Abstract
The virion infectivity factor (Vif) accessory protein of HIV-1 forms a complex with the cellular cytidine deaminase APOBEC3G (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G) to block its antiviral activity. The antiviral property of APOBEC3G is conserved in several mammalian species, but the ability of Vif to block this activity is species-specific. HIV-1 Vif blocks human APOBEC3G but does not block the mouse or African green monkey (AGM) enzyme. Conversely, SIVAGM Vif blocks the antiviral activity of AGM but not human APOBEC3G. We demonstrate that the species specificity is caused by a single amino acid difference in APOBEC3G. Replacement of Asp-128 in human APOBEC3G with the Lys-128 of AGM APOBEC3G caused the enzyme to switch its interaction, becoming sensitive to SIVAGM Vif and resistant to HIV-1 Vif. Conversely, the reciprocal Lys to Asp switch in AGM APOBEC3G reversed its specificity for Vif. The reversal of biological activity was accompanied by the corresponding switch in the species specificity with which the enzyme physically associated with Vif and was excluded from virions. The charge of the amino acid at position 128 was a critical determinant of species specificity. Based on the crystal structure of the distantly related Escherichia coli cytidine deaminase, we propose that this amino acid is positioned on a solvent-exposed loop of APOBEC3G on the same face of the protein as the catalytic site.
Lentiviruses, with the exception of equine infectious anemia virus, encode the accessory protein virion infectivity factor (Vif). HIV-1 Vif is a 24-kDa cytoplasmic protein that is expressed from a partially spliced mRNA late in the replication cycle (1, 2). It lacks obvious homology to previously described proteins. Vif is required for HIV-1 replication in primary cells and in some transformed T cell lines, yet simpler retroviruses such as murine leukemia viruses and avian leucosis viruses lack Vif and are fully replication-competent. HIV-1 that lacks Vif (Δvif) fails to replicate in primary T cells and in transformed T cell lines such as CEM, Hut78, PM1, and MT2, which are termed nonpermissive, but replicates to wild-type levels in cell lines such as HeLa.CD4, SupT1, C8166, and CEMss, which are termed permissive (3-5). Cell:cell fusion experiments suggested that nonpermissive cells contained a dominant inhibitor that interfered with the replication of Δvif but not wild-type HIV-1 (6, 7). The hypothesized inhibitor was identified by Sheehy et al. (8), who used a subtractive cloning approach to identify a cDNA, CEM15 [later named APOBEC3G (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G)], whose expression is restricted to nonpermissive cells. Transfer of the cDNA into the permissive cell line CEMss rendered cells nonpermissive, demonstrating the critical role of APOBEC3G in the phenotype. These findings suggested that the role of Vif is to overcome the antiviral activity of APOBEC3G.
APOBEC3G is a member of the APOBEC family of cytidine deaminases that includes APOBEC1, -2, and -3 and activation-induced deaminase (AID) (9, 10). APOBEC1 is the catalytic subunit of the apoB mRNA-editing enzyme that catalyzes the deamination of C6666 to U in the mRNA-encoding apolipoprotein B, generating a premature termination codon (11-13). APOBEC1 also has DNA mutator activity that induces C → T changes when expressed in Escherichia coli (10). In rodents, APOBEC3 is encoded by a single gene. In humans, the APOBEC3 gene has been expanded to include seven members, APOBEC3A to -G, that lie in tandem on chromosome 22 (9). AID acts to hypermutate Ig variable genes and deaminates class switch DNA sequences to activate Ig isotype class switch recombination (9, 14-16). APOBEC3G contains a signature Cys-His motif that forms a Zn2+ coordination domain characteristic of cytidine deaminases (9). The enzyme consists of an amino-terminal catalytic domain fused to a pseudocatalytic domain, suggesting that it arose through an internal duplication of the coding sequence. APOBEC genes are expressed with characteristic tissue and cell-type specificity. APOBEC1 is primarily expressed in the intestine; APOBEC3G is expressed in lymphoid and myeloid lineage cells (9).
In cells infected with Δvif HIV-1, APOBEC3G is encapsidated into the assembling virions, causing a two to three order of magnitude reduction in infectivity of the newly formed virus. The defective virions are competent to enter target cells and to synthesize full-length double stranded viral DNA; however, most of this DNA fails to integrate and few proviruses are generated (8, 17, 18). The viral DNA contained numerous G → A mutations caused by deamination of minus-strand cytosines to uracil (18-22). In cells infected with wild-type HIV-1, little APOBEC3G was encapsidated into the virions, and G → A mutations in the viral reverse transcripts were rare (18, 23, 24). Vif physically associates with APOBEC3G (18, 23-25), inducing its rapid ubiquitination and degradation by a proteasome-dependent pathway (23, 24, 26, 27).
The ability of Vif to block the antiviral activity of APOBEC3G is species-specific (18). HIV-1, SIVAGM, and SIVMac Vif block the APOBEC3G of the species from which they are derived but generally do not block those of other species. HIV-1 Vif failed to neutralize the antiviral activity of African green monkey (AGM) or rhesus APOBEC3G (18). Similarly, SIVAGM Vif was unable to neutralize human or macaque APOBEC3G. Only SIVMac Vif, which blocked human and AGM APOBEC3G, was able to block nonhomologous APOBEC3G. Mouse APOBEC3G, the sequence of which is diverged from its primate homologues, was not blocked by any of the Vifs tested, making it a potent inhibitor of lentiviral replication. The species specificity of Vif function correlated with the ability of Vif to physically associate with APOBEC3G. HIV-1 Vif formed a complex with human APOBEC3G but not with the mouse enzyme (18), suggesting that its species specificity results from its ability to physically associate with APOBEC3G.
Here, we explored the mechanism of Vif:APOBEC3G species specificity. We localized the specificity-determining region of APOBEC3G to a single amino acid. This amino acid specified the functional interaction, the physical association with Vif, and the exclusion of the enzyme from virions. These data provide an indication of the Vif interaction site on APOBEC3G and demonstrate that species specificity results from the ability of Vif to physically associate with individual APOBEC3Gs.
Methods
APOBEC3G Expression Vectors. Human and AGM APOBEC3G chimeras and point mutants were generated by overlapping extension PCR by using human and AGM APOBEC3G expression plasmids (described in ref. 18) as templates. 5′ and 3′ fragments were amplified separately by using a primer specific for the overlap region and a 5′ external primer, CEM15-CM1, containing an EcoRI site (5′-TAA GCG GAA TTC GGC CCT GGG AGG TCA CTT TAG GG) or a 3′ primer, CEM15 HA-C, containing an XhoI site (5′-TAG AAG CTC GAG TCA AGC GTA ATC TGG AAC ATC GTA TGG ATA GTT TTC CTG ATT CTG GAG). The 5′ and 3′ fragments were then mixed and amplified with the two external primers. The resulting full-length APOBEC3G gene was cleaved with restriction enzymes to generate sticky ends and cloned into pcDNA3.1 (Invitrogen) at the EcoRI and XhoI sites. All constructs were sequenced.
Luciferase Reporter Virus Assay of Vif/APOBEC3G Function. Vif and APOBEC3G function was measured by using VSV-G pseudotyped single-cycle luciferase reporter virus as described (18). Briefly, 293T cells were cotransfected by lipofection with 2 μg of wild-type or Δvif NL4-3-based single-cycle luciferase reporter virus plasmid pNL-LucR-E- or the SIVAGM Tantalus plasmid pSIVAGM-Luc, 2 μg of pcAPOBEC3G-HA expression vector or pcDNA3.1 vector control and 1 μg of pcVSV-G. Virus-containing supernatant was harvested 2 days posttransfection, filtered, and quantitated by p24 or p27 ELISA. To measure the infectivity of the viruses, 1 × 10-4 HOS.CD4 cells were infected (1.0 ng of p24 or p27) in 96-well culture dishes, in triplicate. Three days later, intracellular luciferase activity was quantitated by using Luc-Lite Plus reagent (Packard) and a Topcount luminometer (Perkin-Elmer). The data are presented as mean cps ± SE of the triplicates. The amount of APOBEC3G in 20 μg of the transfected cell lysate was quantitated on immunoblots probed with anti-hemagglutinin (HA) mAb 16B12 (Covance, Princeton) as described (18).
Virion Encapsidation. Virion-encapsidated APOBEC3G was quantitated as described (18). Briefly, 5.0 ml of culture supernatant from transfected 293T cells was pelleted by ultracentrifugation in an SW41 rotor (Beckman) at 30,000 rpm for 1.5 h through a layer of 20% sucrose. The pellet was solubilized in 100 μl of 1% Triton-containing buffer. p24 or p27 content in the lysed virions was measured by ELISA, and an amount containing 100 ng was analyzed on immunoblots probed with anti-HA mAb 16B12.
Detection of Vif:APOBEC3G Complexes. 293T cells were cotransfected with viral plasmid and APOBEC3G-HA expression vector at a ratio of 1:1 and lysed 2 days posttransfection in CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) buffer (5 mM CHAPS/50 mM NaCl/20 mM Tris, pH 7.5). The lysates containing 200 μg of protein were precleared by incubation with 50 μl of protein-A agarose beads for 1 h at 37°C. Complexes were immunoprecipitated with 1 μg of 16B12 anti-HA mAb (Covance) for 1 h, followed by addition of 20 μl of protein A-agarose beads for 1 h. These steps were carried out at 37°C to maximize differences in stability of the complexes. The beads were washed in CHAPS buffer three times and analyzed on immunoblots with anti-HA mAb or rabbit antiserum to HIV-1 Vif no. 2221, contributed to the AIDS Research and Reference Reagent Program by D. Gabuzda (Dana-Farber Cancer Center).
Results
Amino Acid 128 Determines the Vif Interaction Specificity of Human, AGM, and Macaque APOBEC3G. To identify a region of APOBEC3G that determines the species specificity of the interaction with Vif, we constructed a panel of human/AGM APOBEC3G chimeras containing a C-terminal HA epitope tag (Fig. 1A). Chimeras A1 to -5 contained increasing lengths of 5′ AGM sequence joined to human APOBEC3G sequence. Reciprocal chimeras H1 to -5 contained increasing lengths of 5′ human APOBEC3G sequence fused to 3′ AGM sequence. The chimeras were tested by using a single-cycle luciferase reporter virus assay to determine their antiviral activity and functional interaction with Vif (18, 28). Wild-type and Δvif NL4-3-based reporter virus stocks were produced in 293T cells that were cotransfected with APOBEC3G expression vector. The viruses were normalized for p24 concentration, and their infectivity was measured on HOS.CD4 cells by quantitation of intracellular luciferase activity 3 days postinfection. APOBEC3G expression vector and reporter virus plasmid were transfected at a 1:1 ratio, which provided a sensitive measure of Vif function. This ratio almost completely suppressed Δvif virus whereas wild-type virus infectivity was reduced only ≈2-fold. In the absence of APOBEC3G, Δvif virus was generally 30-50% more infectious than wild-type (Fig. 1B). This relatively small difference probably reflects a weak negative inhibitory activity of Vif.
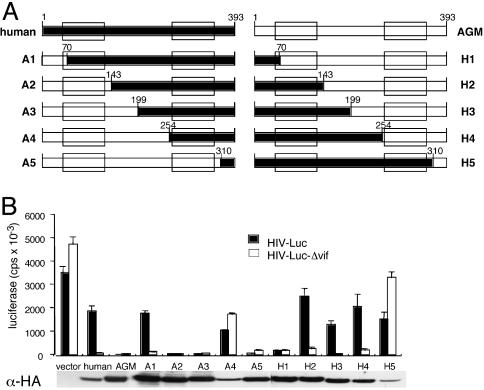
Fig. 1.
Analysis of human:AGM APOBEC3G chimeras maps the Vif specificity-determining region to residues 1-143. (A) Structure of human:AGM APOBEC3G chimeras. Filled bars denote human sequence; open bars denote AGM sequence. The Zn2+-coordination domains are boxed. (B) Luciferase reporter virus assay of chimeric APOBEC3Gs. Wild-type and Δvif luciferase reporter viruses were produced in 293T cells cotransfected with chimeric APOBEC3G expression. vector, Empty vector control. Reporter virus infectivity was measured on HOS.CD4 cells infected with viruses normalized to 1.0 ng of p24. The data are the average of triplicates ± SD. Chimeric APOBEC3G expression in the transfected 293T producer cells detected on immunoblots is shown in Lower.
Analysis of the chimeras using the luciferase reporter virus assay defined a region of APOBEC3G that determines the specificity of the Vif interaction (Fig. 1B). Chimera A1 containing the 70 N-terminal amino acids of AGM APOBEC3G, was, like human APOBEC3G, sensitive to HIV-1 Vif. In contrast, chimera A2, in which the AGM sequence was extended to amino acid 143, was resistant. Chimeras A3 and A5, in which the length of AGM sequence was further extended, were also HIV-1 Vif-resistant. Chimera A4 was only weakly active on wild-type and Δvif virus. Immunoblot analysis of this protein showed that it was poorly expressed. Of the reciprocal chimeras, H1 containing the N-terminal 70 aa of human APOBEC3G was, like AGM APOBEC3G, HIV-1 Vif-resistant. In contrast, chimera H2, in which the human sequence was extended to amino acid 143, was sensitive. Chimeras H3 and H4, in which the human sequence was further extended, were also HIV-1 Vif sensitive. Chimera H5 was inactive and poorly expressed. Taken together, these findings mapped the Vif species specificity determining domain to amino acids 1-143, most likely to within amino acids 71-143 (Fig. 2A).
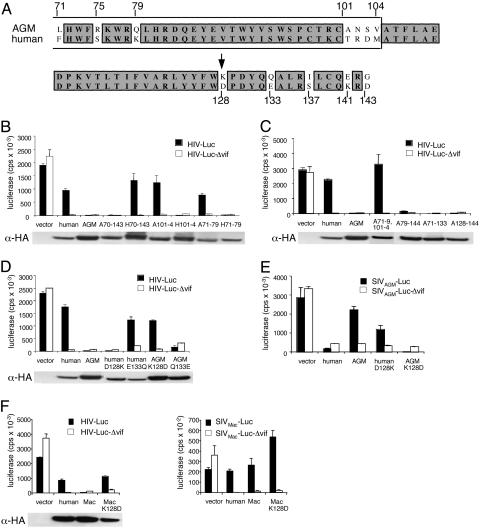
Fig. 2.
Amino acid 128 of APOBEC3G determines the species specificity of the functional interaction with Vif. (A) Alignment of amino acids 71-143 of human and AGM APOBEC3G. The Zn2+-coordination domain is boxed. (B-D) Chimeric and point mutant APOBEC3G activity was determined in the luciferase reporter virus assay as described in Methods. vector, Empty vector control. (E) Amino acid 128 APOBEC3G mutants were tested for their functional interaction with SIVAGM Vif by using SIVAGM reporter virus. (F) The role of amino acid 128 in macaque APOBEC3G was tested by using HIV-1 and SIVMac luciferase reporter viruses. APOBEC3G in the transfected 293T producer cells was detected on immunoblots (Lower, each histogram). Representative results from one of three repetitions of the experiment are shown.
Comparison of the human and AGM APOBEC3G sequence within the region 71-143 showed that there are two conserved stretches (79-100 and 105-127), a patch of four nonconserved residues (101-104) and eight isolated single amino acid changes (71, 75, 79, 128, 133, 137, 141, and 143) (Fig. 2A). To determine which of these differences was important for the interaction with Vif, additional chimeras and point mutants were constructed. A pair of reciprocal chimeras A70-143 (human sequence with amino acids 70-143 of AGM APOBEC3G) and H70-143 (AGM sequence with region 70-143 of human APOBEC3G) narrowed the active domain to 70-143 (Fig. 2B). The reciprocal chimeras A101-104 (human sequence with amino acids 101-104 from AGM APBEC3G) and H101-104 (AGM sequence with amino acids 101-104 from human APOBEC3G) demonstrated that the patch from 101-104 was not critical (Fig. 2B). Chimeras A71-79 and H71-79 showed that the single amino acid differences between 71-79 have no influence for the interaction with HIV-1 Vif. Indeed, exchange of the two patches together (A71-79,101-104) showed that the combination did not affect the interaction with Vif (Fig. 2C). In contrast, chimeras A79-143 (human APOBEC3G with AGM 79-143) and A71-133 (human APOBEC3G with AGM 71-133) were Vif-resistant, mapping the determinant to within amino acids 79-133 (Fig. 2C). Chimera A128-143 was HIV-1 Vif-resistant, narrowing the active region to 128-133. The human and AGM enzymes differed in this region at only two positions, 128 (Asp in human and Lys in AGM) and 133 (Glu in human and Gln in AGM). Substitution of Asp-128 in human APOBEC3G with the corresponding Lys (D128K) in AGM APOBEC3G resulted in resistance to HIV-1 Vif (Fig. 2D). The reciprocal substitution in AGM APOBEC3G (K128D) caused the enzyme to become sensitive to HIV-1 Vif. In contrast, the exchange at position 133 (human E133Q and AGM Q133E) had no effect.
The role of amino acid 128 in determining the functional interaction of APOBEC3G with SIVAGM Vif was tested by using SIVAGM luciferase reporter viruses (Fig. 2E). Substitution of Asp with Lys in human APOBEC3G (D128K) caused the enzyme to become sensitive to SIVAGM Vif. Conversely, substitution of Lys with Asp in AGM APOBEC3G (K128D) caused the enzyme to become resistant to SIVAGM Vif. Taken together, these data demonstrated that position 128 determines the species specificity of the functional interaction of APOBEC3G with Vif.
Macaque APOBEC3G, like the AGM enzyme, is resistant to HIV-1 Vif (18). This resistance poses a block to replication of HIV-1 in macaque cells. Interestingly, macaque APOBEC3G, like the AGM enzyme, has a Lys at position 128. When Lys-128 was changed to Asp, macaque APOBEC3G became sensitive to HIV Vif (Fig. 2F). SIVmac Vif is relatively permissive in its ability to interact with different APOBEC3Gs (18). Consistent with this finding, macaque K128D APOBEC3G retained its sensitivity to SIVmac Vif.
In these analyses, the interaction specificity of the mutant APOBEC3Gs was tested with a single ratio of Vif:APOBEC3G. To determine whether the exchange of amino acid 128 completely changed the specificity of the functional interaction with Vif, we titrated APOBEC3G levels in the reporter virus assay. As the amount of human APOBEC3G increased, the infectivity of the wild-type virus decreased (Fig. 6 Top Left, which is published as supporting information on the PNAS web site). This effect was more pronounced with AGM APOBEC3G, which is resistant to HIV-1 Vif. In contrast, AGM K128D APOBEC3G was as resistant to Vif as the wild-type human enzyme. Against Δvif reporter virus, the three APOBEC3Gs were equally active. This result demonstrated that the amino acid exchange had not affected their enzymatic or antiviral activities (Fig. 6 Lower Left). Conversely, on SIVAGM, human D128K APOBEC3G showed a similar ability to resistant Vif (Fig. 6 Upper Right). The three enzymes were similarly active on Δvif virus (Fig. 6 Lower Right).
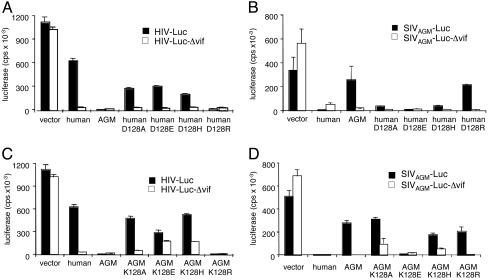
The Charge at Amino Acid 128 Determines the Functional Interaction with Vif. The Asp to Lys exchange changed the overall charge of the enzyme by 2. To determine the effect of charge at this position on the interaction with Vif, AGM and human APOBEC3G were substituted at position 128 with Ala, Glu, His, or Arg. Substitution in human APOBEC3G of Asp-128 with Ala, Glu, or His had only a minor effect (≈2-fold) on the sensitivity to HIV-1 Vif (Fig. 3A). In contrast, substitution with Arg caused resistance to HIV-1 Vif. These findings demonstrated that the interaction of human APOBEC3G with HIV-1 Vif requires a negative or neutral charge at position 128. The Ala, Glu, and His substitutions had no effect on resistance to SIVAGM Vif (Fig. 3B). In contrast, substitution with Arg (D128K) caused the enzyme to become sensitive to SIVAGM. Thus, a strong positive charge at this position is incompatible for the interaction with SIVAGM Vif.
Fig. 3.
The charge of amino acid 128 determines the specificity of the functional interaction with Vif. The functional interaction of human (A and B) and AGM (C and D) APOBEC3G-containing charge changes at amino acid 128 were measured on HIV-1 (A and C) and SIVAGM (B and D) luciferase reporter viruses. The experiment was repeated in three experiments with similar results.
Substitution of Lys-128 in AGM APOBEC3G with Ala caused the enzyme to become sensitive to HIV-1 Vif (Fig. 3C). Glu and His substitutions caused a reduction in the overall activity of the enzyme as judged by their reduced ability to block Δvif HIV-1, and, as a result, a definitive conclusion regarding their functional interaction with HIV-1 Vif could not be drawn. The conservative substitution of Lys-128 with Arg had no effect on resistance to HIV-1 Vif and remained the wild-type AGM APOBEC3G phenotype. Ala- and His-substituted AGM APOBEC3G remained sensitive to SIVAGM Vif (Fig. 3D). In contrast, substitution with Glu resulted in resistance to SIVAGM Vif. These findings led to the conclusion that, to functionally interact with SIVAGM Vif, amino acid 128 of AGM APOBEC3G must be positive or neutral but cannot be negative. In contrast, to interact with HIV-1 Vif, this position must be negatively charged or neutral. Thus, a neutral charge at amino acid 128 in AGM APOBEC3G results in a “dual-tropic” APOBEC3G that is sensitive to both Vifs.
Amino Acid 128 Determines the Specificity with Which Vif Excludes APOBEC3G from Virions. Vif dramatically reduces the amount of APOBEC3G encapsidated into virions (18, 23, 24). Because mouse APOBEC3G does not form a complex with HIV-1 Vif, it is not excluded from virions by HIV-1 Vif and as a result the enzyme is Vif-resistant (18). To determine whether the sensitivity of the amino acid 128 mutants to Vif was associated with the ability of Vif to exclude APOBEC3G from virions, APOBEC3G was quantitated in pelleted virions and in transfected cell lysates on immunoblots. As expected, human APOBEC3G, but not AGM APOBEC3G, was excluded from wild-type HIV-1 (Fig. 4A). HIV-1 Vif failed to exclude human APOBEC3G containing the amino acid 128 Asp to Lys substitution (D128K). In contrast, SIVAGM Vif excluded this mutant from SIVAGM virions (Fig. 4B). Conversely, AGM APOBEC3G in which Lys-128 was replaced with Asp (K128D) was excluded from virions by HIV-1 Vif (Fig. 4A). This mutant was not excluded from SIVAGM virions by SIVAGM Vif (Fig. 4B). Thus, amino acid 128 determined the specificity with which APOBEC3G is excluded from virions by Vif.
Fig. 4.
Amino acid 128 determines the specificity with which Vif excludes APOBEC3G from virions and the specificity of Vif:APOBEC3G complex formation. (A-F) Wild-type and Δvif HIV-1 and SIVAGM virions produced in 293T cells cotransfected with APOBEC3G expression vector were pelleted and normalized for p24 or p27. The encapsidated APOBEC3G was detected on immunoblots probed with anti-HA mAb (Upper). APOBEC3G expression in the transfected cells was confirmed on immunoblots of the cell lysates (Lower). Human and AGM APOBEC3G amino acid 128 exchange mutants were encapsidated in HIV-1 virions (A) and SIVAGM virions (B). mock, Control transfection in which the reporter virus plasmid was omitted. Ala-, Glu-, His-, and Arg-substituted human APOBEC3G were encapsidated in HIV-1 (C) and SIVAGM (D) virions. Ala-, Glu-, His-, and Arg-substituted AGM APOBEC3G was encapsidated in HIV-1 virions (E) or SIVAGM virions (F). The sensitivity of each virus to Vif as determined in Fig. 3 is summarized above: R, Vif resistant; S, Vif sensitive; s, intermediate. (G-H) Vif:APOBEC3G complexes were immunoprecipitated from 293T cells cotransfected with APOBEC3G expression vector and HIV-1 wild-type or Δvif HIV-1 luciferase reporter virus plasmid. The lysates were immunoprecipitated with anti-HA mAb and protein-A beads, and the complexes were analyzed on immunoblots. The filters were probed with anti-HA mAb to detect APOBEC3G and with rabbit anti-Vif antiserum to detect coimmunoprecipitated Vif. Efficient pull-down of APOBEC3G was confirmed on duplicate blots probed with anti-HA mAb. Equivalent expression of Vif in the transfected cells was confirmed on immunoblots of the cell lysates probed with rabbit anti-Vif antiserum. All of the experiments were repeated three to five times, and representative results are shown.
Substitution of human APOBEC3G at amino acid Asp-128 with Ala, Glu, or His did not affect the ability of HIV-1 Vif to exclude the enzyme from virions (Fig. 4C). In contrast, substitution with Arg blocked the ability of Vif to exclude APOBEC3G from virions. The Ala-, Glu-, and His-substituted human APOBEC3G remained resistant to the effects of SIVAGM Vif. Substitution with Arg caused the protein to be excluded by Vif (Fig. 4D). Conversely, AGM APOBEC3G substituted with Ala was excluded from HIV-1 (Fig. 4E) and SIVAGM virions (Fig. 4F). The His substitution in AGM APOBEC3G was partially excluded from SIVAGM virions. Partial exclusion is in accord with the luciferase reporter virus results in Fig. 3, which showed that the enzyme was partially resistant to SIVAGM Vif. Arg-substituted AGM APOBEC3G was efficiently excluded. On HIV virions, the Ala, Glu, and His AGM APOBEC3G mutants were excluded, but the Arg mutant was not (Fig. 4E). The ability of HIV-1 Vif to exclude the Glu-substituted enzyme from virions seemed to contradict the finding in Fig. 3C, which showed that it inhibited wild-type particle infectivity. However, the inhibition was relatively weak. In summary, these findings showed that HIV-1 Vif excludes APOBEC3G from virions if amino acid 128 is neutral or negatively charged. Conversely, SIVAGM Vif excludes APOBEC3G from virions if amino acid 128 is positively charged. This pattern of interaction corresponds to the sensitivity of each APOBEC3G to Vif in the functional assay (as summarized above each lane in Fig. 4).
Amino Acid 128 Determines the Specificity with Which APOBEC3G Physically Associates with Vif. To determine whether the species specificity of Vif function results from the ability of Vif to form a complex with APOBEC3G, we tested whether HIV-1 Vif coimmunoprecipitated from transfected 293T cell lysates with the amino acid 128 mutants. Cells transfected with wild-type or Δvif reporter virus and APOBEC3G expression vector were lysed in mild detergent. Vif:APOBEC3G complexes were immunoprecipitated with anti-HA mAb to pull down the epitope-tagged APOBEC3G. Associated Vif was detected on immunoblots probed with anti-Vif serum. Control human APOBEC3G efficiently pulled down HIV-1 Vif. In addition, Vif reduced the amount of APOBEC3G detected, consistent with its previously reported ability to degrade the enzyme (Fig. 4G) (23, 24, 27). AGM APOBEC3G, although expressed at considerably higher quantities than human APOBEC3G, coimmunoprecipitated much less HIV-1 Vif (≈10-fold by densitometry). A small amount of HIV-1 Vif reproducibly coimmunoprecipitated with AGM APOBEC3G. Substitution of Asp-128 with Lys in human APOBEC3G (D128K) reduced the association with Vif to control levels (Fig. 4G). Conversely, substitution of Lys-128 with Asp in AGM APOBEC3G (K128D) caused it to efficiently form a complex with HIV-1 Vif. Human APOBEC3G with Ala, Glu, and His substitutions associated with HIV-1 Vif, but Arg substitution significantly reduced complex formation (Fig. 4H). In AGM APOBEC3G, Ala, Glu, and His but not Arg interacted with Vif. We concluded that amino acid 128 controls the species-specific association with Vif. HIV-1 Vif can form a complex with APOBEC3G if amino acid 128 is negative or neutral, but not if it is positively charged.
Discussion
We show here that the species specificity of Vif for APOBEC3G is determined by a single amino acid. Human APOBEC3G containing an exchange of Asp-128 with the Lys of AGM APOBEC3G became resistant to HIV-1 Vif and sensitive to SIVAGM Vif. Conversely, AGM APOBEC3G containing the reciprocal Lys-128 to Asp exchange became resistant to SIVAGM Vif and sensitive to HIV-1 Vif. In addition, the exchange at position 128 switched the specificity with which APOBEC3G was excluded from virions and formed a complex with Vif. These findings offered a definitive confirmation that the association of APOBEC3G with Vif causes it to exclude the enzyme from virions and to rescue viral infectivity.
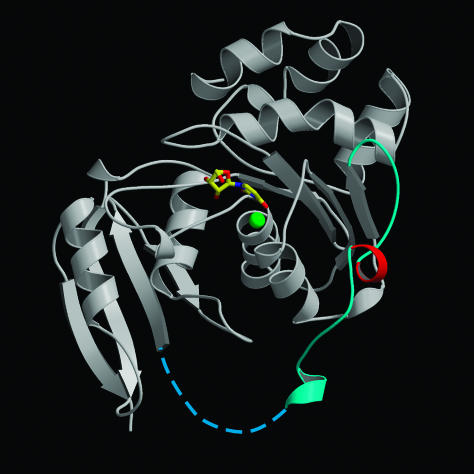
The three dimensional structure of APOBEC3G has not yet been determined; however, the crystal structure of E. coli cytidine deaminase has been solved to high resolution (29). Although only distantly related in primary sequence, the enzyme has features that suggest conservation of structure. The E. coli enzyme, like APOBEC3G, is composed of a catalytic domain followed by a topologically similar (αββαβαββ) pseudocatalytic domain and conserves the signature Cys-His Zn2+ coordination motif (15). To determine the approximate position of amino acid 128 on APOBEC3G, we mapped this position onto the E. coli cytidine deaminase structure. Structure-based sequence alignment of cytidine deaminases placed Asp-128 on the loop that connects the catalytic domain with its pseudocatalytic domain (Fig. 5). In the E. coli cytidine deaminase, this loop connects β5, the last β-strand of the five-stranded β-sheet in the catalytic domain, to the first α-helix of the pseudocatalytic domain. By comparison, APOBEC3G seems to lack the first α-helix of the pseudocatalytic domain. The loop containing Asp-128 most likely connects β5 of the catalytic domain to the first β-strand of the pseudocatalytic domain. Nevertheless, the assignment of this charged residue to a loop suggests that it is exposed to solvent and available to play a direct role in binding Vif. In addition, the loop is on the same face of the protein as the active site such that binding of Vif to APOBEC3G could block with deaminase activity.
Fig. 5.
Ribbon representation of E. coli cytidine deaminase. Asp-128 maps to a helical turn (red) situated on a loop (cyan and dotted line) connecting the catalytic and pseudocatalytic domains of the E. coli enzyme. Uridine at the active site is shown in bonds representation. The green sphere represents Zn2+.
Amino acid 128 could either be a site of direct contact with Vif or could influence global conformation of the enzyme. However, our data are suggestive of a contact site. First, structural modeling placed this amino acid on a solvent-exposed loop that would be accessible for protein:protein interaction. Second, positioned on a loop, this amino acid would be unlikely to affect global conformation of the protein. Third, switching the polarity at this position in human or AGM APOBEC3G had no effect on the catalytic activity of the enzyme as judged by their strong antiviral activity, further arguing that this position does not play a structural role in the enzyme.
Whether the Vif:APOBEC3G complex results from direct contact of the two proteins or involves an intermediary (most likely RNA or protein) is not yet known; however, our findings suggest a direct interaction. The finding that amino acid exchange between APOBEC3Gs caused them to switch their interaction with Vif suggests that they interact directly. If APOBEC3G and Vif bound a common intermediary, there would be no species specificity to the association.
Because amino acid 128 seems to be a contact site for Vif, we speculate that the interaction is mediated by electrostatic interactions. The negatively charged Asp-128 of human APOBEC3G may interact with a positively charged amino acid of HIV-1 Vif. Conversely, the positively charged Lys-128 of AGM APOBEC3G would interact with a negative charge in SIVAGM Vif. HIV-1 and SIVAGM Vif are only 27% similar; however, their isoelectric points are similar (pI 9.97 for HIV-1 Vif and pI 10.04 for SIVAGM Vif). Sequence alignment shows four positions in which a charge is changed from positive in HIV-1 Vif to negative in SIVAGM Vif (R15E, R94E, R123E, and K168D). Preliminary testing of Vif mutants containing charge changes at these positions did not show conclusive effects on the interaction with APOBEC3G (data not shown).
These results provide an interesting insight into the mechanism by which viruses evolve to adapt to interact with the topology of specific cellular cofactors in their host species. Because mammalian genomes encode proteins that are polymorphic between species, plasticity of viral genomes is required for viruses to expand their host range. The lentiviral Tat gene is similarly adapted to a species-specific interaction with its cellular cofactor cyclin-T1 (30). For SIV to replicate in the various primate species, Vif had to accommodate the differences at amino acid 128 of APOBEC3G. The ability of Vif to adapt to the Asp-128 of chimpanzee APOBEC3G (18) was a prerequisite for the zoonosis of the virus to humans. Understanding the molecular basis of these species-specific interactions with host cellular factors will be instrumental for development of novel animal models for AIDS.
Supplementary Material
Acknowledgments
We thank Partho Ghosh for structural modeling and Richard Barnard for critical reading of the manuscript. This work was supported by National Institutes of Health Grants DA14494 and AI 58864 and the University-wide AIDS Research Program. N.R.L. is an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Vif, virion infectivity factor; APOBEC3G, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G; HA, hemagglutinin; AGM, African green monkey; SIV, simian immunodeficiency virus.
See Commentary on page 3725.
References
- 1.Trono, D. (1995) Cell 82, 189-192. [DOI] [PubMed] [Google Scholar]
- 2.Emerman, M. & Malim, M. H. (1998) Science 280, 1880-1884. [DOI] [PubMed] [Google Scholar]
- 3.Strebel, K., Daugherty, D., Clouse, K., Cohen, D., Folks, T. & Martin, M. A. (1987) Nature 328, 728-730. [DOI] [PubMed] [Google Scholar]
- 4.von Schwedler, U., Song, J., Aiken, C. & Trono, D. (1993) J. Virol. 67, 4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabuzda, D. H., Lawrence, K., Langhoff, E., Terwilliger, E., Dorfman, T., Haseltine, W. A. & Sodroski, J. (1992) J. Virol. 66, 6489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon, J. H., Miller, D. L., Fouchier, R. A., Soares, M. A., Peden, K. W. & Malim, M. H. (1998) EMBO J. 17, 1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madani, N. & Kabat, D. (1998) J. Virol. 72, 10251-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. (2002) Nature 418, 646-650. [DOI] [PubMed] [Google Scholar]
- 9.Jarmuz, A., Chester, A., Bayliss, J., Gisbourne, J., Dunham, I., Scott, J. & Navaratnam, N. (2002) Genomics 79, 285-296. [DOI] [PubMed] [Google Scholar]
- 10.Harris, R. S., Petersen-Mahrt, S. K. & Neuberger, M. S. (2002) Mol. Cell 10, 1247-1253. [DOI] [PubMed] [Google Scholar]
- 11.Anant, S., MacGinnitie, A. J. & Davidson, N. O. (1995) J. Biol. Chem. 270, 14762-14767. [PubMed] [Google Scholar]
- 12.Lau, P. P., Villanueva, H., Kobayashi, K., Nakamuta, M., Chang, B. H. & Chan, L. (2001) J. Biol. Chem. 276, 46445-46452. [DOI] [PubMed] [Google Scholar]
- 13.Yang, Y., Sowden, M. P. & Smith, H. C. (2000) J. Biol. Chem. 275, 22663-22669. [DOI] [PubMed] [Google Scholar]
- 14.Muto, T., Muramatsu, M., Taniwaki, M., Kinoshita, K. & Honjo, T. (2000) Genomics 68, 85-88. [DOI] [PubMed] [Google Scholar]
- 15.Wedekind, J. E., Dance, G. S., Sowden, M. P. & Smith, H. C. (2003) Trends Genet. 19, 207-216. [DOI] [PubMed] [Google Scholar]
- 16.Petersen-Mahrt, S. K., Harris, R. S. & Neuberger, M. S. (2002) Nature 418, 99-103. [DOI] [PubMed] [Google Scholar]
- 17.Gaddis, N. C., Chertova, E., Sheehy, A. M., Henderson, L. E. & Malim, M. H. (2003) J. Virol. 77, 5810-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mariani, R., Chen, D., Schröfelbauer, B., Navarro, F., Konig, R., Bollman, B., Munk, C., Nymark-McMahon, H. & Landau, N. R. (2003) Cell 114, 21-31. [DOI] [PubMed] [Google Scholar]
- 19.Lecossier, D., Bouchonnet, F., Clavel, F. & Hance, A. J. (2003) Science 300, 1112. [DOI] [PubMed] [Google Scholar]
- 20.Zhang, H., Yang, B., Pomerantz, R. J., Zhang, C., Arunachalam, S. C. & Gao, L. (2003) Nature 424, 94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L. & Trono, D. (2003) Nature 424, 99-103. [DOI] [PubMed] [Google Scholar]
- 22.Harris, R. S., Bishop, K. N., Sheehy, A. M., Craig, H. M., Petersen-Mahrt, S. K., Watt, I. N., Neuberger, M. S. & Malim, M. H. (2003) Cell 113, 803-809. [DOI] [PubMed] [Google Scholar]
- 23.Marin, M., Rose, K. M., Kozak, S. L. & Kabat, D. (2003) Nat. Med. 9, 1398-1403. [DOI] [PubMed] [Google Scholar]
- 24.Stopak, K., de Noronha, C., Yonemoto, W. & Greene, W. C. (2003) Mol. Cell 12, 591-601. [DOI] [PubMed] [Google Scholar]
- 25.Kao, S., Khan, M. A., Miyagi, E., Plishka, R., Buckler-White, A. & Strebel, K. (2003) J. Virol. 77, 11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu, X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P. & Yu, X. F. (2003) Science 302, 1056-1060. [DOI] [PubMed] [Google Scholar]
- 27.Sheehy, A. M., Gaddis, N. C. & Malim, M. H. (2003) Nat. Med. 9, 1404-1407. [DOI] [PubMed] [Google Scholar]
- 28.Deng, H., Liu, R., Ellmeier, W., Choe, S., Unutmaz, D., Burkhart, M., Di Marzio, P., Marmon, S., Sutton, R. E., Hill, C. M., et al. (1996) Nature 381, 661-666. [DOI] [PubMed] [Google Scholar]
- 29.Betts, L., Xiang, S., Short, S. A., Wolfenden, R. & Carter, C. W., Jr. (1994) J. Mol. Biol. 235, 635-656. [DOI] [PubMed] [Google Scholar]
- 30.Bieniasz, P. D., Grdina, T. A., Bogerd, H. P. & Cullen, B. R. (1998) EMBO J. 17, 7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.