Su(H) site: CGTGGGAAAWBCSM. The five additional nucleotides sometimes have the appearance of a 3′ Dorsal half-site, so that the entire 14-bp sequence often resembles overlapping Su(H) and Dorsal sites (Fig. 2B). There is a single copy of this extended motif in each of the four neurogenic enhancers.
The second most significant shared sequence motif that was identified, SGGAAANYCSS, is related to Dorsal consensus sequences, GGGW4-5CCM and GGGW4CYS. Each enhancer contains at least one copy of a consensus Dorsal-binding site, and a copy of the recently identified Dorsal-like motif. In some cases, the Dorsal-like motif corresponds to the consensus sequence, as seen in the rho enhancer (GGGAAATTCCC; Fig. 2 A). However, in most cases the specialized motif represents a weak Dorsal site that is distinct from the optimal site. We hereafter refer to this sequence as the “Dorsal-like” motif. Some variants of this motif represent very poor Dorsal-binding sites, as seen in the case of the vnd enhancer.
The third shared sequence motif that was identified, CTGRCCBKSMM, is related to a shared motif that was previously identified in the rho and vnd enhancers, as well as in the mesoderm enhancer of the Mes3 gene: RGGNCAG (or CTGNCCY; ref. 6). There is a single copy of a more extended version of this sequence in the rho, vnd, brk, and vn enhancers: CTGRCCBKSMM. This extended sequence is hereafter called the μ motif. It is related to the consensus recognition sequence for a ubiquitous transcription factor called Dorsal interacting protein 3 (Dip3) (Fig. 2C; see Discussion). The Drosophila genome contains only four clusters of tightly linked Dorsal-like, extended Su(H), and μ-binding sites, and these correspond to the rho, vnd, brk, and vn enhancers (see Table 2). These binding sites, along with the Twist site, appear to mediate transcriptional activation. All four NEEs also contain putative Snail repressor sites (MMMCWTGY), which block expression in the ventral mesoderm (9).
Conserved Organizational Features. The three aforementioned specialized sequence motifs appear to exhibit similar arrangements in all four enhancers (see summary diagrams in Fig. 1). First, the 14-bp extended Su(H) site is located on the same strand of the DNA double helix in each enhancer (Fig. 2B). Second, a Twist-binding site, CACATGT, maps between 108 and 168 bp 5′ of an oriented μ motif (Fig. 2C). The exact separation of the two sequences exhibits 15-bp periodicity among the different enhancers (Figs. 1 and 2C). Finally, the specialized Dorsal-like motif is closely linked to the CACATGT that is positioned 5′ of μ (Fig. 2 A). There are two potentially interesting aspects of this linkage. The Dorsal-like and Twist-binding sites map between 5 and 20 bp, with a 5-bp periodicity among the different enhancers (Fig. 2 A). Moreover, the linked Twist site is oriented toward the Dorsal-like sequence, which may reflect specific protein-protein interactions (see Discussion). All three potential organizational features, oriented Su(H), Twist-μ phasing, and Twist/Dorsal-like linkage, occur within a core domain of ≈300 bp in each enhancer (Fig. 1).
Identification of the Anopheles vnd Enhancer. If the organization is significant, then it might be retained in an evolutionarily divergent neurogenic enhancer, such as one from Anopheles. Because we were not able to identify corresponding Anopheles enhancers by using blast-based alignments to Drosophila NEE sequences, we scanned the entire Anopheles genome for clusters of relaxed versions of the four shared sequence motifs.
In one whole-genome query (Table 3), 13 composite clusters of 300 bp or less were identified that contain at least one copy of the Twist site (CACATGT), a degenerate Dorsal-like motif (SGSAARDYYSC), the Su(H) core consensus sequence GTGGGAA, and the core μ (mystery) motif, CTGRCC. Only 5 of the 13 clusters possess a Twist site located upstream of an oriented μ core (Table 3), and just two of these five clusters conform to phased map distances seen in the Drosophila enhancers (206 bp, 236 bp). The first such cluster is located <10 kb upstream of the fred (friend of echinoid) ortholog, corresponding to a Drosophila neurogenic gene involved in Delta/Notch signaling (11). The second cluster maps within the first intron of the Anopheles vnd gene (Fig. 3A), as determined by a conserved N-terminal coding sequence that spans exons 1 and 2 (Fig. 3B). Despite the similar intronic locations of the two clusters, the fly and mosquito enhancers lack simple sequence similarity above random levels (Fig. 3C).
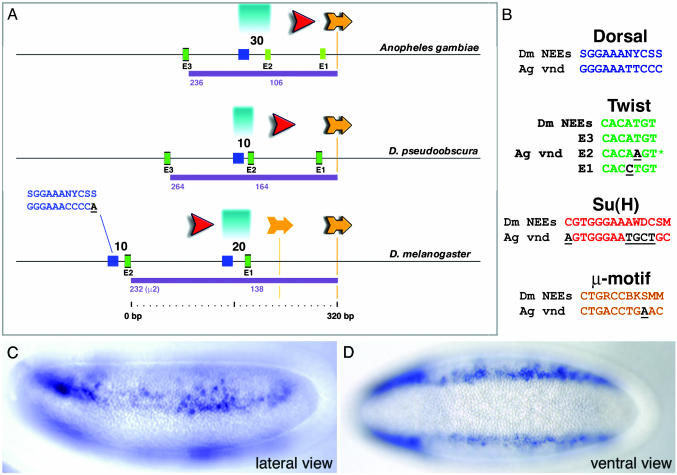
The mosquito vnd cluster appears to retain many of the features seen for the fly NEEs (Fig. 4A). The Su(H) site is positioned in the same orientation as those contained in the Drosophila enhancers. This site shares seven of eight matches with the simple Su(H) consensus sequence (YGTGDGAA), and 9 of 14 matches with the extended sequence (Fig. 4B). The Anopheles Dorsal-like site (SGGAAANYCSS) is an exact match to the optimal Dorsal consensus sequence, GGGW4-5CCC. As in Drosophila, the Dorsal-like site maps ≈160 bp upstream of μ (Fig. 4A). Dorsal-Twist linkage may be somewhat relaxed in the mosquito vnd enhancer as compared with the Drosophila NEEs. The closest oriented Twist motif (CACATGT) maps nearly 90 bp from the Dorsal site (E3, Fig. 4A); they are located within 30 bp in the Drosophila NEEs. There is a properly oriented Twist-like motif, CACAAGT, located just 30 bp from the Dorsal site (E2, Fig. 4A), but it is not clear that it represents an authentic Twist-binding site because it does not conform to the general E-box consensus sequence, CANNTG. The reverse complement of this Twist-like motif is an E-box, CACTTGT, but positioned in the “wrong” orientation relative to the Dorsal site. The conversion of the E2 Twist-like motif into an optimal sequence might be expected to augment expression from the Anopheles vnd-lacZ fusion gene (see below). Finally, there is a single μ motif that contains 10 of 11 matches with the Drosophila sequence. (Fig. 4 A and B). It exhibits the same type of distance (≈110 bp) and orientation from the Twist-like site as seen for the Twist-μ linkages in the fly NEEs (Fig. 4 A and B).
Fig. 4.
Anopheles vnd enhancer retains conserved NEE structure and function. (A) The mosquito vnd enhancer displays many of the organizational features seen for all of the Drosophila neurogenic enhancers. Comparison with the Drosophila pseudoobscura NEE sequence reveals an interesting intermediate compared to the former two. Both the Twist/Dorsal-like and Twist/μ distances are indicated for each sequence. For discussion of salient aspects of this organization, see text. (B) Alignments of Anopheles vnd sites to Drosophila NEE consensus sequences are depicted with mismatches underlined. The E2 sequence (asterisk) shown here overlaps a CACTTGT E-box in the opposite orientation. Lateral (C) and ventral (D) views of transgenic Drosophila embryos carrying the Anopheles vnd enhancer attached to the lacZ reporter gene are shown. Staining is detected in ventral regions of the neurogenic ectoderm.
The Anopheles vnd Enhancer Works in Transgenic Drosophila Embryos. The conservation and organization of the neurogenic regulatory elements in the Anopheles vnd intronic cluster suggested that it might be able to activate gene expression in response to intermediate levels of the Dorsal gradient. To test this possibility, a ≈900-bp genomic DNA fragment that encompasses the Anopheles vnd cluster was attached to a minimal eve-lacZ fusion gene and expressed in transgenic Drosophila embryos (Fig. 4 C and D). The Anopheles enhancer directs lateral stripes of lacZ expression within ventral regions of the presumptive neurogenic ectoderm. The staining pattern is somewhat weak and erratic, but is nonetheless similar to the expression profiles observed for the different Drosophila NEEs. These observations suggest that the Anopheles vnd intronic cluster corresponds to the orthologous Drosophila vnd enhancer, even though they lack sequence similarity (see Discussion).
Discussion
The systematic comparison of the rho, vnd, brk, and vn, enhancers led to the identification of specialized Dorsal, Su(H), and mystery (μ) sites. Simple versions of these binding motifs were identified in previous studies that focused on a two-way comparison of the rho and vnd enhancers (6), or a three-way comparison of rho, vnd, and brk (9). The current use of all four coregulated neurogenic enhancers permitted a more refined search for shared motifs. The focused attention to specialized sites revealed a shared arrangement of regulatory elements within all four enhancers. There are three major features of this organization: tightly linked Dorsal-like and Twist sites, fixed phasing between Twist and an oriented 3′ μ motif, and a common orientation of the extended Su(H) sequence.
These organizational constraints are reminiscent of the IFN-β enhanceosome, which contains linked and oriented binding sites for two sequence-specific transcriptional activators, IRF and the ATF2/c-Jun heterodimer (1). Protein-protein interactions between the two protein complexes are essential for full activation of the IFN-β gene. These interactions are impaired by a variety of manipulations, including inverting the ATF2/c-Jun-binding site relative to the IRF site. The recent analysis of Drosophila immunity gene regulation identified linked and oriented REL and GATA sites in a number of the 5′ regulatory DNAs (13). Similarly, all four Drosophila NEEs contain linked Dorsal and Twist sites, with the Twist site oriented toward the Dorsal site. This organization may be required for protein-protein interactions that foster cooperative occupancy of the linked sites. These Twist sites are located within a 34-bp window centered 125 bp upstream of μ. It is conceivable that this fixed linkage is required for interactions between the Dorsal-Twist complex and whatever regulatory protein binds the μ motif. The μ sequence shares 10 of 11 matches with the consensus sequence for a regulatory protein called Dip3 (14, 15), which augments transcriptional activation by Dorsal and Twist (15). Dip3 is a member of the MADF-BESS family of DNA-binding proteins. MADF contains a helix-turn-helix DNA-binding domain, whereas the C-terminal BESS domain mediates protein-protein interactions. The BESS domain in Dip3 specifically interacts with the Rel homology domain in the Dorsal protein (15). In a recent survey of the Drosophila proteome (16), Dip3 was found to interact with Ubc9, which was independently isolated as “Dip4” in the same screen that identified Dip3 and Twist (“Dip5”) (14). Ubc9, a nuclear ubiquitin-like conjugating enzyme, was found to mediate Dorsal-SMT3 (SUMO-1) conjugation with a corresponding synergistic effect on Dorsal target activation (14). Perhaps the formation of appropriate Dorsal-Twist-Dip3-Ubc9 complexes depends on the specific arrangement of Dorsal, Twist, and μ sites seen in the rho, vnd, brk, and vn enhancers.
An implication of this study is that metazoan enhancers possess higher-order structures. The organizational constraints described in this study are not as stringent as the arrangement of regulatory elements seen in the IFN-β or MHCII enhanceosomes (1, 17), which are located in promoter-proximal regions. In contrast, the NEEs considered in this study represent distal enhancers that map as far as 10 kb 5′ of the brk promoter and >7 kb 3′ of the vn promoter. The typical distal enhancer might possess a structure somewhere between enhanceosomes and a random distribution of regulatory elements. It will be important to test the functional significance of NEE organization by manipulating the distance between linked Dorsal and Twist sites, and inverting the orientation of the extended Su(H) site. Previous attempts to disrupt enhancer organization provide mixed results. Inversion of a GATA site relative to the linked REL site impairs the expression of the cecropin regulatory DNA in transgenic Drosophila larvae (13). In contrast, the relocation of the critical Bicoid-1 activator site to a new position within the eve stripe 2 enhancer had only a modest effect on expression (18).
The organization of binding sites seen in the Drosophila NEEs appears to be largely retained in the Anopheles vnd enhancer, even though the fly and mosquito enhancers have diverged for 230 million years. There is little doubt that the mosquito and fly enhancers are orthologous because they direct similar patterns of gene expression in transgenic Drosophila embryos and are located in the same relative position within the vnd locus. The maintenance of organized binding sites would impose specific constraints on enhancer evolution. Short insertion sequences (indels) could impair enhancer function by disrupting phasing of linked sites. The acquisition of novel activator-binding sites might not alter enhancer function if improperly positioned. We suggest that the simplest way to change gene activity is through the acquisition of binding sites for localized transcriptional repressors, which can work in a dominant fashion to alter enhancer function regardless of orientation and location (e.g., ref. 19).
Supplementary Material
Acknowledgments
We thank Robert Zinzen and Michele Markstein for sharing unpublished results. This work was funded by National Institutes of Health Grants GM46638 and P01 HD37105 (to M.L.).
Abbreviations: NEE, neurogenic ectoderm enhancer; Su(H), Suppressor of Hairless; Dip3, Dorsal interacting protein 3.
References
- 1.Thanos, D. & Maniatis, T. (1995) Cell 83, 1091-1100. [DOI] [PubMed] [Google Scholar]
- 2.Belvin, M. P. & Anderson, K. V. (1996) Annu. Rev. Cell Dev. Biol. 12, 393-416. [DOI] [PubMed] [Google Scholar]
- 3.Rusch, J. & Levine, M. (1996) Curr. Opin. Genet. Dev. 6, 416-423. [DOI] [PubMed] [Google Scholar]
- 4.Drier, E. A. & Steward, R. (1997) Semin. Cancer Biol. 8, 83-92. [DOI] [PubMed] [Google Scholar]
- 5.Stathopoulos, A. & Levine, M. (2002) Dev. Biol. 246, 57-67. [DOI] [PubMed] [Google Scholar]
- 6.Stathopoulos, A., Van Drenth, M., Erives, A., Markstein, M. & Levine, M. (2002) Cell 111, 687-701. [DOI] [PubMed] [Google Scholar]
- 7.Ip, Y. T., Park, R. E., Kosman, D., Bier, E. & Levine, M. (1992) Genes Dev. 6, 1728-1739. [DOI] [PubMed] [Google Scholar]
- 8.Markstein, M., Markstein, P., Markstein, V. & Levine, M. S. (2002) Proc. Natl. Acad. Sci. USA 99, 763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markstein, M., Zinzen, R., Markstein, P., Yee, K.-P., Erives, A., Stathopoulous, A. & Levine, M. (2004) Development (Cambridge, U.K.), in press. [DOI] [PubMed]
- 10.Sosinsky, A., Bonin, C. P., Mann, R. S. & Honig, B. (2003) Nucleic Acids Res. 31, 3589-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey, A. M. & Posakony, J. W. (1995) Genes. Dev. 9, 2609-2622. [DOI] [PubMed] [Google Scholar]
- 12.Chandra, S., Ahmed, A. & Vaessin, H. (2003) Dev. Biol. 256, 302-316. [DOI] [PubMed] [Google Scholar]
- 13.Senger, K., Armstrong, G. W., Rowell, W. J., Kwan, J. M., Markstein, M. & Levine, M. (2004) Mol. Cell 13, 19-32. [DOI] [PubMed] [Google Scholar]
- 14.Bhaskar, V., Valentine, S. A. & Courey, A. J. (2000) J. Biol. Chem. 275, 4033-4040. [DOI] [PubMed] [Google Scholar]
- 15.Bhaskar, V. & Courey, A. J. (2002) Gene 299, 173-184. [DOI] [PubMed] [Google Scholar]
- 16.Giot, L., Bader, J. S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y. L., Ooi, C. E., Godwin, B., Vitols, E., et al. (2003) Science 302, 1727-1736. [DOI] [PubMed] [Google Scholar]
- 17.Masternak, K. & Reith, W. (2002) EMBO J. 21, 1378-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnosti, D. N., Barolo, S., Levine, M. & Small, S. (1996) Development (Cambridge, U.K.) 122, 205-214. [DOI] [PubMed] [Google Scholar]
- 19.Gray, S. & Levine, M. (1996) Genes Dev. 10, 700-710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.