Abstract
Background
Microalbuminuria (MA) has been questioned as a predictor of progressive renal dysfunction in patients with type 1 diabetes (T1D). Consequently, new clinical end points are needed that identify or predict patients that are at risk for early renal function decline (ERFD). The potential clinical utility of differential scanning calorimetry (DSC) analysis of blood plasma and other biofluids has recently been reported. This method provides an alternate physical basis with which to study disease-associated changes in the bulk plasma proteome.
Methods
DSC analysis of blood plasma was applied to identify unique signatures of ERFD in subjects enrolled in the 1st Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes, a prospective cohort study of T1D patients. Recent data suggests differences in the plasma peptidome of these patients correlate with longitudinal measures of renal function. Differences in DSC profile (thermogram) features were evaluated between T1D MA individuals exhibiting ERFD (n=15) and matched control subjects (n=14).
Results
The average control group thermogram resembled a previously defined healthy thermogram. Differences were evident between ERFD and control individuals. Heat capacity values of the main two transitions were found to be significant discriminators of patient status.
Conclusions
Results from this pilot study suggests the potential utility of DSC proteome analysis to prognostic indicators of renal disease in T1D.
General significance
DSC shows sensitivity to changes in the bulk plasma proteome that correlate with clinical status in T1D providing additional support for the utility of DSC profiling in clinical diagnostics.
Keywords: Plasma proteome, differential scanning calorimetry, thermogram, clinical diagnostics, type 1 diabetes, renal function decline
1. INTRODUCTION
For over twenty years urinary albumin levels or urinary albumin excretion rates have been used to identify patients with type 1 diabetes (T1D) at risk for future renal dysfunction [1, 2]. However, the predictive value of microalbuminuria (MA) has been questioned as only 20% of T1D patients with MA will progress to proteinuria, patients with MA can revert to normoalbuminuria and an equal number experience early renal function decline (ERFD) before development of MA [3–6]. Therefore, development of new laboratory methods that identify T1D patients at risk of progressive renal dysfunction is important.
Currently the plasma/serum proteome is routinely interrogated by clinical laboratory procedures such as serum protein electrophoresis or immunochemical assays as an important indicator of patient health [7]. Human plasma is a complex fluid containing over 3000 proteins and peptides ranging in concentration from picograms to tens of milligrams per liter [8–10]. The majority of these proteins and peptides are present in extremely small quantities and the plasma proteome is dominated by only a few proteins: ten proteins make up 90 % of the plasma proteome and an additional 12 proteins constitute a total of 99 % of the total mass concentration of plasma. It is the interrelationship between the most abundant plasma proteins that are monitored by clinical assays as indicators of infection, inflammation, or the progression of diseases and pathophysiologic processes [11].
The low abundance region of the plasma proteome has been targeted in recent years in the search for biomarkers of disease [12–18]. High affinity bead based methods and mass spectrometry have been successfully applied to characterize the low abundance, low molecular weight region of the plasma proteome known as the “peptidome” [19]. Although these techniques have successfully identified a number of previously unknown peptides in the circulation it has not yet been possible to validate the identified peptides as reliable diagnostic markers for specific diseases. Moreover, these approaches have been complicated by the realization of the “interactome” where components of the peptidome are complexed with the more abundant plasma proteins and by problems in appropriate study design [20, 21].
We have recently developed a calorimetric approach to study the plasma/serum proteome that has been described in detail in recent publications [22–24]. Briefly, differential scanning calorimetry (DSC) is used to measure the thermal properties of dilute protein solutions as a function of temperature. For a pure protein, DSC provides a unique temperature-induced denaturation profile, or thermogram, with a characteristic melting temperature and melting enthalpy. In a protein mixture, such as plasma, we have shown that the observed thermogram is a composite of the denaturation behavior of each of the component proteins weighted according to their concentration within the mixture [23]. DSC analysis of blood plasma is therefore a new and useful approach with which to examine the plasma proteome, one based on the thermal properties of the plasma components rather than their mass and charge which is exploited by protein electrophoresis and mass spectrometry.
Our earlier reports of DSC blood plasma analysis have shown that thermograms of plasma from healthy individuals are highly reproducible with characteristic melting temperatures and a well-defined shape [23]. The thermograms obtained from plasma or serum from individuals with various diseases is strikingly different from the “normal” thermogram from healthy individuals. Moreover, the striking differences observed with our DSC assay were not observable using serum protein electrophoresis, a standard clinical tool. To-date, the thermograms for each disease are markedly different from one other and this leads to our current focus in developing this assay as a clinical diagnostic tool for early disease screening. The general applicability of DSC analysis with potential for clinical diagnostics is further underlined by reports from other investigators indicating promising preliminary results for DSC analysis of a number of diseases [25–30]. Here we report the application of DSC to examine the plasma proteome of selected samples obtained during the 1st Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes [31].
2. MATERIALS AND METHODS
2.1 Plasma samples
Patients who were enrolled in the first Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes were eligible for this pilot study. The study protocol and patient consent procedures were approved by the Committee on Human Studies of the Joslin Diabetes Center and the Institutional Review Board of the University of Louisville. Thirty-three plasma samples were selected from the sample repository maintained at the Joslin Diabetes Center for patients enrolled in the 1st Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes. All plasma samples (100 µL) were dialyzed for 24 h at 4 °C against 10 mM potassium phosphate, 150 mM NaCl, 15 mM (w/v) sodium citrate, pH 7.5, to ensure complete solvent exchange and then diluted 25-fold with the same buffer. All samples (0.45 µm, cellulose acetate or polyethersulfone) and buffers (0.22 µm, polyethersulfone) were filtered before use.
2.2 DSC analysis
Scans were performed using an automated VP-Capillary DSC system (MicroCal, Northampton, MA, now part of GE Healthcare). Electrical calibration of the differential power signal and temperature calibration using hydrocarbon temperature standards were performed as part of the manufacturer periodic instrument maintenance. Samples and dialysate were stored in 96-well plates at 5 °C until being loaded into the calorimeter by the robotic attachment. Scans were recorded from 20 °C to 110 °C at 1 °C/min using the mid feedback mode, a filtering period of 2 s, and with a prescan thermostat of 15 min. Data were analyzed using Origin version 7.0 (OriginLab Corporation, Northampton, MA). Sample scans were first corrected for the instrument baseline by subtracting an appropriate buffer scan. Scans were normalized for the gram concentration of protein. Total protein concentrations of the plasma samples were measured by the bicinchoninic acid method (Pierce, Rockford, IL). Nonzero baselines were then corrected by applying a linear baseline fit. Thermograms were plotted as excess specific heat capacity (J/K․g) versus temperature for subsequent analysis.
3. RESULTS
3.1 Study sample characteristics
Plasma samples from thirty-three participants in the 1st Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes were selected for study with our DSC assay. The Joslin study is a prospective cohort study of patients with T1D attending the Joslin Clinic. Subsequent to enrollment, these patients have been followed for 10 to 15 years, thus allowing us to know if their renal function changed over time. Samples from the Joslin study are therefore very valuable in terms of understanding the natural history of early renal function decline in T1D and identifying biomarkers to diagnose it. Recent data suggests differences in the plasma peptidome of these patients correlate with longitudinal measures of renal function [32]. Samples for DSC analysis were selected from individuals with varying renal function, with 17 individuals retaining good renal function and 16 individuals exhibiting ERFD. The threshold for ERFD was defined as having renal function loss of 3.3 % or more per year (−3.3 to −16.1); representing the 2.5th percentile of age-standardized estimated glomerular filtration rate (eGFR) decline among non-diabetic participants of the Baltimore Aging Study [6]. Plasma from patients with less renal function decline (−3.2 to +1.9) were considered to be control samples. All patients selected were Caucasian, and patient groups were matched for eGFR, age, sex, duration of diabetes, blood pressure and medication, as well as several other clinical indices [6].
3.2 Differences between clinical group thermograms
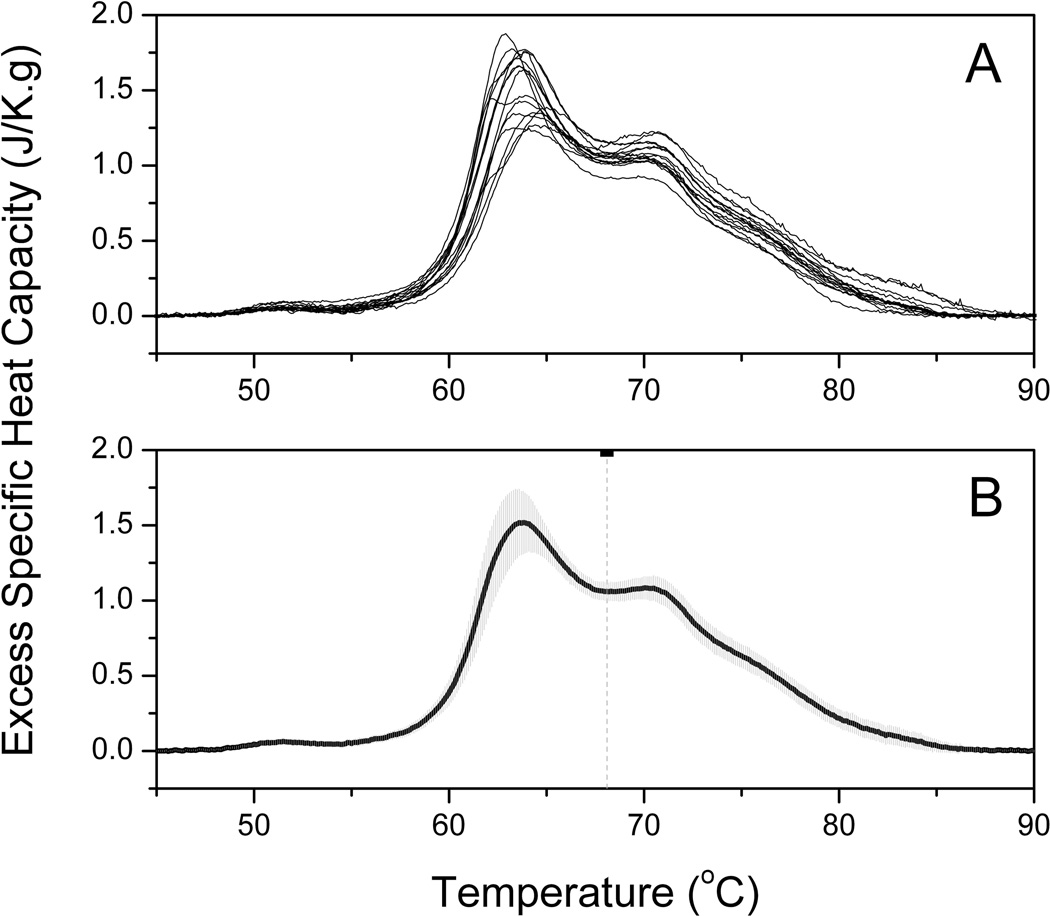
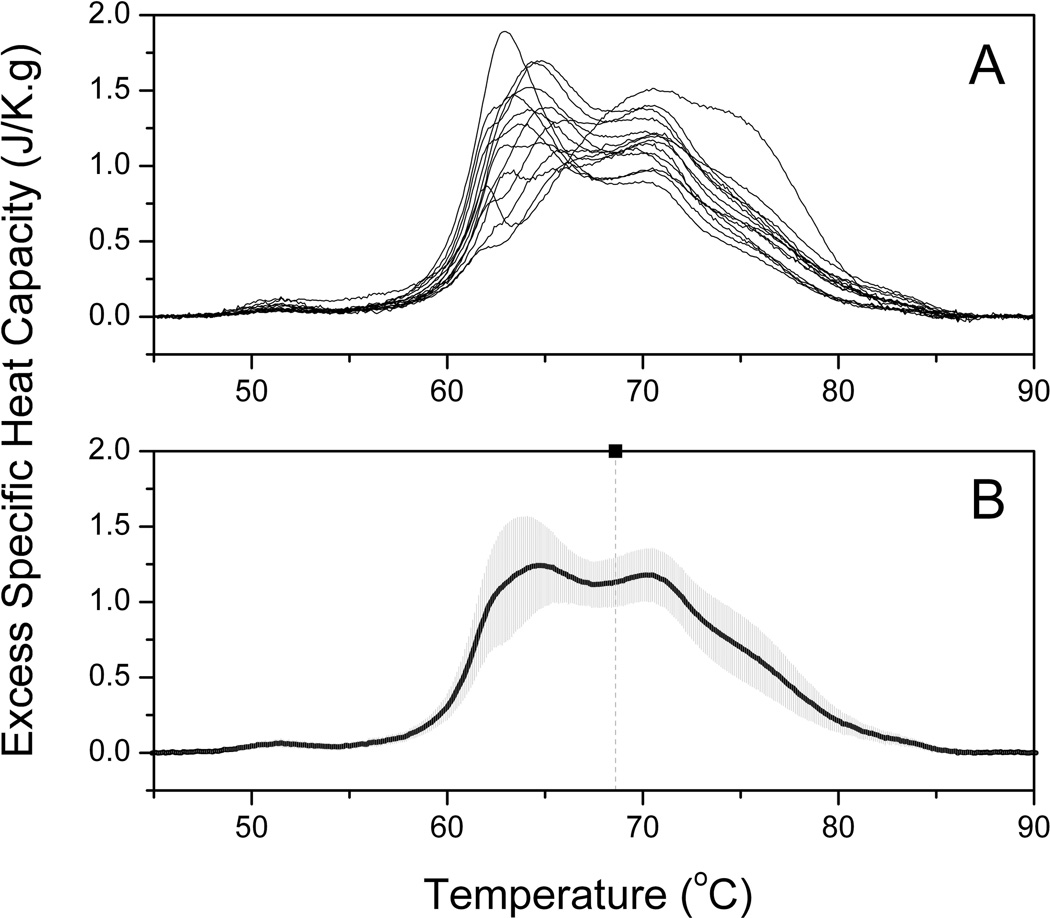
Plasma samples were analyzed in duplicate by DSC and grouped according to ERFD status. Four samples were excluded from further analysis where significant variance was observed between duplicate runs. A composite plot of thermograms for 15 individuals in the control group is shown by Panel A of Figure 1 and the average thermogram and standard deviation shown by Panel B. The standard deviation is modest and within acceptable clinical ranges for human plasma protein concentrations. A composite plot and average thermogram for 14 individuals in the ERFD group is shown by Figure 2. It is immediately apparent by simple inspection of the average control thermogram and the average ERFD thermogram that there are significant differences between these groups. The peak centered ~ 63 °C is diminished while that ~ 70 °C is increased and the entire thermogram is shifted to higher temperatures. While there are clear qualitative differences, a more quantitative comparison of the groups can be made using quantile-quantile plots [33].
Figure 1. Average DSC thermogram from individuals in the control group.
(A) Composite plot of 15 thermograms of plasma samples from 15 individuals in the control group. Each thermogram shown is the average of duplicate measurements.
(B) The solid black line is the average of the thermograms shown in Panel A. The shaded gray area is the standard deviation at each temperature. The vertical dashed line is the first moment of the thermogram at 68.1 °C (see Discussion).
Figure 2. Average DSC thermogram from individuals in the ERFD group.
(A) Composite plot of 14 thermograms of plasma samples from 14 individuals in the ERFD group. Each thermogram shown is the average of duplicate measurements.
(B) The solid black line is the average of the thermograms shown in Panel A. The shaded gray area is the standard deviation at each temperature. The vertical dashed line is the first moment of the thermogram at 68.6 °C (see Discussion).
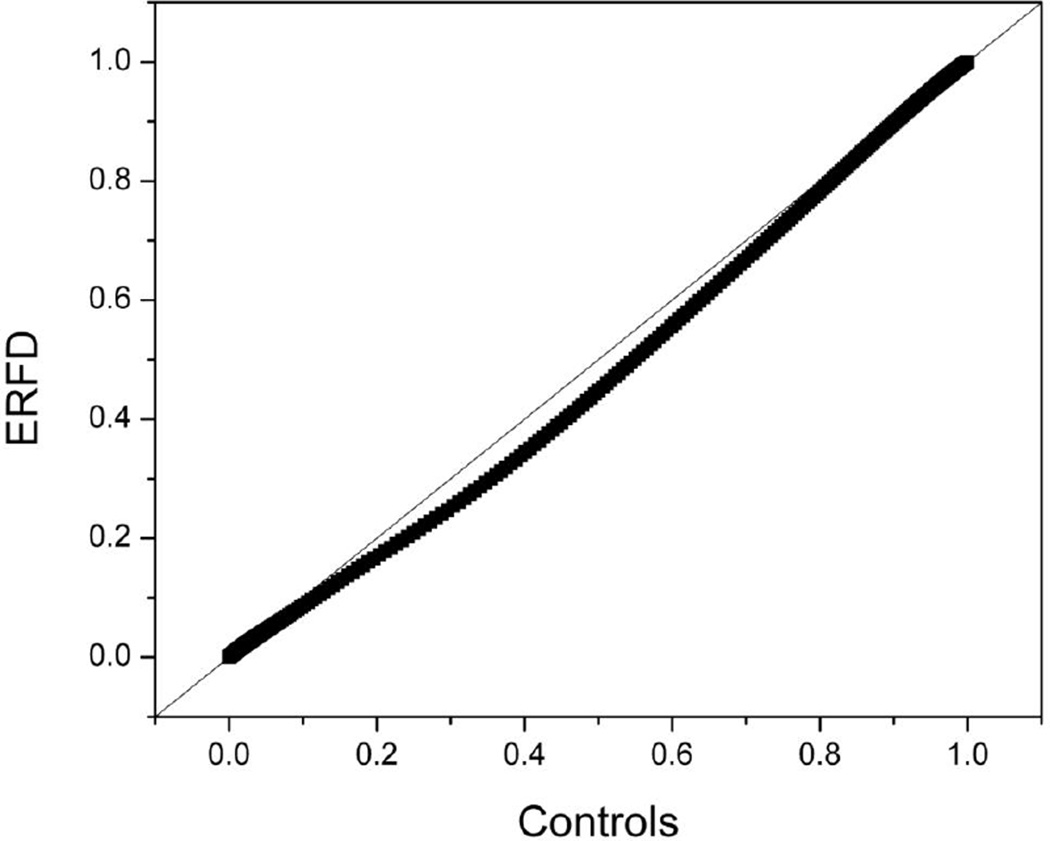
In a quantile-quantile plot, two normalized data sets are plotted against each other to discern if the two groups are different from each other. The thermograms for the two groups are normalized against a common temperature axis and then plotted in a pairwise fashion. The quantile-quantile plot for the control and ERFD groups is shown in Figure 3. If there is no difference between the groups, then the data will fall on the 45° reference line indicated in the plot. Figure 3 demonstrates there to be a clear deviation from this line, indicating that there are differences between the distributions. A quantile-quantile plot is a useful graphical representation that can potentially provide more insight into the nature of the difference than an overall statistical measure such as a P-value. For instance, the pairwise comparison of the control and ERFD data sets shows that over the 45 – 90 °C temperature range under consideration, there are only very small differences between the thermograms over the first ~ 13 % of the range (45 – 51 °C) and the final ~ 7 % of the range (87 – 90 °C). The difference between the thermograms then increases from 51 °C to a maximal difference ~ 40 % (63 °C) and then gradually decreases until no difference is discernible ~ 87 °C. This corresponds to the earlier qualitative observations.
Figure 3. Quantile-quantile plot comparing the control group thermogram and the ERFD group thermogram.
The 45° line shows indicates where the data should fall if there is no difference between the groups. The experimental data do not fall on this line, indicating that differences exist between the control and ERFD thermograms.
3.3 Correlation of thermogram features with renal function index
In order to further examine the nature of differences between the control and ERFD group thermograms, we examined a number of characteristic thermogram features and shape metrics: total peak area; peak height; peak width at half height; temperature of the peak maximum (Tmax); first moment temperature (TFM); excess specific heat capacity of the main transition (Cpex [63.8°C]); excess specific heat capacity of the second transition (Cpex [70.3°C]); and the ratio of the excess specific heat capacities of the main and second transitions (Cpex [63.8°C]/Cpex [70.3°C]). As a first analysis, thermogram features were calculated for each thermogram and associated with renal function index. The correlation of each parameter to the renal function index was assessed using Spearman’s correlation coefficient. The results shown by Table 1 indicate that specific thermogram features, Tmax, Cpex [63.8°C], Cpex [70.3°C] and Cpex [63.8°C]/Cpex [70.3°C] are significantly correlated with renal function index. However, parameters describing the entirety of the thermogram profile, peak area, height, width and TFM, showed no significant correlation. These results suggest that certain thermogram parameters could be used to distinguish between clinical groups.
Table 1.
Correlation of thermogram features to renal function index.
| Parameter | Spearman r | p-value |
|---|---|---|
| Area (J/g) | 0.1970 | 0.3056 |
| Width (°C) | −0.2911 | 0.1256 |
| Height (J/K․g) | 0.1946 | 0.3118 |
| Tmaxa (°C) | −0.5493 | 0.0020 |
| TFMb (°C) | −0.2498 | 0.1913 |
| Cpex [63.8°C]c (J/K․g) | 0.4645 | 0.0111 |
| Cpex [70.3°C]d (J/K․g) | −0.3793 | 0.0424 |
| Cpex [63.8°C] / Cpex [70.3°C]e | 0.5081 | 0.0049 |
Tmax, temperature of the peak maximum;
TFM, first moment temperature;
Cpex [63.8°C], excess specific heat capacity of the main transition;
Cpex [70.3°C], excess specific heat capacity of the second transition;
Cpex [63.8°C]/Cpex [70.3°C], ratio of the excess specific heat capacities of the main and second transitions
3.4 Discrimination between ERFD and control groups
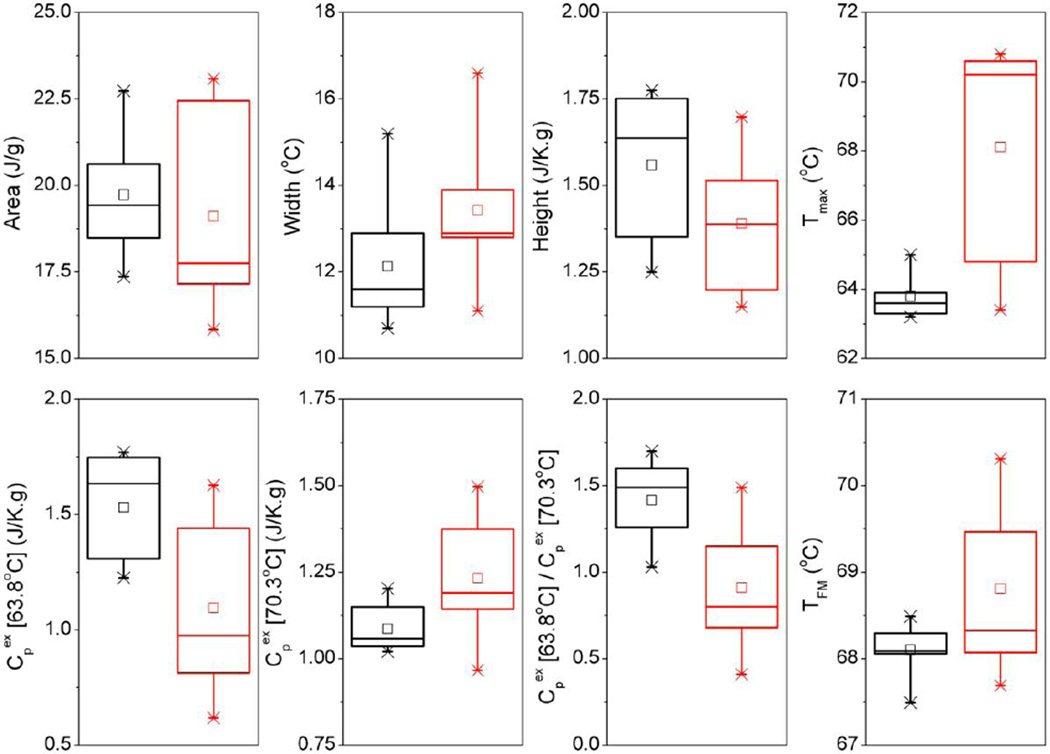
To assess the use of thermogram parameters to distinguish between disease categories, thermogram features were organized according to renal function index into tertiles with a comparison of the 1st and 3rd tertiles representing control patients and those with significant ERFD. The 2nd tertile comprising patients with renal function close to the discriminatory normal function cut-off value was excluded from analysis. Further, it was observed from Figures 1A and 2A that 2-quartile grouping according to the normal renal function cutoff value revealed significant variability in thermogram profiles in the ERFD group in contrast to the control group. Re-grouping into 3-quartiles thus served to reduce the thermogram variability and produce more robust classifications for controls and patients with significant ERFD. Thermogram feature values in the 1st and 3rd tertiles were examined using box plots (Figure 4) and differences between groups tested for significance using the nonparametric Mann-Whitney U test for unequal medians [34] (Table 2). Despite overlap of thermogram feature values between tertiles there were clear trends in most of these parameters. Thermogram area decreased for the ERFD group compared to the control group, characterized by an overall increase in width and decrease of height of the profile. Both Tmax and TFM increased in the ERFD group with significantly greater spread in the Tmax parameter that is reflective of subtle differences in the relative amplitudes of the two major transitions within the ERFD group. Peak heights decreased for the main transition at 63.8 °C and increased for the second transition at 70.3 °C. The ratio of these values was close to 1.5 for the control group and 1.0 for the ERFD group. These reflect significantly decreased main transition amplitude and slightly increased second transition height for the ERFD group compared to the control group. Statistical analyses using the nonparametric Mann-Whitney U test [34] for comparison of medians resulted in high significance of the observed differences for Tmax and all Cpex parameters; all other parameters were less significant. Use of certain characteristic thermogram feature metrics can therefore provide useful quantitative measures of changes in DSC profiles between clinical groups.
Figure 4. Box plots showing key thermogram feature parameters for the 1st (black) and 3rd (red) tertiles of renal function index.
Summary of statistical analyses of these parameters is shown by Table 2.
Table 2.
Comparison of thermogram features for the 1st and 3rd tertiles of renal function index.
| Parameter | 1st Tertile [Mean ± SD]f |
3rd Tertile [Mean ± SD]f |
1st Tertile [Median (LQ,UQ)]g |
3rd Tertile [Median(LQ,UQ)]g |
p-value [Median] |
|---|---|---|---|---|---|
| Area (J/g) | 19.73 ± 1.55 | 19.11 ± 2.75 | 19.82 (18.48,20.63) | 18.05 (17.16,22.45) | 0.3527 |
| Width (°C) | 12.1 ± 1.4 | 13.4 ± 1.5 | 11.8 (11.2,12.9) | 13.3 (12.8,13.9) | 0.0637 |
| Height (J/K․g) | 1.56 ± 0.20 | 1.39 ± 0.21 | 1.65 (1.35,1.75) | 1.39 (1.20,1.51) | 0.1431 |
| Tmaxa (°C) | 63.8 ± 0.5 | 68.1 ± 3.2 | 63.8 (63.3,63.9) | 70.4 (64.8,70.6) | 0.0024 |
| TFMb (°C) | 68.1 ± 0.3 | 68.8 ± 0.9 | 68.1 (68.1,68.3) | 68.7 (68.1,69.5) | 0.1431 |
| Cpex [63.8°C]c (J/K․g) | 1.53 ± 0.23 | 1.10 ± 0.37 | 1.64 (1.31,1.75) | 1.05 (0.81,1.44) | 0.0039 |
| Cpex [70.3°C]d (J/K․g) | 1.09 ± 0.06 | 1.23 ± 0.16 | 1.07 (1.04,1.15) | 1.20 (1.14,1.37) | 0.0232 |
| Cpex [63.8°C] / Cpex [70.3°C]e | 1.42 ± 0.23 | 0.91 ± 0.34 | 1.51 (1.26,1.60) | 0.88 (0.68,1.15) | 0.0032 |
Tmax, temperature of the peak maximum;
TFM, first moment temperature;
Cpex [63.8°C], excess specific heat capacity of the main transition;
Cpex [70.3°C], excess specific heat capacity of the second transition;
Cpex [63.8°C]/Cpex [70.3°C], ratio of the excess specific heat capacities of the main and second transitions;
mean value and standard deviation;
median value, lower quartile and upper quartile
4. DISCUSSION
4.1 Potential clinical utility of DSC proteome analysis
We have recently demonstrated the clinical utility of DSC analysis to study the plasma or serum proteome. The assay design has been described in greater detail in our earlier publications but briefly, small volumes of plasma or serum (50 – 100 µL) are dialyzed against a standard phosphate buffer, diluted 25-fold to yield a suitable working concentration and then directly analyzed by DSC. Our previous studies have defined a distinct and reproducible profile for healthy individuals defined by major peaks at 50.8, 62.8 and 69.8 °C and higher temperature shoulders and by a modest standard deviation [23]. The “normal” thermogram was shown to be represented by a composite of the denaturation behavior of the most abundant plasma proteins Comparison of the “normal” plasma profile with individual thermograms of the 16 most abundant plasma proteins allowed the identification of a number of peaks. The small peak at 50.8 °C corresponds to a transition of fibrinogen, the peak at 62.8 °C is dominated by the denaturation of human serum albumin (HSA) with a smaller contribution from haptoglobin, and the peak at 69.8 °C and higher temperature shoulders can largely be assigned to immunoglobulin G (IgG). We have demonstrated initial proof-of-principle of the clinical utility of DSC plasma profiles through comparing the “normal” thermogram with thermograms obtained from individuals with a number of different diseases. Disease thermograms are found to differ significantly from the “normal” profile and, more importantly, are characterized by unique and distinguishable thermograms. Differences in disease thermograms compared with “normal” correspond with redistribution in thermogram shape with a shift towards higher denaturation temperature. The origin of the thermogram differences is currently under investigation in our laboratories but it is thought not to arise from significant differences in the concentration of the major plasma proteins but from their thermal stabilization resulting from protein modifications or interactions with disease biomarkers, as described by the “interactome” hypothesis [20].
4.2 Application of DSC to profile T1D renal dysfunction
Having established initial clinical utility of DSC plasma analysis our goal in this study was to apply our approach to investigate the nature of the thermograms from subjects enrolled in the 1st Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes. The Joslin study is a unique collection of samples from T1D recruited in 1991 for a prospective study of renal function in these patients, in this way it presents an exceptional collection of specimens for the investigation of biomarker detection of T1D renal function. In this present study we have attempted to define if there are characteristic plasma thermograms that define T1D patients with MA who retain stable renal function from those diabetics with ERFD that develop progressive renal dysfunction. If such characteristic thermograms exist then the DSC assay presents an important clinical assay for the early detection of progressive renal damage in T1D.
Examination of the composite plot of thermograms of plasma from 15 control individuals (Figure 1A) reveals the thermograms to be similar in shape and position to the “normal” thermogram previously observed [23]. In comparison, the individual thermograms of the ERFD group show differences from the “normal” shape with a shift of the thermogram shape towards higher temperature (Figure 2A). The average thermograms from the control (Figure 1B) and ERFD (Figure 2B) groups are visibly different, with an attenuation of the main peak ~ 63 °C and an increase of the second peak ~ 70 °C. The entire thermogram is shifted to higher temperatures with an increase in first moment temperature from 68.1 °C for the control group to 68.6 °C for the ERFD group. These observations are similar to those described for other diseases [23]. The change in first moment temperature is modest but significant. Modulation of the thermogram profile is supported by the quantile-quantile plot (Figure 3). The quantile-quantile plot shows clear differences between the control and ERFD thermograms with the greatest difference corresponding to a decrease in magnitude of the peak ~ 63 °C and an increase in the magnitude of higher temperature peaks. This is consistent with previous observations in our laboratory and our current explanation is that this reflects thermal stabilization of abundant plasma proteins, such as HSA and IgG, through protein post-translational modification or binding of putative biomarker to and structural stabilization of the abundant plasma proteins. We and our collaborators [32, 35] have recently demonstrated using contemporaneous serum and plasma samples that the decline patient samples contain increased abundances of markers of protein oxidation and specifically increased abundances of modified and oxidized high molecular weight kininogen. Given the ability of kininogen to interact with other proteins [36] and its role in blood coagulation, our observations here are consistent more broadly with thermogram changes occurring via an “interactome” hypothesis.
Although the qualitative observations between the control and ERFD thermograms are similar to previous observations between “normal” and diseased thermograms, specific thermogram shapes are associated with different clinical groups. We have begun to develop quantitative approaches to discern thermogram changes which ultimately will allow for accurate clinical classification based on DSC analysis. We have shown here that characteristic thermogram feature metrics can be used to describe differences in profiles between control and ERFD patients. In this study, heat capacity values of the major transitions at 63.8 and 70.3 °C were significant discriminators of the patients. These thermogram parameters were demonstrated to be significant discriminators both through correlation with the rate of renal function decline as well as between discrete control and ERFD clinical groups. We have also recently described a combined shape and distance metric which allows for use of the entire thermogram profile for the purpose of thermogram classification [37]. This latter approach requires a well-defined reference sets for particular disease states to which test thermograms can be compared. While this was beyond the current study this approach could be of significant utility in a future, expanded study.
4.3 Conclusions
The DSC assay does not offer a direct method to identify putative biomarkers or protein modifications associated with early renal function decline but will report with high sensitivity on the alteration in properties of the abundant plasma proteins as a consequence of these disease-specific changes with application for clinical diagnostics. DSC profiles were significantly altered for ERFD, discernibly difference from controls, which suggests the potential utility of DSC proteome analysis in the further investigation of renal disease in T1D. Furthermore, it is important to recognize that the plasma samples analyzed in this study were those collected prospectively from patients in the Joslin study with subsequent determination of ultimate renal function status. The DSC approach may therefore also exhibit sensitivity to prognostic indicators of diabetic renal disease.
HIGHLIGHTS.
DSC profiles of control T1D patients were similar to an established healthy profile
Altered profiles were observed for T1D patients with renal dysfunction
DSC profile feature and shape metrics were correlated with renal function index
Heat capacity values of the major transitions were discriminators of patient status
DSC has potential sensitivity to prognostic indicators of diabetic renal disease
ACKNOWLEDGEMENTS
The authors would like to acknowledge and thank Dr. Andrzej S. Krolewski and the Joslin Diabetes Research Center, Harvard Medical School, Boston, MA for providing the clincial samples used in this study as well as clinical data collected in the Natural History of Type 1 Diabetes study funded through the NIDDK-NIH. This work was also supported by a subcontract awarded to JBC from National Cancer Institute grant R44 CA103437 and by a grant to JBC from the Elsa. U. Pardee Foundation and by NIH 5 R01 DK067638 02 (ASK and JBK) and DOE DE-FG02-05ER6406 (JBK and MLM). The authors want to thank Daniel J. Fish for helpful discussions and critical reading of the manuscript.
Abbreviations
- MA
microalbuminuria
- T1D
type 1 diabetes
- DSC
differential scanning calorimetry
- ERFD
early renal function decline
- eGFR
estimated glomerular filtration rate
- Tmax
temperature of the peak maximum
- TFM
first moment temperature
- Cpex
excess specific heat capacity
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE
NCG and JBC are co-inventors on patent applications describing the DSC plasma thermogram technology for which Louisville Bioscience, Inc. (LBI) holds an exclusive license from the University of Louisville. JBC is a founder and shareholder of LBI; NCG is a founder, shareholder and employee of LBI.
REFERENCES
- 1.Mogensen CE, Keane WF, Bennett PH, Jerums G, Parving HH, Passa P, Steffes MW, Striker GE, Viberti GC. Prevention of diabetic renal disease with special reference to microalbuminuria. Lancet. 1995;346:1080–1084. doi: 10.1016/s0140-6736(95)91747-0. [DOI] [PubMed] [Google Scholar]
- 2.Viberti GC, Jarrett RJ, Keen H. Microalbuminuria as prediction of nephropathy in diabetics. Lancet. 1982;2:611. doi: 10.1016/s0140-6736(82)90688-2. [DOI] [PubMed] [Google Scholar]
- 3.Giorgino F, Laviola L, Cavallo Perin P, Solnica B, Fuller J, Chaturvedi N. Factors associated with progression to macroalbuminuria in microalbuminuric Type 1 diabetic patients: the EURODIAB Prospective Complications Study. Diabetologia. 2004;47:1020–1028. doi: 10.1007/s00125-004-1413-8. [DOI] [PubMed] [Google Scholar]
- 4.Perkins BA, Ficociello LH, Silva KH, Finkelstein DM, Warram JH, Krolewski AS. Regression of microalbuminuria in type 1 diabetes. N.Engl.J.Med. 2003;348:2285–2293. doi: 10.1056/NEJMoa021835. [DOI] [PubMed] [Google Scholar]
- 5.Ficociello LH, Perkins BA, Silva KH, Finkelstein DM, Ignatowska-Switalska H, Gaciong Z, Cupples LA, Aschengrau A, Warram JH, Krolewski AS. Determinants of progression from microalbuminuria to proteinuria in patients who have type 1 diabetes and are treated with angiotensin-converting enzyme inhibitors. Clin J Am Soc Nephrol. 2007;2:461–469. doi: 10.2215/CJN.03691106. [DOI] [PubMed] [Google Scholar]
- 6.Perkins BA, Ficociello LH, Ostrander BE, Silva KH, Weinberg J, Warram JH, Krolewski AS. Microalbuminuria and the risk for early progressive renal function decline in type 1 diabetes. J Am Soc Nephrol. 2007;18:1353–1361. doi: 10.1681/ASN.2006080872. [DOI] [PubMed] [Google Scholar]
- 7.O'Connell TX, Horita TJ, Kasravi B. Understanding and interpreting serum protein electrophoresis. Am. Fam. Physician. 2005;71:105–112. [PubMed] [Google Scholar]
- 8.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 9.Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, Lobley A. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol. Cell. Proteomics. 2004;3:311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 11.Craig W, Ledue T, Ritchie R. Foundation for Blood Research. Scarborough, Maine: 2004. Plasma Proteins: Clinical Utility and Interpretation; p. 152. [Google Scholar]
- 12.Anderson L, Anderson NG. High resolution two-dimensional electrophoresis of human plasma proteins. Proc. Natl. Acad. Sci. U.S.A. 1977;74:5421–5425. doi: 10.1073/pnas.74.12.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson NL, Anderson NG. A two-dimensional gel database of human plasma proteins. Electrophoresis. 1991;12:883–906. doi: 10.1002/elps.1150121108. [DOI] [PubMed] [Google Scholar]
- 14.Adkins JN, Varnum SM, Auberry KJ, Moore RJ, Angell NH, Smith RD, Springer DL, Pounds JG. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell. Proteomics. 2002;1:947–955. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 15.Anderson NL. The roles of multiple proteomic platforms in a pipeline for new diagnostics. Mol. Cell. Proteomics. 2005;4:1441–1444. doi: 10.1074/mcp.I500001-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Gygi SP, Aebersold R. Mass spectrometry and proteomics. Curr. Opin. Chem. Biol. 2000;4:489–494. doi: 10.1016/s1367-5931(00)00121-6. [DOI] [PubMed] [Google Scholar]
- 17.Liotta LA, Kohn EC, Petricoin EF. Clinical proteomics: personalized molecular medicine. JAMA. 2001;286:2211–2214. doi: 10.1001/jama.286.18.2211. [DOI] [PubMed] [Google Scholar]
- 18.Yates I., JR Mass spectrometry. From genomics to proteomics. Trends Genet. 2000;16:5–8. doi: 10.1016/s0168-9525(99)01879-x. [DOI] [PubMed] [Google Scholar]
- 19.Liotta LA, Petricoin EF. Serum peptidome for cancer detection: spinning biologic trash into diagnostic gold. J. Clin. Invest. 2006;116:26–30. doi: 10.1172/JCI27467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou M, Lucas DA, Chan KC, Issaq HJ, Petricoin I, Liotta EFLA, Veenstra TD, Conrads TP. An investigation into the human serum "interactome". Electrophoresis. 2004;25:1289–1298. doi: 10.1002/elps.200405866. [DOI] [PubMed] [Google Scholar]
- 21.Diamandis EP. Validation of breast cancer biomarkers identified by mass spectrometry. Clin Chem. 2006;52:771–772. doi: 10.1373/clinchem.2005.064972. author reply 772. [DOI] [PubMed] [Google Scholar]
- 22.Garbett NC, Miller JJ, Jenson AB, Miller DM, Chaires JB. Interrogation of The Plasma Proteome with Differential Scanning Calorimetry. Clin. Chem. 2007;53:2012–2014. doi: 10.1373/clinchem.2007.091165. [DOI] [PubMed] [Google Scholar]
- 23.Garbett NC, Miller JJ, Jenson AB, Chaires JB. Calorimetry outside the box: a new window into the plasma proteome. Biophys. J. 2008;94:1377–1383. doi: 10.1529/biophysj.107.119453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garbett NC, Mekmaysy CS, Helm CW, Jenson AB, Chaires JB. Differential scanning calorimetry of blood plasma for clinical diagnosis and monitoring. Exp. Mol. Pathol. 2009;86:186–191. doi: 10.1016/j.yexmp.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Ferencz A, Fekecs T, Lőrinczy D. Differential Scanning Calorimetry, as a New Method to Monitor Human Plasma in Melanoma Patients with Regional Lymph Node or Distal Metastases. In: Xi Y, editor. Skin Cancer Overview, InTech. 2011. Available from: http://www.intechopen.com/books/skin-cancer-overview/differential-scanningcalorimetry-as-a-new-method-to-monitor-human-plasma-in-melanoma-patients-with- [Google Scholar]
- 26.Zapf I, Fekecs T, Ferencz A, Tizedes G, Pavlovics G, Kálmán E, Lőrinczy D. DSC analysis of human plasma in breast cancer patients. Thermochim. Acta. 2011;524:88–91. [Google Scholar]
- 27.Todinova S, Krumova S, Gartcheva L, Robeerst C, Taneva SG. Microcalorimetry of blood serum proteome: a modified interaction network in the multiple myeloma case. Anal. Chem. 2011;83:7992–7998. doi: 10.1021/ac202055m. [DOI] [PubMed] [Google Scholar]
- 28.Todinova S, Krumova S, Kurtev P, Dimitrov V, Djongov L, Dudunkov Z, Taneva SG. Calorimetry-based profiling of blood plasma from colorectal cancer patients. Biochim. Biophys. Acta. 2012;1820:1879–1885. doi: 10.1016/j.bbagen.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Michnik A, Drzazga Z, Michalik K, Barczyk A, Santura I, Sozańska E, Pierzchała W. Differential scanning calorimetry study of blood serum in chronic obstructive pulmonary disease. J. Therm. Anal. Calorim. 2010;102:57–60. [Google Scholar]
- 30.Chagovetz AA, Jensen RL, Recht L, Glantz M, Chagovetz AM. Preliminary use of differential scanning calorimetry of cerebrospinal fluid for the diagnosis of glioblastoma multiforme. J Neurooncol. 2011;105:499–506. doi: 10.1007/s11060-011-0630-5. [DOI] [PubMed] [Google Scholar]
- 31.Perkins BA, Ficociello LH, Silva KH, Finkelstein DM, Warram JH, Krolewski AS. Regression of microalbuminuria in type 1 diabetes. N Engl J Med. 2003;348:2285–2293. doi: 10.1056/NEJMoa021835. [DOI] [PubMed] [Google Scholar]
- 32.Merchant ML, Niewczas MA, Ficociello LH, Lukenbill JA, Wilkey DW, Li M, Khundmiri SJ, Warram JH, Krolewski AS, Klein JB. Plasma kininogen and kininogen fragments are biomarkers of progressive renal decline in type 1 diabetes. Kidney Int. 2013 doi: 10.1038/ki.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lodder RA, Hieftje GM. Quantile analysis: a method fo characterizing data distributions. Appl. Spectrosc. 1988;42:1512–1520. [Google Scholar]
- 34.Conover WJ. Practical Nonparametric Statistics. Third Edition ed. New York: Wiley; 1999. [Google Scholar]
- 35.Perkins BA, Rabbani N, Weston A, Ficociello LH, Adaikalakoteswari A, Niewczas M, Warram J, Krolewski AS, Thornalley P. Serum levels of advanced glycation endproducts and other markers of protein damage in early diabetic nephropathy in type 1 diabetes. PLoS ONE. 2012;7:e35655. doi: 10.1371/journal.pone.0035655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ben Nasr AB, Herwald H, Muller-Esterl W, Björck L. Human kininogens interact with M protein, a bacterial surface protein and virulence determinant. Biochem J. 1995;305(Pt 1):173–180. doi: 10.1042/bj3050173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fish DJ, Brewood GP, Kim JS, Garbett NC, Chaires JB, Benight AS. Statistical analysis of plasma thermograms measured by differential scanning calorimetry. Biophys. Chem. 2010;152:184–190. doi: 10.1016/j.bpc.2010.09.007. [DOI] [PubMed] [Google Scholar]