Abstract
Dendritic cells (DCs) play important roles in the initiation and regulation of immune responses. Although several subsets of DCs were identified according to their expression of surface molecules such as CD4, CD8, and CD11b, the regulatory mechanism for the development and homeostasis of these DC subsets remains unclear. Here we show that mice lacking IFN regulatory factor-2 (IRF-2-/- mice) exhibited a marked and selective defect in splenic CD4+CD11b+DCs, instead of CD8α+CD11b-DCs that were reported to be missing in mice lacking the related transcription factor IRF-8. Furthermore, the numbers of epidermal Langerhans cells in IRF-2-/- mice were reduced at least in part because of the lack of the CD4+CD11b+ subset. Studies with radiation bone marrow chimeras as well as in vitro retrovirus-mediated gene transduction showed that IRF-2 was required cell-autonomously for the development of myeloid-related DCs. Notably, these abnormalities in DCs diminished in mice lacking both IRF-2 and the IFN-α/β receptor, indicating that IRF-2 acted through negatively regulating IFN-α/β signals. In contrast, natural killer cells still showed developmental arrest in these double mutant mice, indicating that the mode of action of IRF-2 for CD4+DC development is distinct from that for natural killer cell development. Our current findings thus pointed to a previously unknown unique cell-type-selective multimode function of IRF-2 in the regulation of lymphohematopoiesis.
Dendritic cells (DCs) play pivotal roles not only in the initiation but also in the determination of the direction, toward either type 1 or 2, of T cell-mediated immune responses against infection (1). In response to pathogens, DCs undergo differentiation from immature to mature DCs that act as principal antigen-presenting cells (APCs) in secondary lymphoid organs. Maturation of DCs is induced not only by microbial products acting through Toll-like receptors (2) but also by cytokines produced on infection with pathogens (1). Among such cytokines are type I IFNs (IFN-α/β) produced by cells infected with viruses and in response to microbial products such as lipopolysaccharide (LPS). However, there are other reports showing that IFN-α/β act suppressively on the differentiation of DCs (3, 4). There might therefore be yet-unrecognized regulatory mechanisms operating to control the negative and positive effects of IFN-α/β on the differentiation and functions of DCs.
Murine splenic DCs have been classified into three major subsets based on their surface expression of CD4 and CD8α molecules (5, 6). Although these cells were originally thought to represent distinct cell lineages, reports demonstrating that all three subsets can be generated from either common myeloid or lymphoid progenitors cast skepticism on this lineage hypothesis (7). Nevertheless, there appear to be differences, albeit not necessarily absolute, among these subsets in terms of immunological function such as production of IL-12, crosspriming of CD8+ T cells, and maintenance of self tolerance as well as anatomical localization within lymphoid organs (8, 9). In addition to these three major DC subsets, a rare DC subset with plasmacytoid characteristics has been identified recently (10, 11).
Given the potential importance of DC subset differentiation in the regulation of immune responses, it is critical to understand the molecular nature of the factors regulating murine DC subset differentiation. Studies using gene-disrupted mice have started to shed light on the mechanism of DC subset regulation. Recently, among them, two groups have shown that mice lacking IFN consensus sequence-binding protein (ICSBP), also called IFN regulatory factor (IRF)-8, exhibited a defect in CD8α+, plasmacytoid DCs, and epidermal Langerhans cells (LCs) (12-14). As we have shown previously, another member of the IRF family, IRF-2, attenuates signals evoked by spontaneously produced IFN-α/β, thereby preventing a CD8+ T cell-mediated skin inflammation (15, 16). The function of IRF-2 might not be confined to those as a transcriptional repressor, and direct gene activation was also known to be induced by IRF-2 for several genes, such as those encoding vascular cell adhesion molecule-1 and gp91phox (17, 18). Moreover, IRF-2 was shown to be required for natural killer (NK) cell development (19). In terms of DC biology, IRF-2 is of great interest, because it was reported that IRF-2 and ICSBP/IRF-8 not only formed complexes but also acted cooperatively, for instance, in the expression of the IL-12p40 gene (20). Here we examined the roles of IRF-2 in the development and functions of DCs using mice lacking this transcription factor (IRF-2-/- mice, ref. 21). Contrary to the case in IRF-8-/- mice, we found that IRF-2-/- mice exhibited a selective cell autonomous deficiency in the CD4+ DC subset, including splenic CD4+CD11b+ DCs and epidermal CD4+ LCs. Inactivation of the IFN-α/β receptor restored the development of both CD4+ DCs and epidermal LCs, but not NK cells, in IRF-2-/- mice. Thus, IRF-2 is a unique regulator of lymphohematopoiesis, acting differently in CD4+ DCs and NK cells in terms of its relationship to IFN-α/β signals.
Materials and Methods
Mice. IRF-2-/- mice kindly provided by Tak W. Mak (University of Toronto, Toronto) (21) were backcrossed 6 or 10 times to C57BL/6 (BN6 and BN10, respectively). These two lines of backcrossed mice gave identical results, and we did not discriminate these two series of mice in this work. IFNAR1-/- mice were purchased from B&K Universal (Hull, U.K.) and backcrossed 10 times to C57BL/6. IRF-2-/-IFNAR1-/- double mutant mice were generated by crossing IRF-2-/-BN10 and IFNAR1-/-BN10 mice. IRF-2-/-H-2d mice were established by intercrossing F1 progenies of IRF-2-/-BN10 × B10.D2 (SLC, Shizuoka, Japan) breeding. DO11.10 transgenic (tg) B10.D2 mice were established by backcrossing original DO11.10 tg BALB/c mice at least five times to B10.D2 mice. B6-Ly5.1 mice were purchased from Sankyo (Tsukuba, Japan). All mice were maintained under specific pathogen-free conditions and used at 8-12 wk of age. All experiments were performed according to institutional guidelines.
Antibodies and Reagents. Fluorochrome- and biotin-conjugated mAbs and streptavidins used in this study (FITC-anti-CD11c, FITC-anti-CD43, APC-anti-CD11b, APC-anti-NK1.1, APC-anti-CD8α, APC-anti-B220/CD45R, PE-anti-CD4, PE-anti-CD11b, PE-anti-I-Ab, PE-anti-CD86, PE-Cy7-anti-CD11b, biotin-anti-CD40, biotin-anti-Ly5.1, biotin-anti-CD19, and PerCP-streptavidin) were purchased from BD Pharmingen except for FITC-anti-Ly5.2, which was from e-Bioscience (San Diego). Biotin-labeled antibodies were developed with APC-streptavidin. OptEIA kits for measuring mouse IL-6 and IL-12p40 were from BD Bioscience.
Bone Marrow (BM) Chimeras. Radiation BM chimeras were established by transferring 5-10 × 106 red cell-depleted BM cells i.v. via the tail vein into 8- to 10-wk-old B6-Ly5.1 mice that had been irradiated by 9.0 Gy and analyzed 8-10 weeks later.
BM-Derived and Splenic DCs. BM cells were cultured in vitro in the presence of granulocyte-macrophage colony-stimulating factor (R & D Systems) for 8 days (22). Recovered cells were analyzed directly or purified by using FITC-anti-CD11c antibody and anti-FITC microbeads with MACS columns (Miltenyi Biotec, Auburn, CA). Purified cell preparations contained constantly >90% CD11c+ cells. These cells were stimulated with 1 μg/ml LPS (Escherichia coli O55, Wako Biochemicals, Osaka) or with 1 μM CpG DNA (TCCATGACGTTCCTGATGCTT, completely phosphorothioate-modified; Qiagen, Valencia, CA) for 24 h. The supernatants and cells were collected for ELISA assays and for analyses of the expression of surface markers, respectively. Magnetically purified CD4+ T cells from DO11.10 transgenic B10.D2 mice (1 × 105) were cultured with graded numbers of BM-DCs established from IRF-2-/-H-2d mice and control littermates in the presence of an OVA peptide as described (23), and T cell proliferation was measured by using CellTiter 96 Aqueous One kit (Promega). Splenic CD11c+ cells were prepared by digesting with collagenase D (2.5 mg/ml, Roche Diagnostics).
Retroviral Transduction of IRF-2 cDNA. Mouse IRF-2 cDNA was amplified and cloned into the BamHI-XhoI site of pMX-IRES-EGFP (a kind gift from T. Kitamura, University of Tokyo; ref. 24). The recombinant vector was transfected into a packaging cell phoenix (a kind gift from G. P. Nolan, Stanford University, Stanford, CA). On days 2, 3, and 4 of the granulocyte-macrophage colony-stimulating factor-assisted BM culture, BM cells were incubated in the virus-containing medium in the presence of 8 μg/ml polybrene (Sigma-Aldrich) under centrifugation (300 × g) for 2 h at 32°C. CD11c expression of the cells was analyzed on day 7.
Epidermal Sheets and LCs. Low-density single-cell suspension from epidermal layers was prepared as described (25). Briefly, ears were divided into dorsal and ventral halves with forceps. These halves were trypsinized and then split into epidermal and dermal layers. Single-cell suspension from epidermal layers was prepared by mechanical disaggregation through a stainless steel strainer. Low-density cells were collected by centrifugation by using 30% BSA solution. Epidermal cells were permeabilized by using the cell permeabilization kit FIX & PERM (Caltag, South San Francisco, CA) for intracellular staining because surface CD4 molecules were removed by trypsinization. Epidermal sheets were obtained as described (26), fixed by acetone, and subjected to immunohistochemistry.
Flow Cytometry. Cells were stained with fluorochrome-conjugated antibodies and analyzed by using Cytomics FC500 (Beckman Coulter) for the analyses of NK cells and a FACScalibur cytometer (BD Bioscience) for others. Data analyses were performed by using rxp analysis software (Beckman Coulter) or cellquest software (BD Biosciences). Dead cells were gated out by propidium iodide staining.
Results
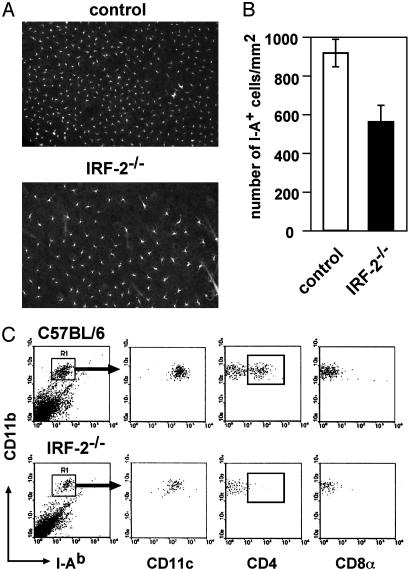
Severe Reduction in CD4+CD11b+ DCs in IRF-2-/- Mice. Flow cytometry showed that the frequencies of CD11chigh cells in IRF-2-/- mice were slightly lower than were those in control littermates (Fig. 1A; 29.0 ± 1.1 × 105 vs. 17.8 ± 9.6 × 105 per spleen for control and IRF-2-/- mice, respectively). It was also found that CD4+CD11b+ DCs, referred thereafter as CD4+ DCs, were reduced in IRF-2-/- mice, whereas the percentage of CD8α+CD11b- DCs (CD8+ DCs) were increased (Fig. 1 B and C). Accordingly, the numbers of CD4+ and CD8+ DCs per spleen were 12.6 ± 1.8 × 105 and 8.4 ± 0.5 × 105, respectively, for control and 2.4 ± 0.7 × 105 and 11.0 ± 7.7 × 105, respectively, for IRF-2-/- mice. The frequencies of CD4-CD8α- DCs were not dramatically altered in these mice compared with control littermates. These results suggested that CD4+ DC development was defective in IRF-2-/- mice. The frequencies of yet another type of DCs, plasmacytoid DCs, defined as CD11cdullCD11b-B220+ in the spleen, were not altered significantly in IRF-2-/- mice (E.I., unpublished data).
Fig. 1.
Impaired splenic CD4+ DC subpopulation in IRF-2-/- mice. (A) The percentages of CD11chigh cells in viable spleen cells, where filled circles represent control littermates and open circles represent IRF-2-/- mice, respectively. (B) CD11chigh cells gated as indicated were analyzed for CD4, CD8α, and CD11b expression (C). The frequencies of CD4+ and CD8α+ DC subsets within total CD11chigh cells were calculated. In C, filled circles denote CD4+CD11chigh cells and open circles denote CD8α+CD11chigh cells. Each dot represents the value obtained from an individual animal (A and C).
Next, cells bearing MHC class II (I-A) in epidermal sheets prepared from the ears were enumerated. As depicted in Fig. 2A and B, the densities of epidermal I-A+ cells representing LCs in IRF-2-/- mice were lower than those in control littermates. Epidermal cells recovered from IRF-2-/- mice contained consistently fewer numbers of CD11b+I-A+CD11c+ cells representing LCs than those from control mice (Fig. 2C). Notably LCs positive for cytoplasmic CD4 (cCD4+ LCs) were almost completely missing in IRF-2-/- mice (Fig. 2C Lower). Thus, the reduction of the density of I-A+ epidermal cells was largely due to the absence of cCD4+ LCs, although the impairment of CD4-CD8- LCs might also have contributed to the reduction.
Fig. 2.
Lack of CD4+ LCs in the epidermis of IRF-2-/- mice. Epidermal sheets were stained for I-A (A), and the numbers of I-A+ cells were counted (B). I-A+CD11b+ cells isolated from epidermis of control littermates (Upper) and IRF-2-/- mice (Lower) were gated as indicated and analyzed for the indicated cell surface markers and cytoplasmic CD4 (C).
Defective CD4+ DC Generation from IRF-2-Deficient BM Cells in Vivo Irradiated B6-Ly5.1 mice were reconstituted with BM cells from Ly5.2-expressing IRF-2-/- mice (IRF-2 chimeras) or control littermates (control chimeras). In these chimeras, >99% of splenic CD11chigh cells were of donor origin, because they expressed surface Ly5.2 (E.I., unpublished data) but not Ly5.1 markers (Fig. 3A). The frequencies of CD11chigh cells within the spleens in IRF-2-/- chimeras were regularly approximately one-third of those in control chimeras (15.2 ± 3.0 × 105 and 5.9 ± 1.0 × 105 per spleen for control and IRF-2 chimeras, respectively). In control chimeras, CD4+ DCs occupied ≈60% of total splenic CD11chigh cells, whereas in IRF-2-/- chimeras, this population was dramatically shrunk (Fig. 3 B and C). When the frequencies of these DC subsets in total spleen cells were compared between these two types of BM chimeras, a remarkable reduction of CD4+ DC frequencies was apparent (Fig. 3D). Consistently, the numbers of CD4+ and CD8+ DCs were 9.0 ± 2.1 × 105 and 1.2 ± 0.3 × 105 per spleen, respectively, for control and 1.1 ± 0.3 × 105 and 3.3 ± 0.4 × 105 per spleen, respectively, for IRF-2 chimeras. Because chimeras generated by transferring wild-type BM cells into irradiated IRF-2-/-RAG-1-/- mice showed splenic DC subsets indistinguishable from those in RAG-1-/- mice received wild-type BM cells (Fig. 6, which is published as supporting information on the PNAS web site), it is clear that the nonhematopoietic environment did not play any role in CD4+ DC development. These results indicated that IRF-2 deficiency affected selectively, if not exclusively, the potential of BM cells to develop into CD4+ DCs, because of the defect intrinsic to BM progenitors.
Fig. 3.
Cell autonomous developmental defects in IRF-2-deficient BM cells. (A) In BM chimeras reconstituted with control BM cells (control chimera) or with IRF-2-deficient BM cells (IRF-2 chimera), >99% of splenic CD11chigh cells gated as indicated were negative for Ly5.1. (B) CD4 vs. CD8α profiles for CD11chigh cells are shown. (C and D) The percentages of either CD4+ or CD8+ DCs within CD11chigh cells (C) or total spleen cells (D) were plotted for control and IRF-2 chimeras. Vertical bars represent the means of the data obtained with seven chimeras generated in three independent transfers.
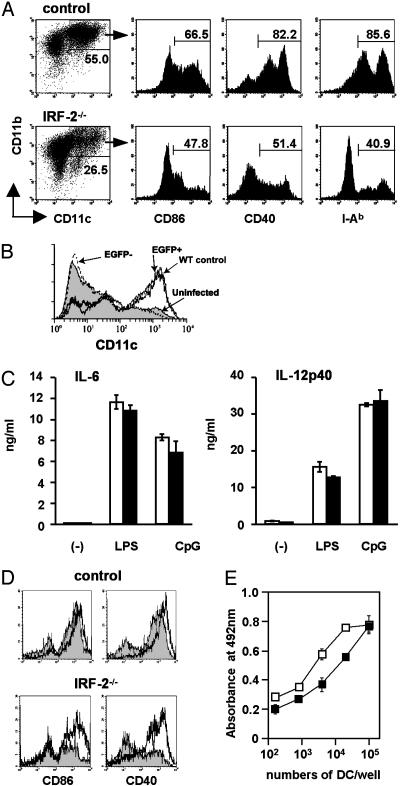
Reduced Frequencies of the Generation of Mature DCs from IRF-2-Deficient BM in Vitro. BM cells isolated from IRF-2-/- mice and control littermates were cultured in vitro in the presence of granulocyte-macrophage colony-stimulating factor, a standard protocol to generate myeloid-related CD11b+ DCs (22). Total cell numbers recovered from the cultures of IRF-2-deficient BM cells ranged from 50% to 100% of those from control cultures. We found that IRF-2-deficient BM cells gave rise to CD11c+CD11b+ DCs (BM-DCs) less efficiently than did control BM cells (Fig. 4A). Moreover, the BM-DCs generated from IRF-2-deficient BM cells contained less CD86+, CD40+, and I-A+ relatively mature DCs than control BM-DCs (Fig. 4A). These observations, together with another result in which retrovirus-mediated transduction of the IRF-2 cDNA restored the development of CD11c+ cells from IRF-2-deficient BM cells (Fig. 4B), confirmed the notion that the developmental potential to myeloid-related DCs was impaired in IRF-2-deficient BM cells in a cell autonomous manner.
Fig. 4.
Inefficient generation of mature DCs from IRF-2-deficient BM cells in vitro. (A) BM-DCs generated in vitro were stained for CD11c and CD11b, together with one of three activation markers, as indicated. Numbers indicate the percentages of cells within the gates (a representative result of more than five independent experiments). (B) BM cells were transduced with an expression vector for IRF-2 and enhanced GFP (EGFP) and cultured in vitro as above. The histograms “WT control,” “uninfected,” “EGFP+,” and “EGFP-” represent control BM-DCs, uninfected IRF-2-deficient BM-DCs, IRF-2-deficient BM-DCs expressing EGFP-IRF-2, and those that failed to express EGFP-IRF-2, respectively. (C) The amounts of IL-6 and IL-12p40 produced by BM-DCs in response to medium alone (-), LPS, or CpG were measured. Open and filled columns represent the means and SD of triplicate cultures of control and IRF-2-deficient BM-DCs, respectively. (D) BM-DCs stimulated with LPS as in C were examined for CD86 and CD40. Shaded histograms were for BM-DCs cultured in medium alone and bold lines for those stimulated with LPS. Note that the CD86dull or CD40dull populations are remaining in IRF-2-deficient BM-DCs even after stimulation. (E) T cell proliferation induced by graded numbers of control (open squares) or IRF-2-deficient (filled squares) BM-DCs. Symbols and error bars represent the means and the SD of duplicate cultures, respectively. Where not seen, error bars were within the symbols.
We stimulated magnetically purified BM-DCs with LPS or with unmethylated CpG DNA. As depicted in Fig. 4C, the amounts of IL-6 and IL-12p40 produced 24 h later did not differ between IRF-2-deficient and control BM-DCs. In contrast, although the up-regulation of CD86 and CD40 expression was observed on the vast majority of control BM-DCs on stimulation with LPS (Fig. 4D) and CpG (E.I., unpublished data), substantial fractions of IRF-2-deficient BM-DCs stayed to become CD86dull and CD40dull (Fig. 4D and E.I., unpublished data). In addition, antigen presentation to CD4+ T cells by unstimulated IRF-2-deficient BM-DCs was less potent than that by control BM-DCs (Fig. 4E). Thus, IRF-2 seemed to be required not only for the generation but also, albeit partially, for the efficient functional maturation of DCs in vitro.
Roles of IFN Signals in Impaired Lymphohematopoietic Development in IRF-2-/- Mice. We next asked whether the attenuator function of IRF-2 on IFN-α/β signals contributed to the efficient development of splenic CD4+ DCs, by generating mice concomitantly deficient for IRF-2 and the IFN-α/β receptor (IRF-2-/-IFNAR1-/- mice). Notably, the frequencies of CD4+ DCs within splenic CD11chigh cells were restored in IRF-2-/-IFNAR1-/- mice to levels comparable to, if slightly lower than, those in control littermates (Fig. 5A and B). In addition, the numbers of I-A+ cells in the epidermis were restored in IRF-2-/-IFNAR1-/- mice to levels seen in control littermates and IFNAR1-/- mice (Fig. 5C). These results together indicated that the function of IRF-2 relevant to the development of splenic CD4+ DCs and epidermal LCs was to attenuate IFN-α/β signals.
Fig. 5.
Roles of IFN-α/β signals in the development of CD4+ DCs and NK cells. (A and B) Splenic CD11chigh cells from IRF-2-/-, IFNAR1-/-, IRF-2-/-IFNAR1-/- (dKO) mice and control littermates were analyzed for CD4+ and CD8+ DC subsets. Numbers indicate the percentages of cells within each quadrant (A). (C) The epidermal sheets of control (open), IRF-2-/- (filled), IFNAR1-/- (dotted), and dKO (hatched) mice were analyzed for I-A+ cells. Data are shown as the mean numbers of I-A+ cells per mm2 with the SD. (D) BM cells isolated from the indicated mice were stained for NK1.1, CD3, CD43, and CD11b. Dot plots are shown for CD11b and CD43 on viable NK lineage cells (CD3-NK1.1+) (representative of three independent analyses).
We next asked whether the mechanisms by which IRF-2 regulates the development of CD4+ DCs and NK cells were the same. As has been proposed recently, NK1.1+ cells in the BM acquire CD11b and CD43 sequentially as they differentiate into mature NK cells (27). As can be seen in Fig. 5D, NK1.1+CD3- cells in the BM of IRF-2-/- mice contained severely reduced numbers of CD11bhigh and CD43+ cells that were present abundantly in control littermates. This developmental arrest appeared to be due to IRF-2 deficiency within BM cells, because BM chimeras receiving IRF-2-deficient BM cells still showed identical developmental arrest (S.T., unpublished observation). Importantly, the developmental arrest of NK cells in the BM was not restored at all in IRF-2-/-IFNAR1-/- mice (Fig. 5D), a contrasting situation to that observed for CD4+ DCs (Fig. 5 A and B). Thus, although IRF-2 is required for the development of both CD4+ DCs and NK cells, its mode of action seems to be different in these two cell types.
Discussion
We showed here that IRF-2-/- mice exhibited a selective cell-autonomous defect in splenic CD4+ DC subset in a fashion that depends on the intact IFN-α/β signaling pathway. Because we did not observe any increase of DC numbers in several other lymphoid organs in IRF-2-/- mice including s.c. and mesenteric lymph nodes and the BM (unpublished observations), we consider that IRF-2 is critical to the development rather than the migration of CD4+ DCs to the spleen. The numbers of CD8+ DCs seemed to be slightly increased in IRF-2-/- mice and IRF-2 chimeras. Although we cannot exclude the possibility that the reduction of the CD4+ DC population vacated the space in the spleen, thereby allowing the expansion of CD8+ DCs, these observations may implicate a relatively minor function of IRF-2 acting negatively in CD8+ DC development. In accordance with the positive role of IRF-2 in DC development in vivo, we observed that the generation of CD11c+CD11b+ DCs from IRF-2-deficent BM cells was also impaired in a cell-autonomous manner. Although BM-DCs hardly express CD4, and their relationship to splenic CD4+ DCs was not clear, both of these DCs are CD11b+ and are regarded as “myeloid-related” DCs. We envisage therefore that IRF-2 is required commonly for the efficient development of “myeloid-related” but not “lymphoid-related” DCs.
That IRF-8 is required for the development of lymphoid-related CD8+ DCs instead of CD4+ DCs (12-14), together with our current findings, indicates that distinct DC subpopulations use different IRF family transcription factors for their development. Notably, we also observed a nearly complete lack of cCD4+ epidermal LCs in IRF-2-/- mice (Fig. 2). This observation, together with the simultaneous restoration of both CD4+ splenic DCs and epidermal LCs in IRF-2-/-IFNAR1-/- mice (Fig. 5), suggests the close relationship between these two types of DCs. It has recently been reported that IRF-8-/- mice showed a reduction by ≈50% in the numbers of epidermal LCs (28). Although epidermal LCs did not contain a CD8+ subset (Fig. 2C), the remaining epidermal LCs in IRF-8-/- mice could be a sister population of splenic CD8+, rather than CD4+, DCs.
Currently, it is not clear how IRF-2 supports CD4+ DC development selectively. One may argue that IRF-2 is selectively expressed in CD4+ DCs by analogy with the observation that IRF-8 expression was restricted to CD8+ DCs (14). However, because IFN-α/β receptors are thought to be expressed ubiquitously, such a simple selective expression model does not explain why CD8+ but not CD4+ DCs could tolerate the up-regulated IFN-α/β signals in IRF-2-/- mice (15), which appear to play a negative role in the development of CD4+ DCs (Fig. 5). It is possible that CD4+ and CD8+ DCs may be different substantially in IFN-α/β-related signaling machinery, including IRF-2 expression. Understanding the mechanism for the cell type specificity of IRF-2 would thus provide a deeper insight into the regulation of DC subset differentiation.
Our current findings, together with the previous observations that RelB, Ikaros C, PU.1, and TRAF6 were required selectively for the development of “myeloid-related” DCs (29-33), implicated crosstalks between IRF-2/IFN-α/β signals and the pathways involving these signaling/transcriptional regulators. In this regard, an interesting report appeared recently in which RelB-/- mice developed an atopic dermatitis-like skin lesion that resembled the IFN-α/β-dependent skin inflammation in IRF-2-/- mice (15); both lesions developed in a T cell-dependent manner and showed several common pathogenic alterations such as thickening of the epidermis, keratinocyte proliferation at the basement membrane, and hair loss (34). On the other hand, NK cells in RelB-/- mice developed normally (35), and IRF-2-/-IFNAR1-/- as well as IRF-2-/- mice exhibited an arrest in NK cell development (Fig. 5D), indicating that the role of IRF-2 in NK cell development was independent of IFN-α/β and RelB pathways. IRF-2 is thus a unique gene regulator that functions with distinct mechanisms in different cell types by attenuating IFN-α/β signals in CD4+ DC development on the one hand and perhaps by directly activating a gene(s) promoting NK cell development on the other. Curiously, Id2-/- mice that lacked CD8+ DCs were also reported to be defective in NK cell development (36). This raised an intriguing possibility that IRF-2 interacted with distinct transcription factors in CD4+ DCs and NK cells.
Contrary to previous findings that IFN-α/β had adjuvant effects on immune responses likely by activating DCs (37, 38), we observed a defective maturation of DCs in IRF-2-/- mice despite the up-regulated IFN-α/β signals (Fig. 5 and ref. 15). Our observation agrees rather with a report showing that IFN-α/β have an inhibitory effect on human DC maturation (3, 4). A speculation would hence be that developing DCs, likely myeloid-related subsets, might be sensitive to IFN-α/β at a certain stage(s) of maturation where IRF-2 normally protects them from maturation arrest as far as the amounts of IFN-α/β not exceeding the limit of its control. In this regard, IRF-2 is a regulator critical for efficient immune responses against pathogens by repressing the harmful effects of IFN-α/β on DC development and allows these cytokines to exert beneficial effects. Importantly, however, the defect in CD4+ DCs was associated not with immunoinsufficiencies but with an autoimmune-like cutaneous inflammation (15). Evidence has accumulated recently for the suppressive activities of DCs (39), and our current and previous (15) findings together raise an interesting possibility that CD4+ DCs suppressed CD8+ T cell-mediated immune responses.
Supplementary Material
Acknowledgments
We thank Drs. Tak W. Mak, Toshio Kitamura, and Garry P. Nolan for IRF-2-/- mice, pMX-IRES-GFP vector, and phoenix cells, respectively. Ms. Namiko Azuta's technical assistance and Dr. Tsutomu Katsuyama's permission for access to the FACS facility at Shinshu University Hospital are also acknowledged. This work has been supported in part by Grants-in-Aid for Scientific Research on Priority Area 15019035 (to S.T.) and 13140202 (to K.I.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; Grants-in-Aid 15390159 (to S.T.) and 14370075 (to K.I.) from the Japan Science Promotion Society; and grants from the Novartis Foundation (Japan) and the Yamanouchi Foundation.
Abbreviations: DC, dendritic cells; LC, Langerhans cells; IRF, IFN regulatory factor; BM, bone marrow; LPS, lipopolysaccharide; APC, antigen-presenting cell; NK, natural killer.
References
- 1.Banchereau, J., Briere, F., Caux, C., Davoust, J., Lebecque, S., Liu, Y. J., Pulendran, B. & Palucka, K. (2000) Annu. Rev. Immunol. 18, 767-811. [DOI] [PubMed] [Google Scholar]
- 2.Takeda, K., Kaisho, T. & Akira, S. (2003) Annu. Rev. Immunol. 21, 335-376. [DOI] [PubMed] [Google Scholar]
- 3.Lehner, M., Felzmann, T., Clodi, K. & Holter, W. (2001) Blood 98, 736-742. [DOI] [PubMed] [Google Scholar]
- 4.McRae, B. L., Nagai, T., Semnani, R. T., van Seventer, J. M. & van Seventer, G. A. (2000) Blood 96, 210-217. [PubMed] [Google Scholar]
- 5.Shortman, K. & Liu, Y. J. (2002) Nat. Rev. Immunol. 2, 151-161. [DOI] [PubMed] [Google Scholar]
- 6.Vremec, D., Pooley, J., Hochrein, H., Wu, L. & Shortman, K. (2000) J. Immunol. 164, 2978-2986. [DOI] [PubMed] [Google Scholar]
- 7.Traver, D., Akashi, K., Manz, M., Merad, M., Miyamoto, T., Engleman, E. G. & Weissman, I. L. (2000) Science 290, 2152-2154. [DOI] [PubMed] [Google Scholar]
- 8.Moser, M. & Murphy, K. M. (2000) Nat. Immunol. 1, 199-205. [DOI] [PubMed] [Google Scholar]
- 9.Ardavin, C. (2003) Nat. Rev. Immunol. 3, 1-9. [DOI] [PubMed] [Google Scholar]
- 10.Asselin-Paturel, C., Boonstra, A., Dalod, M., Durand, I., Yessaad, N., Dezutter-Dambuyant, C., Vicari, A., O'Garra, A., Biron, C., et al. (2001) Nat. Immunol. 2, 1144-1150. [DOI] [PubMed] [Google Scholar]
- 11.Nakano, H., Yanagita, M. & Gunn, M. D. (2001) J. Exp. Med. 194, 1171-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsujimura, H., Tamura, T. & Ozato, K. (2003) J. Immunol. 170, 1131-1135. [DOI] [PubMed] [Google Scholar]
- 13.Schiavoni, G., Mattei, F., Sestili, P., Borghi, P., Venditti, M., Morse, H. C., 3rd, Belardelli, F. & Gabriele, L. (2002) J. Exp. Med. 196, 1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aliberti, J., Schulz, O., Pennington, D. J., Tsujimura, H., Reis e Sousa, C., Ozato, K. & Sher, A. (2003) Blood 101, 305-310. [DOI] [PubMed] [Google Scholar]
- 15.Hida, S., Ogasawara, K., Sato, K., Abe, M., Takayanagi, H., Yokochi, T., Sato, T., Hirose, S., Shirai, T., Taki, S., et al. (2000) Immunity 13, 643-655. [DOI] [PubMed] [Google Scholar]
- 16.Taki, S. (2002) Cytokine Growth Factor Rev. 13, 379-391. [DOI] [PubMed] [Google Scholar]
- 17.Jesse, T. L., LaChance, R., Iademarco, M. F. & Dean, D. C. (1998) J. Cell. Biol. 140, 1265-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo, W. & Skalnik, D. G. (1996) J. Biol. Chem. 271, 23445-23451. [DOI] [PubMed] [Google Scholar]
- 19.Lohoff, M., Duncan, G. S., Ferrick, D., Mittrucker, H. W., Bischof, S., Prechtl, S., Rollinghoff, M., Schmitt, E., Pahl, A. & Mak, T. W. (2000) J. Exp. Med. 192, 325-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, I. M., Contursi, C., Masumi, A., Ma, X., Trinchieri, G. & Ozato, K. (2000) J. Immunol. 165, 271-279. [DOI] [PubMed] [Google Scholar]
- 21.Matsuyama, T., Kimura, T., Kitagawa, M., Pfeffer, K., Kawakami, T., Watanabe, N., Kundig, T. M., Amakawa, R., Kishihara, K., Wakeham, A., et al. (1993) Cell 75, 83-97. [PubMed] [Google Scholar]
- 22.Inaba, K., Inaba, M., Romani, N., Aya, H., Deguchi, M., Ikehara, S., Muramatsu, S. & Steinman, R. M. (1992) J. Exp. Med. 176, 1693-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taki, S., Sato, T., Ogasawara, K., Fukuda, T., Sato, M., Hida, S., Suzuki, G., Mitsuyama, M., Shin, E.-H., Kojima, K., et al. (1997) Immunity 6, 673-679. [DOI] [PubMed] [Google Scholar]
- 24.Onishi, M., Kinoshita, S., Morikawa, Y., Shibuya, A., Phillips, J., Lanier, L. L., Gorman, D. M., Nolan, G. P., Miyajima, A. & Kitamura, T. (1996) Exp. Hematol. 24, 324-329. [PubMed] [Google Scholar]
- 25.Schuler, G. & Steinman, R. (1985) J. Exp. Med. 161, 526-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen, C., Steinman, R., Witmer-Pack, M., Hankins, D., Morris, P. & Austyn, J. (1990) J. Exp. Med. 172, 1483-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, S., Iizuka, K., Kang, H. S., Dokun, A., French, A. R., Greco, S. & Yokoyama, W. M. (2002) Nat. Immunol. 3, 523-528. [DOI] [PubMed] [Google Scholar]
- 28.Schiavoni, G., Mattei, F., Borghi, P., Sestili, P., Venditti, M., Morse, H. C., Belardelli, F. & Gabriele, L. (November 13, 2003) Blood, 10.1182/blood-2003-09-3007.
- 29.Wu, L., Nichogiannopoulou, A., Shortman, K. & Georgopoulos, K. (1997) Immunity 7, 483-492. [DOI] [PubMed] [Google Scholar]
- 30.Wu, L., D'Amico, A., Winkel, K. D., Suter, M., Lo, D. & Shortman, K. (1998) Immunity 9, 839-847. [DOI] [PubMed] [Google Scholar]
- 31.Anderson, K. L., Perkin, H., Surh, C. D., Venturini, S., Maki, R. A. & Torbett, B. E. (2000) J. Immunol. 164, 1855-1861. [DOI] [PubMed] [Google Scholar]
- 32.Guerriero, A., Langmuir, P. B., Spain, L. M. & Scott, E. W. (2000) Blood 95, 879-885. [PubMed] [Google Scholar]
- 33.Kobayashi, T., Walsh, P. T., Walsh, M. C., Speirs, K. M., Chiffoleau, E., King, C. G., Hancock, W. W., Caamano, J. H., Hunter, C. A., Scott, P., et al. (2003) Immunity 19, 353-363. [DOI] [PubMed] [Google Scholar]
- 34.Barton, D., HogenEsch, H. & Weih, F. (2000) Eur. J. Immunol. 30, 2323-2332. [DOI] [PubMed] [Google Scholar]
- 35.Caamano, J., Alexander, J., Craig, L., Bravo, R. & Hunter, C. A. (1999) J. Immunol. 163, 4453-4461. [PubMed] [Google Scholar]
- 36.Yokota, Y., Mansouri, A., Mori, S., Sugawara, S., Adachi, S., Nishikawa, S. & Gruss, P. (1999) Nature 397, 702-706. [DOI] [PubMed] [Google Scholar]
- 37.Montoya, M., Schiavoni, G., Mattei, F., Gresser, I., Belardelli, F., Borrow, P. & Tough, D. F. (2002) Blood 99, 3263-3271. [DOI] [PubMed] [Google Scholar]
- 38.Santini, S. M., Lapenta, C., Logozzi, M., Parlato, S., Spada, M., Di Pucchio, T. & Belardelli, F. (2000) J. Exp. Med. 191, 1777-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinman, R. M., Hawiger, D., Nussenzweig, M. C. (2003) Annu. Rev. Immunol. 21, 685-711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.