Abstract
Adaptive immunity exists in all vertebrates and plays a defense role against microbial pathogens and tumors. T cell responses begin when precursor T cells recognize antigen on specialized antigen-presenting cells and differentiate into effector cells. Currently, dendritic cells are considered the only cells capable of stimulating T lymphocytes. Here, we show that mature naïve B lymphocytes can be genetically programmed by using nonviral DNA and turned into powerful antigen-presenting cells with a dual capacity of synthesis and presentation of antigen to T cells in vivo. A single i.v. injection of transgenic lymphocytes activates T cell responses reproducibly and specifically even at very low cell doses (≈102). We also demonstrate that T cell priming can occur in the absence of dendritic cells and results in immunological memory with protective effector functions. These findings disclose aspects in the regulation of adaptive immunity and indicate possibilities for vaccination against viruses and cancer in humans.
Adaptive immunity is based on the selection and expansion of lymphocytes bearing clonally distributed antigen receptors (1). It is based on genes having common structural features, the Ig and T cell receptor genes, that code for conformationally stable molecules at the surface of B and T lymphocytes where they serve as receptors for antigen. Clonal selection and expansion follow recognition of antigen by T cells, an event that requires the positive effect of coreceptor molecules that stabilize the interaction with the antigen-presenting cell (APC) and costimulatory molecules that provide a jump start to T cell activation (2, 3). The immune system also stores the ability to respond with greater intensity and faster kinetics upon re-encounter of the same antigen, immunological memory (4).
T cells recognize antigen in the form of small peptides in the groove of the MHC molecule at the surface of the APC, dendritic cells (DCs), B lymphocytes, and monocytes/macrophages. Peptides that bind MHC class I molecules consist of 8-9 residues, whereas those that bind class II MHC molecules consist of 13-17 residues (5, 6). Antigenic peptides presented in the context of MHC class I molecules induce cytotoxic T lymphocyte (CTL) responses and are generally the product of degradation of cytosolic proteins via an ATP-dependent, ubiquitin-driven proteolytic system (7). Antigenic peptides presented in the context of MHC class II molecules induce CD4 T cell responses and result typically from the degradation of exogenous antigens internalized by receptor-mediated endocytosis (8). Currently, DCs are viewed as possibly the only APCs able to initiate a T cell adaptive response (9). DCs are endowed with an extraordinary capacity to take up antigen from the extracellular compartment, process it, and display its peptides at the cell surface in conjunction with costimulatory molecules. Owing to these properties, many new approaches to vaccination are based on DCs, notwithstanding the fact that while DC cluster with T cells in lymph nodes rapidly (≈24 h) they also leave lymph nodes rapidly (10). Thus, spatio-temporal factors and considerations on supply of peptides to MHC molecules, which depend on the half-life of the peptide/MHC on the APC (≈4 h for class I and ≈8-22 h class II) (11-13), are relevant to optimize T cell activation in vivo. From the foregoing, the ideal approach is that which uses an APC that is able to actively synthesize and process/present antigen at the same time.
B lymphocytes possess distinct APC function. On a per-cell basis, activated B cells are as effective as DCs in activating naïve T cells (14, 15). In vivo expansion of T cells can be driven by activated B cells (16-18) and resting B lymphocytes depending on the availability of adequate costimulation via B7 and CD40 (19, 20). Resting B lymphocytes also activate naïve T lymphocytes specific for their endogenous light chain variable region peptide in vivo (21). This finding is consistent with the fact that lymphoma cell transfectants process and present to T cells peptides of endogenously synthesized Ig variable region via both MHC class II or class I pathways (22, 23). When plasmid DNA coding for an Ig heavy chain gene controlled by the Ig promoter is injected into the spleen of adult mice, production of antibodies and activation of T cell responses against heterologous epitopes expressed in the variable region are readily induced (24, 25).
The present study sought to determine whether primary B lymphocytes rendered transgenic ex vivo with nonviral DNA can induce antigen-specific responses in vivo. Typically, murine spleen lymphocytes were rendered transgenic for plasmid DNA coding for transgenes under the control of an Ig-specific promoter by incubation in PBS without Ca2+/Mg2+ for 60 min (26). To induce antigen-specific CD4 and CD8 T cell responses, we used heavy chain genes engineered to express heterologous T cell determinants in the complementarity-determining regions (CDRs) (27). Transgenic cells were injected into adult histocompatible recipients i.v. within 24 h from in vitro transgenesis (Fig. 1A).
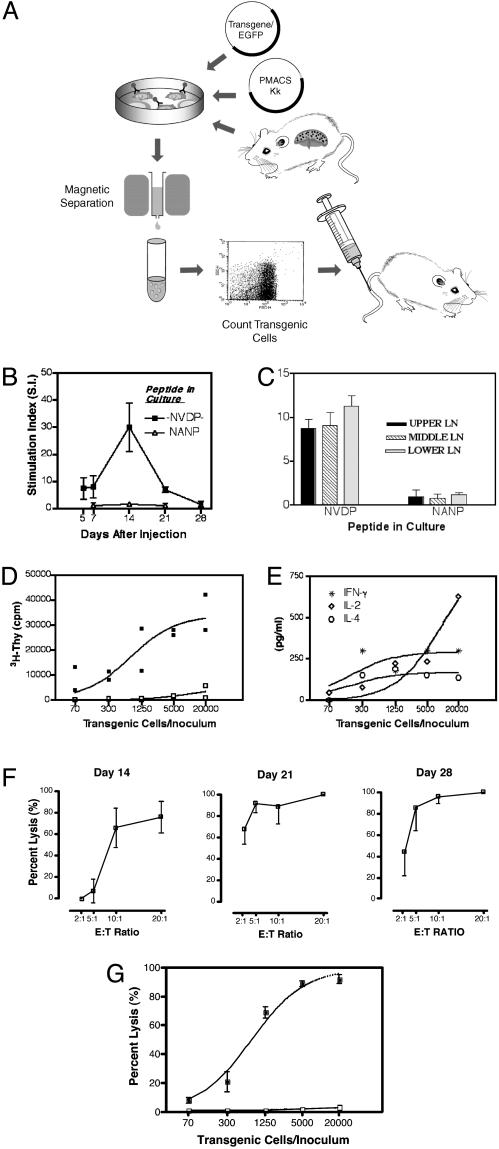
Fig. 1.
(A) The process of spontaneous transgenesis and immunization with transgenic lymphocytes. Details of the two procedures are described in Materials and Methods. (B) The characteristics of the primary CD4 T cell response: kinetics and specificity. C57BL/6 mice were injected with 5 × 103 γ1NV2NA3 transgenic cells. Spleen cells were harvested at different time points and tested in a T cell proliferation assay. Tests were run in triplicate. Values represent the mean stimulation index ± SD of four mice per group (24). The cpm at each time point are as follows: day 5, NVDP peptide (11,581)/medium alone (1,988); day 7, NVDP peptide (40,016)/medium alone (4,932); day 14, NVDP peptide (29,856)/medium alone (1,165); day 21, NVDP peptide (15,788)/medium alone (2,322); and day 28, NVDP peptide (1,014)/medium alone (732). (C) T cell proliferation in lymph nodes (LNs). Fourteen days after injection LNs were collected from the upper, middle, and lower body anatomical regions. The cpm per each group are as follows: upper LN, NVDP peptide (10,576)/NANP peptide (1,201); middle LN, NVDP peptide (10,236)/NANP peptide (1,130); and lower LN, NVDP peptide (9,786)/NANP peptide (887). Spleen cells in the same experiment (data not shown) were NVDP peptide (14,505)/NANP peptide (1,022). (D) Dose-response of CD4 T cell induction. Mice (two per group) were injected with lymphocytes transgenic for γ1NV2NA3 (▪) or γ1NANP (□) as a control, with varying numbers (20,000-70) of transgenic cells. Mice were killed on day 14, and spleen CD4 T cells were restimulated in vitro with the -NVDP- peptide. Proliferation is expressed as cpm. Two independent experiments yielded a similar result. (E) Cytokine production vs. immunizing dose. Culture supernatants from the experiment depicted in D were analyzed for cytokine content (pg/ml). (F) The CD8 T cell response: phase of induction and kinetics after injection of 5 × 103 γ1NV2NP3 transgenic cells. Spleen cells were harvested at the time points indicated, restimulated in vitro with peptide NP366-374 (5 μg/ml), and tested in a standard 51Cr-release assay using EL-4 cells pulsed with NP366-374 as target at various effector-to-target cell ratios. Results are expressed as the percentage of specific lysis. (G) Dose-response of CTL induction. Groups of three mice each received a single injection of varying numbers (20,000-70) of transgenic cells. Mice were killed on day 14, and the CTL response was measured by using EL-4 cells pulsed with NP366-374 (filled symbols) or EL-4 alone (open symbols) as targets.
Materials and Methods
Mice and Generation of Bone Marrow (BM) Chimeras. Eight- to 10-week-old C57BL/6 mice were purchased from The Jackson Laboratory. Homozygous relB (-/-) mice were bred in the animal facility of the University of California, San Diego. BM chimeras were generated as described (28). MHC class I and II (-/-) mice were the kind gift of S. Schoenberger (La Jolla Allergy and Immunology Institute, La Jolla, CA).
Plasmid DNA, Proteins, and Synthetic Peptides. Plasmid γ1NANP, γ1NV2NA3, and γ1NV2NP3 were engineered as described (25, 29). The gene for enhanced GFP (EGFP) was inserted at the C terminus of the human γ1 constant region. Plasmid pSV2Neo is the original plasmid forming the backbone of the pNeoγ1 vector without the human γ1 C region gene and served as a control. Plasmid DNAs were prepared and purified by using a Qiagen Megaprep kit (Qiagen, Chatsworth, CA) and stored at -20°C until used. Synthetic peptides NANP NVDP NANP (-NVDP-), NANP NANP NANP (NANP)3, and ASNENMETM (NP366-374), were synthesized in the Peptide Chemistry Facility of the California Institute of Technology (Pasadena).
Spontaneous Transgenesis and Enumeration of Transgenic Cells. Spleen cells (4 × 106) in 200 μl of PBS without Ca2+ and Mg2+ (CellGro, Herndon, VA) were incubated with 25 μg of plasmid DNA for 1 h at 37°C together with 5 μg of PMACS Kk plasmid (Miltenyi Biotec, Auburn, CA), which codes for a truncated mouse H-2 Kk molecule as a selectable cell surface marker. The cells were then washed and incubated in RPMI medium 1640 (Irvine Scientific) supplemented with Hepes buffer, glutamine, and 10% FCS at 37°C in 5% CO2 atmosphere overnight. Next, the cells were harvested and resuspended in 320 μl of PBS/0.5% BSA/5 mM EDTA (PBE) containing 80 μl of MACSselect Kk microbeads. The suspension was incubated for 15 min at 6-12°C. Transgenic and magnetically labeled cells were enriched by positive selection on a column MS+/RS+ mounted on the magnetic field of a MACS separator. After washing, transgenic (EGFP+) cells were flushed out with PBE by using a plunger and analyzed by flow cytometry on a FACsCalibur (Becton Dickinson). A gate in the forward vs. side scatter map was set to exclude debris and dead cells. Cells collected in the flow-through were similarly analyzed and found to be EGFP-. In dual staining experiments, magnetically sorted cells were stained with a phycoerythrin (PE)-conjugated mAb to CD19 (PharMingen). Mononuclear cells were isolated from peripheral blood lymphocytes on a Lympholyte-M gradient (Cedarlane Laboratories) and then cultured in RPMI medium 1640 and 10% FCS supplemented with 12 mg/ml lipopolysaccharide (Sigma) and 0.7 μg/ml dextran sulfate (Amersham Pharmacia). In some experiments B cells were enriched by using magnetic beads coated with a mAb against mouse CD43 followed by sorting on a MACS magnetic separation column (Miltenyi Biotech). The CD43- population was analyzed for the expression of CD19 by using a mAb anti-CD19 (clone 1D3, PharMingen). The B cell population was then transfected with plasmid DNA together with PMACS Kk plasmid and analyzed as described above. Spleen cells depleted of B cells were used as control.
In Vivo Immunization. Mice were immunized by single injection via the tail vein with 200 μl of a suspension of spleen lymphocytes containing transgenic lymphocytes in a number determined by fluorescence-activated cell sorting. The dose used in each experiment refers to the number of transgenic lymphocytes.
T Cell Assays. Proliferative assays were performed as described (24). Results are expressed as cpm or as stimulation index calculated as the ratio of [cpm of cells cultured in the presence of synthetic peptide]/[cpm of cells cultured in the absence of peptide]. CTLs against NP366-374 were detected in a conventional 51Cr-release assay as described (25). Results are expressed as the percentage of specific lysis and determined as follows: [(experimental cpm - spontaneous cpm)/(maximum cpm - spontaneous cpm)] × 100. IL-2, IL-4, and IFN-γ were detected in culture supernatants harvested 40 h after initial seeding with Opt EIA mouse cytokine sets (PharMingen).
Fluorescence-Activated Cell Sorting Studies. The phenotypic characterization of transgenic lymphocytes was performed by fluorescence-activated cell sorting using PE-conjugated mAb: anti-I-Ab (clone AF6-120.1), anti-CD86 (clone PO.3), anti-CD40 (clone 3/23), anti-Db (clone KH95), and anti-IgD (clone IA6-2) (PharMingen). PE-conjugated Db-NP366-374 tetramer was kindly obtained from the National Institute of Allergy and Infectious Diseases Tetramer Facility (Atlanta). Before staining, mononuclear cells were isolated from peripheral blood lymphocytes on a lympholyte-M gradient (Cedarlane), and CD8+ T cells were labeled and positively sorted on a MS+/RS+ column (Miltenyi Biotech).
Virus Challenge Experiments. Vaccinated mice and their controls were challenged intranasally with 50 μl in each nostril of a stock solution of A/PR8/34 (MA) influenza virus (108 plaque-forming units/ml) (the kind gift of Yoshihiro Kawaoka, University of Wisconsin, Madison). Mice survival was monitored daily and recorded by using the Kaplan-Meier method. In some experiments (tetramer staining) mice were challenged with a sublethal dose of virus corresponding to 1:1,000 dilution of the lethal dose.
Results
A CD4 T cell response was measured in 68 of 68 (100%) mice immunized with spleen cells rendered transgenic for the plasmid γ1NV2NA3 coding for the T helper (Th) cell determinant -NVDP- in CDR2 and the B cell epitope -NANP- in CDR3 (29) irrespective of the dose used (103 to 2 × 104 cells per inoculum). The kinetics of the response against -NVDP- followed canonical characteristics of proliferative T cell responses in mice with a peak on day 14 and a return to undetectable levels by the fourth week (Fig. 1B). No response was detected against the B cell epitope -NANP- used as specificity control. To ensure that immunity was systemic and not merely confined to the spleen, an experiment was performed in which we assessed the proliferative response of T cells from lymph nodes pooled according to their anatomical distribution (upper, middle, and lower body). Proliferation occurred in all lymph nodes, and its magnitude was in the same range as the one induced in the spleen (Fig. 1C). Lymph nodes from mice immunized with cells transgenic for a plasmid lacking the -NVDP- determinant did not proliferate (data not shown). Next, we decided to see whether the level of immunity was proportional to the dose of transgenic cells inoculated. As shown (Fig. 1D), the CD4 T cell response is dose dependent over a wide range of transgenic cells per inoculum (20,000-70 cells), but as few as 70 transgenic cells were sufficient to induce marked proliferation (8,662 cpm vs. 159 cpm in mice immunized with control cells). This response was specific as no proliferation occurred when cultures from the same mice were set with control peptide -NANP- or when spleen cells from mice immunized with cells transgenic for -NANP- only were restimulated in culture with the Th cell determinant -NVDP- (Fig. 1D). A cytokine analysis of culture supernatants showed secretion of IL-2, IFN-γ, and IL-4 (Fig. 1E), indicating no Th1/Th2 bias and the expansion largely of uncommitted Th0 cells. Nonspecific effects such as overexpression of self molecules (e.g., MHC class II molecules or costimulatory molecules) are apparently not at play. It is noteworthy that during the first 72 h after contact with DNA no [3H]Thy incorporation occurred in transgenic cells (data not shown), ruling out activation by contaminant lipopolysaccharide. Collectively, these data demonstrate that a single i.v. injection of transgenic cells yields a systemic CD4 T cell response that is reproducible, dose dependent, and specific.
Activation of CD8 T cells was studied by immunization with cells rendered transgenic for plasmid γ1NV2NP3. This plasmid codes for the Db-restricted CTL epitope of the influenza virus A nucleoprotein (NP366-374) in CDR3 and the Th cell determinant -NVDP- in CDR2, providing help in CD8 T cell activation (25). Within the dose range used (2 × 104 to 103 transgenic cells per inoculum) a CTL response was measured in 39 of 39 (100%) mice. The response started on day 14 and remained elevated through day 28 (Fig. 1F). By day 180, lysis ranged ≈50% at 40:1 effector-to-target cell ratio (not shown). The ability to restimulate CTLs in vitro with the NP366-374 peptide for a period of months demonstrates that a single inoculation of transgenic cells generates long-lived effectors and memory precursor CTLs. The CD8 response was also cell dose dependent over a range of transgenic cells (20,000 to 70 per mouse) with maximal lysis (>90%) reaching a plateau at a dose of 5,000 transgenic cells per inoculum (Fig. 1G). A weak, but specific, CTL activity was still detectable in mice immunized with 70 transgenic cells. The response was specific because no lytic activity was detected on EL-4 cells not pulsed with the NP366-374 peptide. Taken together, these data demonstrate that transgenic lymphocyte immunization is a reproducible and highly effective way to mobilize a CTL response in vivo. As in the case of the CD4 response, a very small number of transgenic cells (70 per inoculum) proved sufficient to trigger a specific CTL response.
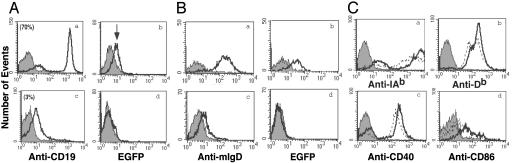
To characterize the cell type serving as target of spontaneous transgenesis, spleen cells from naïve C57BL/6 mice were enriched in B cells by depleting for CD43, an antigen expressed on all leukocytes except B lymphocytes (Fig. 2A). Among the CD43 negative cells, ≈70% expressed CD19 (Fig. 2Aa). This B cell population was then subject to spontaneous transgenesis. Transfected cells were positively sorted and EGFP expression was analyzed by fluorescence-activated cell sorting. An increase in the mean cell fluorescence was seen in the CD43-/CD19+ population (Fig. 2Ab). In a reciprocal experiment spleen cells from naïve mice were depleted of B cells. The flow-through population had a negligible (3%) residual number of CD19+ cells within the total enriched population (Fig. 2Ac) and did not undergo transgenesis (Fig. 2Ad). That B lymphocytes are the target of spontaneous transgenesis in vitro is also supported by the fact that spleen lymphocytes from IgM-/- (B cell deficient) mice could not be rendered transgenic nor immunize (Table 1, which is published as supporting information on the PNAS web site). The B cell subset was characterized by performing spontaneous trangenesis on peripheral blood lymphocytes cultured for 72 h with lipopolysaccharide and dextran sulfate to up-regulate surface IgD expression. Transgenesis occurred in IgD+ B cells (Fig. 2Bb) but not in B cells expressing IgM only (Fig. 2Bd). We conclude that B cells susceptible to spontaneous transgenesis are mature B cells with the IgM+/IgD+ phenotype consistent with what we observed in human B lymphocytes (26). Because T cell activation in vitro requires two signals, a specific signal by the peptide/MHC complex and a second signal through costimulatory molecules (3, 30), we also analyzed the expression of MHC class II and class I, CD40 and CD86, on spontaneously transfected cells. Fig. 2C shows that 18-24 h after contact with plasmid DNA transgenic lymphocytes display a moderate but reproducible up-regulation of these molecules.
Fig. 2.
Phenotypic analysis on transgenic cells. (A) Spleen cells from C57BL/6 mice were enriched for B cells by negative selection on CD43 beads. CD43- and CD43+ populations were stained with anti-CD19 antibody (a and c). These cells were then transfected with plasmid DNA and analyzed for EGFP expression (b and d). Solid histograms refer to background autofluorescence. The experiments were repeated twice with identical results. (B) Murine peripheral blood lymphocytes were cultured for 72 h (a and b) or 10 days (c and d) in the presence of lipopolysaccharide and dextran sulfate (0.7 μg/ml). Cells (85% CD19+ cells) were analyzed for membrane IgD expression (antibody IA6-2) (a and c) and transfected with plasmid DNA. The EGFP expression in each population is shown in b and d. (C) B cells were analyzed before and after transfection with PE-conjugated antibodies against MHC class II (IAb) (a), MHC class I (Db) (b), CD40 (c), and CD86 (d). Dotted lines represent staining on B cells before transfection. The experiment was repeated twice with identical results.
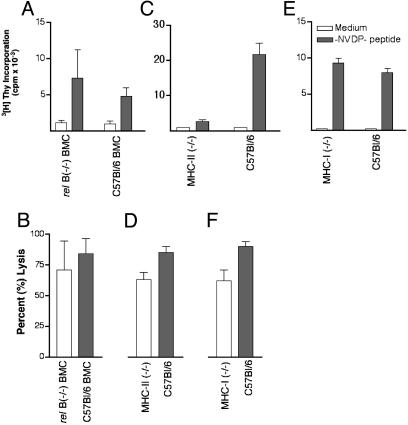
T cell priming in vivo is presently thought to result from antigen presentation by DCs after uptake of soluble antigen or uptake of antigen transferred from another cell (cross-priming). Therefore, experiments were designed to distinguish between T cell priming by the injected transgenic B lymphocytes vs. T cell priming by host DCs. A role by donor DCs in the inoculum was ruled out based on the fact that no CD11c+ cells were present in the cell suspension used for injection (data not shown). In an experiment transgenic lymphocytes were used to immunize mice with a null (-/-) mutation in the relB component of the NF-κB complex. Homozygous relB (-/-) mice lack BM-derived DCs yet possess residual lymphoid (CD8α+) DCs (31, 32). BM chimeras generated by transferring homozygous (-/-) relB BM cells into lethally irradiated recipients carry the same DC defect as relB (-/-) mice. However, we previously showed that residual CD8α+ DCs fail to up-regulate MHC class II and costimulatory molecules after in vitro stimulation and can not prime CD4 and CD8 T cells in vitro and in vivo (28, 33). Immunization with lymphocytes transgenic for γ1NV2NP3 elicited similar CD4 T cell responses in relB (-/-) and control C57BL/6 BM chimeras (Fig. 3A). Similarly, the CD8 T cell response was comparable in both groups of mice (Fig. 3B). Thus, T cell priming is readily achieved in the absence of functional DCs. Whether or not in DC-competent animals host DCs also play a role was addressed in two additional experiments where C57BL/6 mice were immunized with histocompatible transgenic B lymphocytes from either MHC class II (-/-) or class I (Db) (-/-) mice. We reasoned that if T cell priming was solely caused by the injected transgenic lymphocytes, lack of MHC molecules would negatively impact the generation of T cell responses in vivo. The CD4 T cell response in mice given class II (-/-) transgenic lymphocytes was virtually abolished (90% reduction) compared with that of mice immunized with transgenic B lymphocytes from C57BL/6 mice (Fig. 3C). In contrast, the CD8 T cell response was diminished but not abrogated (Fig. 3D), consistent with our previous report that T cell help heightens the CTL response to NP366-374 but is not necessary for T cell priming per se (25). Because MHC class II (-/-) lymphocytes express MHC class I, the result of this experiment indicates that transgenic B lymphocytes are the necessary APCs for in vivo CD4 T cell priming. In a reciprocal experiment, mice were immunized with class I (-/-) transgenic lymphocytes. In these mice the CD4 T cell response was comparable to that of mice given transgenic lymphocytes from C57BL/6 mice (Fig. 3E), indicating efficient presentation to CD4 T cells. CD8 T cells were also induced in both groups of mice even though mice given the class I (-/-) transgenic B lymphocytes had a reduced response (Fig. 3F). This finding suggests that in DC-competent mice cross-priming by means of uptake of secreted transgenic molecules or apoptotic transgenic lymphocytes can contribute to induce a CD8 T cell response. Collectively, the results demonstrate that during transgenic lymphocyte immunization, T cell priming requires direct presentation by transgenic B lymphocytes for CD4 T cell responses but can involve host DCs for CD8 T cell responses. This finding implies that CD8 T cell activation by transgenic B lymphocytes and host DCs complement each other, demonstrating that the immune system possesses intrinsic functional plasticity.
Fig. 3.
Cellular and molecular requirements for CD4 and CD8 T cell responses. (A and B) T cell priming in the absence of DCs. BM chimeras were immunized with 5 × 103 WT cells transgenic for γ1NV2NP3. Spleen cells were harvested on day 21. For the proliferative response (A) cells were cultured with -NVDP- peptide (filled bars) or medium (open bars). Tests were run in triplicate. Results are expressed as cpm ± SD of four mice per group. For CTL induction (B) cells were cultured with peptide NP366-374, and the cytotoxic activity was determined in a 51Cr-release assay. Tests were done in triplicate. Bars refer to the mean percentage of lysis ± SD of four mice per group at an effector-to-target cell ratio of 10:1. (C and D) T cell priming using MHC class II (-/-) transgenic lymphocytes. C57BL/6 mice were immunized with class II (-/-) or WT lymphocytes (5 × 103) transgenic γ1NV2NP3. (E and F) T cell priming using MHC class I (-/-) transgenic lymphocytes. C56Bl/6 mice were immunized with class I (-/-) or WT lymphocytes (5 × 103) transgenic γ1NV2NP3. CD4 (C and E) and CD8 (D and F) T cell responses were tested as above.
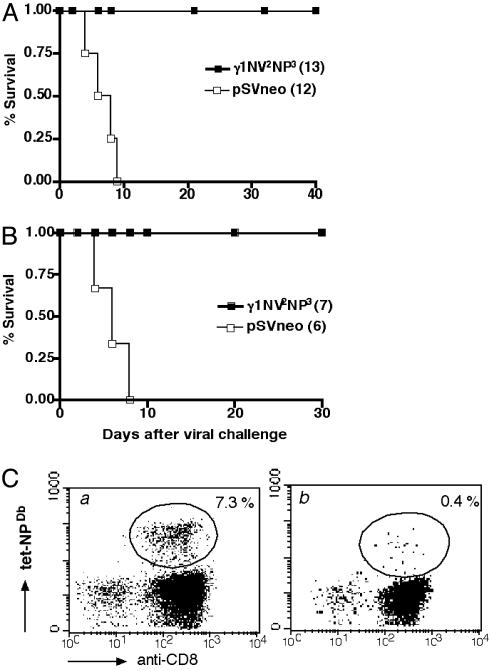
To determine whether the immunity induced by transgenic lymphocyte immunization was protective, we used an accepted model of lethal virus infection, intranasal infection by influenza A/PR8/34 virus, which causes 100% mortality in mice in ≈10 days. In the first experiment, C57BL/6 mice were immunized by a single i.v. injection of lymphocytes rendered transgenic for plasmid γ1NV2NP3. Twenty-one days after vaccination mice were challenged intranasally with a lethal dose of virus. Whereas all control mice (i.e., mice inoculated with lymphocytes rendered transgenic for control plasmid) died by day 9, all vaccinated mice survived through the end of the observation period free of signs of disease (e.g., weight loss) (Fig. 4A). This result was confirmed in a second experiment in which the longevity of protection was probed in mice allowed to rest for 2 months after vaccination. Again, after lethal virus challenge, only vaccinated mice survived (Fig. 4B). Because precursor CTLs against the influenza peptide NP366-374 persist for several months after immunization, the protection data argue that the virus challenge restimulated specific CTLs and mobilized them to the lung in time and in number sufficient to block disease and prevent death. In fact, mice primed by transgenic lymphocyte immunization and subsequently challenged with a sublethal dose of A/PR8/34 virus underwent an expansion of Db/NP366-374 tetramer-positive CD8 T cells, which did not occur in mice given virus challenge only (Fig. 4C).
Fig. 4.
Protection against lethal challenge with influenza virus. C57BL/6 mice were immunized with 5 × 103 transgenic for γ1NV2NP3 (▪) or pSV2neo (□) as a control. All mice were infected by intranasal inoculation with a lethal dose of virus (A/PR8/34) (MA). Mice were challenged 21 (A) or 60 (B) days after priming. Survival was calculated by the Kaplan-Meyer method. The number of mice used is indicated in parentheses. (C) Mice immunized with 5 × 103 transgenic lymphocytes (a) or naïve mice as control (b) were infected on day 21 with a sublethal dose of influenza virus. After 10 days NP366-374-specific CD8 T cells in the blood were identified by staining with the PE-conjugated Db-NP366-374 tetramer. Blood was pooled from four mice. The percentage of CD8+ tetramer+ T cells is shown.
Discussion
The adaptive immune response occurs when precursor T lymphocytes exist at sufficient frequency and their encounter with the APC fulfills certain rules of spatial and structural organization in secondary lymphoid organs where costimulatory signals are present (34). These factors determine the temporal aspects of clonal expansion and kinetics of the immune response. This level of stringency is particularly relevant because for T cell priming the antigen needs to be presented in the form of short peptides in association with MHC molecules at the surface of the APC. Based on these considerations transgenic lymphocyte immunization is a way to program the adaptive immune response, unique with respect to control of the APC involved, synthesis of antigen, and the ability to direct transgenic B lymphocytes to the site of immune induction. In our opinion, several factors contribute to the efficiency of this approach. First is the possibility to program mature naive B lymphocytes into efficient APCs by using nonviral DNA without the need for isolation of the B cell population or for drug selection. Second is the up-regulation of MHC molecules and costimulatory molecules (CD40 and CD86). Third is the transcription and synthesis of transgenic Ig heavy chains and the assumption that quality-control processes regulate the degradation of misfolded transgenic polypeptides in the lumen of the endoplasmic reticulum vs. secretion of intact transgenic Ig molecules. This process guarantees a prolonged supply of peptides to the MHC molecules beyond their short half-life (11-13). Fourth is that upon injection in vivo transgenic lymphocytes localize to secondary lymphoid organs, mainly the spleen, where they persist for several weeks (Fig. 5A, which is published as supporting information on the PNAS web site). We monitored the fate of injected cells by a nested PCR amplifying a 198-bp fragment specific for the transgene. Six hours after i.v. injection of 5 × 103 transgenic lymphocytes, and through the first week, a PCR product was amplified in the spleen (Fig. 5B) and in blood and organs with high perfusion rate. By day 21 the transgene was detected only in the spleen consistent with the notion that B lymphocytes injected i.v. localize to the spleen (16). From days 28-210 all of the samples were negative. Amplification from lymph nodes required the injection of a larger dose (2.5 × 104) of transgenic lymphocytes (Fig. 5C), possibly reflecting different homing characteristics in the spleen and lymph nodes.
Adaptive immune responses can occur only in organized lymphoid tissues where antigen needs to be present for a long enough time and at sufficient levels. Here, we showed that ≈102 transgenic B lymphocytes suffice to induce CD4 and CD8 T cell responses, suggesting that a major factor in the success of transgenic lymphocyte immunization may result from the ability to target the transgenic APC to secondary lymphoid organs. This finding is reminiscent of a view of immunity centered on antigen localization dose and time, a concept termed “geographical immunity” (35). There, a CTL response against the lymphocytic choriomeningitis virus (LCMV) could be induced by intra-spleen immunization with only 500 nonprofessional APCs transgenic for a LCMV glycoprotein (36). The antigen dose is directly linked with the type of resulting immune response, antibody vs. T cells (37, 38) or the type of T cells (37, 38), with small doses of antigen favoring cell-mediated responses and vice versa. However, because too little antigen may be ignored, establishing the lower limit of an antigen dose that induces protective cell-mediated responses remains an empirical procedure. Our experiments show that one injection of a very small number of transgenic lymphocytes activates both CD4 and CD8 T cell responses. Commitment to T cell proliferation and differentiation requires antigen contact for ≈20 h for naïve CD4 T cells (39) and CD8 T cells (40). Thus, the parameters established for immunization with transgenic B lymphocytes are within the requirements for (i) efficient T cell priming (e.g., the duration of contact with naïve T cells even at a low dose of cells), (ii) establishment of immunological memory, and (iii) induction of protective immunity. Further support for our model comes from a recent study showing that the greatest expansion of memory CD8 T cells follows priming with a very low dose of antigen (41). Thus, the model of immunization presented here is optimally designed to address the spatio-temporal prerequisites for a fast synchronization of T cell priming, locally and systemically.
Here, we show that it is easy to capture in naïve B lymphocytes the essence of an effective APC, i.e., prolonged endogenous synthesis of antigen and up-regulation of surface molecules required for efficient T cell priming (e.g., signals 1 and 2), by genetic reprogramming with nonviral DNA. Possibly related to our system is the report on the immunogenicity of hepatoma cells fused with activated B lymphocytes (42), where the latter contribute costimulatory molecules (CD40 and CD86). In our case, B lymphocytes may be turned into APCs when CpG motifs in bacterial DNA signal TLR-9 intracellularly (43), even though spontaneous transgenesis does not induce cell-cycle entry and proliferation (unpublished data). On the other hand, the physiological role of the phenomenon illustrated here remains to be explored. However, because spontaneous transgenesis of B lymphocytes is apparently conserved across species (26), a physiological role is possible. For instance, nuclear DNA released from dying B lymphocytes in germinal centers during a primary response may be internalized by neighboring B lymphocytes that are escaping death or terminal differentiation. Similarly, viral DNA from dying infected cells could also be internalized in B cells and transcribed as the type of promoter is not a limiting factor (26). Because a very low number of transgenic B lymphocytes is sufficient for T cell priming, the mechanism described here could play a role in idiotype regulation of B cells or the amplification of antigen presentation during a primary response with a contribution to the establishment of immunological memory.
Although the characteristics of the system described here have only begun to be delineated, it is already evident that the efficiency of immunization by transgenic lymphocytes depends on the factors discussed above working cooperatively to build immunity within the spatio-temporal geometry of organized secondary lymphoid organs. This paradigm can be exploited to induce adaptive immune responses applicable to strategies of vaccination against viral infections and cancer.
Supplementary Material
Acknowledgments
We thank Drs. J. Hernandez, S. Schoenberger, J. Urbain, and A. Vitiello for critical reading of the manuscript and helpful suggestions and Dr. S. Schoenberger for the gift of MHC (-/-) mice. This work was supported by National Institutes of Health Grants RO1CA77427, R21AI49771, and R01CA92119.
Abbreviations: APC, antigen-presenting cell; DC, dendritic cell; CTL, cytotoxic T lymphocyte; CDR, complementarity-determining region; BM, bone marrow; EGFP, enhanced GFP; PE, phycoerythrin; Th, T helper.
References
- 1.Burnet, F. (1959) The Clonal Selection Theory of Acquired Immunity (Cambridge Univ. Press, London).
- 2.Zamoyska, R. (1998) Curr. Opin. Immunol. 10, 82-87. [DOI] [PubMed] [Google Scholar]
- 3.Bluestone, J. A., Khattri, R. & van Seventer, G. A. (1999) in Fundamental Immunology, ed. Paul, W. E. (Raven, New York), pp. 449-478.
- 4.Ahmed, R. & Gray, D. (1996) Science 272, 54-60. [DOI] [PubMed] [Google Scholar]
- 5.Rotzschke, O., Falk, K., Deres, K., Schild, H., Norda, M., Metzger, J., Jung, G. & Rammensee, H. G. (1990) Nature 348, 252-254. [DOI] [PubMed] [Google Scholar]
- 6.Rudensky, A., Preston-Hurlburt, P., Hong, S.-C., Barlow, A. & Janeway, C., Jr. (1991) Nature 353, 622-627. [DOI] [PubMed] [Google Scholar]
- 7.Yewdell, J. W., Norbury, C. C. & Bennink, J. R. (1999) Adv. Immunol. 73, 1-77. [DOI] [PubMed] [Google Scholar]
- 8.Unanue, E. R. (1992) Curr. Opin. Immunol. 4, 63-69. [DOI] [PubMed] [Google Scholar]
- 9.Banchereau, J. & Steinman, R. M. (1998) Nature 392, 245-252. [DOI] [PubMed] [Google Scholar]
- 10.Ingulli, E., Mondino, A., Khoruts, A. & Jenkins, M. K. (1997) J. Exp. Med. 185, 2133-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellone, M., Iezzi, G., Manfredi, A. A., Protti, M. P., Dellabona, P., Casorati, G. & Rugarli, C. (1994) Eur. J. Immunol. 24, 2691-2698. [DOI] [PubMed] [Google Scholar]
- 12.Adorini, L., Appella, E., Doria, G., Cardinaux, F. & Nagy, Z. A. (1989) Nature 342, 800-803. [DOI] [PubMed] [Google Scholar]
- 13.Muller, K. P., Schumacher, J. & Kyewski, B. A. (1993) Eur. J. Immunol. 23, 3203-3207. [DOI] [PubMed] [Google Scholar]
- 14.Lanzavecchia, A. (1985) Nature 314, 537-539. [DOI] [PubMed] [Google Scholar]
- 15.Cassell, D. J. & Schwartz, R. H. (1994) J. Exp. Med. 180, 1829-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ron, Y. & Sprent, J. (1987) J. Immunol. 138, 2848-2856. [PubMed] [Google Scholar]
- 17.Constant, S., Schweitzer, N., West, J., Ranney, P. & Bottomly, K. (1995) J. Immunol. 155, 3734-3741. [PubMed] [Google Scholar]
- 18.Morris, S. C., Lees, A. & Finkelman, F. D. (1994) J. Immunol. 152, 3768-3776. [PubMed] [Google Scholar]
- 19.Constant, S. L. (1999) J. Immunol. 162, 5695-5703. [PubMed] [Google Scholar]
- 20.Evans, D. E., Munks, M. W., Purkerson, J. M. & Parker, D. C. (2000) J. Immunol. 164, 688-697. [DOI] [PubMed] [Google Scholar]
- 21.Munthe, L. A., Kyte, J. A. & Bogen, B. (1999) Eur. J. Immunol. 29, 4043-4052. [DOI] [PubMed] [Google Scholar]
- 22.Weiss, S. & Bogen, B. (1989) Proc. Natl. Acad. Sci. USA 86, 282-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billetta, R., Filaci, G. & Zanetti, M. (1995) Eur. J. Immunol. 25, 776-783. [DOI] [PubMed] [Google Scholar]
- 24.Gerloni, M., Miner, K. T., Xiong, S., Croft, M. & Zanetti, M. (1999) J. Immunol. 162, 3782-3789. [PubMed] [Google Scholar]
- 25.Langlade-Demoyen, P., Garcia-Pons, F., Castiglioni, P., Garcia, Z., Cardinaud, S., Xiong, S., Gerloni, M. & Zanetti, M. (2003) Eur. J. Immunol. 33, 720-728. [DOI] [PubMed] [Google Scholar]
- 26.Filaci, G., Gerloni, M., Rizzi, M., Castiglioni, P., Chang, H.-D., Wheeler, M., Fiocca, R. & Zanetti, M. (2004) Gene Therapy 11, 42-51. [DOI] [PubMed] [Google Scholar]
- 27.Zanetti, M. (1992) Nature 355, 476-477. [DOI] [PubMed] [Google Scholar]
- 28.Castiglioni, P., Lu, C., Lo, D., Croft, M., Langlade-Demoyen, P., Zanetti, M. & Gerloni, M. (2003) Int. Immunol. 15, 127-136. [DOI] [PubMed] [Google Scholar]
- 29.Xiong, S., Gerloni, M. & Zanetti, M. (1997) Nat. Biotechnol. 15, 882-886. [DOI] [PubMed] [Google Scholar]
- 30.Bretscher, P. & Cohn, M. (1970) Science 169, 1042-1049. [DOI] [PubMed] [Google Scholar]
- 31.Lo, D., Quill, H., Burkly, L., Scott, B., Palmiter, R. D. & Brinster, R. L. (1992) Am. J. Pathol. 141, 1237-1246. [PMC free article] [PubMed] [Google Scholar]
- 32.Wu, L., D'Amico, A., Winkel, K. D., Suter, M., Lo, D. & Shortman, K. (1998) Immunity 9, 839-847. [DOI] [PubMed] [Google Scholar]
- 33.Zanetti, M., Castiglioni, P., Schoenberger, S. & Gerloni, M. (2003) Ann. N.Y. Acad. Sci. 987, 249-257. [DOI] [PubMed] [Google Scholar]
- 34.Zinkernagel, R. M. (2000) Nat. Immunol. 1, 181-185. [DOI] [PubMed] [Google Scholar]
- 35.Zinkernagel, R. M., Ehl, S., Aichele, P., Oehen, S., Kundig, T. & Hengartner, H. (1997) Immunol. Rev. 156, 199-209. [DOI] [PubMed] [Google Scholar]
- 36.Kundig, T. M., Bachmann, M. F., DiPaolo, C., Simard, J. J., Battegay, M., Lother, H., Gessner, A., Kuhlcke, K., Ohashi, P. S., Hengartner, H., et al. (1995) Science 268, 1343-1347. [DOI] [PubMed] [Google Scholar]
- 37.Parish, C. R. (1971) J. Exp. Med. 134, 21-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hosken, N. A., Shibuya, K., Heath, A. W., Murphy, K. M. & O'Garra, A. (1995) J. Exp. Med. 182, 1579-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iezzi, G., Karjalainen, K. & Lanzavecchia, A. (1998) Immunity 8, 89-95. [DOI] [PubMed] [Google Scholar]
- 40.van Stipdonk, M. J., Hardenberg, G., Bijker, M. S., Lemmens, E. E., Droin, N. M., Green, D. R. & Schoenberger, S. P. (2003) Nat. Immunol. 4, 361-365. [DOI] [PubMed] [Google Scholar]
- 41.Badovinac, V. P., Porter, B. B. & Harty, J. T. (2002) Nat. Immunol. 3, 619-626. [DOI] [PubMed] [Google Scholar]
- 42.Guo, Y., Wu, M., Chen, H., Wang, X., Liu, G., Li, G., Ma, J. & Sy, M. S. (1994) Science 263, 518-520. [DOI] [PubMed] [Google Scholar]
- 43.Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., Matsumoto, M., Hoshino, K., Wagner, H., Takeda, K. & Akira, S. (2000) Nature 408, 740-745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.