Abstract
Double-strand break (DSB) repair and DNA replication are tightly linked in the life cycle of bacteriophage T4. Indeed, the major mode of phage DNA replication depends on recombination proteins and can be stimulated by DSBs. DSB-stimulated DNA replication is dramatically demonstrated when T4 infects cells carrying two plasmids that share homology. A DSB on one plasmid triggered extensive replication of the second plasmid, providing a useful model for T4 recombination-dependent replication (RDR). This system also provides a view of DSB repair in T4-infected cells and revealed that the DSB repair products had been replicated in their entirety by the T4 replication machinery. We analyzed the detailed structure of these products, which do not fit the simple predictions of any of three models for DSB repair. We also present evidence that the T4 RDR system functions to restart stalled or inactivated replication forks. First, we review experiments involving antitumor drug-stabilized topoisomerase cleavage complexes. The results suggest that forks blocked at cleavage complexes are resolved by recombinational repair, likely involving RDR. Second, we show here that the presence of a T4 replication origin on one plasmid substantially stimulated recombination events between it and a homologous second plasmid that did not contain a T4 origin. Furthermore, replication of the second plasmid was increased when the first plasmid contained the T4 origin. Our interpretation is that origin-initiated forks become inactivated at some frequency during replication of the first plasmid and are then restarted via RDR on the second plasmid.
T4 Recombination-Dependent Replication
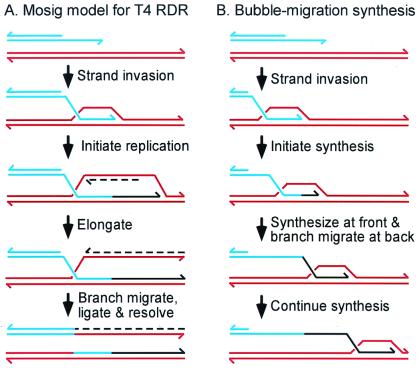
One of the earliest indications of a tight linkage between DNA replication and recombination came from studies of bacteriophage T4, when it was found that mutations in the same genes could substantially reduce both processes (1). This finding and a variety of genetic results led Mosig (2) to propose the first model for recombination-dependent DNA replication (RDR). In this model, most phage DNA replication initiates at the 3′ ends of the invading DNA within D-loops that are formed during the infection (Fig. 1A). A major route for formation of such D-loops involves the ends of the T4 chromosome. When a replication fork reaches a duplex genome end, the 3′ end of the parental strand remains single-stranded (ss) because of the polarity of DNA polymerases and the requirement for a primer. A 3′ ss end can also be generated after 5′ to 3′ exonuclease action at a duplex end. In either case, the 3′ ss end invades homologous duplex DNA either at the other end of the same DNA molecule or in another molecule (T4 DNA is terminally redundant and circularly permuted). A replication fork is then assembled on the D-loop, leading to normal semiconservative replication. The simplest possibility is that replication is unidirectional from the D-loop, leaving behind a Holliday junction (also see ref. 3 for discussion of bidirectional and tridirectional replication).
Figure 1.
Models for T4 RDR. The Mosig (2) model for T4 RDR in A invokes semiconservative replication initiated at a D-loop. Newly replicated DNA is shown in black, with lagging-strand product dashed. During the last step, branch migration and ligation create a Holliday junction (not shown), which can be cleaved in either of two orientations (only one is shown). The Formosa and Alberts (4) model for conservative bubble-migration synthesis is presented in B.
Biochemical studies have elucidated the roles of certain T4 proteins in T4 RDR and have established at least two possible mechanisms, semiconservative replication and bubble-migration synthesis. The first in vitro reaction related to RDR was reconstituted in the pioneering experiments of Formosa and Alberts (4). In this “bubble-migration synthesis,” the 3′ end of the leading strand product is extended by DNA polymerase as the product strand is extruded from the back of the D-loop bubble by a three-stranded branch-migration reaction (Fig. 1B). In these early experiments, the T4 helicase–primase complex (gp41/61) failed to assemble, and thus there was no lagging-strand synthesis. The reaction was formally conservative, because the starting template molecule was recovered unchanged.
In subsequent experiments, the addition of the gp59 protein led to successful loading of the helicase/primase complex onto the displaced strand of the D-loop. Gp59 is a branched-DNA-binding protein (5), and its role in loading gp41/61 qualifies it as a replication/recombination mediator protein (6). When all three proteins were present, semiconservative RDR was achieved (J. Barry and B. Alberts, cited in ref. 7).
In these in vitro reactions, the substrates consist of an ssDNA fragment and homologous duplex template DNA. The strand-exchange protein UvsX, together with UvsY (loads UvsX) and gp32 (ssDNA-binding protein), catalyzes D-loop formation. The inclusion of the ssDNA fragment bypasses whatever step is necessary for generating ssDNA in vivo. The gp46/47 complex probably contributes to this process in vivo by catalyzing resection and/or unwinding at DNA ends (3, 8).
The semiconservative reaction (as in Fig. 1A) is likely to be the major pathway of T4 RDR in a wild-type infection. Nonetheless, bubble-migration synthesis could function in certain mutant infections or in the presence of DNA damage, where template jumping might allow damage bypass (8). In addition, bubble-migration synthesis is central to one of the models for double-strand break (DSB) repair (see below).
One difficulty with studying T4 RDR in vivo is that the process occurs randomly throughout the genome (because T4 DNA is circularly permuted). We have tried to overcome this difficulty by establishing site-specific versions of T4 RDR. The first such version involved the replication of plasmids that contained a segment of T4 DNA but no T4 replication origin. Such plasmids replicated after T4 infection via a pathway that required UvsX, UvsY, gp46/47, and gp59, each of which is also required for phage genomic RDR (9). In a subsequent study (10), we demonstrated that the process of RDR could be stimulated by DSBs, as predicted by the Mosig (2) model. The amount of plasmid DNA replication was approximately doubled when the homologous region of the phage chromosome contained a site for DSBs (generated by an intron endonuclease; see below), and there was no stimulation when the plasmid was homologous to a region of the phage chromosome distant from the DSB.
In a study designed to analyze DSB repair, we demonstrated that a plasmid containing two inverted segments of homology replicated extensively via RDR when one of the two segments sustained a DSB (11). The products consisted of long concatamers that were replicated in their entirety by the T4 replication machinery. As described below, these results led to the extensive chromosome replication (ECR) model for DSB repair, in which repair is achieved simply by starting a new semiconservative replication fork with each of the two broken ends.
Although each of these site-specific versions of T4 RDR revealed the same protein requirements and provided strong evidence for the role of DNA ends in the reaction, they did not clearly define the immediate products of RDR. In the case of plasmid by phage studies, the initial products of RDR were subject to subsequent rounds of replication initiated within the phage chromosome. In the case of the inverted-repeat plasmid, extensive plasmid replication amplified the products. In the experiments below, we have analyzed a two-plasmid system for RDR to follow the fate of the two participating homologous DNA molecules, one with a DSB and one without.
Relationship of T4 RDR to DSB Repair
Early genetic studies on phage T4 provided the first evidence that DNA ends are recombinogenic (12, 13). However, it was difficult to explicitly study DSB repair in T4 until the discovery of mobile group-I introns in the T4 genome (14). Intron mobility depends on a site-specific endonuclease, encoded within the intron, which cleaves intron-free DNA very close to the site where the intron is located in the intron-containing genome. An efficient DSB repair reaction then ensues, with the intron-containing genome serving as the template.
The process of intron mobility itself has been studied as one version of a DSB repair reaction (15), and the intron-encoded endonuclease I-TevI has also been used to introduce site-specific DSBs at artificial sites for the study of DSB repair (11). In all cases, T4 DSB repair was found to be closely linked to RDR, with the same gene product requirements and a tight coupling between repair and DNA replication. However, studies to date have supported three different models for DSB repair within a T4 infection (see below). It is possible that two or even all three models operate, depending on the nature of the substrates and the genotype of the infecting phage. However, we do not believe that conclusive evidence has been obtained for or against any of the three models. Two major problems have impeded this goal. First, the genome of T4 is difficult to manipulate, because it is large (≈170 kb) and because every dC residue is modified with hydroxymethyl and glucosyl groups (making the DNA refractory to most restriction enzymes). Second, most studies have used plasmid substrates to overcome the first problem, but the products of the DSB repair reaction appear to amplify extensively. Because the mechanism of amplification is not understood, and because only a subset of products contain T4 replication origins in some of the studies, it seems likely that amplification has distorted our view of the repair reaction (also see ref. 3).
As mentioned above, we have used a two-plasmid system in this paper to analyze the mechanism of DSB-directed RDR. This system is also quite relevant to understanding the mechanism of DSB repair. Throughout this paper, results from the two-plasmid system will be discussed with respect to both RDR and DSB repair. Indeed, RDR and DSB repair may well be just two different manifestations of the same underlying reaction.
Three Models of DSB Repair
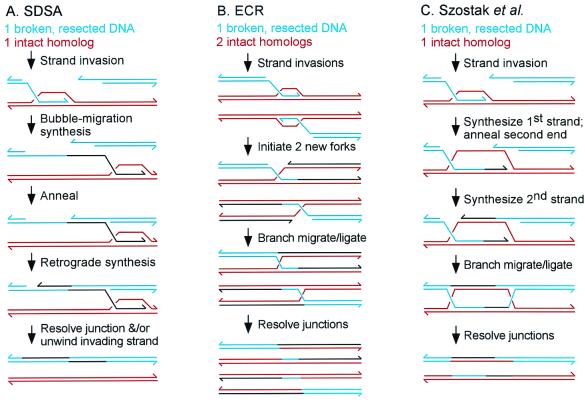
Three models have been proposed for DSB repair in T4-infected cells: synthesis-dependent strand annealing (SDSA), ECR, and the original Szostak et al. (16) model (Fig. 2). All three models begin with the resected 3′ ssDNA from one end of the break invading the intact homologous repair template.
Figure 2.
Three models for DSB repair during phage T4 infections: (A) SDSA (synthesis-dependent strand annealing) (15); (B) ECR (11); and (C) the Szostak et al. (16) model (14, 15). Broken DNA is blue, homologous template red, and newly synthesized DNA black. In each model, only one of several resolution modes is depicted.
In the SDSA model (15), bubble-migration synthesis occurs across the region homologous to the break. After synthesis has extended past the broken region, the newly synthesized strand is complementary to the ss 3′ end from the other broken end, and these anneal. The second strand of newly synthesized DNA is then generated by retrograde synthesis by using the second 3′ end as primer.
The ECR model, an adaptation of the Mosig (2) model for RDR, predicts that each broken end initiates a new semiconservative replication fork after invading homologous DNA (11). Unlike the other two models, the ECR model proposes that the two broken ends are not coordinated with each other, generally invading different homologs, and never actually reconnect to each other. As mentioned above, a Holliday junction might be left behind at each original point of strand invasion, and these would presumably be resolved by T4 endonuclease VII and DNA ligase. When applied to linear DNA, the ECR model predicts that one broken molecule would initiate two new replication forks, generally on two different homologs, leading to four products (assuming unidirectional replication).
In the Szostak et al. (16) model, leading-strand synthesis first crosses the region homologous to the break. The single strand of intact parental DNA exposed during this synthesis is complementary to the 3′ end of the other broken end, and the two anneal. Synthesis of the second strand uses the second 3′ end as primer. With the appropriate ligations and branch migration, the classic “double Holliday” structure is created. Depending on the directions of resolution of the two junctions, the products could be either crossover or noncrossover [an alternative mode of resolution invokes topoisomerase-mediated strand passage (17)].
A Two-Plasmid System for RDR and DSB Repair
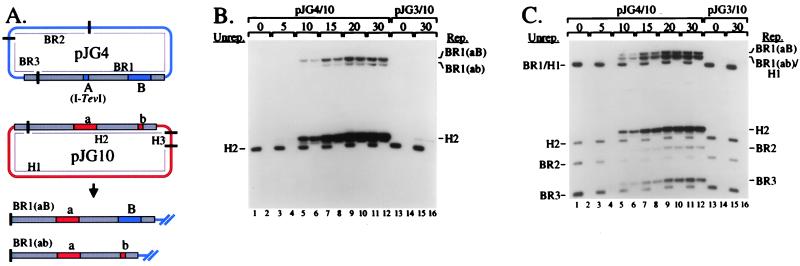
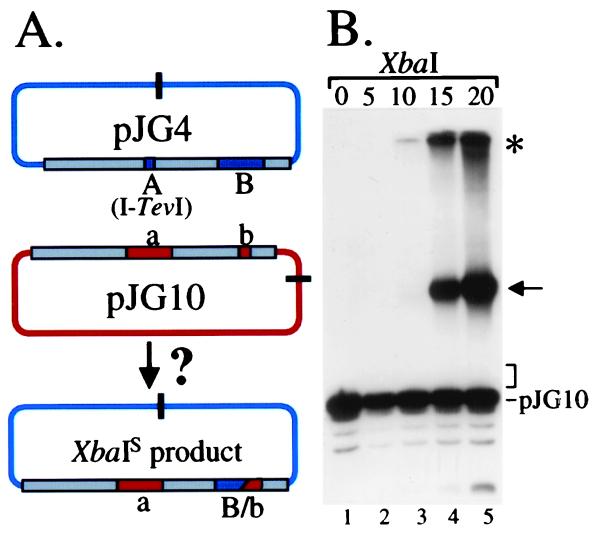
We began by analyzing RDR and DSB repair with two compatible plasmids that are homologous in a region of about 2,000 bp (Fig. 3). Plasmid pJG4 is a pBR322 derivative with a cloned 56-bp I-TevI recognition site (“A”) near the middle of the homologous segment. We refer to pJG4 as the “breakable” (BR) plasmid substrate, and the plasmid restriction fragments are designated BR number (in order of decreasing size; e.g., BR1, BR2, etc.). The second plasmid, pJG10, is a pACYC184 derivative with a 283-bp NaeI fragment (“a”) within the 2,000-bp homologous segment (in place of the I-TevI site). We refer to pJG10 as the “homologous” (H) plasmid substrate and the plasmid restriction fragments as H number.
Figure 3.
DSB repair and DSB-induced replication in a two-plasmid system. Breakable plasmid pJG4 and homologous plasmid pJG10 are depicted in A. The gray boxes represent the homologous regions of the two plasmids, with heterologous insertions (including the cloned I-TevI site) indicated. AseI restriction sites are indicated by bars, and AseI restriction fragments by the BR number and H number designations. Two possible DSB repair products are depicted at the Bottom. Plasmid pJG3 (not shown) is identical to pJG4 except that it lacks an inserted I-TevI recognition site. In B and C, total nucleic acids were prepared as described (11, 27) from a time-course of infection of plasmid-bearing E. coli JG99S (recA; see ref. 11) by T4 strain K10 (wild-type for replication and recombination genes) (11). All digests contained AseI (cleaves T4-modified DNA), with HaeIII also added for even-numbered lanes. The time of infection (min) is indicated above the lanes, with “0” indicating uninfected cells. Positions of unreplicated (Unrep.) bands are indicated to the Left of each gel panel and replicated (Rep.) bands to the Right. All band assignments in this and subsequent gels were verified against molecular size markers (not shown). The probes were radioactive DNA from the “a” segment of pJG10 (B) and from a mix of both plasmids (C).
In the experiments below and in Figs. 8–10 (which are published on the PNAS web site, www.pnas.org), we will introduce several closely related plasmids. In each case, we will refer to the pJG4-related plasmids (pBR322-based) as breakable plasmids, even if the plasmid does not have an inserted I-TevI site. Likewise, the pJG10-related plasmids (pACYC184-based) will be referred to as homologous plasmids.
DSB repair events using the homologous plasmid pJG10 as template would replace the “A” segment originally in pJG4 with “a,” and so the “a” segment provides a convenient molecular probe for DSB repair. A subset of the DSB repair products can be easily visualized after cleavage with restriction enzyme AseI and Southern blotting with the “a” probe, namely those with a crossover between the “a” marker and the blue vector DNA on the right flank. These products generate much larger AseI fragments than the parental pJG10 DNA, which also hybridizes to the “a” probe (see Fig. 3A; AseI sites indicated by solid bars).
The 2,000-bp homologous segment between the breakable and homologous plasmids also has a second heterology, a 248-bp NarI fragment (“B”) that is present in the breakable plasmid but absent (“b”) in the homologous plasmid. The “b” marker undergoes coconversion during some of the selected DSB repair events, with the extent of coconversion presumably reflecting in part the ratio of the “a”–“b” and “b”–vector distances (because we are demanding a crossover between the “a” marker and the blue vector on the right flank).
We have access to a powerful tool to determine whether particular DNA products have been replicated by the T4 machinery during the infection. T4 uses hydroxymethyl-dCTP instead of dCTP as precursor for DNA replication. Newly replicated DNA is therefore marked precisely at the time of replication with this cytosine modification, making it resistant to most restriction enzymes (e.g., HaeIII, which otherwise cleaves these plasmid DNAs frequently). We can therefore ask whether products are T4-replicated by testing their resistance to HaeIII. A few restriction enzymes, such as AseI and PacI, do cleave T4-replicated DNA. Restriction fragments of T4-replicated DNA migrate a bit slower on agarose gels because T4 glucosylates the hydroxymethyl cytosine residues immediately after replication.
In the first experiment, we compared infections of two cell lines, one containing pJG4 (breakable with DSB site) and pJG10 (homologous) and another containing pJG3 (control with no DSB site) and pJG10. Beginning with the uninfected samples (0 time), the “a” probe hybridized to the predicted AseI restriction fragment (H2) from homologous plasmid pJG10, and this fragment was completely destroyed by HaeIII (Fig. 3B; even-numbered lanes contain HaeIII). As the infection progressed, two novel AseI fragments were evident at the size expected for DSB repair events in which the “a” marker is copied into the BR1 restriction fragment [BR1(aB) and BR1(ab)]. The smaller of the repair products is formed when the “b” marker is coconverted during the DSB repair event.
The amounts of these two repair products were not affected by the presence of HaeIII in the digest (Fig. 3B), indicating that most or all of the products had been replicated throughout the length of the AseI restriction fragment (which contains 15 or 16 HaeIII sites). The production of these recombinant products depends strictly on the presence of the I-TevI site in the breakable substrate, because the control infection produced none (lanes 15 and 16). We conclude that DSBs are actively repaired in this two-plasmid system by an intermolecular reaction, and that most or all of the products have been replicated in their entirety by the T4 machinery.
In addition to the predicted repair products, an even larger amount of replicated H2 restriction fragment was observed after the infection in which DSBs occurred within the breakable plasmid (Fig. 3B). An intense band migrated just slower than the starting H2 fragment of pJG10 (because of glucosylation), and this band was completely resistant to HaeIII digestion (i.e., T4-replicated). The generation of this replicated pJG10 DNA depended almost completely on the DSB site in pJG4 (compare lanes 12 and 16). This replicated H2 fragment consists mostly of simple replicated pJG10 DNA, with a small amount of DSB repair products that do not have a crossover between the “a” marker and the blue vector DNA on the right flank (see below and Figs. 8 and 9, which are published as supplemental data on the PNAS web site, www.pnas.org). The major conclusion is that extensive replication of the homologous plasmid is induced by cleavage of the breakable plasmid, dramatically demonstrating DSB-directed DNA replication in phage T4.
We rehybridized the gel with a probe that detects all restriction fragments of both plasmids (Fig. 3C). The AseI fragments from uninfected cells (0 time) were the predicted sizes and, as expected, were completely sensitive to HaeIII (Fig. 3C). As above, large amounts of replicated plasmid DNA were evident when the I-TevI cleavage site was present in pJG4 (Fig. 3C, compare lanes 12 and 16). We also analyzed control infections with each plasmid alone, confirming the restriction fragment assignments and verifying that the plasmids do not replicate when present alone in cells (data not shown).
The relative amounts of replicated plasmids can be judged from the smaller restriction fragments [note that replicated H1 comigrates with BR1(ab)]. The H2 fragment clearly replicated more extensively than the BR2 and BR3 fragments during the T4 infection, demonstrating that the DSB in the breakable plasmid stimulated more replication in the homologous plasmid than in itself.
In summary, DSB repair products are generated at relatively high levels, and these products have been replicated in their entirety by the T4 machinery. In addition, a DSB in one plasmid induces high levels of replication in an unbroken homologous plasmid. We will discuss the nature of the products and how they relate to DSB repair models below.
Requirement for Recombination and Replication Gene Products
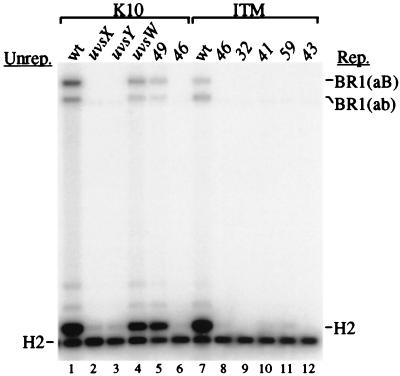
We next asked how mutations in T4 recombination and replication genes affect the DSB repair/replication reaction. One problem with this analysis is that the I-TevI endonuclease is translated from a late phage mRNA (14), but late gene expression depends on DNA replication by the phage (18). Thus, replication mutants are deficient for I-TevI production, and a negative result in the recombination assay may be just the trivial consequence of failing to generate DSBs. We circumvented this problem by constructing a mutant phage (T4-ITM) in which the I-TevI gene is transcribed from a middle-mode promoter, which is independent of DNA replication (19). The Escherichia coli RecBCD enzyme apparently degrades I-TevI-cleaved DNA when it is produced at middle times by the ITM phage (19), and we therefore used a recD− host for this experiment. The host cells contained breakable plasmid pJG4 (with DSB site) and homologous plasmid pJG13 (identical to pJG10 except for an inserted PacI site; see Fig. 8, www.pnas.org), and were infected for 30 min with wild-type or mutant phage. The DNA was analyzed exactly as above, with AseI restriction digests and the “a” probe (as in Fig. 3B).
Consider first the recombination gene mutants, where I-TevI production was from the normal late promoter. Mutations in uvsX (encodes strand-exchange protein), uvsY (UvsX loading protein), and 46 (putative nuclease/helicase) abolished production of the DSB repair products and the replicated H2 band (Fig. 4, lanes 2, 3, and 6). The same three proteins are required for DSB repair and DSB-stimulated replication in the one-plasmid (inverted-repeat) system (11) and also for the major RDR pathway of phage chromosomal DNA replication. In both the one- and two-plasmid systems, DSB repair and replication displayed a partial requirement for the products of genes uvsW (branch-specific helicase) and 49 (branch-specific nuclease) (11, 20) (Fig. 4, lanes 4 and 5).
Figure 4.
Replication and recombination mutations affect DSB repair in the two-plasmid system. The analysis was as in Fig. 3B except: (i) the homologous plasmid was pJG13, which is identical to pJG10 except for an inserted PacI site; (ii) the E. coli host was recD strain DJT1 (also recA and rpsL; derived from Hfr cross of strains CAG12135 and KL16–99S); (iii) all digests contained only AseI; and (iv) all samples were collected at the 30-min time point.
Turning to the replication mutants and the ITM background, we repeated the control with wild-type phage and also the 46-mutant infection (to make sure that the defect was not an indirect consequence of reduced 46-mutant phage DNA replication). The ITM wild-type phage produced somewhat less of the DSB repair products and the replicated H2 fragment than the non-ITM wild type (Fig. 4, compare lanes 1 and 7). We do not understand why earlier I-TevI production leads to less product, but a similar result was seen in our analysis of single-strand annealing (19). The 46 mutation completely abolished the signals, strengthening the conclusion that gp46/47 is required for these reactions (Fig. 4, lane 8).
The DSB repair products and DSB-stimulated replication of homologous plasmid were both essentially abolished by mutations in each of four replication genes (32, encoding the T4 ssDNA-binding protein; 41, replicative helicase; 59, helicase loading protein; and 43, DNA polymerase; Fig. 4, lanes 9–12). We failed to detect unreplicated recombination products in these infections even with an overexposure (data not shown). One possible explanation is that DSB-promoted recombination requires phage replication proteins, perhaps to stabilize DSB-promoted D-loops (i.e., by replication). An alternative interpretation is that the visible DSB repair products are the result of extensive amplification of the original repair products, with the replication gene mutations simply eliminating the amplification (but not the original repair reaction). Further experiments are necessary to distinguish between these two interpretations.
Do Any of the DSB Repair Models Explain the DSB Repair Products?
Both the Szostak and SDSA models predict only localized DNA replication in the region of the repaired DSB, but we found extensive replication throughout the DSB repair products. Furthermore, neither model predicts that the DSB would lead to preferential replication of the homologous plasmid. By tinkering with Holliday junction resolution, one could modify the Szostak model to predict rolling circle replication of a plasmid heterodimer, but this should equally amplify the breakable and homologous plasmids, which was not observed (Fig. 3C; also see Figs. 8 and 9, www.pnas.org).
We looked directly for any repaired pJG4 products that had not been replicated through the vector portion of the plasmid during a time course of an infection. The DNA was cleaved with XbaI, which is blocked by the T4 modifications, and then probed with the “a” segment. If the DSB was repaired with only local DNA synthesis near the break, the products would be linearized by XbaI (Fig. 5A). As expected, the “a” probe lit up the 5,747-bp XbaI-linearized pJG10 from the uninfected cells (Fig. 5B, lane 1). As the infection progressed, no novel bands were detected in the region where the predicted repair products should migrate (Fig. 5B, lanes 2–5, expected products 6,456 and 6,704 bp; position indicated by bracket). Instead, a large amount of slower migrating DNA was detected. The two intense bands presumably contain replicated (XbaI-resistant) plasmid concatamer DNA. The band that does not enter the gel (Fig. 5B, asterisk) is likely branched concatamers, whereas the band that enters the gel (arrow) is at the position of limit migration for very long linear DNA. We conclude that repair products with only a patch of replicated DNA, as predicted by the simplest versions of the SDSA and Szostak models, are not detectable under conditions where fully replicated repair product is easily detected.
Figure 5.
DSB repair products with replicated patches are not detected. The strategy is diagrammed in A, with XbaI sites indicated by bars. DSB repair with only localized DNA replication (and no crossing over) would create the circle at the bottom, with the XbaI site sensitive to cleavage. For the samples in B, DNA was prepared from a T4 K10 infection of plasmid-bearing E. coli JG99S (11) at the indicated times (min). All digests contained XbaI, and the probe was the “a” segment of pJG10. The bracket indicates the expected position for XbaI-linearized DSB repair product.
In its simplest form, the ECR model would predict that the two broken ends of pJG4 would each invade a copy of the homologous plasmid and trigger replication of that plasmid. Thus, ECR does predict massive replication of the homologous plasmid, as observed. However, the ECR model also predicts that the two ends should behave independently, invading two different homologous plasmid circles. This in turn predicts that the pJG4 vector sequences within the DSB repair products should be unreplicated. However, most or all of the BR1 repair products detected above were replicated in their entirety (Fig. 3).
These considerations indicate that none of the three repair models in their simplest form explain the observed products. To further explore the nature of the repair products, we carried out experiments in which one or the other plasmid carried a single site for restriction enzyme PacI (cleaves T4-modified DNA). The analysis is presented in Figs. 8 and 9, www.pnas.org, and the conclusions will be discussed here.
By analyzing repair products in which only the homologous (H; red) plasmid carried the PacI site, we found that most of the repaired breakable (BR; blue) plasmid was present in a single copy surrounded by H plasmid DNA on both sides (Fig. 6A). This structure is consistent with either plasmid heterodimers (H-BR) or multimers in which single copies of breakable plasmid are surrounded by multiple copies of homologous plasmid. This H-BR-H “sandwich” was completely resistant to XbaI, which would cut the BR vector sequence if it were not T4-replicated (Fig. 6A). Again, we find that the repaired products are completely replicated by T4.
Figure 6.
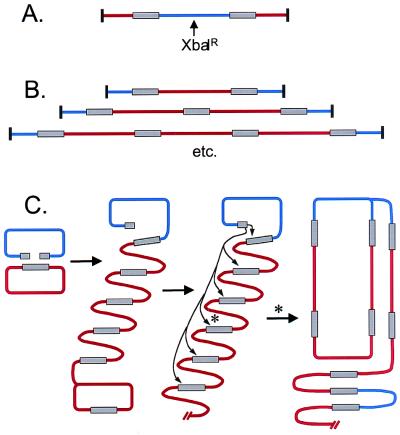
Summary of repair product structures and a revised ECR model. A and B summarize the structure of the DSB repair products, as determined in Figs. 8 and 9, respectively (www.pnas.org). As above, breakable plasmid is depicted in blue and homologous plasmid in red. In C, an adaptation of the ECR model is presented to explain the structure of the products. In the first step, one end of the broken plasmid invades the homologous plasmid and initiates a rolling circle. In the second step, the second broken end invades a homologous segment somewhere in the concatameric product. One particular invasion is indicated by an asterisk; the result of this invasion is depicted in the final step, where a second replication fork is established to create a larger rolling circle.
The relationship between adjacent copies of the plasmids was further investigated by placing the PacI site in only the BR plasmid. In this case, we observed a ladder of increasing multimers on a pulse-field gel, as depicted in Fig. 6B. Increasing numbers of H plasmid copies were found in between BR plasmid copies. The inverting-field gel resolved sandwiches with up to four copies of H plasmid (BR–H4–BR) as individual bands, and there was also an intense band in the region where sandwiches with five or more copies would migrate. These various multimeric products could be from linear and/or circular concatamers.
With this more complete product analysis, we can return to a discussion of DSB repair models. The major repair products, consisting of the series of … Hn-BR-Hn-BR… products, cannot easily be explained by either the Szostak or SDSA model, even assuming some kind of plasmid amplification by rolling circle replication. They can be explained by a derivative of the ECR model (Fig. 6C). In this model, one broken end initiates a replication fork on the H plasmid, triggering rolling circle replication. The second broken end then preferentially invades the product of the first replication event somewhere along the length of the multimeric product. We can rationalize this preference as being because: (i) the product of the first round of ECR is located within the same molecule; or (ii) the product of the first round is T4 modified, which may be preferred by the T4 recombination machinery. The second invasion would set up a retrograde fork that replicates back through the multimeric product toward the original B plasmid. One simple outcome is a “rolling multimer,” in which a multimeric circle serves as template for repeated rounds of replication. This model explains essentially all of the products that we detect, and accounts for their replicated nature.
This modified ECR model intrinsically involves rolling circle replication, which produces many copies of product from a single event. Clearly, the DSB repair products we detect may be extensively amplified, and the amount of these products may exaggerate the actual number of repair events. Another important caveat is that amplification might preferentially amplify some products over others, just as in the previous studies of DSB repair using plasmid substrates in T4. We had hoped that the two-plasmid system would allow us to identify the initial repair products without amplification, but once again the propensity of T4 to couple extensive DNA replication to DSB repair has prevented this identification. In future experiments, we hope to finally overcome this limitation by using prelabeled radioactive DNA substrates.
In summary, the observed repair products can be explained by a modified version of the ECR model, but we have been unable to come up with modifications of the Szostak or SDSA models that explain most of the products. However, we admit that repair may also occur by the Szostak and/or SDSA models, and that without amplification, these products may fall below our level of detection.
Recombination Events Stimulated by Replication Forks Blocked at Topoisomerase Cleavage Complexes
To this point, we have been considering DNA replication triggered by recombination in the process of RDR. We now turn to experiments that analyze the opposite connection, namely the stimulation of recombination by DNA replication. These opposing connections can be understood as two sides of the same coin, namely the rescue of replication forks that have stalled. When a replication fork stalls, recombination is presumably triggered as part of the rescue, and this recombination in turn triggers renewed DNA replication. Indeed, there is a growing belief that this is the major role for recombination at the cellular level (21).
In recent studies, we have directly analyzed replication fork blockage during a T4 infection. T4 encodes a type II DNA topoisomerase that is sensitive to antitumor drugs that also inhibit the mammalian enzyme (22). These drugs trap the covalent enzyme–DNA cleavage complex, which is critical for drug cytotoxicity (23). Furthermore, the process of DNA replication has been implicated in cytotoxicity, suggesting that some critical event occurs when a replication fork meets a cleavage complex.
Using two-dimensional gel electrophoresis and a plasmid that contains both a T4 replication origin and a cloned recognition site for the T4 topoisomerase, we demonstrated that replication forks are blocked in vivo when they run into a drug-stabilized cleavage complex (23). The blocked replication fork may be on the pathway to cytotoxicity, perhaps by leading to a frank DSB (e.g., after cleavage by a recombination endonuclease such as T4 endonuclease VII; see refs. 3 and 24). Whatever the exact nature of the DNA damage caused by the cleavage complex, it is important to note that recombinational repair significantly reduces the toxic effects. Mutations in T4 genes uvsX, uvsY, uvsW, 46, and 59 all increase sensitivity to the topoisomerase-targeting drugs (25, 26).
Because forks become blocked at the cleavage complex and recombinational repair reduces cytotoxicity, we asked whether recombination is stimulated by the presence of a cloned topoisomerase recognition site. Indeed, plasmid–plasmid homologous recombination was significantly increased, with the increase depending on the presence of a T4 origin (on the plasmid with the cloned topoisomerase site), the antitumor drug, the topoisomerase, and the phage-encoded UvsX, UvsY, 32, and 46 proteins (27). The simplest model is that the blocked forks feed into the T4 recombination pathway, perhaps after generation of a DSB. Given the tight linkage between recombination and replication demonstrated above, it seems very likely that the recombination events lead to new replication forks. A cautious note is also in order—we have not directly linked the blocked fork and the stimulated recombination events but have simply shown that both are consequences of the drug-stabilized cleavage complex.
Interplasmid Recombination Events Simulated by a T4 Origin of Replication on One of the Plasmids
Does DNA replication in the absence of any overt DNA damage also lead to stalled replication forks, followed by fork restart via recombination? We came across a strong hint during the above-mentioned studies of recombination induced by drug-stabilized cleavage complexes (27). We observed interplasmid homologous recombination in control infections lacking the drug when one of the two plasmids contained a T4 replication origin.
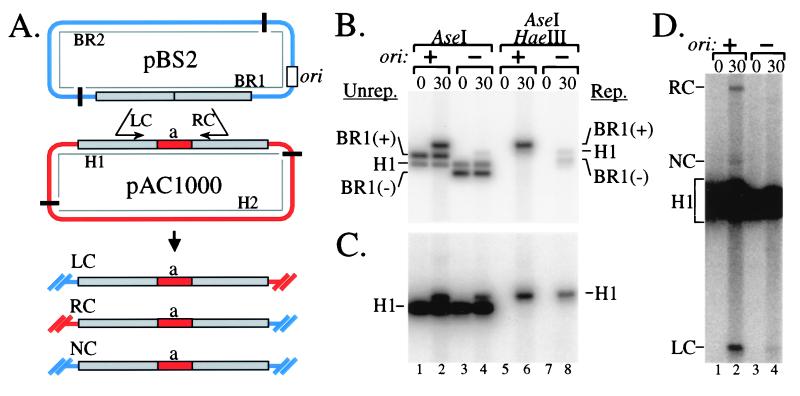
We addressed this issue with the two-plasmid assay outlined in Fig. 7A. The breakable plasmid pBS2 and the homologous plasmid pAC1000 share an ≈1,000-bp region of homology, which is interrupted centrally by the 283-bp “a” fragment in pAC1000. The pBS2 plasmid has a T4 origin of replication (and no I-TevI site; we refer to it as “breakable” for consistency with the above studies, and because origin-directed replication of this plasmid may lead to DSBs). A control plasmid pBS6 is identical to pBS2 except that it lacks the T4 origin of replication.
Figure 7.
T4 replication origin induces interplasmid recombination and RDR. Breakable plasmid pBS2 and homologous plasmid pAC1000 are depicted in A (see ref. 27 for plasmid constructions). The gray boxes represent regions of homology, and ori indicates the cloned T4 replication origin ori(34). The infections, DNA purification, and analysis were as described for Fig. 3B, with all DNA samples isolated at either 0 min (uninfected) or 30 min after infection of plasmid-bearing E. coli JGD1 (27) with T4 strain K10. Control infections (−ori) contained pBS6 (no T4 replication origin) in place of pBS2. In B and C, DNA samples were digested with AseI or AseI/HaeIII, as indicated and the probe was from the region of homology shared by both plasmids (B) or the “a” fragment of pAC1000 (C). The (+) and (−) signs after BR1 indicates the presence or absence of the T4 replication origin. D shows a dark exposure of AseI-digested DNA hybridized with the “a” probe to allow visualization of the three recombination products. Labeling of bands in D is not split on the basis of replication status. The noncrossover (NC) and right crossover (RC) products in the pBS6 lanes, if present, would run slightly faster than the corresponding bands in the pBS2 lanes. LC, left crossover.
We first compared infections of cell lines containing pAC1000 and either pBS2 or pBS6. DNA samples were digested with AseI alone (Fig. 7B, lanes 1–4) or AseI/HaeIII (Fig. 7B, lanes 5–8) and hybridized to a probe for the region of homology shared by the two plasmids. After phage infection, the breakable plasmid replicated extensively because of the cloned T4 origin of replication (Fig. 7B, compare BR1(+) in lane 6 to BR1(−) in lane 8).
To analyze replication of the pAC1000 plasmid, the same blot was hybridized with a probe for the “a” fragment. Replication of pAC1000 was approximately doubled when the breakable plasmid contained the T4 replication origin (Fig. 7C, compare lanes 6 and 8). The significance of this difference was confirmed through four independent repetitions, along with control infections in which E. coli contained pAC1000 alone. The amount of replicated pAC1000 (relative to unreplicated pAC1000 DNA) in the latter control infection was arbitrarily set to 1.0 (measured standard deviation ± 0.1). The relative amount of replicated pAC1000 in the pBS2/pAC1000 samples was measured at 2.0 ± 0.3, compared with 0.9 ± 0.1 for the pBS6/pAC1000 control. We conclude that origin activity on one plasmid causes a significant increase in replication of a second homologous plasmid (which does not itself contain an origin).
To determine whether this increase in pAC1000 replication might result from RDR, we compared the amount of interplasmid recombination in infections with pBS2/pAC1000 vs. the pBS6/pAC1000 control. DNA samples were digested with AseI and hybridized with the “a” probe, which will detect the nonrecombinant H1 fragment and three expected recombination products (see Fig. 7A). The right crossover (RC) and left crossover (LC) products were clearly visible in the pBS2/pAC1000 sample but nearly undetectable in the pBS6/pAC1000 control (Fig. 7D, compare lanes 2 and 4; quantitation revealed approximately 10-fold more LC and RC products in lane 2 than in lane 4). The noncrossover (NC) product was also increased in the pBS2/pAC1000 sample, although accurate quantitation of this difference was not possible. Thus, the presence of an origin on the pBS2 plasmid leads to substantially increased interplasmid recombination, which is likely responsible for the increased replication of the pAC1000 plasmid discussed above. Although the levels of stimulated recombination and RDR are much lower than with I-TevI-generated DSBs, the parallels between the two systems suggest that replication of the breakable plasmid does indeed lead to DSBs that stimulate recombination and RDR.
We also explored the genetic requirements for this interplasmid recombination by using phage with mutations in genes that encode the recombination/repair proteins UvsX, UvsY, gp46, gp32, and gp49. No recombination was detectable with the 46 and 32 mutants, whereas greatly reduced recombination was evident with the uvsX, uvsY, and 49 mutants (Fig. 10, www.pnas.org). Origin-induced interplasmid recombination therefore requires the same proteins as DSB repair, arguing for similar pathways.
The Tight Connections of T4 Replication, Recombination, and DSB Repair
The plasmid model systems described here provide windows on interconnections between DNA replication, recombination, and repair during T4 infections. All of the results can be understood in relation to the general problem of replication fork stalling and the recombinational restart of stalled forks. We infer that replication forks initiated from an origin on one plasmid sometimes stall, leading to an increase in interplasmid recombination and RDR on the homolog. Topoisomerase cleavage complexes further increase replication fork blockage (23) and correspondingly increase interplasmid recombination (27). A recent study of UV-stimulated recombination in T4 similarly argues that recombinational restart of replication forks is important for survival after other forms of DNA damage (28).
Most phage DNA replication initiates by RDR, and even this can be viewed as a special case of replication fork restart. The infecting T4 chromosome is linear, and thus every replication fork initiated at an origin eventually reaches a genome end. RDR initiated from this genome end restarts the replication fork that was lost when the fork reached the end. In this context, the massive stimulation of homologous plasmid replication by a DSB in the breakable plasmid provides a unique model system to study the detailed mechanism of T4 RDR and DSB repair. The results argue that RDR and DSB repair are simply two different ways of viewing the same reaction: the assembly of semiconservative replication forks at D-loops created from double-strand ends.
Supplementary Material
Acknowledgments
This work was supported by Grant GM34622 from the National Institutes of Health, along with the following training grants/fellowships: F32 GM15985 (to J.W.G.), T32 GM07171 (to B.A.S.), and T32 GM07184 (to D.J.T.).
Abbreviations
- DSB
double-strand break
- ECR
extensive chromosome replication
- RDR
recombination-dependent DNA replication
- SDSA
synthesis-dependent strand annealing
- ss
single-stranded
- BR
breakable
- H
homologous
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Epstein R H, Bolle A, Steinberg C M, Kellenberger E, Boy de la Tour E, Chevalley R, Edgar R S, Susman M, Denhardt G H, Lielausis A. Cold Spring Harbor Symp Quant Biol. 1963;28:375–392. [Google Scholar]
- 2.Mosig G. In: Bacteriophage T4. Mathews C K, Kutter E M, Mosig G, Berget P B, editors. Washington, DC: Am. Soc. Microbiol.; 1983. pp. 120–130. [Google Scholar]
- 3.Kreuzer K N. Trends Biochem Sci. 2000;25:165–173. doi: 10.1016/s0968-0004(00)01559-0. [DOI] [PubMed] [Google Scholar]
- 4.Formosa T, Alberts B M. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- 5.Jones C F, Mueser T C, Nossal N G. J Biol Chem. 2000;275:27145–27154. doi: 10.1074/jbc.M003808200. [DOI] [PubMed] [Google Scholar]
- 6.Beernink H T, Morrical S W. Trends Biochem Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- 7.Kreuzer K N, Morrical S W. In: Molecular Biology of Bacteriophage T4. Karam J D, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 28–42. [Google Scholar]
- 8.Bleuit J, Xu H, Ma Y, Wang T, Liu J, Morrical S. Proc Natl Acad Sci USA. 2001;98:8298–8305. doi: 10.1073/pnas.131007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kreuzer K N, Yap W Y, Menkens A E, Engman H W. J Biol Chem. 1988;263:11366–11373. [PubMed] [Google Scholar]
- 10.Kreuzer K N, Saunders M, Weislo L J, Kreuzer H W E. J Bacteriol. 1995;177:6844–6853. doi: 10.1128/jb.177.23.6844-6853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George J W, Kreuzer K N. Genetics. 1996;143:1507–1520. doi: 10.1093/genetics/143.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mosig G. Cold Spring Harbor Symp Quant Biol. 1963;28:35–41. [Google Scholar]
- 13.Womack F C. Virology. 1963;21:232–241. doi: 10.1016/0042-6822(63)90262-9. [DOI] [PubMed] [Google Scholar]
- 14.Belfort M. Annu Rev Genet. 1990;24:363–385. doi: 10.1146/annurev.ge.24.120190.002051. [DOI] [PubMed] [Google Scholar]
- 15.Mueller J E, Clyman T, Huang Y J, Parker M M, Belfort M. Genes Dev. 1996;10:351–364. doi: 10.1101/gad.10.3.351. [DOI] [PubMed] [Google Scholar]
- 16.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 17.Thaler D S, Stahl M M, Stahl F W. Genetics. 1987;116:501–511. doi: 10.1093/genetics/116.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams K P, Kassavetis G A, Herendeen D R, Geiduschek E P. In: Molecular Biology of Bacteriophage T4. Karam J D, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 161–175. [Google Scholar]
- 19.Tomso D J, Kreuzer K N. Genetics. 2000;155:1493–1504. doi: 10.1093/genetics/155.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carles-Kinch K, George J W, Kreuzer K N. EMBO J. 1997;16:4142–4151. doi: 10.1093/emboj/16.13.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Hippel P H. Trends Biochem Sci. 2000;25:155. doi: 10.1016/s0968-0004(00)01571-1. [DOI] [PubMed] [Google Scholar]
- 22.Kreuzer K N. Biochim Biophys Acta Gene Struct Expression. 1998;1400:339–347. doi: 10.1016/s0167-4781(98)00145-6. [DOI] [PubMed] [Google Scholar]
- 23.Hong G, Kreuzer K N. Mol Cell Biol. 2000;20:594–603. doi: 10.1128/mcb.20.2.594-603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michel B. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- 25.Neece S H, Carles-Kinch K, Tomso D J, Kreuzer K N. Mol Microbiol. 1996;20:1145–1154. doi: 10.1111/j.1365-2958.1996.tb02635.x. [DOI] [PubMed] [Google Scholar]
- 26.Woodworth D L, Kreuzer K N. Genetics. 1996;143:1081–1090. doi: 10.1093/genetics/143.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stohr B A, Kreuzer K N. Genetics. 2001;158:19–28. doi: 10.1093/genetics/158.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doan P L, Belanger K G, Kreuzer K N. Genetics. 2001;157:1077–1087. doi: 10.1093/genetics/157.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.