Abstract
Molecular mechanisms of leukemogenesis have been successfully unraveled by studying genes involved in simple rearrangements including balanced translocations and inversions. In contrast, little is known about genes altered in complex karyotypic abnormalities. We studied acute myeloid leukemia (AML) patients with complex karyotypes and abnormal chromosome 21. High-resolution bacterial artificial chromosome (BAC) array-based comparative genomic hybridization disclosed amplification predominantly in the 25- to 30-megabase (MB) region that harbors the APP gene (26.3 MB) and at position 38.7-39.1 MB that harbors the transcription factors ERG and ETS2. Using oligonucleotide arrays, APP was by far the most overexpressed gene (mean fold change 19.74, P = 0.0003) compared to a control group of AML with normal cytogenetics; ERG and ETS2 also ranked among the most highly expressed chromosome 21 genes. Overexpression of APP and ETS2 correlated with genomic amplification, but high APP expression occurred even in a subset of AML patients with normal cytogenetics (10 of 64, 16%). APP encodes a glycoprotein of unknown function previously implicated in Alzheimer's disease, but not in AML. We hypothesize that APP and the transcription factors ERG and ETS2 are altered by yet unknown molecular mechanisms involved in leukemogenesis. Our results highlight the value of molecularly dissecting leukemic cells with complex karyotypes.
Balanced translocations and inversions are recurrent events in acute myeloid leukemia (AML) and usually occur in relatively simple karyotypes (1, 2). Genes involved in these rearrangements have been identified and chimeric fusion genes have been shown to result in deregulation of downstream genes (3). Other mechanisms leading to altered gene function are intragenic molecular rearrangements such as deletions and duplications (4, 5). The discovery of these genomic changes have had a profound influence on our understanding of the molecular mechanisms contributing to leukemogenesis. Moreover, the altered pathways uncovered in this way are beginning to be targeted in the design of novel therapies (6).
In contrast, few genes involved in complex karyotypes are known, and their impact on leukemogenesis research has been modest (7). AML patients with complex karyotypes have a very poor prognosis (2); therefore, the identification of recurrent genomic changes might provide insights into the pathways involved. In a recent study we used spectral karyotyping to refine the interpretation of complex karyotypes in AML (8). A striking finding was the unanticipated amplification of chromosomal material from several chromosomes, the most frequent one being chromosome 21 in 8 of 29 cases. Amplification of 21q has also been observed by others (7, 9), supporting the notion that gain of chromosome 21 material appears to be a nonrandom event implicated in AML.
We reasoned that the recurring amplification of chromosome 21 seen in AML patients with complex karyotypes might be related to the function of a specific gene or set of genes. It had already been shown that RUNX1 (AML1, CBFA2) was not the likely target (8). Our strategy was to use a bacterial artificial chromosome (BAC) array to obtain a high-resolution genomic dosage map of chromosome 21 followed by expression analyses in search of overexpressed genes within amplified regions. We report here how these studies led to the detection of two regions frequently amplified. Candidate genes located in these overrepresented regions showing considerable overexpression include the transcription factors ERG and ETS2 (genes implicated in leukemia) (10, 11), as well as the APP gene [amyloid β (A4) precursor protein], a gene not previously related to AML. Furthermore, we show that subsets of patients with AML greatly overexpress APP even in the absence of amplification.
Materials and Methods
Patients. We studied 12 AML patients with complex karyotypes and abnormalities of chromosome 21 by BAC array comparative genomic hybridization (CGH; referred to as AML patients with complex karyotypes). See Table 2 and Supporting Text, which are published as supporting information on the PNAS web site, for a detailed description. Additionally, BAC array CGH was performed on DNA samples from two AML patients who had a noncomplex karyotype with +13 and -21 as the only chromosomal aberrations present, as well as from five AML patients with a normal karyotype (AML CN), and from five healthy donors.
For gene expression analyses using oligonucleotide arrays, we studied bone marrow aspirates from 6 of the above 12 patients, and 23 de novo AML patients with normal cytogenetics. Additionally, blood samples from 64 AML patients with normal karyotypes were included in the real-time RT-PCR studies. These samples were obtained from the Cancer and Leukemia Group B Leukemia Tissue Bank (CALGB protocols 9665 and 8461).
Cytogenetic Analyses. G-banding, spectral karyotyping, and fluorescence in situ hybridization (FISH) analyses were preformed as described in Supporting Text. To validate DNA copy number changes of the APP, ETS2, and ERG genes, we performed FISH assays using the BAC clones RP11-44F3 (containing the APP locus), RP11-830D9 (containing the ETS2 locus), and RP11-66C14 (containing the ERG locus) in seven, four, and three patients (Table 2). This was of particular importance for the APP and the ETS2 genes, which, unlike ERG, were not contained in any of the BAC clones used for the array construction.
BAC Array-Based CGH. A BAC array providing high-resolution mapping of chromosome 21 dosage was constructed (Supporting Text). Thirty-six RPCI-11 BAC clones were selected covering the q arm of chromosome 21 from position 15.1 megabases (MB, close to the centromere) to the telomeric position 46.9 MB, with an average gap between clones of 800 kb (range, 346-1,593 kb). In addition, 23 randomly selected clones, each representing one of the other chromosomes, were included as controls (Table 3, which is published as supporting information on the PNAS web site). Positions of genes and BAC clones were determined according to the NCBI database (www.ncbi.nlm.nih.gov/mapview) based on sequence information available on April 10th, 2003 (build 33).
For BAC array-based CGH pooled blood DNA from five healthy donors (three females and two males) was used as reference. Genomic DNA from tester (AML) and reference samples were differentially labeled and cohybridized to the arrays.
Expression Analyses Using Oligonucleotide Arrays. Total RNA was extracted from AML samples by using Trizol reagent (Invitrogen) following the manufacturer's directions. RNA integrity was assessed by electrophoresis on the Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA). Microarray analysis was performed as described at www.dnaarrays.org. In brief, 8 μg of high-quality total RNA was used for the cRNA synthesis and hybridized to U133A Affymetrix oligonucleotide arrays and scanned according to the manufacturer's protocols (Affymetrix, Santa Clara, CA). Scanned microarray images were analyzed with dchip software (www.dchip.org) to obtain model-based gene expression estimates using the PM Only Model (12).
Quantitative RT-PCR. Comparative real-time RT-PCR assays were performed for each sample in duplicate in a final reaction volume of 25 μl. APP, ERG, and ETS2 expression levels were estimated relative to the expression levels of the housekeeping gene Glucose-phosphate isomerase (GPI). The comparative cycle threshold (CT) cycles for the targets and GPI were determined, and the cycle number difference (ΔCT = GPI - target) was calculated for each replicate. Relative target expression values were calculated by using the mean of ΔCT from the two replicates, that is, μ(ΔCT) = (ΣΔCT)/2, and expressed as 2μ(ΔCT). A detailed protocol is provided in Supporting Text.
Statistical Analyses
BAC Arrays. The analysis of the BAC array CGH experiments is outlined in Supporting Text. In brief, fluorescence intensity ratios were measured in four control experiments (reference versus reference hybridizations) to weight the DNA amplifications (fluorescence ratio >1) and deletions (fluorescence ratio <1). The normal variation at each clone was measured by the standard deviation (SD) of the four control experiments at each clone (13). Fluorescence ratios >1 + 2 SD were considered to be amplified, and the ratios <1 - 2 SD were considered to be deleted if they showed at least a 10% change in the copy number from the base line.
Oligonucleotide Arrays. Model based gene expression estimates obtained by dchip software were used to compare the expression levels of the complex karyotype AML group and the AML CN group. To identify differentially expressed genes, unpaired two-sample t tests between the two groups and the fold change between their means were performed for each gene on the chip. A gene was said to be differentially expressed between complex karyotype AML cases and AML CN cases if the P value was <0.05 (one-sided t test) with a fold change >1.5 with an absolute mean difference between the two groups >100. The initial study included hybridization experiments of six AML samples with complex karyotypes and a reference group of five AML CN (data set 1). Differences in gene expression patterns between the two groups were further validated by comparison of expression estimates of the six AML with complex karyotypes to a second set of AML CN (n = 23; data set 2), which were studied in the same way.
To compare the DNA copy number change and the RNA expression ratio for genes on chromosome 21, we obtained the RNA expression ratios for each of the six AML patients with complex karyotypes individually by comparing the expression estimate with the pooled AML CN (data set 1). Then each gene was mapped to the nearest BAC clone, and the corresponding DNA copy number change was obtained. The Spearman rank correlation between RNA expression ratios and the DNA copy number change was calculated and considered important if the associated P value was <0.05.
Results
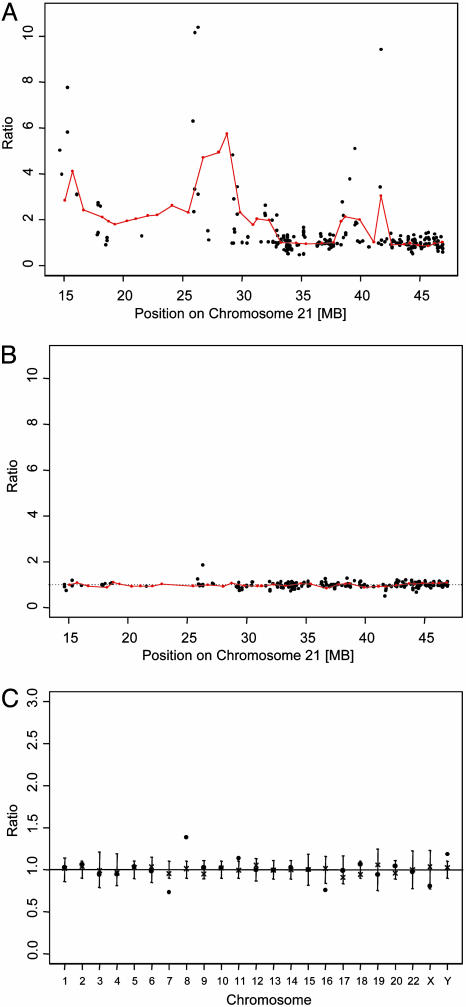
Characterization of DNA Copy Number Changes of Chromosome 21 by BAC Array CGH. We identified patterns of genomic imbalances with over- and underrepresentation of specific regions of chromosome 21 that were unique to each of the 12 AML patients with complex karyotypes and abnormal chromosome 21 studied by BAC array CGH (Figs. 1 and 2A). Amplification was present in all but one patient and varied in degree (range, up to 5.7-fold) and amplicon size (range, 0.5-31.8 MB). The proximal gene poor half of 21q, which corresponds mainly to the Giemsa dark band 21q21, showed frequent gain of chromosomal material. Loss of chromosome 21 material was less extensive and was found in eight patients, involving segments in the distal half of 21q in five patients and in the proximal half of 21q in four patients. In contrast to these AML patients with complex karyotypes, BAC array CGH confirmed loss of the entire 21q in 2 AML patients with monosomy 21 in a noncomplex karyotype. Furthermore, analysis of five AML patients with AML CN as well as blood DNA from four healthy donors showed no changes in DNA copy number of chromosome 21 material or any of the loci on other chromosomes (Fig. 2B).
Fig. 1.
Gains (red) and losses (green) of 21q material detected by BAC array CGH in 12 AML patients with complex karyotype. Genes appearing in boldface are contained in the corresponding BAC clone; the remaining genes are located in the vicinity of the BAC clone used here but are not present in this BAC. The asterisk indicates cases studied for gene expression by oligonucleotide array.
Fig. 2.
Ratios of DNA copy number of chromosome 21 clones (red dots connected with red line) and array-derived gene expression estimates (black dots) of 230 probe sets for AML patient 5 (A) and for a representative AML case with a normal karyotype (B) (expression ratios for the AML CN case in B were obtained by the comparison to the four remaining AML CN of the control group). (C) DNA copy number ratios for the clones from chromosomes other than chromosome 21 are shown for patient 11 (dots) and the reference DNA (× for normal blood), illustrating loss of material from chromosomes 7 and 16 and gain of material from chromosomes 8 and 11 in patient 11. These changes were confirmed by cytogenetic analyses. The ratios for the X and Y chromosomes are typical for males. Ratios were calculated as described in Materials and Methods and were plotted as a function of their position; SD for each clone in reference DNA is depicted.
Two regions predominantly amplified in AML with complex karyotypes were of particular interest: the region spanning positions 25-30 MB included the highest degree of amplification (26.7-28.7 MB, 4.7- to 5.7-fold; patient 5) and was amplified in 8 of the 12 patients. Genes implicated in leukemia have not been found in this region that was noted to contain the APP gene (located at position 26.3 MB). The second chromosomal region (MB position 38.7) was the most frequently amplified region, overrepresented in all but three patients. This region contained the ERG gene (38.7 MB) and was close to the ETS2 locus (39.1 MB). In contrast, the region of the RUNX1 locus (in 21q22.12, MB position 35.2) was only amplified in three patients, deleted in one, and unchanged in eight patients.
Correlation of Dosage Estimates Obtained by Cytogenetic Analyses and BAC Array CGH. To validate the accuracy of the BAC array CGH in the detection of copy number changes in the 12 AML patients with complex karyotypes, we compared results of BAC array CGH with cytogenetic findings and found a high concordance between results of BAC array CGH and cytogenetics (Fig. 2C).
To further confirm the variation in copy number changes across 21q, we performed FISH using different probes specific for the APP, ERG, ETS2, and RUNX1 genes. FISH assays confirmed the accuracy with which DNA copy numbers were detected by BAC array CGH for these loci, and highlighted the substantial amplification of the regions encompassing the APP and the ERG/ETS2 loci (Fig. 4 and Table 2, which are published as supporting information on the PNAS web site).
DNA Copy Number and Expression of Genes on Chromosome 21. We performed gene expression analyses using oligonucleotide arrays on samples from those six AML cases with complex karyotypes that had high-quality RNA available (Fig. 1). The U133A oligonucleotide array includes 22,000 probe sets, of which 230 recognize 139 of the 225 different genes predicted on chromosome 21 (14). Concordance between RNA expression level and DNA copy number was identified for genes located on chromosome 21 (Fig. 2 A and B), and a significant correlation was observed for 30 chromosome 21 genes that were overexpressed and amplified (Fig. 5, which is published as supporting information on the PNAS web site).
We then analyzed differences in gene expression regardless of the DNA copy number and found that levels of 10 genes located on chromosome 21 were significantly higher in the group of AML with complex karyotypes compared to the two control groups of AML CN (data sets 1 and 2; Table 1). However, only one gene was consistently overexpressed in all six AML samples with complex karyotype: the APP gene. Compared to five AML CN cases (mean expression estimate, 52; range, 28-96), significantly higher APP expression was found in three patients with amplification of the region containing the APP locus [DNA copy number ranging from 1.48- to 4.70-fold; mean expression estimate, 1,248; range, 1,082-1,568; P value (t test), 0.008]. Significantly higher expression was also detected in the two cases without increased copy number and one case with deletion of the APP locus (mean expression estimate, 803; range, 693-967) compared to the five AML CN cases [P value (t test), 0.005]. Thus, expression of APP correlated with DNA dosage, but overexpression was not restricted to cases with an increased copy number.
Table 1. Significantly up-regulated chromosome 21 genes in AML with complex karyotypes (n = 6).
| Mean fold change (range) compared to AML CN data set 1 | P value (t test) compared to AML CN data set 1 | Mean fold change (range) compared to AML CN data set 2 | P value (t test) compared to AML CN data set 2 | Gene | Gene name | Unigene cluster | Position, MB |
|---|---|---|---|---|---|---|---|
| 19.74 (13.3-30.17) | 0.0003 | 7.20 (5.31-10.77) | 0.0002 | APP | Amyloid β (A4) precursor protein | Hs. 177486 | 26.3 |
| 2.61 (0.76-5.02) | 0.02 | 1.83 (0.93-2.46) | 0.009 | ERG | v-ets erythroblastosis virus E26 oncogene like (avian) | Hs. 45514 | 38.7 |
| 2.55 (1.08-5.80) | 0.04 | 2.32 (1.23-4.50) | 0.02 | NRIP1 | Nuclear receptor interacting protein 1 | Hs. 155017 | 15.3 |
| 2.44 (0.83-3.77) | 0.02 | 2.33 (0.96-3.42) | 0.01 | ETS2 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | Hs. 292477 | 39.1 |
| 2.10 (0.57-3.98) | 0.04 | 1.86 (0.69-3.20) | 0.04 | SAMSN1 | SAM domain, domain and nuclear localization signals 1 | Hs. 221851 | 14.8 |
| 1.91 (0.71-3.43) | 0.04 | 2.16 (1.02-3.07) | 0.01 | BACH1 | BTB and CNC homology 1, basic leucine zipper transcription factor | Hs. 154276 | 29.6 |
| 1.81 (0.74-2.90) | 0.03 | 2.39 (1.23-3.46) | 0.007 | USP16 | Ubiquitin specific protease 16 | Hs. 99819 | 29.3 |
| 1.76 (1.03-3.02) | 0.04 | 1.88 (1.27-2.81) | 0.01 | TTC3 | Tetratricopeptide repeat domain 3 | Hs. 132605 | 37.4 |
| 1.62 (0.94-2.01) | 0.01 | 1.77 (1.11-2.88) | 0.0009 | DYRK1A | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | Hs. 75842 | 37.7 |
| 1.52 (0.90-2.10) | 0.03 | 1.59 (1.19-2.06) | 0.004 | U2AF1 | U2 (RNU2) small nuclear RNA auxiliary factor 1 | Hs. 365116 | 43.4 |
Significant overexpression was also observed for the ERG and ETS2 genes located in the second region of frequent amplification (Table 1). There was a strong correlation between DNA copy number and RNA amount for ETS2, whereas the expression of ERG did not significantly correlate with DNA copy number (Fig. 5).
Comparison of expression estimates of all 22,000 probe sets (regardless of chromosomal assignment) recognized by the U133A array identified 47 genes whose expression in the six AML cases with complex karyotypes was significantly greater compared to AML CN. Overexpression of all these genes was also confirmed by using data set 2 that included AML CN (n = 23) as an additional control cohort. In this comparison, APP remained by far the most overexpressed gene [mean fold change, 7.2; P value (t test), 0.00017] as compared to AML CN. This analysis further confirmed the significant overexpression of the ETS2 gene (Table 4, which is published as supporting information on the PNAS web site).
Eight genes were significantly underexpressed in the group of AML with complex karyotypes (Table 5, which is published as supporting information on the PNAS web site). Interestingly, four of these genes were located on chromosome 17 (at 17p13.1, 17q11.2, 17q12, and 17q21.3), and cytogenetic analysis revealed loss of 17p13.1 in five of the six cases. TP53 was significantly underexpressed in the five AML cases with complex karyotypes and deletion of 17p13.1 (range of expression estimates, 76.6-215.9) as compared to the control group of AML CN (range, 111-915; P = 0.0001).
APP, ETS2, and ERG Expression in AML by Quantitative RT-PCR. To confirm and validate our findings, we analyzed APP, ETS2, and ERG mRNA expression in patients that had been studied by oligonucleotide arrays as well as in patients that had only been evaluated for DNA copy number changes by BAC array CGH.
Quantitative RT-PCR confirmed increased APP expression in all 11 AML cases with complex karyotypes with RNA available [patients 1-11; median APP expression, 0.48 2μ(ΔCT); range, 0.1-3.1]. In contrast, APP expression was significantly lower in AML CN cases included in the gene array expression studies [9 of 23 cases had RNA available for quantitative RT-PCR; median APP expression, 0.0002 2μ(ΔCT); range, 0.0001-0.0039; P value, 0.01]. We then explored further the implications of APP in AML by determining expression levels in an independent set of 64 AML CN cases. Significantly lower APP expression values [median APP expression 0.0001 2μ(ΔCT); range, 0-1.3; P value, 0.012] were found. Levels within the range found in the 11 AML cases with complex karyotypes were defined as high APP expression. Using this definition, high APP expression was observed in 10 of 64 (16%) AML CN cases (Fig. 3).
Fig. 3.
APP mRNA expression determined by real-time RT-PCR in AML. Bracket indicates samples that were included in gene expression studies using oligonucleotide arrays.
ETS2 expression detected by quantitative RT-PCR showed significantly higher values for AML with complex karyotypes [median ETS2 expression, 3.543 2μ(ΔCT); range, 1.25-8.31) compared to the group of AML CN included in the gene array expression studies [n = 9; median, 2.4 2μ(ΔCT); range, 0.63-3.18; P value, 0.027). In contrast, no significant difference in expression values was observed for ERG measurements between the two groups, in concert with our data showing that DNA amplification of ERG appeared not to confer overexpression by oligonucleotide expression array analysis (see above).
Discussion
Amplification is a genomic alteration typically resulting in overexpression of genes, in particular targeting oncogenes located within the amplicon. Among candidate genes located on chromosome 21 implicated in leukemogenesis is the RUNX1 gene located at 21q22. Amplification of RUNX1 is infrequent in AML and mainly occurs in childhood acute lymphoblastic leukemia (15). In support of previous observations (8), we found underrepresentation of the RUNX1 locus relative to the amount of 21q gained in most patients, and did not see significant overexpression of RUNX1, making it unlikely to be a relevant target of amplification in AML. Other potential candidate genes include two members of the ETS transcription factor family, ETS2 and ERG, located at 21q22. Little is known about the roles of these genes in leukemogenesis; however, ETS2 amplification has been reported in one AML patient who, interestingly, had a hypodiploid karyotype with a complex translocation t(6;18;21), and ERG is involved in the rare t(16;21)(p11;q22), leading to the chimeric FUS/ERG gene fusion (10, 11).
We applied array CGH, which is becoming the method of choice for high-resolution screening of genomic imbalances (16), in a larger series of AML patients with complex karyotypes and involvement of chromosome 21 and identified two main regions showing significant amplification. One region mapped to the gene poor middle region of 21q21 (25-30 MB). Genes implicated in leukemogenesis have not yet been identified in this region. Global gene expression analyses revealed the APP gene located in 21q21 (position 26.3 MB) as a likely candidate within this amplicon, because it was by far the most highly expressed gene in AML patients with complex karyotypes compared to AML CN. APP was located in the region with the highest degree of amplification, and the highest mRNA expression was associated with the highest copy number. Nevertheless, overexpression was not restricted to cases with an increased copy number of the APP locus. Therefore, the increased DNA dosage might contribute to but is not the only factor leading to overexpression of APP.
The APP gene encodes a transmembrane glycoprotein that has been implicated in the pathogenesis of Alzheimer's disease, and in dementia in adults with Down syndrome (17). APP has hitherto been believed to be predominantly expressed in the brain. The specific function of the APP protein in neural cells is not fully characterized, and remains unknown in nonneural cells; however, studies have provided evidence that APP promotes proliferation in colon cancer and various epithelial cells (18, 19). Very recently, APP was shown to be overexpressed in pancreatic cancer cells, and soluble APP signaling increased proliferation in a pancreatic cancer cell line (20). Although APP has not been implicated in AML previously, careful scrutiny of recently published expression profiling studies suggest its possible involvement in hematological malignancy. Profiling identified APP as one of the most differentially expressed genes in lymphoid malignancies, showing high expression levels in Epstein-Barr virus-negative Burkitt lymphoma and low expression levels in acute lymphoblastic leukemia with MLL translocations (21, 22) Our study demonstrates that APP is highly expressed in subsets of patients with AML. Protein expression analyses by Western blotting and flow cytometry confirmed that high APP transcript levels correlate with high APP protein expression and identified the localization of the APP protein to be intracellular (data not shown). Further RNA expression analyses of a larger series of AML patients revealed that high expression levels are found in a subset, ≈16%, of AML with normal karyotype. We found low-level APP expression in AML with 11q23 rearrangements (data not shown), consistent with the observed low-level APP expression in acute lymphoblastic leukemia with 11q23 translocations (22).
Several lines of evidence support the notion that increased APP expression is not a random change, but may relate to an altered pathway implicated in AML. First, genes known to increase APP expression, such as ETS2 and HGF (23, 24), were among the genes overexpressed in the cohort of AML with complex karyotype (Table 4). These genes have already been associated with AML. Second, in our study, several hematopoietic stem cell genes, such as CD34, BAALC, MDR1, and CD133, were also highly expressed with expression profiles similar to APP, suggesting that APP might be part of a leukemic stem cell signature (Table 4). In fact, gene expression profiling has identified APP to be among the most highly expressed genes in CD34-positive hematopoietic progenitor cells, thus further supporting the notion that APP may be involved in hemato-poiesis (25).
Interestingly, it has been reported that wild-type APP blocks the proapoptotic activation of p53 in neuronal cells by controlling p53 activation at the posttranslational level and that overexpression of the ETS2 gene induces apoptosis in the presence of normal p53 (26, 27). We observed frequent loss of the 17p13.1 region in five of the six AML with complex karyotypes resulting in significant decrease of TP53 expression (Table 5). Overexpression of APP and/or ETS2 in conjunction with loss of TP53 might result in disruption of the apoptotic pathways involved in leukemia.
The second region showing significant amplification included the loci of the transcription factors ERG and ETS2. Alterations of these genes, which are critical for the control of proliferation, differentiation, and apoptosis, might have a substantial impact on cellular processes. We observed a significant correlation between DNA amount and RNA expression of ETS2, making it a likely target of amplification resulting in substantial overexpression. In contrast, expression of ERG did not seem to be influenced by DNA copy number, so it must be up-regulated by other mechanisms.
We conclude that amplification of two regions (25-30 MB and 38.7-39.1 MB) of chromosome 21 is frequently found in AML with complex karyotypes and abnormal chromosome 21. Amplification of at least one of these regions was observed in 10 of the 12 patients analyzed (83%). There was significant overexpression of the APP and ETS2 genes located in these regions. Overexpression of APP also occurred in some cases with complex karyotypes without amplification of the region and in a subset of AML with normal karyotype. We hypothesize that high APP expression relates to a yet unknown or incompletely dissected pathway implicated in leukemia, and that overexpression of the ETS2 transcription factor may lead to deregulation of critical processes in leukemogenesis. Future characterization of these changes may help to uncover molecular mechanisms that could guide the development of novel treatment strategies in AML.
Supplementary Material
Acknowledgments
We thank Danilo Perrotti for helpful discussions. We thank the physicians and patients who participated in the sample collection for the Cancer and Leukemia Group B leukemia tissue bank. This work was supported by National Cancer Institute Grants CA101140, CA16058, CA31946, and CA77658 and the Leukemia Clinical Research Foundation. C. D. Baldus was supported by a grant from the Deutsche Krebshilfe.
Abbreviations: AML, acute myeloid leukemia; MB, megabase(s); BAC, bacterial artificial chromosome; CGH, comparative genomic hybridization; FISH, fluorescence in situ hybridization.
References
- 1.Rowley, J. D. (1999) Semin. Hematol. 36, Suppl. 7, 59-72. [PubMed] [Google Scholar]
- 2.Byrd, J. C., Mrózek, K., Dodge, R. K., Carroll, A. J., Edwards, C. G., Arthur, D. C., Pettenati, M. J., Patil, S. R., Rao, K. W., Watson, M. S., et al. (2002) Blood 100, 4325-4336. [DOI] [PubMed] [Google Scholar]
- 3.Look, A. T. (1997) Science 278, 1059-1064. [DOI] [PubMed] [Google Scholar]
- 4.Song, W.-J., Sullivan, M. G., Legare, R. D., Hutchings, S., Tan, X., Kufrin, D., Ratajczak, J., Resende, I. C., Haworth, C., Hock, R., et al. (1999) Nat. Genet. 23, 166-175. [DOI] [PubMed] [Google Scholar]
- 5.Steudel, C., Wermke, M., Schaich, M., Schäkel, U., Illmer, T., Ehninger, G. & Thiede, C. (2003) Genes Chromosomes Cancer 37, 237-251. [DOI] [PubMed] [Google Scholar]
- 6.Kelly, L. M., Yu, J.-C., Boulton, C. L., Apatira, M., Li, J., Sullivan, C. M., Williams, I., Amaral, S. M., Curley, D. P., Duclos, N., et al. (2002) Cancer Cell 1, 421-432. [DOI] [PubMed] [Google Scholar]
- 7.Schoch, C., Haferlach, T., Bursch, S., Gerstner, D., Schnittger, S., Dugas, M., Kern, W., Löffler, H. & Hiddemann, W. (2002) Genes Chromosomes Cancer 35, 20-29. [DOI] [PubMed] [Google Scholar]
- 8.Mrózek, K., Heinonen, K., Theil, K. S. & Bloomfield, C. D. (2002) Genes Chromosomes Cancer 34, 137-153. [DOI] [PubMed] [Google Scholar]
- 9.Hilgenfeld, E., Padilla-Nash, H., McNeil, N., Knutsen, T., Montagna, C., Tchinda, J., Horst, J., Ludwig, W.-D., Serve, H., Büchner, T., et al. (2001) Br. J. Haematol. 113, 305-317. [DOI] [PubMed] [Google Scholar]
- 10.Santoro, A., Maggio, A., Carbone, P., Mirto, S., Caronia, F. & Acuto, S. (1992) Cancer Genet. Cytogenet. 58, 71-75. [DOI] [PubMed] [Google Scholar]
- 11.Ichikawa, H., Shimizu, K., Hayashi, Y. & Ohki, M. (1994) Cancer Res. 54, 2865-2868. [PubMed] [Google Scholar]
- 12.Li, C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98, 31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veltman, J. A., Schoenmakers, E. F. P. M., Eussen, B. H., Janssen, I., Merkx, G., van Cleef, B., van Ravenswaaij, C. M., Brunner, H. G., Smeets, D. & van Kessel, A. G. (2002) Am. J. Hum. Genet. 70, 1269-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hattori, M., Fujiyama, A., Taylor, T. D., Watanabe, H., Yada, T., Park, H.-S., Toyoda, A., Ishii, K., Totoki, Y., Choi, D.-K., et al. (2000) Nature 405, 311-319. [DOI] [PubMed] [Google Scholar]
- 15.Roumier, C., Fenaux, P., Lafage, M., Imbert, M., Eclache, V. & Preudhomme, C. (2003) Leukemia 17, 9-16. [DOI] [PubMed] [Google Scholar]
- 16.Snijders, A. M., Nowak, N., Segraves, R, Blackwood, S., Brown, N., Conroy, J., Hamilton, G., Hindle, A. K., Huey, B., Kimura, K., et al. (2001) Nat. Genet. 29, 263-264. [DOI] [PubMed] [Google Scholar]
- 17.Teller, J. K., Russo, C., DeBusk, L. M., Angelini, G., Zaccheo, D., Dagna-Bricarelli, F., Scartezzini, P., Bertolini, S., Mann, D. M. A., Tabaton, M., et al. (1996) Nat. Med. 2, 93-95. [DOI] [PubMed] [Google Scholar]
- 18.Meng, J.-Y., Kataoka, H., Itoh, H. & Koono, M. (2001) Int. J. Cancer 92, 31-39. [PubMed] [Google Scholar]
- 19.Schmitz, A., Tikkanen, R., Kirfel, G. & Herzog, V. (2002) Histochem. Cell Biol. 117, 171-180. [DOI] [PubMed] [Google Scholar]
- 20.Hansel, D. E., Rahman, A., Wehner, S., Herzog, V., Yeo, C. J. & Maitra, A. (2003) Cancer Res. 63, 7032-7037. [PubMed] [Google Scholar]
- 21.Maesako, Y., Uchiyama, T. & Ohno, H. (2003) Cancer Sci. 94, 774-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armstrong, S. A., Staunton, J. E., Silverman, L. B., Pieters, R., den Boer, M. L., Minden, M. D., Sallan, S. E., Lander, E. S., Golub, T. R. & Korsmeyer, S. J. (2002) Nat. Genet. 30, 41-47. [DOI] [PubMed] [Google Scholar]
- 23.Wolvetang, E. W., Bradfield, O. M., Tymms, M., Zavarsek, S., Hatzistavrou, T., Kola, I. & Hertzog, P. J. (2003) Biochim. Biophys. Acta 1628, 105-110. [DOI] [PubMed] [Google Scholar]
- 24.Liu F., Su, Y., Li, B. & Ni, B. (2003) Exp. Cell Res. 287, 387-396. [DOI] [PubMed] [Google Scholar]
- 25.Steidl, U., Kronenwett, R., Rohr, U.-P., Fenk, R., Kliszewski, S., Maercker, C., Neubert, P., Aivado, M., Koch, J., Modlich, O., et al. (2002) Blood 99, 2037-2044. [DOI] [PubMed] [Google Scholar]
- 26.Xu, X., Yang, D., Wyss-Coray, T., Yan, J., Gan, L., Sun, Y. & Mucke, L. (1999) Proc. Natl. Acad. Sci. USA 96, 7547-7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolvetang, E. J., Wilson, T. J., Sanij, E., Busciglio, J., Hatzistavrou, T., Seth, A., Hertzog, P. J. & Kola, I. (2003) Hum. Mol. Genet. 12, 247-255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.