Abstract
Over the past decade, the role of the ubiquitin–proteasome system (UPS) has been the subject of numerous studies to elucidate its role in cardiovascular physiology and pathophysiology. There have been many advances in this field including the use of proteomics to achieve a better understanding of how the cardiac proteasome is regulated. Moreover, improved methods for the assessment of UPS function and the development of genetic models to study the role of the UPS have led to the realization that often the function of this system deviates from the norm in many cardiovascular pathologies. Hence, dysfunction has been described in atherosclerosis, familial cardiac proteinopathies, idiopathic dilated cardiomyopathies, and myocardial ischemia. This has led to numerous studies of the ubiquitin protein (E3) ligases and their roles in cardiac physiology and pathophysiology. This has also led to the controversial proposition of treating atherosclerosis, cardiac hypertrophy, and myocardial ischemia with proteasome inhibitors. Furthering our knowledge of this system may help in the development of new UPS-based therapeutic modalities for mitigation of cardiovascular disease.
Keywords: Ubiquitin–proteasome system, Heart, Vascular, Atherosclerosis, Ubiquitin protein ligases, Protein quality control, Heart failure, Cardiomyopathy, Myocardial ischemia, Preconditioning
I. Introduction
The ubiquitin-proteasome system (UPS) is the major nonlysosomal pathway for intracellular degradation of proteins and plays a major role in regulating many cellular processes. Many of these processes have already been or will be described in the accompanying chapters. For the most part, the structure of the 26S proteasome, the immunoproteasome, and the 11S-activated proteasome have been described in detail in earlier chapters of this book. To avoid overlap, the sections in this chapter deal exclusively with UPS function and regulation as they pertain to the cardiovascular system. The chapter first examines regulation of the cardiac proteasome followed by an in-depth discussion of the role of the UPS and the controversial use of proteasome inhibitors in atherosclerosis. A description of the ubiquitin protein (E3) ligases follows with a discussion of their potential roles in cardiac physiology and pathophysiology. This leads up to the entire issue of protein quality control (PQC) in the cardiovascular system and how the UPS functions and malfunctions in both familial proteinopathies and idiopathic dilated cardiomyopathies (heart failure). The chapter ends with a discussion of proteasome dysfunction in myocardial ischemia, the potential role of the UPS in ischemic preconditioning (IPC), and a revisit of the controversial proposed use of proteasome inhibitors to treat some aspects of ischemia.
II. Regulation of the Cardiac Proteasome
Given the importance of the UPS in cardiovascular physiology and pathology, it was only natural that the cardiac proteasome would attract the attention of one of the premier proteomics groups who have conducted an extensive characterization to provide some clues to the regulation of proteasome function. The following section is a discussion of the regulation of the UPS as it applies to the cardiac proteasome although it may be applicable to proteasomes in other cell types as well.
A. Heterogeneity as a Basis for Regulation
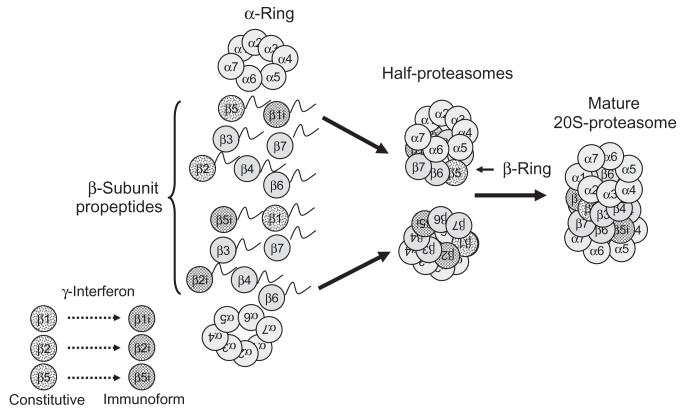
Prior to 2005, the prevailing hypothesis was that both β rings of the 20S proteasome were homologous to each other, that is, both rings had the same distribution of constitutive versus immunoforms of β1, β2, or β5 subunits. Since then, studies from the Ping group at UCLA have revealed a rather surprising heterogeneity in at least the β rings.1-4 These investigators have shown that the total cardiac 20S proteasome population can consist of multiple subpopulations containing different proportions of the catalytic β subunits, that is, the β1, β2, and β5 subunits may be replaced by the immunoform on one β ring, but not the other, or perhaps even a mixture of immunoform versus nonimmunoform subunits. As predicted, the different subpopulations of the 20S proteasome exhibit unique proteolytic activities, and specificity and selectivity for various substrates, suggesting a role in regulation of intracellular proteolysis.3-5 A potential scheme illustrating how proteasomes may be regulated by heterogeneous configurations is presented in Fig. 1. In regard to the 19S regulatory particle, alternate splicing of Rpn10 has been reported2 and a recent study describes isolation of two distinct populations of the 19S regulatory particle from the murine heart.7 The presence of multiple proteasome subpopulations suggests the potential for directing assembly to effect selective degradation (or not) of a protein or class of proteins, possibly altering the outcome of a disease process.
Fig. 1.
Regulation of 20S proteasomes by assembly of heterogeneous configurations. Under the influence of γ-interferon, select β-subunits are replaced by their immunoforms, which are directed by the complete α-rings to form half-proteasomes with specific configurations. In the example shown earlier, the top half-proteasome has constitutive β2 and β5 subunits and the β1 subunit has been replaced by the immunoform. The bottom half-proteasome has a constitutive β1 subunit, but the β2 and β5 subunits have been replaced by their immunoforms. These two distinctly different half-proteasomes mate to form a mature 20S proteasome that has a heterogeneous configuration. For the sake of clarity, the involvement of the chaperones PAC1, PAC2, PAC3, and PAC4, and the maturation factor POMP, are not depicted here. Modified and reproduced from Ref. 6 with the permission of Walters Kluwer Health.
B. Posttranslational Modification and Associating Partners as a Basis for Regulation
These two topics are intimately related and are discussed together. Often, as is the case with phosphorylation, it is the associating partner of the proteasome that results in the posttranslational modification. Studies have suggested a dynamic link between the activity of the proteasome and metabolic demand as glycosylation of the 19S regulatory particle ATPase, Rpt2, can inhibit ATPase activity and shut down proteolytic activity.8 Phosphorylation of the 19S regulatory particle ATPase, Rpt6, at ser120 by cAMP-dependent protein kinase A (PKA) is associated with an increase in proteasome proteolytic activities. Dephosphorylation by the serine threonine protein phosphatase, PP1γ, reverses this.9 Taken together, these findings suggest that hormones using the PKA system can act in concert with glucose metabolism to regulate proteasome function.
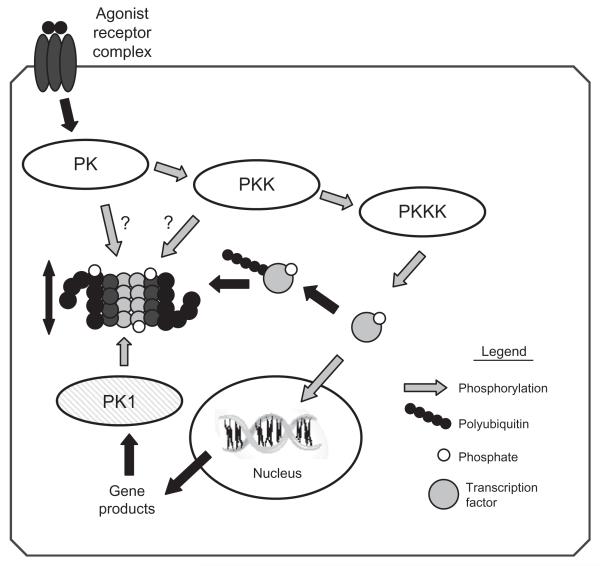
In regard to the heart, Ping and coworkers have published a series of studies that have characterized both the associating partners and posttranslational modifications of the murine cardiac proteasome and have provided much of the evidence linking PKA-mediated phosphorylation of multiple proteasome subunits with changes in proteolytic activity.2,7,10-13 Multiple posttranslational modifications have been described, including N-terminal acetylation of Rpn1, Rpn5, Rpn6, Rpt3, and Rpt6 of the 19S regulatory particle, and α2, α5, α7, β3, and β4 subunits of the 20S proteasome; and myristolation of Rpt2. In addition, phosphorylation of the α1, α2, α3, α6, α7, β2, and β5 subunits was described and linked to the previously unknown proteasome-associating partner, PKA. Incubation with PKA increased all three proteolytic activities, which was reversed by incubation with the serine threonine protein phosphatase, PP2A.2,11-13 Taken as a whole, these studies suggest that cAMP-dependent PKA plays a central role in regulating proteasome function and activity, which may be altered in the development of certain cardiac diseases, such as hypertrophy.10 Based largely on these studies, it was proposed that the cardiac proteasome may actually be regulated by multiple kinases and may be a component of signal transduction pathways6 (Fig. 2 presents one possible generic scenario).
Fig. 2.
Regulation of proteasome activity by kinase cascades. In this scenario, binding of an agonist with a cell surface receptor initiates a phosphorylation cascade with an end effector protein kinase kinase kinase (PKKK) phosphorylating some transcription factor which can then enter the nucleus and interact with DNA resulting in gene transcription. Phosphorylation of signaling intermediates, in this case the transcription factor, is often a signal for ubiquitination and targeting to the 26S proteasome for degradation. To amplify or dampen the effect of the signaling intermediate, any one of the protein kinases within the cascade might phosphorylate proteasome subunits resulting in increased or decreased peptidase activity, which would lessen or enhance availability of the signaling molecule accordingly. Another possibility would be for the gene product to activate a different protein kinase (PK1), which would phosphorylate proteasome, resulting in activation and thus decreased availability of the transcription factor, essentially a form of feedback control. Modified and reproduced from Ref. 6 with the permission of Walters Kluwer Health.
For the most part, the cardiac 19S regulatory particle remains a mystery. However, as pointed out earlier, a recent study has reported isolation of two distinct populations of cardiac 19S regulatory particles, one of which associates with heat shock protein 90 (HSP90) and has a decreased functional potency.7 Inhibition of the associating HSP90 led to an enhanced activating potential of this subpopulation of 19S regulatory particles on 20S proteasomes. While much work needs to be done on regulation of both the 20S proteasome and 19S regulatory particle, as well as other regulatory particles, it is becoming clearer that the cardiac proteasome is a dynamic structure under dynamic control, which probably allows it to respond to rapid changes in cardiac metabolism.
III. Involvement of the UPS in atherosclerosis
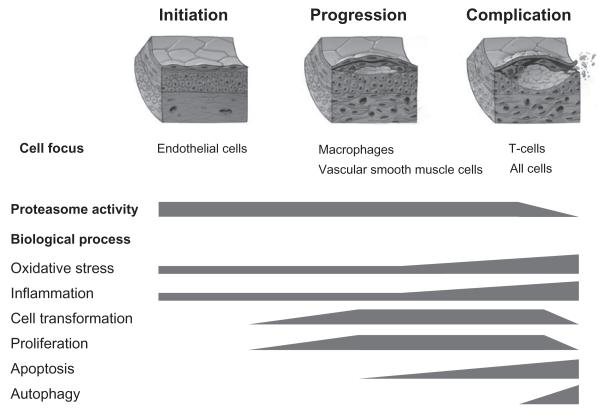
Beyond rhetoric, one may therefore wonder about an involvement of the leading cellular degenerative machinery in the leading human degenerative disease process. Over the past decade, a number of studies have explored this question and an integrative view is provided in this section. As the influence of the UPS varies by disease stage, its role in atherosclerosis is reviewed accordingly (Fig. 3).
Fig. 3.
Focus overview of the role of the ubiquitin–proteasome system in atherosclerosis. In general, the ubiquitin system is less vulnerable to changes and its activity remains preserved throughout the stages of atherosclerosis. On the contrary, proteasome activity seems to be increased in the initiation and progression stage to meet the increased substrate demand generated by oxidative stress, heightened metabolic activity, and cell transformation. Accumulation of oxidative stress and aging of the system likely lead to a decline in proteasome activity, which is notable in the complication stage along with apoptosis and autophagy.
A. The UPS in the Initiation Stage of Atherosclerosis
It has been said that atherosclerosis is a disease of the intima of large to medium-sized arteries with hardening plaque formation. While it has now been realized that the entire arterial wall undergoes remodeling, traditional teaching still holds true in the sense that the disease process starts with the lining of the intima, that is, the endothelium. Endothelial dysfunction and activation presents the first and still reversible event in the atherosclerotic sequence.14,15 On a molecular level, this disease stage is closely related to reduced bioavailability of nitric oxide (NO), which can be due to a reduction of NO production and/or an increase in NO consumption. Over the last few years, exciting discoveries have been made concerning the influence of the UPS, both under normal and pathological conditions, on the activity of the enzyme that generates NO in the vascular wall: endothelial NO synthase (eNOS).
Under normal conditions, eNOS is kept in an inactive state at the cell membrane by interaction with caveolin-1. Importantly, the posttranscriptional level of eNOS and hence the relative ratio to caveolin-1 is influenced by the proteasome and possibly favorably altered by proteasome inhibition.16 In addition to release from caveolin-1 and dimerization, normal eNOS activity requires Akt-induced phosphorylation, particularly of its serine residue 1179, and dephosphorylation of this residue by protein phosphatase 2A leads to eNOS inactivation.17 Interestingly, both Akt and protein phosphatase 2A are influenced in their activity by the UPS but seemingly in a different manner. While ubiquitination of Akt leads to its degradation by the proteasome complex, ubiquitination of protein phosphatase 2A leads to its translocation from the cytosol to the cell membrane, where it can then interact with eNOS.17 These differences also highlight the various regulatory influences of the ubiquitin system on intracellular proteins.18 NO generated by eNOS exerts a broad range of effects within endothelial and neighboring cells. On the level of the vascular smooth muscle cells (VSMCs) of the media, NO binds and activates soluble guanyl cyclase (sGC). This enzyme then generates cGMP from GTP, which mediates VSMC relaxation by various mechanisms. Importantly, cellular levels of sGC are negatively controlled by proteasomal degradation following ubiquitination by the co-chaperone/ubiquitin ligase carboxyl terminus of Hsc70 interacting protein (CHIP).19 For this reason, the effect of proteasome inhibition on protein phosphatase 2A can prevail over other outlined effects, leading to a decrease in eNOS activity on endothelial cell level, whereas the overall scenario might be different when testing organs or organisms.16,17
Cardiovascular risk factors can downregulate eNOS activity and efficacy quite potently. One key mechanism is depletion of tetrahydro-l-biopterin (BH4), which serves as an essential cofactor for normal eNOS function. Very intriguingly, under hyperglycemic conditions, BH4 can become depleted by UPS-dependent degradation of guanosine 5′-triphosphate cyclohydrolase I.20 Oxidative stress products such as 4-hydroxynonenal (4-HNE) generated in the setting of exposure to hypercholesterolemia or hypertension can accomplish the same.21 In the absence of BH4, eNOS generates superoxide anions instead of NO (so-called eNOS uncoupling).22 By this mechanism, the bioavailability of NO can be reduced quite quickly as NO is no longer produced and any remaining NO is consumed by superoxide anions to generate the potent cytotoxin, peroxynitrite. While superoxide anions are the epitome of oxidative stress in the arterial wall, a number of reactive oxygen and nitrate species are generated and insufficiencies of antioxidant defense mechanisms have to be considered as well. One of these involves nuclear erythroid 2-related factor 2 (Nrf2). As long as the level of oxidative stress is low, this transcription factor is marked for proteasomal degradation by constitutive ubiquitination, which involves Keap-1 as a substrate adaptor for a Cul3-dependent E3 ubiquitin ligase complex.23 Under circumstances of increased cellular oxidative stress, Keap-1 itself undergoes ubiquitination and proteasomal degradation, which stabilizes Nrf2 then on and allows it to bind to genome sequences with an antioxidant response element. This leads to the expression of genes encoding for proteins which have been linked to the amelioration of oxidative stress. Of note, Nrf2 also leads to an upregulation of proteasome subunits, and their overexpression via the Nrf2 pathway has been found to increase cellular resistance even against toxic misfolded proteins.24 Likewise, proteasome inhibitors can “shield” against oxidative injury in an Nrf2-dependent manner.25 However, this potential is not infinite and, at higher dosages, proteasome inhibitors can in fact worsen cellular oxidative stress.26 Vice versa, a dual effect on proteasome function has been shown for oxidative stress products such as oxLDL and HNE.27,28
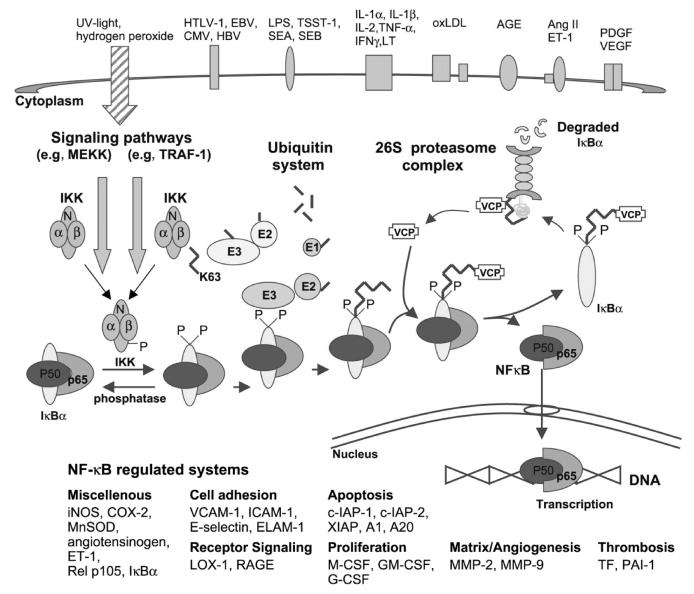
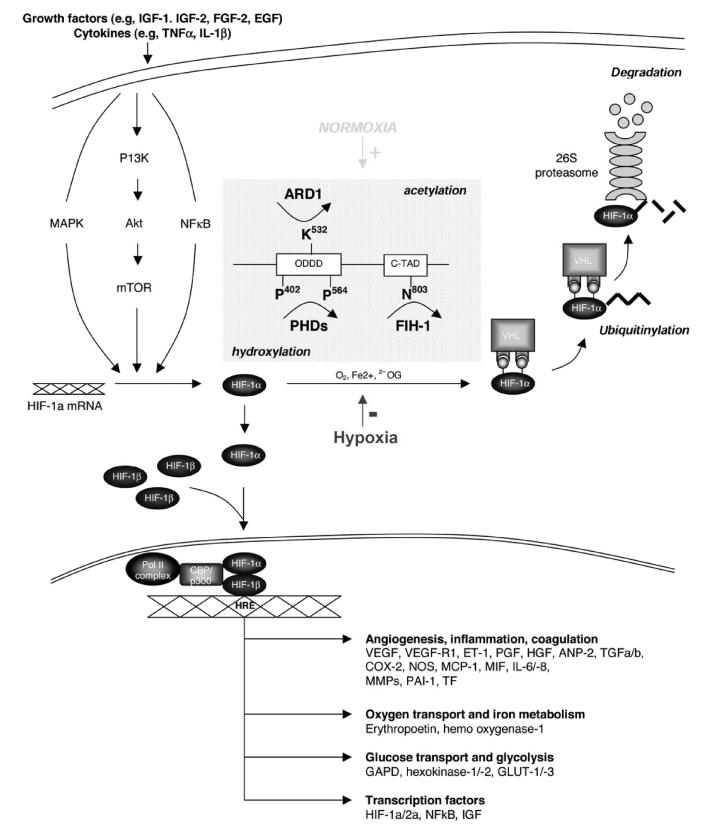
Interestingly, there are two other transcription factors whose activation pathways are stimulated by elevated intracellular levels of oxidative stress with involvement of the UPS.29 The first one is nuclear factor kappa-B (NFκB), and gene sequences controlled by NFκB include those that encode for vascular cell adhesion molecule-1, E-selectin, and monocyte chemoattractant protein-1 (Fig. 4).30 These molecules lead to the attraction and accumulation of inflammatory cells to and in the arterial wall, and hence NFκB is a pivotal factor for the development of atherosclerosis as an inflammatory disease from its early stages onward. NFκB also adds to the cell proliferation aspects of atherosclerosis, for instance, by regulating the expression of endothelin-1. The second transcription factor to be mentioned in this context is hypoxia-inducible factor (HIF)-1α (Fig. 5).31,32 Importantly, HIF-1α adds to and stimulates NFκB activity but remains best known for plaque facilitating vasa vasorum and then neovascularization.
Fig. 4.
The classical example of NFκB is the heterodimer of p50 and p65 and the classical activation pathway entails the release from its association with an inhibitory molecule of the IκB family. Upon exposure of the cell to various stimuli such as increase in oxidative stress, two specific serine residues are rapidly phosphorylated by the IKK1/2 kinases, whose activity is coupled to upstream signaling cascades by their association with a third protein called NFκB essential modulator (NEMO). Once phosphorylated, IκBs undergo degradation via the ubiquitin–proteasome pathway in the main route of NFκB activation. This is mediated by initial recognition of phosphorylated IκBs by the β-TrCP component of the ubiquitin ligase complex Skp1/Cul1/ROC1/F-box protein FWD 1 and subsequent covalent attachment of multiple moities of ubiquitin to the IκBs. The multi-ubiquitin chain allows subsequent association with the amino-terminal domain of valosin-containing protein (VCP), and thereby the targeting of IκBs to the 26S proteasome and the translocation of the transcriptionally active dimers to the nucleus. Worth noticing is the fact that the very same IKK/E3 combination is also involved in the generation of p50 (NFκB1) from its precursor molecule p105 by limited proteolysis, adding to its NFκB activating function. Modified and reproduced with permission from the European Society of Cardiology.30
Fig. 5.
Illustration of the hypoxia-inducible factor (HIF) activation pathway, highlighting hydroxylation and acetylation of HIF-1α depending on cellular oxygen tension, that is, normoxia and hypoxia. Hydroxylation of the proline residues 402 and 564 of HIF-1α allows recognition by the ubiquitin system, and thereby labeling for degradation by the proteasome complex. Modified and reproduced with permission from the American Heart Association.31
B. The UPS and Progression Stage of Atherosclerosis
The dysfunctional endothelium with reduced barrier function allows cholesterol, especially in the form of so-called low-density lipoprotein (LDL) particles, to enter the subintimal space. In the environment marked by oxidative stress, these particles then undergo various modifications that lead to their entrapment and uptake by macrophages. An imbalance of cholesterol uptake and efflux along with suppression of apoptosis leads to the conversion of macrophages to foam cells. The expression of CD36, a member of the family of scavenger receptors that mediate the uptake of modified LDL particles, is regulated by protein kinase C alpha (PKCα). Importantly, ubiquitination of PKCα leads to the downregulation of this enzyme, CD36, cholesterol accumulation, and development of atherosclerosis.33 On the other side of the input–output equation, cholesterol efflux from macrophages depends on ATP-binding cassette transporter molecules (such as ABCA1 and G1), and their expression level is likewise negatively controlled by the UPS.34 As such, inhibition of the UPS can yield two opposing effects: increase of cholesterol uptake via CD36 and increase in reverse cholesterol transport via ABCA1 and G1. A pathway by which the UPS may contribute to foam cell maintenance is via adipose differentiation-related protein. This protein is associated with lipid droplets in various cell types including foam cells, and a functional UPS is required for the regression of these cells.35 Finally, and in fact the very first link between the UPS and foam cells was provided by the observation that aggregated LDL particles stimulate the expression of a ubiquitin-conjugating enzyme that mediates the ubiquitination and degradation of p53, and thereby facilitates the suppression of apoptosis of lipid-bearing macrophages contributing to foam cell formation.36
As yet another reflection of the evolving disease process, VSMCs invade the subendothelial space from the media and transform into a proliferating phenotype. They also become metabolically active and generate the collagen-rich matrix that eventually overlies an atheromatous lipid pool. This transformation of smooth muscle cells involves extensive intracellular remolding and is prevented by proteasome inhibitors.30 Myocardin has been identified as one specific molecular mediator of this process. This transcriptional coactivator controls the expression of genes for a contractile smooth muscle cell phenotype but is downregulated by CHIP-mediated ubiquitination and proteasomal degradation.37 Likely mediated by upregulation of the p21 cyclin-dependent kinase inhibitor, proteasome inhibitors also inhibit migration and proliferation of VSMCs.38,39 A shift in the balance in favor of proapoptotic molecules may also contribute to the antiproliferation effects of proteasome inhibitors.39,40 Under normal conditions, NO may recapture these effects by inhibitory S-nitrosylation of proteolytic core proteins and modification of proteasome composition.41 Furthermore, NO inhibits activation of NFκB in VSMCs by interfering with phosphorylation and thereby ubiquitination and proteasomal degradation of IκBα.42 Importantly, the activation sequence of NFκB as a pivotal contributor to the proliferative response of the arterial wall can also be modified by deubiquitinating enzymes. Ubiquitin C-terminal hydroxylase L1 is a prominent example, expressed not only in endothelial cells but also especially in neointimal VSMCs in an effort to downregulate NFκB activation by IκBα deubiquitination.43 Cylindromatosis, another deubiquitinating enzyme expressed strongly in VSMCs, accomplishes the same by acting on TRAF2, a central molecule in the intracellular tumor necrosis factor (TNF)-α-receptor signaling cascade upstream from IκBs.44
C. The UPS in the Complication Stage of Atherosclerosis
The developing atherosclerotic lesions can impair blood flow to the extent that it does not meet the demand of an end organ, leading to recurrent episodes of ischemia. While this in itself has clinical implications, the greater concern relates to acute (near-) closure of arteries as a consequence of acute plaque rupture causing acute ischemic events in various vascular territories. In a simplified equation, the vulnerability of a plaque toward this complication stage is determined by the relative content of collagen, produced by transformed VSMCs, and the volume of the lipid pool, generated by degenerating macrophages. Interestingly enough, the ratio of fibrous cap to necrotic core is influenced by inflammation and neovascularization at their very shoulder points.
T cells are of utmost importance for these tissue dynamics, and the UPS is, for one, uniquely involved in the regulation of NFκB activity in these cells.15 Upon T cell stimulation, covalent attachment of regulatory ubiquitin chains to regulatory protein Malt1 allows interaction with the IκB kinase (IKK) complex and its paracaspase activity cleaves A20, which catalyzes the removal of K63-linked ubiquitin and its paracaspase activity cleaves A20, which catalyzes the removal of K63-chains from Malt1. As such, the balance between these two proteins regulates the strength and duration of the IKK/NFκB response upon T cell receptor (TCR)/CD28 costimulation.45 The significance of this activation mode for atherosclerotic cardiovascular disease has been questioned by the fact that atherosclerotic plaque T cells often lack the costimulatory receptor CD28.46 However, the significance of A20 for the attenuation of atherosclerosis has been confirmed in diabetic mice even though this was not specifically related to the TCR.47 Another negative feedback loop in T cells involves Bcl10, which promotes activation of the IKK complex that then will phosphorylate Bcl10 after T cell antigen receptor stimulation and causes its proteolysis via the beta-TrCP ubiquitin ligase/proteasome pathway.48
T cell activity is also potently regulated by proteasome-mediated activation and nuclear translocation of the nuclear factor of activated T cells (NFAT). This transcription factor controls the expression of the activation-associated cell surface receptors CD25, CD28, CD120b, and CD134 as well as production of interferon (IFN)-γ, TNF-α, and interleukin (IL)-4 and IL-5.49 Furthermore, T cells can be arrested in the G(1) phase by accumulation of cyclin-dependent kinase inhibitors p21(WAF1/CIP1) and p27(KIP1) and the disappearance of cyclin A, cyclin D2, and proliferating cell nuclear antigen.49 These observations were made in CD4(+) T cells activated by dendritic cells (DCs), which is very relevant as DCs are particularly prevalent in the neovascularization-rich shoulder areas of complicated and symptomatic atherosclerotic plaques.50 Importantly, the proteasome is crucial for the maturation and function of DCs. As a consequence, DCs with impaired proteasome function fail to stimulate allogeneic CD4(+) and CD8(+) T cells and autologous CD4(+) T cells sufficiently enough to regulate innate and adaptive immunity.51,52 Moreover, inhibition of proteasome function also induces apoptosis of DCs via the mitochondrial pathway.53 Similarly, with prolonged inhibition of proteasome function, T cells undergo apoptosis via the mitochondrial p53 pathway.54,55 Of utmost significance, oxLDL, highly prevalent in human atherosclerotic plaques, can induce apoptosis of CD4+/CD25+ regulatory T cells in a time- and concentration-dependent manner.56 Apoptosis of this regulatory T cell population removes a vital stabilizing force from the atherosclerotic plaque as these cells downregulate T cell responses to foreign and self-antigens.57
Recently, the immunoproteasome was identified as a potential link between inflammation and apoptosis of plaque cells.58 In the presence of IFN-γ, fibrous cap VSMCs are sensitized to apoptosis via the Fas/Fas ligand pathway, which is prevented by interference with the inducible β5 subunit of the immunoproteasome. As it was subsequently realized, a switch to the immunoproteasome allowed proteolytic processing of myeloid cell leukemia (Mcl)-1, thereby removing a potent inhibitor (sequester) of proapoptotic molecules.59 Intriguingly, this mode of regulation is in addition to the possible degradation of Mcl-1 via the conventional UPS.60 These observations indicate that even subtle nuances in proteasome composition and activity may be important for the phenotype of biological and pathobiological processes.
In addition to apoptosis, autophagic cell death has been reported in atherosclerotic plaques with potentially detrimental consequences.61 Autophagy is activated under circumstances of impaired proteasome function likely as a consequence of endoplasmic reticulum (ER) stress and as a compensatory effort to remove polyubiquitinated protein aggregates.62 However, a number of other mechanisms can induce autophagy in the atherosclerotic plaque, not related to impairment of proteasome function.61 Furthermore, autophagy can also have cytoprotective effects and the detrimental effects prevail only with excessive stimulation of autophagic activity.61
D. Proteasome Inhibition and Atherosclerosis: For Better or for Worse?
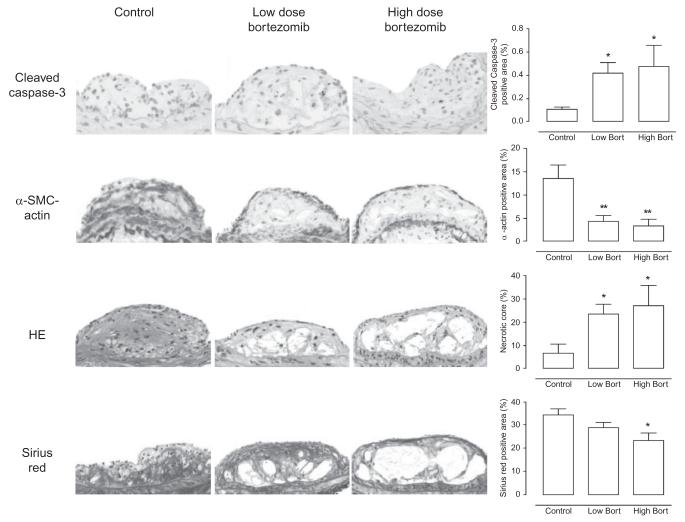
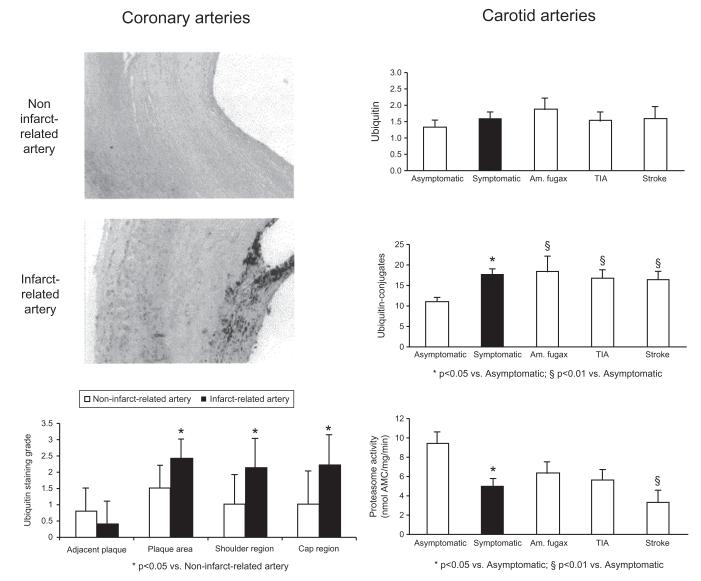
Utilizing a murine carotid collar of atherosclerosis, a 4-week treatment course with the proteasome inhibitor bortezomib, given once every 3 days at either a low- or high-dose regimen (corresponding to nearly 40% and 60% proteasome inhibition in the aorta, respectively), did not influence the composition of early plaques but led to a significant increase in necrotic core area and apoptotic cells as well as a decrease in VSMCs and collagen content in advanced atherosclerotic plaques (Fig. 6).63 Most intriguingly, compared with carotid artery plaques from patients who did not have symptoms of focal cerebral ischemia within 6 months of carotid endarterectomy, plaques from symptomatic patients were characterized by lower collagen content and increased rate of apoptosis.64 In correlation with the latter, the level of ubiquitination was higher and the level of proteasome activity was lower in symptomatic plaques (Fig. 7). Demonstrating that proteasome function can intrinsically decline in atherosclerosis with the potential to propagate plaque dynamics toward an unstable phenotype, these observations provided further insight into the functional significance of the original observation of elevated ubiquitin levels in advanced and complicated coronary artery plaques of patients with acute myocardial infarction (Fig. 8).66 It has to be acknowledged though that some studies demonstrated an increase in proteasome activity in high-risk plaques, macrophages extracted from these plaques, and even peripheral monocytes.67-69 These contradictory results may be explained by differences in the composition of the study population, as subsequent studies did indicate that proteasome activity was lower in atherosclerotic plaques of patients older than 60 years of age.70 Intriguingly, an age-dependent decrease of proteasome function has been shown in other organs including the heart.71,72 Hence, a decline in proteasome function may become a prevailing phenomenon over time with the consequences outlined above.
Fig. 6.
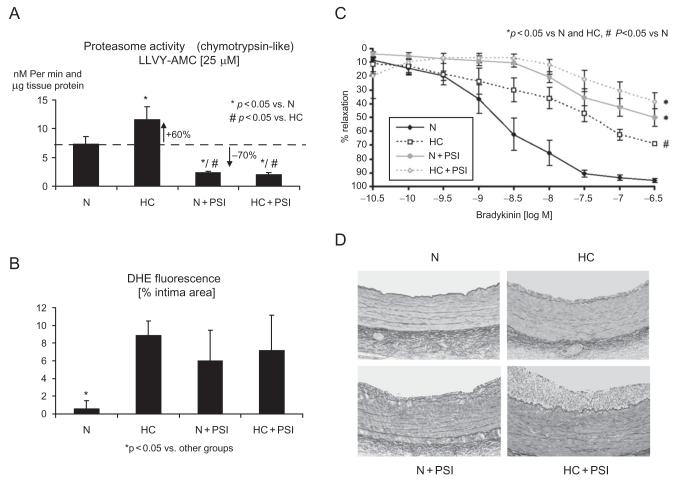
Illustration of the effects of boronate-type proteasome inhibitor treatment (PSI) in addition to normal (N) or high-cholesterol (HC) for 12 weeks in domestic pigs. Compared with N, a 60% increase in coronary artery chymotrypsin-like proteasome activity is notable in HC and a 70% decrease in N+PSI and HC+PSI (panel A). This level of proteasome inhibition causes oxidative stress on the level of the intima, measurable, for instance, by DHE fluorescence, indicating in situ superoxide anion production similar to the degree observed in HC (panel B). Concomitantly, there is a decrease in the vasorelaxation response to the endothelium-dependent agonist bradykinin although more pronounced in PSI pigs (panel C). Along with endothelial dysfunction, intimal thickening develops in N+PSI similar to HC (panel B). Yet, even with a similar level of endothelial dysfunction, intimal thickening is greatly aggravated in HC+PSI. These observations suggest that, while chronic proteasome inhibition can be harmful under otherwise normal conditions, the consequences are much worse when it entails blunting a compensatory increase in proteasome activity. Modified and reproduced with permission from the American Heart Association.18
Fig. 7.
Illustration of the effects of the boronate-type proteasome inhibitor, bortezomib, at low and high dose on the composition of advanced atherosclerotic plaques in ApoE-deficient mice including an increase in immunoreactivity for cleaved caspase-3 and a decrease for α-SMC-actin+cells, a decrease in collagen content by Sirius red staining, and an increase in necrotic core size by hematoxylin eosin (HE) staining; *p<0.05, **p<0.01 versus control; scale bar=50 μm. Modified and reproduced with permission from Springer Verlag.63
Fig. 8.
In patients with fatal myocardial infarction, an increase in ubiquitin immunoreactivity is evident in the complicated plaques of the infarction-related coronary arteries compared with advanced plaques in the noninfarction-related coronary arteries. The difference in ubiquitin immunoreactivity is notable in the shoulder and fibrous cap areas (left panels). In patients undergoing carotid endarterectomy, immunoblotting-determined levels of ubiquitin–protein conjugates, but not free ubiquitin, are higher and chymotrypsin-like proteasome activity is lower in plaques from patients with symptoms of transient ischemic attack (TIA), stroke, or amaurosis (Am.) fugax within 6 months of surgery (right panel). Reproduced with permission from Elsevier.65
Similar to the debate in advanced atherosclerotic plaques, experimental studies in early atherosclerosis have yielded conflicting results. In agreement with the detrimental effect of the UPS on BH4 levels and NFκB activation, proteasome inhibitors or drugs that have proteasome-inhibitory potential such as aspirin were found to improve endothelial function and structural alterations of atherosclerosis in hypercholesterolemic and uremic rabbits as well as diabetic mice.16,20,73,74 On the contrary, we noted a remarkable deterioration endothelial function and aggravation of atherosclerosis in pigs on a normal or a high-cholesterol diet undergoing chronic proteasome inhibitor administration (Fig. 8).18 One readily notable difference between these studies is that boronate-type proteasome inhibitor used in our study is more proteasome-specific and selectively inhibits chymotrypsin-like activity whereas the peptide aldehyde MG132 is not as proteasome and proteasome subunit specific. conclusions on the functional significance (beneficial or detrimental) of upregulation of proteasome activity under pathological conditions which been consistently outlined in all of these studies therefore remains confounded by the inherent limitations of using various proteasome inhibitors. Currently, there is agreement that proteasome inhibitors can be “poisons or remedies” depending on the context in which they are used.75
E. Summary: The UPS and Atherosclerosis
This section reviews the current and most up-to-date knowledge of the UPS in atherosclerosis with the following key points: First, the UPS is variably involved in the regulation of eNOS activity and modulation of oxidative stress, which is highly relevant for the initiation stage of atherosclerosis and beyond. Second, the UPS plays a central role in the activation pathway of NFκB in various cells, mediating adhesion molecule expression, cytokine release, and proliferation. Moreover, it can influence foam cell formation and maintenance, both of which influence atherosclerosis progression. Third, contrary to the initiation and progression stage, the complication stage of atherosclerosis seems to be linked to a reduction of proteasome activity mediated by accumulating levels of oxidative stress in aging cells over time. In this, atherosclerosis shares similarities with neurodegenerative disorders and protein quality diseases. Indeed, the UPS and particularly the proteasome complex is a vital component of PQC mechanisms as it prevents the accumulation of dysfunctional proteins, which can be toxic to the cell and its environment (Fig. 9).36 As such, preservation rather than inhibition of proteasome function should be the goal while acknowledging some clear-cut exceptions and nuances. For instance, transient and reversible low-level proteasome inhibition has clearly been shown to exert beneficial effects such as increasing the resistance of cells against oxidative stress. The key is that this type of proteasome inhibition can be understood as a conditioning rather than an inhibition regimen similar to the protective effects of cardiac preconditioning.26 Otherwise, a more local and focal mode of proteasome inhibition is to be favored such as the use in drug-eluting stents. Particularly advanced plaques may become vulnerable with systemic general proteasome inhibitor administration. In the coming years, further details of the function and significance of the UPS in atherosclerosis will likely be revealed that will hopefully lead to the development of novel strategies.
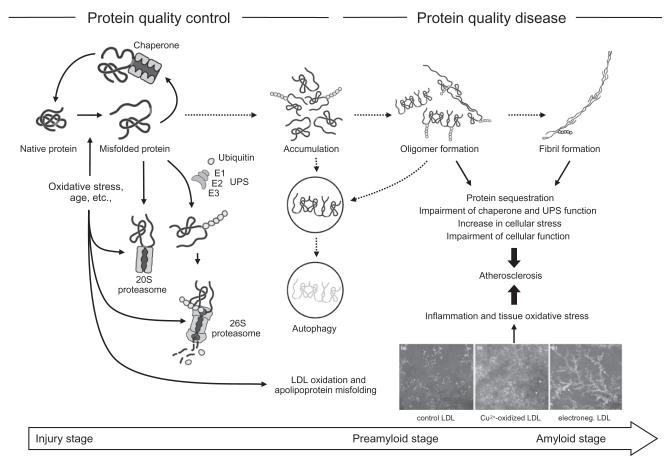
FIG. 9.
Illustration of the role of the ubiquitin–proteasome system in protein quality aspects of atherosclerosis. In the initial phase of cardiovascular risk factor exposure, compensatory upregulation of chaperones and the ubiquitin–proteasome system prevents the overwhelming intracellular accumulation of damaged and dysfunctional proteins. The ubiquitin–proteasome system also contributes to the classical activation pathway of nuclear factor kappa-B and thereby to inflammation and cell proliferation. With the formation and growth of a metabolically active atherosclerotic plaque, there is further production of misfolded and damaged proteins in the progression phase. Once the classical protein quality mechanisms are overwhelmed and fail, these dysfunctional proteins accumulate (and aggregate) and autophagy remains the final clearance pathway. The accumulating proteins can undergo further oxidation, ubiquitination, and cross-linking. Furthermore, beta-pleated sheets can be formed and hence amyloid fibrils. In addition to intracellular proteins, proteins in the extracellular matrix can undergo conformational changes. For instance, oxidation and phospholipid hydrolysis of low-density lipoprotein (LDL) produces oxidatively modified and electronegative particles with unfolding of the apolipoprotein components. The generation of amyloid-like structures in this process serves as a potent “key” to the uptake of these modified LDLs by macrophages via scavenger receptors. Recognition of amyloid-like fibrils by CD36 leads to the production of reactive oxygen species, chemokines, and cytokines, which contributes further to the atherosclerotic disease process, including its complication phase. Reproduced with permission from Elsevier.65
IV. The Role of Cardiac E3 Ligases
A. Pathologic Cardiac Hypertrophy
The role of the UPS, and specifically cardiac E3 ligases, in the development of pathologic cardiac hypertrophy is now well appreciated. Pathologic left ventricular hypertrophy is induced by a variety of factors (such as ischemia and volume or pressure overload) and results in a concomitant increase in both protein synthesis and proteolysis, the latter of which is regulated in large part by the UPS.76,77 Following the induction of pathologic cardiac hypertrophy, the mRNA levels of ubiquitin, E1, E2, and E3s (specifically atrogin-1 and MuRF1 (muscle ring finger 1)) increase.78 A similar increase in transcriptional upregulation of atrogin-1 and MuRF1 occurs in an in vivo model of chronic heart failure (CHF), indicating that cardiac E3 ligases are involved in both the development and end-stage pathology associated with pathologic cardiac hypertrophy. Although there is still some doubt as to whether or not these increases in expression of atrogin-1 and MuRF1 are beneficial in the defense against pathologic cardiac hypertrophy, as will be detailed in the following sections, the evidence suggesting that these cardiac E3 ligases are pivotal in regulating the development of cardiac hypertrophy is accumulating.
1. Atrogin-1
Cardiac atrogin-1 (also known as MAFbx1) is a cardiac and skeletal muscle-specific F-box protein first identified as a crucial participant in skeletal muscle atrophy programs.79,80 The expression level of atrogin-1 is tightly regulated at the transcriptional level in both skeletal myocytes and cardiomyocytes by members of the Forkhead family of transcription factors.81,82 As with all cardiac E3 ligases elucidated thus far, atrogin-1 has generalized antihypertrophic activity, and is able to inhibit cardiac hypertrophy induced by either pathologic or physiologic stimuli. In the case of pathologic cardiac hypertrophy, atrogin-1 works as a cardiac E3 ligase and targets the “pro-hypertrophy” protein calcineurin for proteasomal degradation.83 Calcineurin is a calcium/calmodulin-activated serine/threonine phosphatase that is a critical component of the signaling pathway that promotes pathologic hypertrophic growth of cardiomyocytes.84 Calcineurin initiates nuclear signaling by dephosphorylating the NFAT transcription factor, which in turn activates transcription of hypertrophy-associated genes. Atrogin-1 inhibits this calcineurin-dependent transactivation and downstream NFATc4 translocation by serving as an adaptor for calcineurin to associate with Skp1, Cul1, and Roc1 (the common components of SCF ubiquitin ligase complexes) to assemble an SCF (atrogin-1) E3 complex that ubiquitinates calcineurin and targets it for proteasome-dependent degradation.85 By targeting calcineurin for proteasomal degradation (and thereby inhibiting NFAT transcriptional activation of hypertrophy-associated genes), atrogin-1 disrupts a critical signaling pathway involved in pathologic hypertrophy.83,86 In addition to its ability to inhibit pathologic cardiac hypertrophy, atrogin-1 can also decrease cardiac hypertrophy induced by physiologic stimuli. In this case, however, atrogin-1 uses a completely different mechanism of action. In response to the induction of physiologic cardiac hypertrophy, atrogin-1 targets the transcription factors Foxo1 and Foxo3 for ubiquitination.87 Surprisingly though, instead of targeting the Foxo proteins for proteasomal degradation, atrogin-1-mediated ubiquitination results in enhanced Forkhead transcriptional activity and subsequent inhibition of Akt-dependent induction of hypertrophy.88 Atrogins’ ability to stimulate Foxo activity through ubiquitination is by virtue of the fact that atrogin-1 mediates the noncanonical addition of ubiquitin molecules on the Foxo proteins via lysine-63-linked chains, as opposed to the traditional canonical lysine-48.87 Previous reports have documented the ability of lysine-63-linked chains to modulate protein function89,90; however, the atrogin-1-mediated noncanonical ubiquitination of the Forkhead transcription factors is the first report of this form of regulatory mechanism working within the heart.
2. MuRF Proteins
The MuRF family of proteins comprises three members: MurF1, MuRF2, and MuRF3. All these proteins have ubiquitin ligase activity and localize to the M-line and Z-disc of the sarcomere91-93 where they function, in part, as mechanical stress sensors via their direct and indirect interactions with the giant protein titin.85,94,95 Although all three MuRF proteins appear to play an important role in various aspects of myogenic responses and contractile regulation,96 only MuRF1 appears to be involved in the regulation of skeletal muscle atrophy and inhibition of pathologic cardiac hypertrophy.97 Although the mechanism by which MuRF1 mediates cardiac atrophy has not yet been fully elucidated, recent evidence suggests that MuRF1 may be instrumental in the initial proteolytic breakdown of the myofibril,98 which then exposes the individual myofibrillar component proteins to subsequent proteolytic degradation. In cardiac muscle in vitro, MuRF1 exerts E3 ligase activity by ubiquitinating the sarcomeric protein cardiac troponin I, thereby contributing to its proteasomal degradation.99 However, there are other ways in which MuRF1 may also contribute to the regulation of cardiac atrophy. For example, in cultured cardiomyocytes, MuRF1 inhibits PKCε activity through interactions with RACK1, the receptor for activated protein kinase C protein, which in turn suppresses focal adhesion kinase and ERK1/2 in cardiomyocytes, resulting in the suppression of cardiomyocyte hypertrophy.100 It has also been suggested that MuRF1, through its association with the transcription serum response factor, is involved in a circuit that links mechanical force generation to transcriptional responses in cardiomyocytes, resulting in hypertrophic growth.97 In vivo, MuRF1 has been linked to the regulation of cardiac hypertrophy in both mouse models and human disease.101 MuRF1 protein levels are increased in human cardiac tissue isolated from patients undergoing therapeutic atrophy following placement of a left ventricular assist device (LVAD) as a treatment to decrease pressure overload associated with heart failure.101
B. Protection Against Ischemic Heart Disease
The cardiac pathology profile associated with ischemic injury centers around the induction of cardiomyocyte apoptosis. A major player in this ischemia-related cardiomyocyte apoptosis is the transcription factor p53. p53 target genes include PUMA, Bax, and Apaf1, each of which plays a distinct yet complementary role in the apoptotic signaling pathway. Numerous reports have now demonstrated that p53 is a target substrate of multiple cardiac E3 ligases such as CHIP, murine double minute (MDM) 4, and MDM2. These cardiac E3 ligases ubiquitinate p53 and target it for proteasomal degradation, thereby decreasing cardiomyocyte apoptosis and resulting in an overall cardioprotective effect in response to ischemic injury.
1. CHIP and MDM2
The E3 ligase/co-chaperone CHIP (carboxyl terminus of Hsp70-interacting protein) is highly expressed in cardiac muscle where it ubiquitinates specific proteins, targeting them for degradation by the 26S proteasome. CHIP is also characterized by its ability to bind the HSPs Hsc70–Hsp70 and Hsp90. Much of what is known about CHIP’s cardioprotective function during ischemia/reperfusion injury comes from studies carried out in CHIP−/−mice. CHIP−/−mice are far more susceptible to cardiac damage following an ischemic insult compared to wild-type mice, exhibiting a 50% larger infarct area compared to wild-type controls.102 In addition, CHIP−/−mice exhibit a marked increase in frequency of arrhythmias following reperfusion, indicating an increased degree of cardiac damage compared to wild-type mice.102 CHIP-deficient cardiomyocytes also undergo enhanced apoptosis following ischemia/reperfusion injury, indicating that CHIP inhibits apoptosis during cardiac stress. The mechanism through which CHIP exerts this antiapoptotic effect could be linked to the fact that CHIP interacts with both wild-type and mutant forms of p53. In response to CHIP suppresses the aggregation and stress, ubiquitinates both forms of p53, leading to their 26S proteasome-dependent degradation.103 This in turn would decrease the ability of p53 to stimulate proapoptotic pathways.
MDM2 is another E3 ligase present in cardiac muscle that regulates the activity and degradation of proteins such as p53 and Apoptosis Repressor with Caspase recruitment domain (ARC), both of which are involved in cell growth and apoptosis. Similar to CHIP, MDM2 inhibits p53 transcriptional activity through ubiquitination and subsequent proteasomal degradation.91,104,105 In cardiomyocytes, the decrease in p53 levels as a result of MDM2 ubiquitination leads to an inhibition of apoptosis during pathologic stress.100 Conversely, if MDM2 is inactivated in cardiomyocytes, the level of p53 increases, resulting in enhanced I/R-induced apoptosis and a subsequent reduction in left ventricular function.100 In addition to targeting p53, MDM2 also regulates ARC protein levels in cardiomyocytes.106 MDM2-mediated ubiquitination of ARC leads to proteasomal degradation of ARC and subsequent inhibition of cardiomyocyte apoptosis. Recently, the MDM2 homologue MDM4 has been shown to also inhibit p53 in cardiomyocytes. Interestingly, a conditional MDM4 knock-out mouse model demonstrated that decreased cardiac MDM4 levels result in the development of a dilated cardiomyopathy.107 The cause of this dilated cardiomyopathy was linked back to a p53-dependent decrease in cardiomyocyte number, indicating the importance of MDM4 inhibition of p53 activity in fully differentiated cardiomyocytes. While MDM4 has been shown to have overlapping functions with MDM2, its role in ischemia/reperfusion injury has not yet been elucidated.
2. MuRF3
MuRF3 is another muscle-specific RING finger protein that was originally identified by its interaction with microtubules and its role in skeletal myoblast differentiation.93 However, more recent work has expanded the role of MuRF3 to include activities within the heart. Mice lacking MuRF3 exhibit abnormal sarcomere structure and have a dramatically increased response to ischemia/reperfusion injury compared to wild-type mice. When these mice are challenged with ischemia/reperfusion injury, rapid left ventricular dilation, cardiac rupture, and myocyte degeneration occur within days of the initial insult.108 Two substrates of MuRF3 have been identified as being γ-filamin and the four-and-a-half LIM domain protein (FHL2) and the expression of both these proteins is increased in the hearts of mice lacking MuRF3. Given the dramatic response of MuRF3-deficient mice to myocardial infarction, it has been suggested that MuRF3-dependent turnover of γ-filamin and FHL2 is essential to the integrity and function of the heart following acute ischemic events.108 MuRF3 also plays a role in the regulation and turnover of sarcomeric contractile proteins including MHCIIa and beta/slow myosin heavy chain, a function that MuRF3 shares with MuRF1.108 This cooperative relationship between MuRF1 and MuRF3 is illustrated by the pathological phenotype expressed by mice deficient in both proteins. MuRF1/MuRF3 knockout mice develop a skeletal muscle myopathy and hypertrophy cardiomyopathy in which myofiber fragmentation, MHC accumulation, and diminished muscle performance are also seen.109 These findings reveal a critical role for MuRF1 and MuRF3 in proteasomal-dependent turnover of sarcomeric proteins and suggest a potential mechanism for the development of myosin storage myopathies.108
C. The UPS in Familial Hypertrophic Cardiomyopathy: Proteasomal Involvement Without an Identified Cardiac E3 Ligase
While cardiac E3 ligases have been definitely linked to the regulation of cardiac hypertrophy and ischemia/reperfusion injury, there are other cardiovascular conditions that are known to involve the UPS, but for which a candidate ubiquitin ligase has yet to be discovered. One example is familial hypertrophy cardiomyopathy (FHC). FHC is an inherited cardiac disease that is transmitted as an autosomal-dominant trait. It is estimated that FHC affects 0.2% of the population and is the major cause of sudden death in young, healthy individuals.110 Over 200 mutations associated with FHC have been identified in 14 different genes, with the majority of them involving genes encoding sarcomeric proteins.111 Genetic analysis of families with FHC has revealed that most of the FHC-related mutations occur in the myosin-binding protein-C (cMyBP-C).112 cMyBP-C is a critical constituent of sarcomeric thick filaments and has both regulatory and structural roles within the sarcomere. cMyBP-C interacts with myosin, titin, and actin, which, once again, are essential components of the sarcomeric apparatus.110-113 Recently it has been demonstrated that the UPS rapidly targets naturally occurring mutations in cMyBP-C that result in a truncated protein for degradation. The increase in proteasomal workload associated with clearing of these mutated forms of cMyBP-C results in impairment of proteolytic capacity.114 Frameshift mutant cMyBP-C proteins are expressed at markedly lower levels in muscle, despite the fact that the mRNA levels of these mutant forms of cMyBP-C match the wild-type cMyBP-C mRNA levels,114 in both humans115,116 and transgenic mice.117,118 Experimentally, if the 26S proteasome is inhibited, expression of the truncated cMyBP-C mutant protein increases to that of wild-type cMyBP-C levels, supporting a role for proteasomal degradation in the control of cMyBP-C protein expression levels. Paradoxically, although the UPS is activated to remove mutant cMyBP-C proteins from the cell, over time, the increased proteasomal workload associated with this quality control mechanism results in a breakdown in activity of the proteasome, ultimately leading to the accumulation of mutated cMyBP-C proteins.114 As such, truncated cMyBP-Cs act as both substrates of the UPS and obstructers of the proteolytic process, a dichotomy that is shared with certain protein mutants that accumulate in Alzheimer’s disease and other storage-related neurological conditions.119 The specific E3 ligases that target either wild-type or mutant cMyBP-C are currently unknown. If these E3 ligases could be identified, however, it would be possible to use them as therapeutic targets to alter the severity and progression of FHCs. The role of the UPS in PQC as it relates to cardiac proteinopathies is covered in detail in the next section.
D. Summary: Cardiac E3 Ligases
As highlighted in this section, cardiac E3 ligases such as atrogin-1, CHIP, MDM2/4, and MuRF proteins play a pivotal role in regulating the molecular pathways underlying cardiovascular conditions such as cardiac hypertrophy and ischemic heart disease. Furthermore, proteasomal involvement has been reported in a number of other cardiovascular conditions, such as familial cardiac hypertrophy, although the identity of the participating cardiac E3 ligases remains unknown. However, despite the potential that cardiac E3 ligases have as therapeutic agents, more data need to be gathered in order to understand the extent to which these ligases control various cellular processes, both beneficial and detrimental. Such focus will allow identification of which cardiac E3 ligases might serve as therapeutic targets in the future.
V. The UPS in PQC and Cardiac Proteinopathy
A. Cardiac PQC
For a polypeptide to fulfill its normal duty in the cell, it must attain and maintain its native conformation via proper folding. A misfolded or unfolded protein is not only unable to perform its normal function, but it is also likely to be detrimental to the cell. PQC is a set of cellular mechanisms to keep the level of misfolded/unfolded proteins in the cell to the minimum. All proteins targeted to the secretory pathway go through the ER and are folded in the ER, while the folding of other proteins, such as cytoskeletal and cytosolic proteins, does not involve the ER. Hence, PQC is often divided into ER-associated PQC with which the unfolded protein response is associated, and ER-independent PQC. Given that myofibrils take the majority of space in the cardiomyocyte and that myofibrillar proteins are not folded via ER, ER-independent PQC takes care of more proteins in cardiomyocytes than the ER-associated PQC. Unfortunately, ER-independent PQC is much less understood than ER-associated PQC. Nevertheless, ER-associated and ER-independent PQC share the same general principle. Both are carried out by an intricate collaboration between molecular chaperones and targeted protein degradation. The latter is primarily performed by the UPS.113 Both genetic mutations and environmental stress can give rise to misfolded protein molecules. In the cell, misfolded/unfolded proteins, which inevitably expose hydrophobic surfaces, will be sequestered by molecular chaperones to prevent aberrant aggregation and, in some cases, to help the misfolded/unfolded proteins to refold. If the refolding is unsuccessful, the misfolded proteins will then be known as terminally misfolded proteins and removed by proteasome-mediated degradation. Misfolded proteins that escape from the surveillance of chaperones and the UPS form aberrant aggregates. Recent evidence suggests that the autophagic–lysosomal pathway may also contribute to the removal of misfolded proteins, especially the aggregated form of misfolded proteins (Fig. 9).120
With a few exceptions, misfolded polypeptides are polyubiquitinated before they can be recognized and degraded by the proteasome. A mature degradation signal, known as a degron, on the protein molecule is required to trigger the polyubiquitination and subsequent proteasome-mediated degradation of the protein. For a native protein, the maturation of its degradation signal is highly regulated and determines when it will be ubiquitinated; therefore, the degradation of a native protein is primarily determined by the ubiquitination step. Protein misfolding can either expose encrypted degrons or form a degron de novo (e.g., hydrophobic surfaces)121; therefore, it is conceivable that terminally misfolded proteins “automatically” activate the ubiquitination system and the rate-limiting step likely resides in the proteasome-mediated degradation step. Consistent with this notion, the accumulation of ubiquitinated proteins appears to be a common feature in the cells or tissues with increased production of misfolded proteins, and enhancing proteasome function facilitates the degradation of misfolded, but not native, proteins in the cell.122,123 Therefore, measures to evaluate proteasome functional sufficiency, as well as means to enhance proteasome function, in a cell or organ are vital to the investigation into the role of the UPS in PQC and in the genesis of disease caused by misfolded proteins.
Proteasome peptidase activity assays provide a simple method to estimate changes in the proteolytic activity intrinsic to either the 20S or arguably the 26S proteasome in a crude protein extract, in a proteasome-enriched preparation from cells or tissue specimens, or in intact cells or tissues.123 However, these assays do not assess whether the proteasome proteolytic activity is sufficient or not for adequate PQC in a given cell, tissue, or organ. To circumvent this deficit, full-length fluorescence proteins modified by fusion with a degron have been successfully adopted as surrogate substrates for the proteasome. Stable cell lines and stable transgenic mouse models that constitutively express such surrogate protein substrates have been established and employed to probe UPS functional status in the cell or organ in situ.124-126 Regarding the usefulness of these surrogate protein substrates in the research into PQC and proteinopathy, GFPu/GFPu and GFPdgn are arguably the most interesting and widely used ones. Both GFPu and GFPdgn were engineered via carboxyl fusion of degron CL1 to the enhanced green fluorescence protein (GFP). Conventional GFP is not an effective substrate of the UPS, with a long half-life when expressed in mammalian cells. The fusion of degron CL1 renders GFP a specific substrate for the UPS.124 Amino acid replacement experiments showed that degron CL1, a consensus ubiquitination signal sequence from fission yeast,127 triggers ubiquitination likely by forming a hydrophobic surface in its helical structure.128 Hence, GFPu and GFPdgn are surrogates for misfolded proteins because surface exposure of hydrophobic residues is a conformational signature of misfolded proteins.120 Changes in GFPu/GFPdgn protein levels in the absence of alterations in their synthesis inversely reflect the capacity of the UPS to remove misfolded proteins.
Emerging evidence suggests that proteasomes with a certain composition may be more efficiently equipped to degrade misfolded proteins. For instance, it is well known that viral infection or IFN stimulation triggers the replacement of conventional peptidase subunits (β1, β2, and β5) in 20S proteasomes with the inducible subunits (β1i, β2i, and β5i) and thereby upregulates immunoproteasomes.129 A recent study has demonstrated that immunoproteasomes help the cell to more effectively remove oxidized proteins formed by increased oxidative stress induced by IFN.130 It was also reported at almost the same time that upregulation of 11S proteasomes via overexpression of PA28α enhances proteasome-mediated removal of misfolded proteins and protects against oxidative stress in cultured cardiomyocytes.131
B. Proteasome Functional Insufficiency in Cardiac Proteinopathy
Proteinopathies are a family of diseases caused by misfolded proteins and featured by the presence of aberrant protein aggregates in the cell. The best-studied cardiac proteinopathy is perhaps desmin-related cardiomyopathy (DRC).132 DRC is the cardiac manifestation of desmin-related myopathy (DRM). DRM is a group of heterogeneous myopathies with a hallmark of the presence of desmin-positive aberrant protein aggregates in muscle cells. Mutations in the desmin gene, the aB-crystallin gene (CryAB), and a few other genes whose products interact with the desmin filaments have been linked to DRM.132 DRM can affect all three types of muscle: skeletal, cardiac, and smooth muscle. DRC is the main cause of death of DRM patients. In the past decade, a few DRC mouse models have been created through cardiac-specific transgenic expression of a DRM-linked mutated gene,133,134 such as a missense (R120G) mutant of CryAB (CryABR120G) and a 7-amino-acid (R173 through E179) deletion mutant of the desmin gene (D7-Des).135,136 Both CryABR120G and D7-des transgenic mice recapitulate the main aspects of pathology observed in human DRC, including the presence of desmin-positive protein aggregates in cardiomyocytes and cardiac dysfunction133,134; hence, these mouse models provide useful platforms for studying DRC pathogenesis and exploring therapeutic strategies.113,132
Studies with cross-breeding GFPdgn mice with the mouse models of DRC showed marked increases of GFPdgn protein levels but not GFPdgn transcript levels in mouse hearts overexpressing either CryABR120G or D7-des,137,138 indicating UPS functional insufficiency in DRC hearts. The abundance of total ubiquitinated proteins was significantly increased in the DRC hearts, suggesting that UPS insufficiency is primarily caused by proteasome functional insufficiency (PFI). Further analyses revealed that the insufficiency resides mainly in the step delivering ubiquitinated proteins into the 20S proteasome as decreased protein abundance of critical 19S proteasome subunits, namely Rpt3 and Rpt5, is concurrently associated with the PFI while the peptidase activities of the 20S proteasome are not decreased. Further studies have determined that aberrant protein aggregation is in part responsible for PFI in DRC hearts.137,139 These are highly significant discoveries because although DRC per se is not a common disease, the DRC mice represent valuable models for pathogenic elucidation and therapeutic exploration of cardiac proteinopathies. It is increasingly suggested that a large subset of common heart diseases share likely pathogenic mechanisms with cardiac proteinopathy. Aberrant protein aggregates are also observed and considered the trigger of autophagy activation in the pressure-overloaded mouse heart.140 Significant increases in myocardial ubiquitinated proteins have invariably been reported in studies using this experimental method, although some of the reports dispute the direction of changes in the proteasome peptidase activities as well as the effect of systemic proteasome inhibition on the hypertrophic responses and function of pressure-overloaded hearts.141,142 Both the accumulation of aberrant protein aggregates and elevated levels of ubiquitinated proteins are consistent with PFI but the sufficiency of proteasome function in a pressure-overloaded or ischemic heart has not been directly assessed yet. A very high prevalence of aberrant protein aggregates in the form of preamyloid oligomers in human hearts with end-stage heart failure of common etiologies has been recently reported.143,144 The coexistence of increased ubiquitinated proteins with aberrant protein aggregation signifies PFI and serves potentially as a cause of proteasome impairment. Hence, these observations place a large subset of CHF into the category of cardiac proteinopathy. A better understanding of the molecular pathogenesis of cardiac proteinopathy should shed light on the molecular mechanisms by which these heart diseases progress to CHF.
C. PFI Contributes to the Genesis of Proteinopathy
PFI has been observed or implicated in a variety diseases, of diseases, such as neural degenerative cardiac proteinopathy, and more recently, a large subset of CHF. However, until very recently, a proteasome activator was unavailable, so it was difficult to directly test whether PFI plays a role in the development of any of these diseases. The recent discovery of a benign measure to enhance proteasome function has instantly changed this situation. Using cultured neonatal rat ventricular myocytes and human embryonic kidney cells, it was found that overexpressing PA28α (PA28αOE) stabilizes PA28α and thereby upregulates 11S proteasomes, which destabilizes GFPu, a surrogate misfolded protein substrate for the UPS,120 without affecting the protein levels of endogenous and ectopically overexpressed native proteins.122 Therefore, this marks the establishment of the first benign measure to enhance proteasome proteolytic function in cultured cells. A biological relevance of this exciting new measure was further demonstrated by testing the effect of PA28αOE on oxidative stress. Enhancing proteasome function by PA28 OE decreased the abundance of protein carbonyls, a widely used marker of oxidized proteins, and the prevalence of apoptosis in cultured cardiomyocytes triggered by the treatment of hydrogen peroxide.131 These findings suggest that PFI plays an important role in injury by oxidative stress. The latter represents a major cause of myocardial I/R injury. Additionally, PA28α overexpression and knockdown were shown to enhance and delay the degradation of CryABR120G in cultured neonatal rat ventricular myocytes.
To verify whether PA28αOE enhances proteasome function in intact animals, transgenic mouse lines were created to achieve temporally controllable cardiomyocyte-restricted (CR-PA28αOE),122 using the “Tet-off” system reengineered by Sanbe et al.145 Mice with CR-PA28αOE initiated at the perinatal period do not display remarkable changes in the expression of the fetal gene program, heart function, or cardiac morphology till at least 1 year of age, the longest time observed. Introducing GFPdgn into the CR-PA28αOE mice via cross-breeding, it was validated that CR-PA28αOE facilitates the degradation of GFPdgn, a surrogate misfolded protein.126 This is evidenced by a marked decrease in GFPdgn protein level and the unaltered abundance and unchanged translational efficiency of GFPdgn transcripts. The effect on GFPdgn protein degradation disappears upon turning off CR-PA28αOE via administration of doxycycline.122 These experiments mark the establishment of the first genetic method and the first animal model to benignly enhance proteasome function in the heart, or any other organs. Taking advantage of this model, Li et al. have further demonstrated that overexpression of PA28α significantly attenuates aberrant protein aggregation and cardiac hypertrophy and delays the premature death of mice with transgenic overexpression of CryABR120G in cardiomyocytes, in a bona fide mouse model of cardiac proteinopathy.122 Moreover, we demonstrated that CR-PA28αOE protects against myocardial I/R injury induced by coronary ligation and release in intact mice.122 These studies prove for the first time that PFI plays an important pathogenic role in myocardial I/R injury and cardiac proteinopathy in both cell culture and intact animals. It is anticipated that these exciting discoveries, including the invention of new ways to benignly enhance proteasome function in a cell and an organ, will facilitate research into the pathogenic roles of UPS dysfunction and promote the development of new therapeutic strategies to treat a large subset of diseases, including heart diseases.
D. Potential Mechanisms Underlying the Pathogenic Role of PFI
Conceivably, PFI can impair the heart via a number of mechanisms, such as compromising PQC and disturbing signaling pathways. Sarcomeres are the most fundamental units of mechanical function in cardiac and skeletal muscles. An extremely vigorous stoichiometry among different sarcomeric proteins is followed to form sarcomeres and maintained for proper function of the sarcomere. The replacement of a damaged sarcomeric protein molecule with a new one must be accurately coupled with the removal of the damaged one. The UPS is responsible for the degradation of virtually all myofibrillar proteins. Hence, PFI in cardiomyocytes will leave defective proteins in the sarcomere that are not replaced in a timely fashion, and which will undoubtedly reduce the functioning of the sarcomere and ultimately the mechanical performance of the heart. In this regard, PFI may yield the same consequence as the genetic mutations of the sarcomeric proteins that are known to cause inherited cardiomyopathies,146 a major cause of CHF. PFI inevitably accumulates misfolded proteins and permits aberrant protein aggregation. In both humans and mice, expression of misfolded proteins and resultant aberrant protein aggregation are sufficient to cause cardiomyopathy and CHF.147-149 In addition to physical impedance to contraction and relaxation and structural disruption, aberrant protein aggregates can also be detrimental to multiple cellular processes. Aggregation-prone proteins can directly interact and inhibit proteasome proteolytic subunits.150,151 In both cultured cardiomyocytes and intact mouse hearts, aberrant protein aggregation has been shown to impair proteasome proteolytic function,117,137-139,152,153 thereby forming a vicious cycle.
Pathological changes in mitochondrial morphology and function as well as programmed cell death are often associated with CHF. Mitochondrial malfunction and increased cell death were observed in the mouse hearts of CryABR120G-based DRC that displays severe PFI before other cardiac pathology becomes discernible.137,154 Pharmacologically induced proteasome inhibition has been shown to induce cell death in cultured cardiomyocytes.125,155 The calcineurin–NFAT pathway is an important signaling pathway that transcriptionally regulates pathological remodeling of the heart.156 Interestingly, proteasome inhibition can activate the calcineurin–NFAT pathway and promote maladaptive growth of cultured cardiomyocytes. NFAT transactivation is increased in the D7-des mouse heart and proteasome inhibition facilitates the progression of a stressed heart to heart failure.142 Similar to PFI, the loss of an important chaperone (e.g., HSPB2/HSPB5 double knockout) can also activate the calcineurin–NFAT pathway in the heart,142,157 suggesting that PQC inadequacy may play a role in the activation of this pathway during pathological cardiac remodeling.
E. Improving PQC as a Strategy to Treat Cardiac Proteinopathy
No effective therapies are currently available for treating cardiac proteinopathies. However, by targeting the molecular pathogenesis of proteinopathy, recent experimental studies have shown some promise. As described earlier, enhancing proteasome function can protect against cardiac proteinopathy in mice.122 Additionally, pharmacological agents that behave as chaperones (e.g., high dose of doxycycline) or as inducers of HSPs (e.g., geranylgeranylacetone) have been shown to suppress aberrant protein aggregation in CryABR120G-based DRC mouse hearts, to improve heart function, and to delay premature death of the DRC mice,158,159 providing strong evidence that pharmacological chaperones are promising therapeutic candidates for clinical trials to treat cardiac proteinopathies.
VI. The UPS in Heart Failure
Heart failure is defined “as a complex clinical syndrome that can result from any structural or functional cardiac disorder that impairs the ability of the ventricle to fill with, or eject, blood.160” The causes and risk factors of heart failure are many, and include hypertension, ischemic heart disease, diabetes, obesity, valvular heart disease, and exposure to cardiotoxic agents.160 As discussed in the previous section, some cases of heart failure are familial and associated with mutations in various genes encoding for various myocardial proteins (cardiac proteinopathies), leading to issues with PQC. However, the majority of cases are idiopathic, where no one cause can be identified. This section describes alterations in the UPS as it relates to this latter group. As will become apparent, changes in activity of the proteasome are biphasic and seem to correlate with the severity of the process and thus can be described as early, studied primarily in animal models, versus late, studied using explants from human hearts.
A. Alterations in UPS in Early Failure
One of the early manifestations of heart failure is development of cardiac hypertrophy secondary to ventricular remodeling. This early phase of heart failure development has been modeled in experimental animals using a variety of initiating events, including transverse aortic constriction (TAC), permanent left anterior descending artery (LAD) occlusion, and chronic exposure to hypertrophic agents, such as isoproteronol (ISO). Perhaps the earliest study was by Depre et al.,161 who used a canine TAC model to show that, as the heart hypertrophies, the UPS is activated. Increased proteasome activity was associated with increased expression of proteasome subunits. This result was confirmed in subsequent studies in a murine ISO-induced hypertrophy model162 and in cardiomyocytes exposed to hypertrophic agents.163 This is not to say that these results are not without some controversy, as at least one study observed that proteasome activities are depressed prior to the onset of cardiac dysfunction in pressure-overloaded hearts of mice.155 An interesting observation of these and other studies was that inhibition of the proteasome with any one of an assortment of proteasome inhibitors decreased the amount of hypertrophy associated with the initiating event.161-164 Activation of the proteasome was associated with alterations in the stress-response chaperone H11 Kinase/Hsp22.165 In addition, in mice treated with ISO, cardiac hypertrophy was associated with increased sensitivity of the proteasome to PKA-mediated activation, as well as an altered proteasome phosphoproteome.10
While the exact mechanism of proteasome activation is unclear at this time, the involvement of the UPS in cardiac hypertrophy and remodeling cannot be denied. This can be inferred from the skeletal muscle literature where it is clear that the proteasome and associated ubiquitination machinery mediate the breakdown of muscle protein (remodeling) necessary for exercise-induced physiologic hypertrophy.166-169 Although not studied to the same extent, it is becoming increasingly clear from experimental models that pathological cardiac remodeling is also associated with changes in the ubiquitin protein ligases that target myofibrillar and other cardiac proteins, including atrogin-1 (MAFbx) and Murf-1.94,170,171 A recent study has shown that knockout of atrogin-1 is protective in both TAC and β-adrenergic-induced cardiac hypertrophy.171 There is a key question that remains to be answered: What is the difference between the remodeling associated with cardiac hypertrophy that becomes pathologic and leads to heart failure and the physiologic remodeling associated with exercise that leads to strengthening and optimized function?
B. Alterations in UPS Function in Late Failure
The availability of human heart tissues to study alterations in UPS function in early heart failure is extremely limited. This or is not the case when studying late, so-called end-stage, heart failure, where unfortunately there are ample supplies of explanted human hearts harvested at transplant. Often, these studies are limited by the availability of control human heart tissues, which generally are donor hearts originally destined for transplant but not used for a variety of reasons. Two of the earliest studies investigated for the presence of ubiquitinated proteins or substrates, indirect measures of UPS function at best, and observed these to be increased, suggesting dysfunction.172,173 Subsequently, we published the first comprehensive examination of proteasome function in human end-stage heart failure and confirmed that proteasome activity is diminished in end-stage failure associated with increases in ubiquitinated proteins.174 Unloading the heart via the use of an LVAD partially reversed the proteasome dysfunction.174 This is an observation also reported by another pre- and post-LVAD study which showed enhanced expression of proteasome subunits.175 It is of interest that similar results were obtained with markers of autophagy which were also downregulated following mechanical unloading via LVAD.176 Autophagic and proteasome-mediated degradation are often considered to be parallel proteolytic systems that respond inversely to each other, that is, when proteasome activity is decreased, autophagy is increased, or vice versa.123,177 However, in end-stage heart failure, both appear to be diminished.174,176
As part of our study, increased protein oxidation products that prompted an examination for oxidative damage to proteasome subunits were observed.174 Protein carbonylation of at least one subunit of the 19S regulatory particle, Rpt5, was observed, suggesting that the dysfunction may be secondary to oxidative damage to the proteasome itself, although one rather intriguing study suggests that humoral auto-antibodies to proteasomes may be increased in dilated cardiomyopathy.178 In all fairness, not all studies of human heart explants report the same dysfunction. One study reports increased expression of proteasomes in samples from patients with dilated cardiomyopathy referred for left ventriculoplasty179; however, proteasome activity was not assessed. Nonetheless, if one extrapolates from the animal and human studies, a relationship between progression of heart failure and function of the proteasome can be constructed in which activity initially increases during remodeling but then diminishes below baseline in end-stage heart failure.
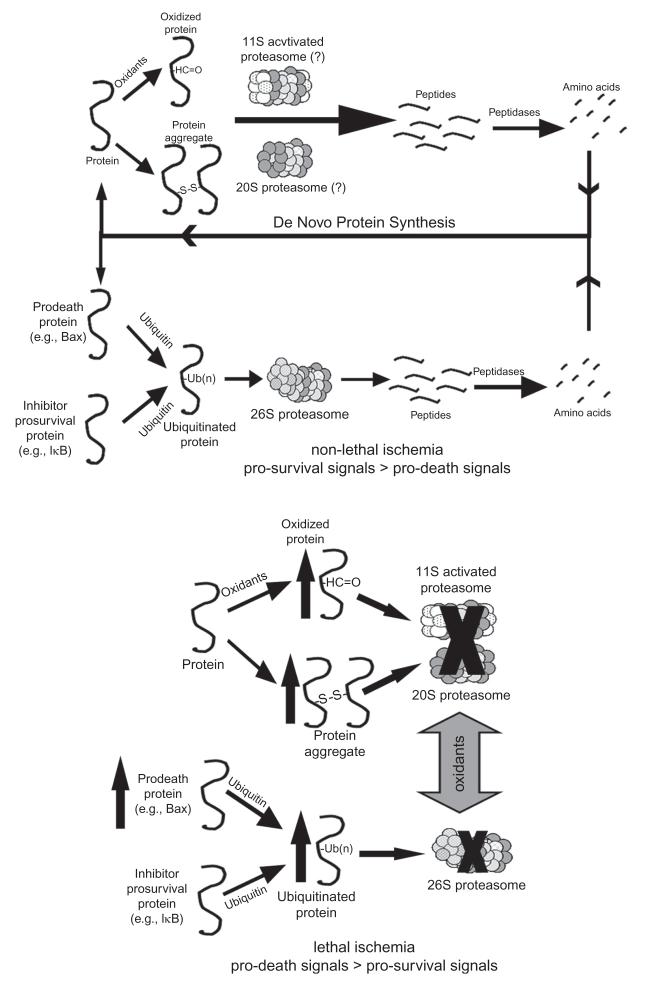
VII. The UPS in Myocardial Ischemia/Reperfusion Injury
Ischemia is defined as reduced tissue perfusion resulting in oxygen deprivation accompanied by diminished removal of metabolites. From a historical perspective, many of the earliest studies suggesting that ischemia may result in dysfunction of the UPS actually derive from the brain literature. These earlier studies report increases in insoluble ubiquitin conjugates in the mitochondria and decreased 26S proteasome activity in the gerbil cortex and hippocampus following transient forebrain ischemia.180,181 Other studies have confirmed these results and provide a link between UPS dysfunction and oxidative stress.182,183 Since the publication of earlier brain studies, there have been several studies examining UPS dysfunction following myocardial ischemia.
Dysfunctional postischemic proteasome in the heart was first described by Bulteau et al.,184 who reported decreased proteasome chymotryptic, caspase-like, and trypsin-like activity accompanied by increased ubiquitinated proteins following 30 min of in vivo LAD artery occlusion. Despite the decrease in tissue proteasome activity, following purification only the decrease in trypsin-like activity was observed. In a series of studies, Powell et al.185,186 confirmed this observation in the isolated perfused heart model and also demonstrated that ATP-dependent proteasome activity is preferentially affected. This suggested defects in 26S proteasome function which were consistent with increases in myocardial ubiquitinated proteins. One rather intriguing study187 suggested that short periods of ischemia/reperfusion may result in selective proteasome dysfunction affecting turnover of specific proteins. This study took the indirect approach of assessing proteasome function by examining the effects of proteasome inhibitors on turnover of signaling proteins known to be subject to UPS-mediated degradation.187 This study must be interpreted with caution as lack of studies of the ubiquitinated homologues of these signaling proteins makes it difficult to determine the role of the proteasome. Nonetheless, given the earlier discussion on regulation of the cardiac UPS and presence of proteasome subpopulations, the differential regulation of protein degradation proposed in this study is conceivable.
A. Potential Mechanisms Involved in Postischemic Proteasome Dysfunction
Various mechanisms have been proposed to explain the dysfunction; however, two potential explanations have recently received credible experimental support.
1. Ischemia-Induced ATP Depletion
This mechanism is based on the known requirement for ATP for 20S proteasome and 19S regulatory particle docking and activation of the proteasome, as well as for protein ubiquitination.188,189 A recent study suggests that intracellular ATP levels regulate proteasome activity190 in a biphasic manner; thus, it is certainly conceivable that depletion of ATP during ischemia191 could be partially responsible for decreased proteasome activity in the ischemic heart. In fact, this mechanism has been invoked to explain proteasome dysfunction in both an in vivo canine and ex vivo isolated rat models of ischemia.192,193 Since both of these studies measured proteasome activity in the presence of ATP, essentially adding back the nucleotide, the dysfunction should have been reversed, yet this was not the observed result. At least one study has shown that, even when measured in the presence of optimal concentrations of ATP, post-ischemic proteasome activity is diminished.186 Taken as a whole, these studies suggest that the dysfunction measured using the standard fluorogenic peptide assays is not related to ATP depletion; however, this does not rule out the possibility that ATP depletion can still be a contributing factor to proteasome dysfunction during the ischemic period. It does, however, suggest a defect in the utilization of ATP by the postischemic proteasome. The presence of oxidative stress and oxidized cytosolic, myofibrillar, and mitochondrial proteins in the postischemic heart194-197 has prompted us and others to hypothesize that the many subunits of the 20S proteasome and its regulatory particles may be targets for oxidative attack.
2. Oxidative Damage to Proteasome and/or Regulatory Subunits
Oxidative modification of proteins affects their secondary and tertiary structures resulting in unfolding, which can lead to loss of function and exposure of hydrophobic patches, which in turn can lead to increased susceptibility to degradation.198,199 Studies of vulnerability of different populations of proteasomes (20S vs. 26S) to oxidative damage have shown that exposure of purified proteasome to oxidants, including peroxynitrite,200,201 hypochlorite, hydrogen peroxide,202 and 4-hydroxynonenal,203,204 leads to inactivation with the 26S configuration being approximately 10-fold more sensitive.202 In their original study, Bulteau et al.184 report that several α-type subunits of the 20S proteasome are subject to 4-hydroxynonelation following ischemia/reperfusion though it was not clear that the dysfunction was related to the modification of these subunits. Subsequently, they reported that proteasome purified from rat heart seems to be somewhat resistant to inactivation requiring concentrations of 4-hydroxynonenal in excess of 100 μM.27 In collaboration with the Das group,205,206 we have shown that pre-treatment of α-tocotrienol, a Vitamin E analogue, can preserve postischemic proteasome function with preferential effects on the ATP-dependent activity. This predilection for the preferential effect of ischemia/reperfusion on the ATP-dependent activity of the proteasome186,193 coupled with the reported higher vulnerability of the 26S proteasome to oxidative inactivation207 has led to a suggestion that subunits of the 19S regulatory particle, which utilizes ATP, may be more sensitive to oxidative damage than those in the 20S proteasome. In fact, the Rpt3 (S6 ATPase) subunit has been reported to be sensitive to carbonylation reactions in SH-SY5Y cells exposed to an oxidative environment.208 Consistent with this report, we have identified Rpt3/Rpt5 as 26S proteasome subunits significantly carbonylated during myocardial ischemia/reperfusion.209 As indicated in the section on heart failure, this was the same subunit(s) that appeared to be oxidized in explants from human hearts from patients with end-stage heart failure.174
B. The 11S-Activated Proteasome and Myocardial Ischemia
So far, this section has primarily dealt with the role of the 26S proteasome in myocardial ischemia. However, as described in an earlier chapter in this book as well as earlier in this chapter, the 20S proteasome can dock with and be regulated by an additional regulatory particle, the 11S activator ring. This regulatory particle can dock at either end or in combination with a 19S regulatory particle at one end (hybrid proteasome). Various forms of this alternate “zome” exist. If it contains one or more immunoforms of the catalytic subunits (β1i, β2i, β5i), it is often referred to as the immunoproteasome, which as described earlier is primarily involved with antigen presentation. If it is docked with a 20S that does not contain immunoforms, it is called the 11S-activated proteasome, which can remove oxidized proteins as studies have suggested.210 This suggestion is given credence by the recent report that forced overexpression of the PA28α subunit in cardiomyocytes results in increased activity of the proteasome associated with enhanced resistance to an oxidative stress.131 Moreover, a recent study has shown that cardiac-specific overexpression of the same subunit increases proteasome activity, improves postischemic function, and decreases infarct size.132 Previously, we had shown that the proteasome was responsible for removal of proteins oxidized during myocardial ischemia and that this appeared not to require ubiquitin.211 Given the oxidative stress associated with myocardial ischemia197,212 as well as the reported upregulation of this form of the proteasome in a related cardiomyopathy also associated with oxidative stress,213 it is conceivable that the 11S-activated proteasome plays a role in the removal of proteins oxidized during myocardial ischemia. A scheme describing the proposed functions of the 26S- and 11S-activated proteasomes is presented in Fig. 10.
Fig. 10.
Potential roles for the UPS and 11S-activated proteasome in short- and long-duration ischemia. In the nonischemic heart, oxidized, misfolded, and ubiquitinated proteins are degraded through both ubiquitin- and nonubiquitin-mediated pathways, recycling the constituent amino acids, and maintaining a dynamic balance between pro-survival and pro-death signals. During an ischemic insult resulting in cell death or dysfunction, UPS function is inhibited leading to accumulation of oxidized and ubiquitinated proteins. In addition, a condition known as dysregulation may occur in which normal UPS-mediated degradation of pro-death proteins is depressed. Reproduced with permission from Oxford University Press.214
C. The UPS in IPC
As IPC is usually associated with myocardial ischemia, it is discussed in this section. IPC is a procedure to decrease vulnerability of the myocardium to ischemia/reperfusion injury via the prior exposure to short ischemic bursts. IPC tends to result in improved postischemic hemodynamic function and reduced markers of myocardial injury. The215 current thinking is that IPC involves preischemic signaling changes that open the inward mitochondrial KATP channels216 and prevent opening of the mitochondrial permeability transition pore.217 Three studies192,193,209 have suggested that perhaps the UPS plays a role in IPC by facilitating some of the associated pre- or postischemic signaling changes.
Since the UPS may become dysfunctional during ischemia, by necessity IPC must in some way preserve postischemic proteasome function. As was the case with the ischemia studies, earlier studies of IPC protocols to protect the brain from ischemia/reperfusion had already examined this. These studies showed less accumulation of protein aggregates and proapoptotic proteins in the postischemic brain.218-220 One of the earliest studies of the heart suggested that pharmacologic preconditioning of the myocardium with nicorandil affords some protection of postischemic function of the proteasome.185 Nicorandil is an agent thought to open the inward mitochondrial KATP channels221 and thus mimic the effects of IPC. While all three of convincing these recent studies provide evidence that IPC preserves postischemic dysfunction of the UPS including improved peptidase activity and less accumulation of ubi-quitinated or misfolded proteins,192,193,209 they differ in their proposed mechanisms.
1. Preserved Function of the UPS Improves Degradation of δ Protein Kinase C During Reperfusion
Inhibition of the pro-death kinase, δ Protein Kinase C (δPKC), is reported to protect the heart from ischemic injury.222 The pro-survival kinase, εPKC, is activated and translocated to cardiac mitochondria as a result of IPC.223 Churchill et al.193 have shown that IPC improves postischemic UPS function and alters the ratios of these kinases to favor tissue survival. They have proposed that the UPS regulates the ratio between these two kinases because a proteasome inhibitor prevents the effects of IPC on postischemic function and the ratios of δPKC/εPKC. This is an attractive hypothesis; however, IPC alters the levels of many pro-survival and pro-death proteins thought to be regulated by the UPS including PTEN,224,225 IκB,187 and Bax.209 As many diverse pathways are regulated by the UPS, however important it may be, it is likely that UPS-mediated changes in δPKC account for only a portion of the protective effects of IPC.
2. Increased PKA-mediated activation of proteasome
Given the regulation of the cardiac proteasome by PKA (Section II.B above), this theory is highly credible. PKA phosphorylates several subunits of the 20S proteasome and the 19S regulatory particle9,13 which activates proteasome peptidase activity. Asai et al.192 present convincing evidence that PKA can enhance docking of the 20S proteasome and 19S regulatory particle(s), and suggest that IPC results in transient preischemic increases in PKA, which enhance assembly of the intact 26S proteasome. This then carries over to the postischemic period to explain the higher peptidase activity of the proteasome observed in the immediate postischemic period. Unfortunately, because assembly of intact proteasome at this time point was not assessed, the relationship between the improved function and assembly is not clear.
3. Decreased postischemic oxidation of 19S regulatory particle subunits
As described earlier, the Rpt5 subunit, one of the ATPase subunits of the 19S regulatory particle, was recently described as subject to oxidative modification during myocardial ischemia.209 One of the observations leading up to an examination of the vulnerability of 19S regulatory particle subunits to oxidative damage was the preferential effect of ischemia/reperfusion on the ATP-dependent activity of the proteasome.193,209 Divald et al.209 report decreased postischemic carbonylation of the Rpt5 subunit when the heart was subjected to an IPC protocol. As the Rpt5 subunit plays pivotal roles in the attachment of the “base” of the 19S regulatory particle to 20S proteasome α-rings226 and the binding of the 19S particle “lid” to the base,227 it is conceivable that by decreasing oxidation, IPC improves docking of these two complexes. However, as this was not examined, it is not clear whether the improved peptidase activity is related to the decreased oxidation of just this one subunit. Given the complexity of IPC, it is likely that no one single mechanism accounts for the protective effect of this procedure on postischemic proteasome function.
D. Proteasome Inhibition in Myocardial Ischemia: for Better or for Worse?
The same controversy that surrounds the proposed use of proteasome inhibitors to treat atherosclerosis applies to myocardial ischemia. There have been a handful of studies in the literature suggesting that this strategy may be beneficial. As the use of proteasome inhibitors in myocardial ischemia has recently been discussed in depth,228 little time will be spent on it here. Suffice it to say that given the proteasome dysfunction that may be present following myocardial ischemia, this strategy raises grave concerns229 particularly in light of several studies230-232 reporting unexpected cardiac toxicity associated with administration of the proteasome inhibitor bortizomeb (Velcade®) for the treatment of multiple myeloma. This is not to suggest that proteasome inhibitors could not be of value in treating some of the problems associated with myocardial ischemia. The problems with the first generation of proteasome inhibitors centers around their nonspecificity. However, it is conceivable that second-generation proteasome inhibitors with more selective effects, such as the immunoproteasome inhibitor ONYX-0914,233 may be of value due to their potential anti-inflammatory properties. Likewise, we foresee altering degradation of specific proteins through targeting of specific ubiquitin protein ligases as a viable therapeutic intervention in the future.
VIII. Concluding Statement
The last 15 years have witnessed a veritable explosion in research surrounding the various roles and functions of the UPS in cellular physiology and pathophysiology. While often described as simply a means for the removal of unwanted, damaged, or otherwise unneeded proteins, nothing could be further from the truth. In reality, in removing proteins the UPS plays a critical role in regulating many of the intracellular processes necessary for cell function and survival. Given the absolute requirement for the UPS, it was only a matter of time before this system was examined for its potential roles in cardiovascular physiology and pathophysiology. This chapter presents a synopsis of this research. Over the past decade, there have been many advances in our understanding of how the UPS functions in vascular and myocardial tissues and we have come to realize that many of the cardiovascular pathologies are associated with some degree of deviation of UPS function from the norm (increased or decreased). In many of these pathologies, it is not clear whether UPS dysfunction caused the primary disease process or is secondary to it, but it is clear that once the UPS becomes dysfunctional, deterioration is enhanced. However, in the familial proteinopathies, the dysfunctional proteasome plays a role not only in progression of the disease, but also, at least in experimental models, in the genesis of the proteinopathy. Although there is still a relative lack of knowledge, efforts to mitigate cardiovascular pathologies by modifying the function of the UPS are promising as recent studies with genetic models and development of activator and second-generation inhibitors have produced encouraging results. It is hoped that this chapter will spur research to further our knowledge of this system and lead to the development of new UPS-based therapeutic modalities for the mitigation of cardiovascular disease.
Acknowledgments
This work was supported by National Institute of Health grants HL 92954, AG 31750 (to A. L.), HL 68936 (to S. R. P. and X. W.), HL72166 and HL85629 (to X. W.), GM061728 and HL065619 (to C. P.), and American Heart Asociation grant 0740025N (to X. W.).
References
- 1.Gomes AV, Zong C, Edmondson RD, Berhane BT, Wang GW, Le S, et al. The murine cardiac 26S proteasome: an organelle awaiting exploration. Ann N Y Acad Sci. 2005;1047:197–207. doi: 10.1196/annals.1341.018. [DOI] [PubMed] [Google Scholar]
- 2.Gomes AV, Zong C, Edmondson RD, Li X, Stefani E, Zhang J, et al. Mapping the murine cardiac 26S proteasome complexes. Circ Res. 2006;99:362–71. doi: 10.1161/01.RES.0000237386.98506.f7. [DOI] [PubMed] [Google Scholar]
- 3.Drews O, Wildgruber R, Zong C, Sukop U, Nissum M, Weber G, et al. Mammalian proteasome subpopulations with distinct molecular compositions and proteolytic activities. Mol Cell Proteomics. 2007;6:2021–31. doi: 10.1074/mcp.M700187-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Gomes AV, Young GW, Wang Y, Zong C, Eghbali M, Drews O, et al. Contrasting proteome biology and functional heterogeneity of the 20 S proteasome complexes in mammalian tissues. Mol Cell Proteomics. 2009;8:302–15. doi: 10.1074/mcp.M800058-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young GW, Wang Y, Ping P. Understanding proteasome assembly and regulation: importance to cardiovascular medicine. Trends Cardiovasc Med. 2008;18:93–8. doi: 10.1016/j.tcm.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powell SR. The cardiac 26S proteasome. Regulating the regulator. Circ Res. 2006;99:342–5. doi: 10.1161/01.RES.0000239412.40685.61. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Zong C, Koag MC, Wang Y, Drews O, Fang C, et al. Proteome dynamics and proteome function of cardiac 19S proteasomes. Mol Cell Proteomics. 2011;10:M110. doi: 10.1074/mcp.M110.006122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, Su K, Yang X, Bowe DB, Paterson AJ, Kudlow JE. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell. 2003;115:715–25. doi: 10.1016/s0092-8674(03)00974-7. [DOI] [PubMed] [Google Scholar]
- 9.Zhang F, Hu Y, Huang P, Toleman CA, Paterson AJ, Kudlow JE. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. J Biol Chem. 2007;282:22460–71. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]
- 10.Drews O, Tsukamoto O, Liem D, Streicher J, Wang Y, Ping P. Differential regulation of proteasome function in isoproterenol-induced cardiac hypertrophy. Circ Res. 2010;107:1094–101. doi: 10.1161/CIRCRESAHA.110.222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu H, Zong C, Wang Y, Young GW, Deng N, Souda P, et al. Revealing the dynamics of the 20 S proteasome phosphoproteome: a combined CID and electron transfer dissociation approach. Mol Cell Proteomics. 2008;7:2073–89. doi: 10.1074/mcp.M800064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zong C, Young GW, Wang Y, Lu H, Deng N, Drews O, et al. Two-dimensional electrophoresis-based characterization of post-translational modifications of mammalian 20S proteasome complexes. Proteomics. 2008;8:5025–37. doi: 10.1002/pmic.200800387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zong C, Gomes AV, Drews O, Li X, Young GW, Berhane B, et al. Regulation of murine cardiac 20S proteasomes: role of associating partners. Circ Res. 2006;99:372–80. doi: 10.1161/01.RES.0000237389.40000.02. [DOI] [PubMed] [Google Scholar]
- 14.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–95. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 15.Weber C, Zernecke A, Libby P. The multifaceted contributions of leukocyte subsets to atherosclerosis: lessons from mouse models. Nat Rev Immunol. 2008;8:802–15. doi: 10.1038/nri2415. [DOI] [PubMed] [Google Scholar]
- 16.Stangl V, Lorenz M, Meiners S, Ludwig A, Bartsch C, Moobed M, et al. Long-term up-regulation of eNOS and improvement of endothelial function by inhibition of the ubiquitin-proteasome pathway. FASEB J. 2004;18:272–9. doi: 10.1096/fj.03-0054com. [DOI] [PubMed] [Google Scholar]
- 17.Wei Q, Xia Y. Proteasome inhibition down-regulates endothelial nitric-oxide synthase phosphorylation and function. J Biol Chem. 2006;281:21652–9. doi: 10.1074/jbc.M602105200. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann J, Saguner AM, Versari D, Peterson TE, Chade A, Olson M, et al. Chronic proteasome inhibition contributes to coronary atherosclerosis. Circ Res. 2007;101:865–74. doi: 10.1161/CIRCRESAHA.107.152959. [DOI] [PubMed] [Google Scholar]
- 19.Xia T, Dimitropoulou C, Zeng J, Antonova GN, Snead C, Venema RC, et al. Chaperone-dependent E3 ligase CHIP ubiquitinates and mediates proteasomal degradation of soluble guanylyl cyclase. Am J Physiol Cell Physiol. 2007;293:H3080–7. doi: 10.1152/ajpheart.00579.2007. [DOI] [PubMed] [Google Scholar]
- 20.Xu J, Wu Y, Song P, Zhang M, Wang S, Zou MH. Proteasome-dependent degradation of guanosine 5′-triphosphate cyclohydrolase I causes tetrahydrobiopterin deficiency in diabetes mellitus. Circulation. 2007;116:944–53. doi: 10.1161/CIRCULATIONAHA.106.684795. [DOI] [PubMed] [Google Scholar]
- 21.Whitsett J, Picklo MJ, Sr., Vasquez-Vivar J. 4-Hydroxy-2-nonenal increases superoxide anion radical in endothelial cells via stimulated GTP cyclohydrolase proteasomal degradation. Arterioscler Thromb Vasc Biol. 2007;27:2340–7. doi: 10.1161/ATVBAHA.107.153742. [DOI] [PubMed] [Google Scholar]
- 22.Forstermann U, Munzel T. Endothelial nitric oxide synthase in vascular disease: from marvel to menace. Circulation. 2006;113:1708–14. doi: 10.1161/CIRCULATIONAHA.105.602532. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. 2009;48:91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park HM, Kim JA, Kwak MK. Protection against amyloid beta cytotoxicity by sulforaphane: role of the proteasome. Arch Pharm Res. 2009;32:109–15. doi: 10.1007/s12272-009-1124-2. [DOI] [PubMed] [Google Scholar]
- 25.Dreger H, Westphal K, Wilck N, Baumann G, Stangl V, Stangl K, et al. Protection of vascular cells from oxidative stress by proteasome inhibition depends on Nrf2. Cardiovasc Res. 2010;85:395–403. doi: 10.1093/cvr/cvp279. [DOI] [PubMed] [Google Scholar]
- 26.Bieler S, Meiners S, Stangl V, Pohl T, Stangl K. Comprehensive proteomic and transcriptomic analysis reveals early induction of a protective anti-oxidative stress response by low-dose proteasome inhibition. Proteomics. 2009;9:3257–67. doi: 10.1002/pmic.200800927. [DOI] [PubMed] [Google Scholar]
- 27.Farout L, Mary J, Vinh J, Szweda LI, Friguet B. Inactivation of the proteasome by 4-hydroxy-2-nonenal is site specific and dependant on 20S proteasome subtypes. Arch Biochem Biophys. 2006;453:135–42. doi: 10.1016/j.abb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 28.Vieira O, Escargueil-Blanc I, Jurgens G, Borner C, Almeida L, Salvayre R, et al. Oxidized LDLs alter the activity of the ubiquitin-proteasome pathway: potential role in oxidized LDL-induced apoptosis. FASEB J. 2000;14:532–42. doi: 10.1096/fasebj.14.3.532. [DOI] [PubMed] [Google Scholar]
- 29.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med. 2010;49:1603–16. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herrmann J, Ciechanover A, Lerman LO, Lerman A. The ubiquitin-proteasome system in cardiovascular diseases-a hypothesis extended. Cardiovasc Res. 2004;61:11–21. doi: 10.1016/j.cardiores.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 31.Herrmann J, Lerman LO, Mukhopadhyay D, Napoli C, Lerman A. Angiogenesis in atherogenesis. Arterioscler Thromb Vasc Biol. 2006;26:1948–57. doi: 10.1161/01.ATV.0000233387.90257.9b. [DOI] [PubMed] [Google Scholar]
- 32.Kietzmann T, Gorlach A. Reactive oxygen species in the control of hypoxia-inducible factor-mediated gene expression. Semin Cell Dev Biol. 2005;16:474–86. doi: 10.1016/j.semcdb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Bradshaw EL, Li XA, Guerin T, Everson WV, Wilson ME, Bruce-Keller AJ, et al. Nucleoside reverse transcriptase inhibitors prevent HIV protease inhibitor-induced atherosclerosis by ubiquitination and degradation of protein kinase C. Am J Physiol Cell Physiol. 2006;291:C1271–8. doi: 10.1152/ajpcell.00211.2006. [DOI] [PubMed] [Google Scholar]
- 34.Ogura M, Ayaori M, Terao Y, Hisada T, Iizuka M, Takiguchi S, et al. Proteasomal inhibition promotes ATP-binding cassette transporter A1 (ABCA1) and ABCG1 expression and cholesterol efflux from macrophages in vitro and in vivo. Arterioscler Thromb Vasc Biol. 2011;31:1980–7. doi: 10.1161/ATVBAHA.111.228478. [DOI] [PubMed] [Google Scholar]
- 35.Masuda Y, Itabe H, Odaki M, Hama K, Fujimoto Y, Mori M, et al. ADRP/adipophilin is degraded through the proteasome-dependent pathway during regression of lipid-storing cells. J Lipid Res. 2006;47:87–98. doi: 10.1194/jlr.M500170-JLR200. [DOI] [PubMed] [Google Scholar]
- 36.Kikuchi J, Furukawa Y, Kubo N, Tokura A, Hayashi N, Nakamura M, et al. Induction of ubiquitin-conjugating enzyme by aggregated low density lipoprotein in human macrophages and its implications for atherosclerosis. Arterioscler Thromb Vasc Biol. 2000;20:128–34. doi: 10.1161/01.atv.20.1.128. [DOI] [PubMed] [Google Scholar]
- 37.Xie P, Fan Y, Zhang H, Zhang Y, She M, Gu D, et al. CHIP represses myocardin-induced smooth muscle cell differentiation via ubiquitin-mediated proteasomal degradation. Mol Cell Biol. 2009;29:2398–408. doi: 10.1128/MCB.01737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meiners S, Laule M, Rother W, Guenther C, Prauka I, Muschick P, et al. Ubiquitin-proteasome pathway as a new target for the prevention of restenosis. Circulation. 2002;105:483–9. doi: 10.1161/hc0402.102951. [DOI] [PubMed] [Google Scholar]
- 39.Barringhaus KG, Matsumura ME. The proteasome inhibitor lactacystin attenuates growth and migration of vascular smooth muscle cells and limits the response to arterial injury. Exp Clin Cardiol. 2007;12:119–24. [PMC free article] [PubMed] [Google Scholar]
- 40.Ying WZ, Zhang HG, Sanders PW. EGF receptor activity modulates apoptosis induced by inhibition of the proteasome of vascular smooth muscle cells. J Am Soc Nephrol. 2007;18:131–42. doi: 10.1681/ASN.2006040333. [DOI] [PubMed] [Google Scholar]
- 41.Kapadia MR, Eng JW, Jiang Q, Stoyanovsky DA, Kibbe MR. Nitric oxide regulates the 26S proteasome in vascular smooth muscle cells. Nitric Oxide. 2009;20:279–88. doi: 10.1016/j.niox.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 42.Katsuyama K, Shichiri M, Marumo F, Hirata Y. NO inhibits cytokine-induced iNOS expression and NF-kappaB activation by interfering with phosphorylation and degradation of IkappaB-alpha. Arterioscler Thromb Vasc Biol. 1998;18:1796–802. doi: 10.1161/01.atv.18.11.1796. [DOI] [PubMed] [Google Scholar]
- 43.Takami Y, Nakagami H, Morishita R, Katsuya T, Cui TX, Ichikawa T, et al. Ubiquitin carboxyl-terminal hydrolase L1, a novel deubiquitinating enzyme in the vasculature, attenuates NF-kappaB activation. Arterioscler Thromb Vasc Biol. 2007;27:2184–90. doi: 10.1161/ATVBAHA.107.142505. [DOI] [PubMed] [Google Scholar]
- 44.Takami Y, Nakagami H, Morishita R, Katsuya T, Hayashi H, Mori M, et al. Potential role of CYLD (Cylindromatosis) as a deubiquitinating enzyme in vascular cells. Am J Pathol. 2008;172:818–29. doi: 10.2353/ajpath.2008.070312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duwel M, Welteke V, Oeckinghaus A, Baens M, Kloo B, Ferch U, et al. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J Immunol. 2009;182:7718–28. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- 46.Weyand CM, Younge BR, Goronzy JJ. T cells in arteritis and atherosclerosis. Curr Opin Lipidol. 2008;19:469–77. doi: 10.1097/mol.0b013e32830bfdc2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shrikhande GV, Scali ST, da Silva CG, Damrauer SM, Csizmadia E, Putheti P, et al. O-glycosylation regulates ubiquitination and degradation of the anti-inflammatory protein A20 to accelerate atherosclerosis in diabetic ApoE-null mice. PLoS One. 2010;5:e14240. doi: 10.1371/journal.pone.0014240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lobry C, Lopez T, Israel A, Weil R. Negative feedback loop in T cell activation through IkappaB kinase-induced phosphorylation and degradation of Bcl10. Proc Natl Acad Sci USA. 2007;104:908–13. doi: 10.1073/pnas.0606982104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berges C, Haberstock H, Fuchs D, Miltz M, Sadeghi M, Opelz G, et al. Proteasome inhibition suppresses essential immune functions of human CD4+ T cells. Immunology. 2008;124:234–46. doi: 10.1111/j.1365-2567.2007.02761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawahara I, Kitagawa N, Tsutsumi K, Nagata I, Hayashi T, Koji T. The expression of vascular dendritic cells in human atherosclerotic carotid plaques. Hum Pathol. 2007;38:1378–85. doi: 10.1016/j.humpath.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 51.Naujokat C, Berges C, Hoh A, Wieczorek H, Fuchs D, Ovens J, et al. Proteasomal chymotrypsin-like peptidase activity is required for essential functions of human monocyte-derived dendritic cells. Immunology. 2007;120:120–32. doi: 10.1111/j.1365-2567.2006.02487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Straube C, Wehner R, Wendisch M, Bornhauser M, Bachmann M, Rieber EP, et al. Bortezomib significantly impairs the immunostimulatory capacity of human myeloid blood dendritic cells. Leukemia. 2007;21:1464–71. doi: 10.1038/sj.leu.2404734. [DOI] [PubMed] [Google Scholar]
- 53.Nencioni A, Garuti A, Schwarzenberg K, Cirmena G, Dal Bello G, Rocco I, et al. Proteasome inhibitor-induced apoptosis in human monocyte-derived dendritic cells. Eur J Immunol. 2006;36:681–9. doi: 10.1002/eji.200535298. [DOI] [PubMed] [Google Scholar]
- 54.van der Heijden JW, Oerlemans R, Lems WF, Scheper RJ, Dijkmans BA, Jansen G. The proteasome inhibitor bortezomib inhibits the release of NFkappaB-inducible cytokines and induces apoptosis of activated T cells from rheumatoid arthritis patients. Clin Exp Rheumatol. 2009;27:92–8. [PubMed] [Google Scholar]
- 55.Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta. 2009;1787:414–20. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meier P, Golshayan D, Blanc E, Pascual M, Burnier M. Oxidized LDL modulates apoptosis of regulatory T cells in patients with ESRD. J Am Soc Nephrol. 2009;20:1368–84. doi: 10.1681/ASN.2008070734. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Taleb S, Tedgui A, Mallat Z. Regulatory T-cell immunity and its relevance to atherosclerosis. J Intern Med. 2008;263:489–99. doi: 10.1111/j.1365-2796.2008.01944.x. [DOI] [PubMed] [Google Scholar]
- 58.Yang Z, Gagarin D, Laurent G, III, Hammell N, Toma I, Hu CA, et al. Cardiovascular inflammation and lesion cell apoptosis: a novel connection via the interferon-inducible immunoproteasome. Arterioscler Thromb Vasc Biol. 2009;29:1213–9. doi: 10.1161/ATVBAHA.109.189407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang-Yen HF. Mcl-1: a highly regulated cell death and survival controller. J Biomed Sci. 2006;13:201–4. doi: 10.1007/s11373-005-9064-4. [DOI] [PubMed] [Google Scholar]
- 60.Cuconati A, Mukherjee C, Perez D, White E. DNA damage response and MCL-1 destruction initiate apoptosis in adenovirus-infected cells. Genes Dev. 2003;17:2922–32. doi: 10.1101/gad.1156903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martinet W, De Meyer GR. Autophagy in atherosclerosis. Curr Atheroscler Rep. 2008;10:216–23. doi: 10.1007/s11883-008-0034-y. [DOI] [PubMed] [Google Scholar]
- 62.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–24. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Herck JL, De Meyer GR, Martinet W, Bult H, Vrints CJ, Herman AG. Proteasome inhibitor bortezomib promotes a rupture-prone plaque phenotype in ApoE-deficient mice. Basic Res Cardiol. 2010;105:39–50. doi: 10.1007/s00395-009-0054-y. [DOI] [PubMed] [Google Scholar]
- 64.Versari D, Herrmann J, Gossl M, Mannheim D, Sattler K, Meyer FB, et al. Dysregulation of the ubiquitin-proteasome system in human carotid atherosclerosis. Arterioscler Thromb Vasc Biol. 2006;26:2132–9. doi: 10.1161/01.ATV.0000232501.08576.73. [DOI] [PubMed] [Google Scholar]
- 65.Herrmann J, Soares SM, Lerman LO, Lerman A. Potential role of the ubiquitin-proteasome system in atherosclerosis: aspects of a protein quality disease. J Am Coll Cardiol. 2008;51:2003–10. doi: 10.1016/j.jacc.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 66.Herrmann J, Edwards WD, Holmes DR, Jr., Shogren KL, Lerman LO, Ciechanover A, et al. Increased ubiquitin immunoreactivity in unstable atherosclerotic plaques associated with acute coronary syndromes. J Am Coll Cardiol. 2002;40:1919–27. doi: 10.1016/s0735-1097(02)02564-0. [DOI] [PubMed] [Google Scholar]
- 67.Marfella R, D’Amico M, Di Filippo C, Baldi A, Siniscalchi M, Sasso FC, et al. Increased activity of the ubiquitin-proteasome system in patients with symptomatic carotid disease is associated with enhanced inflammation and may destabilize the atherosclerotic plaque: effects of rosiglitazone treatment. J Am Coll Cardiol. 2006;47:2444–55. doi: 10.1016/j.jacc.2006.01.073. [DOI] [PubMed] [Google Scholar]
- 68.Marfella R, D’Amico M, Esposito K, Baldi A, Di Filippo C, Siniscalchi M, et al. The ubiquitin-proteasome system and inflammatory activity in diabetic atherosclerotic plaques: effects of rosiglitazone treatment. Diabetes. 2006;55:622–32. doi: 10.2337/diabetes.55.03.06.db05-0832. [DOI] [PubMed] [Google Scholar]
- 69.Marfella R, Di Filippo C, Baldi A, Siniscalchi M, Sasso FC, Crescenzi B, et al. The vascular smooth muscle cells apoptosis in asymptomatic diabetic carotid plaques: role of glycemic control. J Am Coll Cardiol. 2006;47:2118–20. doi: 10.1016/j.jacc.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 70.Marfella R, Di Filippo C, Laieta MT, Vestini R, Barbieri M, Sangiulo P, et al. Effects of ubiquitin-proteasome system deregulation on the vascular senescence and atherosclerosis process in elderly patients. J Gerontol A Biol Sci Med Sci. 2008;63:200–3. doi: 10.1093/gerona/63.2.200. [DOI] [PubMed] [Google Scholar]
- 71.Friguet B, Bulteau AL, Chondrogianni N, Conconi M, Petropoulos I. Protein degradation by the proteasome and its implications in aging. Ann N Y Acad Sci. 2000;908:143–54. doi: 10.1111/j.1749-6632.2000.tb06643.x. [DOI] [PubMed] [Google Scholar]
- 72.Bulteau AL, Szweda LI, Friguet B. Age-dependent declines in proteasome activity in the heart. Arch Biochem Biophys. 2002;397:298–304. doi: 10.1006/abbi.2001.2663. [DOI] [PubMed] [Google Scholar]
- 73.Tan C, Li Y, Tan X, Pan H, Huang W. Inhibition of the ubiquitin-proteasome system: a new avenue for atherosclerosis. Clin Chem Lab Med. 2006;44:1218–25. doi: 10.1515/CCLM.2006.209. [DOI] [PubMed] [Google Scholar]
- 74.Feng B, Zhang Y, Mu J, Ye Z, Zeng W, Qi W, et al. Preventive effect of a proteasome inhibitor on the formation of accelerated atherosclerosis in rabbits with uremia. J Cardiovasc Pharmacol. 2010;55:129–38. doi: 10.1097/FJC.0b013e3181c87f8e. [DOI] [PubMed] [Google Scholar]
- 75.Meiners S, Ludwig A, Stangl V, Stangl K. Proteasome inhibitors: poisons and remedies. Med Res Rev. 2008;28:309–27. doi: 10.1002/med.20111. [DOI] [PubMed] [Google Scholar]
- 76.Bao J, Sato K, Li M, Gao Y, Abid R, Aird W, et al. PR-39 and PR-11 peptides inhibit ischemia-reperfusion injury by blocking proteasome-mediated I kappa B alpha degradation. Am J Physiol Heart Circ Physiol. 2001;281:H2612–8. doi: 10.1152/ajpheart.2001.281.6.H2612. [DOI] [PubMed] [Google Scholar]
- 77.Gao Y, Lecker S, Post MJ, Hietaranta AJ, Li J, Volk R, et al. Inhibition of ubiquitin-proteasome pathway-mediated I kappa B alpha degradation by a naturally occurring antibacterial peptide. J Clin Invest. 2000;106:439–48. doi: 10.1172/JCI9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luss H, Schmitz W, Neumann J. A proteasome inhibitor confers cardioprotection. Cardiovasc Res. 2002;54:140–51. doi: 10.1016/s0008-6363(02)00232-8. [DOI] [PubMed] [Google Scholar]
- 79.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–8. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 80.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA. 2001;98:14440–5. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ni YG, Berenji K, Wang N, Oh M, Sachan N, Dey A, et al. Foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation. 2006;114:1159–68. doi: 10.1161/CIRCULATIONAHA.106.637124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pye J, Ardeshirpour F, McCain A, Bellinger DA, Merricks E, Adams J, et al. Proteasome inhibition ablates activation of NF-kappa B in myocardial reperfusion and reduces reperfusion injury. Am J Physiol Heart Circ Physiol. 2003;284:H919–26. doi: 10.1152/ajpheart.00851.2002. [DOI] [PubMed] [Google Scholar]
- 84.Wilkins BJ, Dai YS, Bueno OF, Parsons SA, Xu J, Plank DM, et al. Calcineurin/NFATcoupling participates in pathological, but not physiological, cardiac hypertrophy. Circ Res. 2004;94:110–8. doi: 10.1161/01.RES.0000109415.17511.18. [DOI] [PubMed] [Google Scholar]
- 85.Li HH, Kedar V, Zhang C, McDonough H, Arya R, Wang DZ, et al. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Invest. 2004;114:1058–71. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stansfield WE, Moss NC, Willis MS, Tang R, Selzman CH. Proteasome inhibition attenuates infarct size and preserves cardiac function in a murine model of myocardial ischemia-reperfusion injury. Ann Thorac Surg. 2007;84:120–5. doi: 10.1016/j.athoracsur.2007.02.049. [DOI] [PubMed] [Google Scholar]
- 87.Johnson SA, Hunter T. Kinomics: methods for deciphering the kinome. Nat Methods. 2005;2:17–25. doi: 10.1038/nmeth731. [DOI] [PubMed] [Google Scholar]
- 88.Li HH, Willis MS, Lockyer P, Miller N, McDonough H, Glass DJ, et al. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J Clin Invest. 2007;117:3211–23. doi: 10.1172/JCI31757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coleman PS, Parmacek MS, Lesch M, Samarel AM. Protein synthesis and degradation during regression of thyroxine-induced cardiac hypertrophy. J Mol Cell Cardiol. 1989;21:911–25. doi: 10.1016/0022-2828(89)90759-1. [DOI] [PubMed] [Google Scholar]
- 90.Wilkinson KD, Ventii KH, Friedrich KL, Mullally JE. The ubiquitin signal: assembly, recognition and termination. Symposium on ubiquitin and signaling. EMBO Rep. 2005;6:815–20. doi: 10.1038/sj.embor.7400506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Centner T, Yano J, Kimura E, McElhinny AS, Pelin K, Witt CC, et al. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J Mol Biol. 2001;306:717–26. doi: 10.1006/jmbi.2001.4448. [DOI] [PubMed] [Google Scholar]
- 92.McElhinny AS, Kakinuma K, Sorimachi H, Labeit S, Gregorio CC. Muscle-specific RING finger-1 interacts with titin to regulate sarcomeric M-line and thick filament structure and may have nuclear functions via its interaction with glucocorticoid modulatory element binding protein-1. J Cell Biol. 2002;157:125–36. doi: 10.1083/jcb.200108089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spencer JA, Eliazer S, Ilaria RL, Jr., Richardson JA, Olson EN. Regulation of microtubule dynamics and myogenic differentiation by MURF, a striated muscle RING-finger protein. J Cell Biol. 2000;150:771–84. doi: 10.1083/jcb.150.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adams V, Linke A, Wisloff U, Doring C, Erbs S, Krankel N, et al. Myocardial expression of Murf-1 and MAFbx after induction of chronic heart failure: effect on myocardial contractility. Cardiovasc Res. 2007;73:120–9. doi: 10.1016/j.cardiores.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 95.Razeghi P, Baskin KK, Sharma S, Young ME, Stepkowski S, Essop MF, et al. Atrophy, hypertrophy, and hypoxemia induce transcriptional regulators of the ubiquitin proteasome system in the rat heart. Biochem Biophys Res Commun. 2006;342:361–4. doi: 10.1016/j.bbrc.2006.01.163. [DOI] [PubMed] [Google Scholar]
- 96.Parmacek MS, Magid NM, Lesch M, Decker RS, Samarel AM. Cardiac protein synthesis and degradation during thyroxine-induced left ventricular hypertrophy. Am J Physiol. 1986;251:C727–36. doi: 10.1152/ajpcell.1986.251.5.C727. [DOI] [PubMed] [Google Scholar]
- 97.Willis MS, Ike C, Li L, Wang DZ, Glass DJ, Patterson C. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res. 2007;100:456–9. doi: 10.1161/01.RES.0000259559.48597.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, et al. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185:1083–95. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- 100.Arya R, Kedar V, Hwang JR, McDonough H, Li HH, Taylor J, et al. Muscle ring finger protein-1 inhibits PKC{epsilon} activation and prevents cardiomyocyte hypertrophy. J Cell Biol. 2004;167:1147–59. doi: 10.1083/jcb.200402033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Willis MS, Rojas M, Li L, Selzman CH, Tang RH, Stansfield WE, et al. Muscle ring finger 1 mediates cardiac atrophy in vivo. Am J Physiol Heart Circ Physiol. 2009;296:H997–H1006. doi: 10.1152/ajpheart.00660.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang C, Xu Z, He XR, Michael LH, Patterson C. CHIP, a cochaperone/ubiquitin ligase that regulates protein quality control, is required for maximal cardioprotection after myocardial infarction in mice. Am J Physiol Heart Circ Physiol. 2005;288:H2836–42. doi: 10.1152/ajpheart.01122.2004. [DOI] [PubMed] [Google Scholar]
- 103.Hoshijima M. Mechanical stress-strain sensors embedded in cardiac cytoskeleton: Z disk, titin, and associated structures. Am J Physiol Heart Circ Physiol. 2006;290:H1313–25. doi: 10.1152/ajpheart.00816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gregorio CC, Perry CN, McElhinny AS. Functional properties of the titin/connectin-associated proteins, the muscle-specific RING finger proteins (MURFs), in striated muscle. J Muscle Res Cell Motil. 2005;26:389–400. doi: 10.1007/s10974-005-9021-x. [DOI] [PubMed] [Google Scholar]
- 105.Mrosek M, Labeit D, Witt S, Heerklotz H, von Castelmur E, Labeit S, et al. Molecular determinants for the recruitment of the ubiquitin-ligase MuRF-1 onto M-line titin. FASEB J. 2007;21:1383–92. doi: 10.1096/fj.06-7644com. [DOI] [PubMed] [Google Scholar]
- 106.Foo RS, Chan LK, Kitsis RN, Bennett MR. Ubiquitination and degradation of the anti-apoptotic protein ARC by MDM2. J Biol Chem. 2007;282:5529–35. doi: 10.1074/jbc.M609046200. [DOI] [PubMed] [Google Scholar]
- 107.Xiong S, Van Pelt CS, Elizondo-Fraire AC, Fernandez-Garcia B, Lozano G. Loss of Mdm4 results in p53-dependent dilated cardiomyopathy. Circulation. 2007;115:2925–30. doi: 10.1161/CIRCULATIONAHA.107.689901. [DOI] [PubMed] [Google Scholar]
- 108.Fielitz J, van Rooij E, Spencer JA, Shelton JM, Latif S, van der Nagel R, et al. Loss of muscle-specific RING-finger 3 predisposes the heart to cardiac rupture after myocardial infarction. Proc Natl Acad Sci USA. 2007;104:4377–82. doi: 10.1073/pnas.0611726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fielitz J, Kim MS, Shelton JM, Latif S, Spencer JA, Glass DJ, et al. Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J Clin Invest. 2007;117:2486–95. doi: 10.1172/JCI32827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Maron BJ. Triggers for sudden cardiac death in the athlete. Cardiol Clin. 1996;14:195–210. doi: 10.1016/s0733-8651(05)70273-3. [DOI] [PubMed] [Google Scholar]
- 111.Bonne G, Carrier L, Richard P, Hainque B, Schwartz K. Familial hypertrophic cardiomyopathy: from mutations to functional defects. Circ Res. 1998;83:580–93. doi: 10.1161/01.res.83.6.580. [DOI] [PubMed] [Google Scholar]
- 112.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–32. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 113.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–28. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 114.Flavigny J, Robert P, Camelin JC, Schwartz K, Carrier L, Berrebi-Bertrand I. Biomolecular interactions between human recombinant beta-MyHC and cMyBP-Cs implicated in familial hypertrophic cardiomyopathy. Cardiovasc Res. 2003;60:388–96. doi: 10.1016/j.cardiores.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 115.Freiburg A, Gautel M. A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur J Biochem. 1996;235:317–23. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- 116.Gruen M, Gautel M. Mutations in beta-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J Mol Biol. 1999;286:933–49. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 117.Sarikas A, Carrier L, Schenke C, Doll D, Flavigny J, Lindenberg KS, et al. Impairment of the ubiquitin-proteasome system by truncated cardiac myosin binding protein C mutants. Cardiovasc Res. 2005;66:33–44. doi: 10.1016/j.cardiores.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 118.Squire JM, Luther PK, Knupp C. Structural evidence interaction and for the of C-protein (MyBP-C) with actin sequence identification of a possible actin-binding domain. J Mol Biol. 2003;331:713–24. doi: 10.1016/s0022-2836(03)00781-2. [DOI] [PubMed] [Google Scholar]
- 119.Moolman JA, Reith S, Uhl K, Bailey S, Gautel M, Jeschke B, et al. A newly created splice donor site in exon 25 of the MyBP-C gene is responsible for inherited hypertrophic cardiomyopathy with incomplete disease penetrance. Circulation. 2000;101:1396–402. doi: 10.1161/01.cir.101.12.1396. [DOI] [PubMed] [Google Scholar]
- 120.Wang X, Su H, Ranek MJ. Protein quality control and degradation in cardiomyocytes. J Mol Cell Cardiol. 2008;45:11–27. doi: 10.1016/j.yjmcc.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–90. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121:3689–700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang X, Li J, Zheng H, Su H, Powell SR. Proteasome functional insufficiency in cardiac pathogenesis. Am J Physiol Heart Circ Physiol. 2011;301:H2207–19. doi: 10.1152/ajpheart.00714.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–5. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 125.Dong X, Liu J, Zheng H, Glasford JW, Huang W, Chen QH, et al. In situ dynamically monitoring the proteolytic function of the ubiquitin-proteasome system in cultured cardiac myocytes. Am J Physiol Heart Circ Physiol. 2004;287:H1417–25. doi: 10.1152/ajpheart.01233.2003. [DOI] [PubMed] [Google Scholar]
- 126.Kumarapeli RKA, Horak KM, Glasford JW, Li J, Chen Q, Liu J, et al. A novel transgenic mouse model reveals deregulation of the ubiquitin-proteasome system in the heart by doxorubicin. FASEB J. 2005;19:2051–3. doi: 10.1096/fj.05-3973fje. [DOI] [PubMed] [Google Scholar]
- 127.Gilon T, Chomsky O, Kulka RG. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 1998;17:2759–66. doi: 10.1093/emboj/17.10.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gilon T, Chomsky O, Kulka RG. Degradation signals recognized by the Ubc6p-Ubc7p ubiquitin-conjugating enzyme pair. Mol Cell Biol. 2000;20:7214–9. doi: 10.1128/mcb.20.19.7214-7219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Griffin TA, Nandi D, Cruz M, Fehling HJ, Kaer LV, Monaco JJ, et al. Immunoproteasome assembly: cooperative incorporation of interferon gamma (IFN-gamma)-inducible subunits. J Exp Med. 1998;187:97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroter F, et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142:613–24. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 131.Li J, Powell SR, Wang X. Enhancement of proteasome function by PA28alpha; overexpression protects against oxidative stress. FASEB J. 2011;25:883–93. doi: 10.1096/fj.10-160895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McLendon PM, Robbins J. Desmin-related cardiomyopathy: an unfolding story. Am J Physiol Heart Circ Physiol. 2011;301:H1220–8. doi: 10.1152/ajpheart.00601.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang X, Osinska H, Klevitsky R, Gerdes AM, Nieman M, Lorenz J, et al. Expression of R120G-alphaB-crystallin causes aberrant desmin and alphaB-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89:84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 134.Wang X, Osinska H, Dorn GW, Nieman M, Lorenz JN, Gerdes AM, et al. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103:2402–7. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- 135.Munoz-Marmol AM, Strasser G, Isamat M, Coulombe PA, Yang Y, Roca X, et al. A dysfunctional desmin mutation in a patient with severe generalized myopathy. Proc Natl Acad Sci USA. 1998;95:11312–7. doi: 10.1073/pnas.95.19.11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vicart P, Caron A, Guicheney P, Li Z, Prevost MC, Faure A, et al. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–5. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 137.Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, et al. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97:1018–26. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 138.Liu J, Chen Q, Huang W, Horak KM, Zheng H, Mestril R, et al. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. FASEB J. 2006;20:362–4. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- 139.Liu J, Tang M, Mestril R, Wang X. Aberrant protein aggregation is essential for a mutant desmin to impair the proteolytic function of the ubiquitin-proteasome system in cardiomyocytes. J Mol Cell Cardiol. 2006;40:451–4. doi: 10.1016/j.yjmcc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 140.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, et al. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–8. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hedhli N, Depre C. Proteasome inhibitors and cardiac cell growth. Cardiovasc Res. 2010;85:321–9. doi: 10.1093/cvr/cvp226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tang M, Li J, Huang W, Su H, Liang Q, Tian Z, et al. Proteasome functional insufficiency activates the calcineurin-NFAT pathway in cardiomyocytes and promotes maladaptive remodelling of stressed mouse hearts. Cardiovasc Res. 2010;88:424–33. doi: 10.1093/cvr/cvq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gianni D, Li A, Tesco G, McKay KM, Moore J, Raygor K, et al. Protein aggregates and novel presenilin gene variants in idiopathic dilated cardiomyopathy. Circulation. 2010;121:1216–26. doi: 10.1161/CIRCULATIONAHA.109.879510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, et al. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci USA. 2004;101:10132–6. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sanbe A, Gulick J, Hanks MC, Liang Q, Osinska H, Robbins J. Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ Res. 2003;92:609–16. doi: 10.1161/01.RES.0000065442.64694.9F. [DOI] [PubMed] [Google Scholar]
- 146.Marian AJ. Hypertrophic cardiomyopathy: from genetics to treatment. Eur J Clin Invest. 2010;40:360–9. doi: 10.1111/j.1365-2362.2010.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Maloyan A, Gulick J, Glabe CG, Kayed R, Robbins J. Exercise reverses preamyloid oligomer and prolongs survival in alphaB-crystallin-based desmin-related cardiomyopathy. Proc Natl Acad Sci USA. 2007;104:5995–6000. doi: 10.1073/pnas.0609202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pattison JS, Sanbe A, Maloyan A, Osinska H, Klevitsky R, Robbins J. Cardiomyocyte expression of a polyglutamine preamyloid oligomer causes heart failure. Circulation. 2008;117:2743–51. doi: 10.1161/CIRCULATIONAHA.107.750232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sanbe A, Osinska H, Villa C, Gulick J, Klevitsky R, Glabe CG, et al. Reversal of amyloid-induced heart disease in desmin-related cardiomyopathy. Proc Natl Acad Sci USA. 2005;102:13592–7. doi: 10.1073/pnas.0503324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Goldberg AL. On prions, proteasomes, and mad cows. N Engl J Med. 2007;357:1150–2. doi: 10.1056/NEJMcibr073962. [DOI] [PubMed] [Google Scholar]
- 151.Kristiansen M, Deriziotis P, Dimcheff DE, Jackson GS, Ovaa H, Naumann H, et al. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol Cell. 2007;26:175–88. doi: 10.1016/j.molcel.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 152.Bennett EJ, Bence NF, Jayakumar R, Kopito RR. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell. 2005;17:351–65. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 153.Sanbe A, Yamauchi J, Miyamoto Y, Fujiwara Y, Murabe M, Tanoue A. Interruption of CryAB-amyloid oligomer formation by HSP22. J Biol Chem. 2007;282:555–63. doi: 10.1074/jbc.M605481200. [DOI] [PubMed] [Google Scholar]
- 154.Maloyan A, Sanbe A, Osinska H, Westfall M, Robinson D, Imahashi K, et al. Mitochondrial dysfunction and apoptosis underlie the pathogenic process in alpha-B-crystallin desmin-related cardiomyopathy. Circulation. 2005;112:3451–61. doi: 10.1161/CIRCULATIONAHA.105.572552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tsukamoto O, Minamino T, Okada K, Shintani Y, Takashima S, Kato H, et al. Depression of proteasome activities during the progression of cardiac dysfunction in pressure-overloaded heart of mice. Biochem Biophys Res Commun. 2006;340:1125–33. doi: 10.1016/j.bbrc.2005.12.120. [DOI] [PubMed] [Google Scholar]
- 156.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–28. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kumarapeli AR, Su H, Huang W, Tang M, Zheng H, Horak KM, et al. Alpha B-crystallin suppresses pressure overload cardiac hypertrophy. Circ Res. 2008;103:1473–82. doi: 10.1161/CIRCRESAHA.108.180117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sanbe A, Daicho T, Mizutani R, Endo T, Miyauchi N, Yamauchi J, et al. Protective effect of geranylgeranylacetone via enhancement of HSPB8 induction in desmin-related cardiomyopathy. PLoS One. 2009;4:e5351. doi: 10.1371/journal.pone.0005351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zheng H, Tang M, Zheng Q, Kumarapeli AR, Horak KM, Tian Z, et al. Doxycycline attenuates protein aggregation in cardiomyocytes and improves survival of a mouse model of cardiac proteinopathy. J Am Coll Cardiol. 2010;56:1418–26. doi: 10.1016/j.jacc.2010.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hunt SA, Abraham WT, Chin MH, Feldman AM, Francis GS, Ganiats TG, et al. 2009 Focused update incorporated into the ACC/AHA 2005 Guidelines for the Diagnosis and Management of Heart Failure in Adults a Report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines Developed in Collaboration With the International Society for Heart and Lung Transplantation. J Am Coll Cardiol. 2009;53:e1–e90. doi: 10.1016/j.jacc.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 161.Depre C, Wang Q, Yan L, Hedhli N, Peter P, Chen L, et al. Activation of the cardiac proteasome during pressure overload promotes ventricular hypertrophy. Circulation. 2006;114:1821–8. doi: 10.1161/CIRCULATIONAHA.106.637827. [DOI] [PubMed] [Google Scholar]
- 162.Stansfield WE, Tang RH, Moss NC, Baldwin AS, Willis MS, Selzman CH. Proteasome inhibition promotes regression of left ventricular hypertrophy. Am J Physiol Heart Circ Physiol. 2008;294:H645–50. doi: 10.1152/ajpheart.00196.2007. [DOI] [PubMed] [Google Scholar]
- 163.Meiners S, Dreger H, Fechner M, Bieler S, Rother W, Gunther C, et al. Suppression of cardiomyocyte hypertrophy by inhibition of the ubiquitin-proteasome system. Hypertension. 2008;51:302–8. doi: 10.1161/HYPERTENSIONAHA.107.097816. [DOI] [PubMed] [Google Scholar]
- 164.Hedhli N, Lizano P, Hong C, Fritzky LF, Dhar SK, Liu H, et al. Proteasome inhibition decreases cardiac remodeling after initiation of pressure overload. Am J Physiol Heart Circ Physiol. 2008;295:H1385–93. doi: 10.1152/ajpheart.00532.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hedhli N, Wang L, Wang Q, Rashed E, Tian Y, Sui X, et al. Proteasome activation during cardiac hypertrophy by the chaperone H11 Kinase/Hsp22. Cardiovasc Res. 2008;77:497–505. doi: 10.1093/cvr/cvm054. [DOI] [PubMed] [Google Scholar]
- 166.Urso ML, Clarkson PM, Hittel D, Hoffman EP, Thompson PD. Changes in ubiquitin proteasome pathway gene expression in skeletal muscle with exercise and statins. Arterioscler Thromb Vasc Biol. 2005;25:2560–6. doi: 10.1161/01.ATV.0000190608.28704.71. [DOI] [PubMed] [Google Scholar]
- 167.Reid MB. Response of the ubiquitin-proteasome pathway to changes in muscle activity. Am J Physiol Regul Integr Comp Physiol. 2005;288:R1423–31. doi: 10.1152/ajpregu.00545.2004. [DOI] [PubMed] [Google Scholar]
- 168.Nedergaard A, Vissing K, Overgaard K, Kjaer M, Schjerling P. Expression patterns of atrogenic and ubiquitin proteasome component genes with exercise: effect of different loading patterns and repeated exercise bouts. J Appl Physiol. 2007;103:1513–22. doi: 10.1152/japplphysiol.01445.2006. [DOI] [PubMed] [Google Scholar]
- 169.Murton AJ, Constantin D, Greenhaff PL. The involvement of the ubiquitin proteasome system in human skeletal muscle remodelling and atrophy. Biochim Biophys Acta. 2008;1782:730–43. doi: 10.1016/j.bbadis.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 170.Adams V, Linke A, Gielen S, Erbs S, Hambrecht R, Schuler G. Modulation of Murf-1 and MAFbx expression in the myocardium by physical exercise training. Eur J Cardiovasc Prev Rehabil. 2008;15:293–9. doi: 10.1097/HJR.0b013e3282f3ec43. [DOI] [PubMed] [Google Scholar]
- 171.Usui S, Maejima Y, Pain J, Hong C, Cho J, Park JY, et al. Endogenous muscle atrophy F-box mediates pressure overload-induced cardiac hypertrophy through regulation of nuclear factor-kappaB. Circ Res. 2011;109:161–71. doi: 10.1161/CIRCRESAHA.110.238717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hein S, Arnon E, Kostin S, Schonburg M, Elsasser A, Polyakova V, et al. Progression from compensated hypertrophy to failure in the pressure-overloaded human heart: structural deterioration and compensatory mechanisms. Circulation. 2003;107:984–91. doi: 10.1161/01.cir.0000051865.66123.b7. [DOI] [PubMed] [Google Scholar]
- 173.Weekes J, Morrison K, Mullen A, Wait R, Barton P, Dunn MJ. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics. 2003;3:208–16. doi: 10.1002/pmic.200390029. [DOI] [PubMed] [Google Scholar]
- 174.Predmore JM, Wang P, Davis F, Bartolone S, Westfall MV, Dyke DB, et al. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation. 2010;21:997–1004. doi: 10.1161/CIRCULATIONAHA.109.904557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wohlschlaeger J, Sixt SU, Stoeppler T, Schmitz KJ, Levkau B, Tsagakis K, et al. Ventricular unloading is associated with increased 20s proteasome protein expression in the myocardium. J Heart Lung Transplant. 2010;29:125–32. doi: 10.1016/j.healun.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 176.Kassiotis C, Ballal K, Wellnitz K, Vela D, Gong M, Salazar R, et al. Markers of autophagy are downregulated in failing human heart after mechanical unloading. Circulation. 2009;120:S191–7. doi: 10.1161/CIRCULATIONAHA.108.842252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zheng Q, Su H, Tian Z, Wang X. Proteasome malfunction activates macroautophagy in the heart. Am J Cardiovasc Dis. 2011;1:214–26. [PMC free article] [PubMed] [Google Scholar]
- 178.Voigt A, Bartel K, Egerer K, Trimpert C, Feist E, Gericke C, et al. Humoral anti-proteasomal autoimmunity in dilated cardiomyopathy. Basic Res Cardiol. 2010;105:9–18. doi: 10.1007/s00395-009-0061-z. [DOI] [PubMed] [Google Scholar]
- 179.Otsuka K, Terasaki F, Shimomura H, Tsukada B, Horii T, Isomura T, et al. Enhanced expression of the ubiquitin-proteasome system in the myocardium from patients with dilated cardiomyopathy referred for left ventriculoplasty: an immunohistochemical study with special reference to oxidative stress. Heart Vessels. 2010;25:474–84. doi: 10.1007/s00380-010-0006-3. [DOI] [PubMed] [Google Scholar]
- 180.Hayashi T, Takada K, Matsuda M. Post-transient ischemia increase in ubiquitin conjugates in the early reperfusion. Neuroreport. 1992;3:519–20. doi: 10.1097/00001756-199206000-00016. [DOI] [PubMed] [Google Scholar]
- 181.Kamikubo T, Hayashi T. Changes in proteasome activity following transient ischemia. Neurochem Int. 1996;28:209–12. doi: 10.1016/0197-0186(95)00071-2. [DOI] [PubMed] [Google Scholar]
- 182.Keller JN, Huang FF, Zhu H, Yu J, Ho YS, Kindy TS. Oxidative stress-associated impairment of proteasome activity during ischemia-reperfusion injury. J Cereb Blood Flow Metab. 2000;20:1467–73. doi: 10.1097/00004647-200010000-00008. [DOI] [PubMed] [Google Scholar]
- 183.Asai A, Tanahashi N, Qiu JH, Saito N, Chi S, Kawahara N, et al. Selective proteasomal dysfunction in the hippocampal CA1 region after transient forebrain ischemia. J Cereb Blood Flow Metab. 2002;22:705–10. doi: 10.1097/00004647-200206000-00009. [DOI] [PubMed] [Google Scholar]
- 184.Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, et al. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276:30057–63. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 185.Powell SR, Wang P, Katzeff HL, Shringarpure R, Teoh C, Khaliulin I, et al. Oxidized and ubiquitinated proteins may predict recovery of postischemic cardiac function. Essential role of the proteasome. Antioxid Redox Signal. 2005;7:538–46. doi: 10.1089/ars.2005.7.538. [DOI] [PubMed] [Google Scholar]
- 186.Powell SR, Davies KJA, Divald A. Optimal determination of heart tissue 26S Proteasome activity requires maximal stimulating concentrations of ATP. J Mol Cell Cardiol. 2007;42:265–9. doi: 10.1016/j.yjmcc.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gurusamy N, Goswami S, Malik G, Das DK. Oxidative injury induces selective rather than global inhibition of proteasomal activity. J Mol Cell Cardiol. 2008;44:419–28. doi: 10.1016/j.yjmcc.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 188.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 189.Horwitz AA, Navon A, Groll M, Smith DM, Reis C, Goldberg AL. ATP-induced structural transitions in PAN, the proteasome-regulatory ATPase complex in Archaea. J Biol Chem. 2007;282:22921–9. doi: 10.1074/jbc.M702846200. [DOI] [PubMed] [Google Scholar]
- 190.Huang H, Zhang X, Li S, Liu N, Lian W, McDowell E, et al. Physiological levels of ATP negatively regulate proteasome function. Cell Res. 2010;20:1372–85. doi: 10.1038/cr.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Jennings RB, Steenbergen C., Jr. Nucleotide metabolism and cellular damage in myocardial ischemia. Annu Rev Physiol. 1985;47:727–49. doi: 10.1146/annurev.ph.47.030185.003455. [DOI] [PubMed] [Google Scholar]
- 192.Asai M, Tsukamoto O, Minamino T, Asanuma H, Fujita M, Asano Y, et al. PKA rapidly enhances proteasome assembly and activity in in vivo canine hearts. J Mol Cell Cardiol. 2009;46:452–62. doi: 10.1016/j.yjmcc.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 193.Churchill EN, Ferreira JC, Brum PC, Szweda LI, Mochly-Rosen D. Ischaemic preconditioning improves proteasomal activity and increases the degradation of deltaPKC during reperfusion. Cardiovasc Res. 2010;85:385–94. doi: 10.1093/cvr/cvp334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Canton M, Neverova I, Menabo R, Van Eyk J, Di Lisa F. Evidence of myofibrillar protein oxidation induced by postischemic reperfusion in isolated rat hearts. Am J Physiol Heart Circ Physiol. 2004;286:H870–7. doi: 10.1152/ajpheart.00714.2003. [DOI] [PubMed] [Google Scholar]
- 195.Khaliulin I, Schwalb H, Wang P, Houminer E, Grinberg L, Katzeff H, et al. Preconditioning improves postischemic mitochondrial function and diminishes oxidation of mitochondrial proteins. Free Radic Biol Med. 2004;37:1–9. doi: 10.1016/j.freeradbiomed.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 196.Park Y, Kanekal S, Kehrer JP. Oxidative changes in hypoxic rat heart tissue. Am J Physiol Heart Circ Physiol. 1991;260:H1395–405. doi: 10.1152/ajpheart.1991.260.5.H1395. [DOI] [PubMed] [Google Scholar]
- 197.Powell SR, Gurzenda EM, Wahezi SE. Actin is oxidized during myocardial ischemia. Free Radic Biol Med. 2001;30:1171–6. doi: 10.1016/s0891-5849(01)00514-7. [DOI] [PubMed] [Google Scholar]
- 198.Davies KJ, Lin SW, Pacifici RE. Protein damage and degradation by oxygen radicals. IV. Degradation of denatured protein. J Biol Chem. 1987;262:9914–20. [PubMed] [Google Scholar]
- 199.Davies KJ, Delsignore ME. Protein damage and degradation by oxygen radicals. III. Modification of secondary and tertiary structure. J Biol Chem. 1987;262:9908–13. [PubMed] [Google Scholar]
- 200.Amici M, Lupidi G, Angeletti M, Fioretti E, Eleuteri AM. Peroxynitrite-induced oxidation and its effects on isolated proteasomal systems. Free Radic Biol Med. 2003;34:987–96. doi: 10.1016/s0891-5849(02)01369-2. [DOI] [PubMed] [Google Scholar]
- 201.Osna NA, Haorah J, Krutik VM, Donohue TM., Jr. Peroxynitrite alters the catalytic activity of rodent liver proteasome in vitro and in vivo. Hepatology. 2004;40:574–82. doi: 10.1002/hep.20352. [DOI] [PubMed] [Google Scholar]
- 202.Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJ, Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem J. 1998;335:637–42. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Friguet B, Szweda LI. Inhibition of the multicatalytic proteinase (proteasome) by 4-hydroxy-2-nonenal cross-linked protein. FEBS Lett. 1997;405:21–5. doi: 10.1016/s0014-5793(97)00148-8. [DOI] [PubMed] [Google Scholar]
- 204.Okada K, Wangpoengtrakul C, Osawa T, Toyokuni S, Tanaka K, Uchida K. 4-Hydroxy-2-nonenal-mediated impairment of intracellular proteolysis during oxidative stress. Identification of proteasomes as target molecules. J Biol Chem. 1999;274:23787–93. doi: 10.1074/jbc.274.34.23787. [DOI] [PubMed] [Google Scholar]
- 205.Das S, Powell SR, Wang P, Divald A, Nesaretnam K, Tosaki A, et al. Cardioprotection with palm tocotrienol: antioxidant activity of tocotrienol is linked with its ability to stabilize proteasomes. Am J Physiol Heart Circ Physiol. 2005;289:361–7. doi: 10.1152/ajpheart.01285.2004. [DOI] [PubMed] [Google Scholar]
- 206.Das S, Lekli I, Das M, Szabo G, Varadi J, Juhasz B, et al. Cardioprotection with palm oil tocotrienols: comparision of different isomers. Am J Physiol Heart Circ Physiol. 2008;294:H970–8. doi: 10.1152/ajpheart.01200.2007. [DOI] [PubMed] [Google Scholar]
- 207.Reinheckel T, Ullrich O, Sitte N, Grune T. Differential impairment of 20S and 26S proteasome activities in human hematopoietic K562 cells during oxidative stress. Arch Biochem Biophys. 2000;377:65–8. doi: 10.1006/abbi.2000.1717. [DOI] [PubMed] [Google Scholar]
- 208.Ishii T, Sakurai T, Usami H, Uchida K. Oxidative modification of proteasome: identification of an oxidation-sensitive subunit in 26 s proteasome. Biochemistry. 2005;44:13893–901. doi: 10.1021/bi051336u. [DOI] [PubMed] [Google Scholar]
- 209.Divald A, Kivity S, Wang P, Hochhauser E, Roberts B, Teichberg S, et al. Myocardial ischemic preconditioning preserves postischemic function of the 26S proteasome through diminished oxidative damage to 19S regulatory particle subunits. Circ Res. 2010;106:1829–38. doi: 10.1161/CIRCRESAHA.110.219485. [DOI] [PubMed] [Google Scholar]
- 210.Pickering AM, Koop AL, Teoh CY, Ermak G, Grune T, Davies KJ. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem J. 2010;432:585–94. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Divald A, Powell SR. Proteasome mediates removal of proteins oxidized during myocardial ischemia. Free Radic Biol Med. 2006;40:156–64. doi: 10.1016/j.freeradbiomed.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 212.Powell SR, Hall D. Use of salicylate as a probe for •OH formation in isolated ischemic rat hearts. Free Radic Biol Med. 1990;9:133–41. doi: 10.1016/0891-5849(90)90116-z. [DOI] [PubMed] [Google Scholar]
- 213.Powell SR, Samuel SM, Wang P, Divald A, Thirunavukkarasu M, Koneru S, et al. Upregulation of myocardial 11S-activated proteasome in experimental hyperglycemia. J Mol Cell Cardiol. 2008;44:618–21. doi: 10.1016/j.yjmcc.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 214.Powell SR, Divald A. The ubiquitin-proteasome system in myocardial ischaemia and preconditioning. Cardiovasc Res. 2010;85:303–11. doi: 10.1093/cvr/cvp321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Murry CE, Jennings RB, Reimer KA. New insights into potential mechanisms of ischemic preconditioning. Circulation. 1991;84:442–5. doi: 10.1161/01.cir.84.1.442. [DOI] [PubMed] [Google Scholar]
- 216.Gross GJ, Fryer RM. Sarcolemmal versus mitochondrial ATP-sensitive K+ channels and myocardial preconditioning. Circ Res. 1999;84:973–9. doi: 10.1161/01.res.84.9.973. [DOI] [PubMed] [Google Scholar]
- 217.Korge P, Honda HM, Weiss JN. Protection of cardiac mitochondria by diazoxide and protein kinase C: implications for ischemic preconditioning. Proc Natl Acad Sci USA. 2002;99:3312–7. doi: 10.1073/pnas.052713199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Meller R, Cameron JA, Torrey DJ, Clayton CE, Ordonez AN, Henshall DC, et al. Rapid degradation of Bim by the ubiquitin-proteasome pathway mediates short-term ischemic tolerance in cultured neurons. J Biol Chem. 2006;281:7429–36. doi: 10.1074/jbc.M512138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pradillo JM, Romera C, Hurtado O, Cardenas A, Moro MA, Leza JC, et al. TNFR1 upregulation mediates tolerance after brain ischemic preconditioning. J Cereb Blood Flow Metab. 2005;25:193–203. doi: 10.1038/sj.jcbfm.9600019. [DOI] [PubMed] [Google Scholar]
- 220.Liu C, Chen S, Kamme F, Hu BR. Ischemic preconditioning prevents protein aggregation after transient cerebral ischemia. Neuroscience. 2005;134:69–80. doi: 10.1016/j.neuroscience.2005.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Sato T, Sasaki N, O’Rourke B, Marban E. Nicorandil, a potent cardioprotective agent, acts by opening mitochondrial ATP-dependent potassium channels. J Am Coll Cardiol. 2000;35:514–8. doi: 10.1016/s0735-1097(99)00552-5. [DOI] [PubMed] [Google Scholar]
- 222.Inagaki K, Chen L, Ikeno F, Lee FH, Imahashi K, Bouley DM, et al. Inhibition of delta-protein kinase C protects against reperfusion injury of the ischemic heart in vivo. Circulation. 2003;108:2304–7. doi: 10.1161/01.CIR.0000101682.24138.36. [DOI] [PubMed] [Google Scholar]
- 223.Baines CP, Song CX, Zheng YT, Wang GW, Zhang J, Wang OL, et al. Protein kinase Cepsilon interacts with and inhibits the permeability transition pore in cardiac mitochondria. Circ Res. 2003;92:873–80. doi: 10.1161/01.RES.0000069215.36389.8D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Cai Z, Semenza GL. PTEN activity is modulated during ischemia and reperfusion: involvement in the induction and decay of preconditioning. Circ Res. 2005;97:1351–9. doi: 10.1161/01.RES.0000195656.52760.30. [DOI] [PubMed] [Google Scholar]
- 225.Cai Z, Shen Z, Kaer LV, Becker LC. Ischemic preconditioning-induced cardioprotection is lost in mice with immunoproteasome subunit low molecular mass polypeptide-2 deficiency. FASEB J. 2008;22:4248–57. doi: 10.1096/fj.08-105940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential role of the C-termini of AAA subunits of PA700 (19S regulator) in asymmetric assembly and activation of the 26S proteasome. J Biol Chem. 2008;283:31813–22. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.DeMartino GN. Purification of PA700, the 19S regulatory complex of the 26S proteasome. Methods Enzymol. 2005;398:295–306. doi: 10.1016/S0076-6879(05)98024-5. [DOI] [PubMed] [Google Scholar]
- 228.Yu X, Kem DC. Proteasome inhibition during myocardial infarction. Cardiovasc Res. 2010;85:312–20. doi: 10.1093/cvr/cvp309. [DOI] [PubMed] [Google Scholar]
- 229.Powell SR. Proteasome inhibitors in myocardial ischemia, some concerns. Ann Thorac Surg. 2008;85:1503–4. doi: 10.1016/j.athoracsur.2007.10.049. [DOI] [PubMed] [Google Scholar]
- 230.Voortman J, Giaccone G. Severe reversible cardiac failure after bortezomib treatment combined with chemotherapy in a non-small cell lung cancer patient: a case report. BMC Cancer. 2006;6:129. doi: 10.1186/1471-2407-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Enrico O, Gabriele B, Nadia C, Sara G, Daniele V, Giulia C, et al. Unexpected cardiotoxicity in haematological bortezomib treated patients. Br J Haematol. 2007;138:396–7. doi: 10.1111/j.1365-2141.2007.06659.x. [DOI] [PubMed] [Google Scholar]
- 232.Hacihanefioglu A, Tarkun P, Gonullu E. Acute severe cardiac failure in a myeloma patient due to proteasome inhibitor bortezomib. Int J Hematol. 2008;88:219–22. doi: 10.1007/s12185-008-0139-7. [DOI] [PubMed] [Google Scholar]
- 233.Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15:781–7. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]