Background: Polysialylated neuropilin-2 mediates CCL21-driven chemotactic migration of dendritic cells.
Results: Deletion of either ST8SiaIV or O-glycosylation sites located between b2 and c domain abrogates polysialylation of neuropilin-2.
Conclusion: Polysialylation of neuropilin-2 occurs in the same linker region as GAGylation of neuropilin-1.
Significance: Defining enzyme and acceptor site requirements is crucial for understanding how polysialylation of neuropilin-2 is regulated.
Keywords: Dendritic Cells, Glycoprotein, Glycosylation, Glycosyltransferases, Post-translational Modification, O-Glycan Analysis, Polysialic Acid
Abstract
Neuropilin-2 (NRP2) is well known as a co-receptor for class 3 semaphorins and vascular endothelial growth factors, involved in axon guidance and angiogenesis. Moreover, NRP2 was shown to promote chemotactic migration of human monocyte-derived dendritic cells (DCs) toward the chemokine CCL21, a function that relies on the presence of polysialic acid (polySia). In vertebrates, this posttranslational modification is predominantly found on the neural cell adhesion molecule (NCAM), where it is synthesized on N-glycans by either of the two polysialyltransferases, ST8SiaII or ST8SiaIV. In contrast to NCAM, little is known on the biosynthesis of polySia on NRP2. Here we identified the polySia attachment sites and demonstrate that NRP2 is recognized only by ST8SiaIV. Although polySia-NRP2 was found on bone marrow-derived DCs from wild-type and St8sia2−/− mice, polySia was completely lost in DCs from St8sia4−/− mice despite normal NRP2 expression. In COS-7 cells, co-expression of NRP2 with ST8SiaIV but not ST8SiaII resulted in the formation of polySia-NRP2, highlighting distinct acceptor specificities of the two polysialyltransferases. Notably, ST8SiaIV synthesized polySia selectively on a NRP2 glycoform that was characterized by the presence of sialylated core 1 and core 2 O-glycans. Based on a comprehensive site-directed mutagenesis study, we localized the polySia attachment sites to an O-glycan cluster located in the linker region between b2 and c domain. Combined alanine exchange of Thr-607, -613, -614, -615, -619, and -624 efficiently blocked polysialylation. Restoration of single sites only partially rescued polysialylation, suggesting that within this cluster, polySia is attached to more than one site.
Introduction
The neuropilins, neuropilin-2 (NRP2)3 and its close homologue NRP1, function as co-receptors for class 3 semaphorins and several vascular endothelial growth factors (VEGFs) and are crucial for repulsive axon guidance, vascularization, and angiogenesis (1–5). Both neuropilins are cell surface glycoproteins with identical domain structure and 44% homology at the amino acid sequence level (6–8). Their extracellular part is composed of five domains, namely two CUB domains, termed a1 and a2, sharing homology with the complement binding factors C1r/C1s, two coagulation factor V/VIII (FV/VIII) homology domains, termed b1 and b2, and a MAM (meprin/A5-protein/phosphotyrosine phosphatase μ) domain, termed c. The extracellular part is followed by a single transmembrane domain (TMD) and a short cytosolic part (9, 10). As neuropilins lack intracellular signaling motifs, they form co-receptor complexes with plexins and VEGF receptors to mediate signal transduction (9, 10).
In addition to their well studied functions during nervous system development, vascularization, and angiogenesis, novel roles emerged for neuropilins in the immune system. NRP1 was found to play a role in the immunological synapse (11, 12) and the transmigration of dendritic cells (DCs) into the lymphatics (13). NRP2 was shown to be involved in chemotactic migration of human monocyte-derived DCs toward the CC-motif chemokine CCL21 (14–16). After pathogen recognition, DC traffic to secondary lymphoid organs to activate naïve T cells through antigen presentation. This migration depends on the G-protein-coupled Cys-Cys-chemokine receptor 7 (CCR7), which has two known ligands, CCL19 and CCL21 (17, 18). Although migration of maturing DCs into peripheral lymphatic vessels is regulated by CCL21 expressed by these vessels, migration from the afferent lymphatics into the lymph node is mediated by CCL19 and CCL21 expressed by stromal cells in the T cell areas of the lymph node (19, 20). In vitro and in vivo studies revealed that NRP2 enhances the chemotaxis of DCs toward CCL21 but not toward CCL19 (16). Notably, this NRP2 function was found to be dependent on the posttranslational modification of NRP2 by polysialic acid (polySia) and was lost by specific removal of this glycan polymer (15, 16).
PolySia represents a linear polysaccharide composed of α2,8-linked sialic acids. In vertebrates, polySia is found predominantly as a developmentally regulated posttranslational modification of the neural cell adhesion molecule NCAM, a member of the immunoglobulin super family (21, 22). Polysialylation of NCAM starts on complex N-glycans at two particular N-glycosylation sites and is mediated by the two Golgi-resident polysialyltransferases, ST8SiaII and ST8SiaIV (23–25). Both enzymes share 59% amino acid sequence identity and are independently able to modify NCAM by polySia chains of up to 90 α2,8-linked sialic acid residues (26–28). Modification of NCAM by these highly hydrated and negatively charged polySia chains not only masks the underlying protein core, preventing NCAM-mediated interactions, but also increases the intermembrane space, resulting in a general negative regulation of cell adhesion (29–31). Accordingly, the polysialylated form of NCAM is involved in dynamic processes such as migration of neural precursor cells, brain wiring, and synaptic plasticity (21, 22, 32). The crucial role of the carbohydrate polymer polySia was highlighted by the lethal phenotype of polySia-negative mice that were generated by genetic ablation of both polysialyltransferases (24).
In contrast to most other glycosyltransferases, the polysialyltransferases are highly protein-specific, and only a few polysialylated proteins have been identified in mammals. Apart from NCAM and NRP2, these are the synaptic cell adhesion molecule SynCAM 1 on NG2 cells of the developing mouse brain (33), the α-subunit of a voltage-gated sodium channel in adult rat brain (34), and the scavenger receptor CD36 in human and murine milk (35). Moreover, the two polysialyltransferases have the ability to modify themselves in a process termed autopolysialylation (36, 37). However, our knowledge on the biosynthesis of polySia on proteins other than NCAM is still limited.
So far, polysialylation of NRP2 has been exclusively found on human monocyte-derived DCs (38). In these cells, transcripts of both polysialyltransferases, ST8SiaII and ST8SiaIV, were detected (38). During maturation, however, only ST8SiaIV mRNA levels were up-regulated, coincident with an increased appearance of polySia-NRP2 on the cell surface (14, 38). Moreover, siRNA knockdown of ST8SiaIV resulted in decreased CCL21-driven chemotaxis (15), suggesting a prominent role for ST8SiaIV in the polysialylation of NRP2. Although previous studies indicated that O-glycans may serve as preferred attachment sites of polySia (14, 38), nothing is known on the localization of these sites.
In the present study we defined the polySia attachment sites of NRP2 and investigated whether ST8SiaII and ST8SiaIV differ in their ability to polysialylate NRP2. Our study revealed that NRP2 is an acceptor molecule exclusively for ST8SiaIV that acts selectively on mucin-type O-glycans that are clustered in a short stretch between b2 and c domain of NRP2.
EXPERIMENTAL PROCEDURES
Materials
Mouse anti-polySia monoclonal antibody (mAb) 735 (39) and endoneuraminidase (endoN) were purified as described previously (28). Mouse anti-Myc mAb 9E10 was purchased from Roche Applied Science, and mouse anti-actin mAb was from Millipore. The rabbit polyclonal antibody directed against human NRP2 (H-300) was obtained from Santa Cruz Biotechnology, and rabbit monoclonal anti-mouse NRP2 (D39A5) was from Cell Signaling. Allophycocyanin-labeled anti-mouse CD11c and phycoerythrin-labeled anti-MHC class II antibodies were obtained from BD Biosciences. Secondary antibodies Alexa568-conjugated anti-rabbit IgG was obtained from Molecular Probes, and horseradish peroxidase-conjugated anti-mouse IgG was purchased from Southern Biotech. The O-glycosylation inhibitor α-benzyl-N-acetylgalactosamine (α-benzyl-GalNAc) was obtained from Merck. The plasmids encoding human ST8SiaIV and ST8SiaII in the vector pcDNA1 (Invitrogen) were gifts from Drs. Minoru Fukuda and Paul Scheidegger, respectively.
Expression Plasmids
For the expression of different human NRP2 isoforms, constructs encoding full-length proteins with a C-terminal Myc/hexahistidine epitope were generated. The coding sequence of NRP2a(17) was amplified by PCR using the primers MR23 and MR24 and a NRP2a(17) cDNA clone (Ultimate ORF Clone, Invitrogen, accession no. NM_003872.2) as template. The resulting PCR product was digested with HindIII and XbaI and ligated into the corresponding sites of pcDNA3.1/myc-His (Invitrogen) leading to the plasmid pNRPa(17)-Myc. Transcript variant a(22), which encodes for the longest isoform, was generated based on pNRP2a(17)-Myc by insertion of an oligonucleotide adapter encoding the amino acid sequence GENFK using the primers MR121 and MR122. Accordingly, the plasmid encoding isoform a(0) was obtained by deletion of the sequence encoding the 17-amino acid stretch VDIPEIHEREGYEDEID using the primers MR124 and MR125. For the generation of a construct encoding isoform b(0), the coding sequence was amplified by PCR with the primer pair MR23 and MR25 and an NRP2b(0) cDNA clone (Ultimate ORF Clone, Invitrogen, accession no. NM_201267.1) as template. After digestion with HindIII/XbaI, the obtained PCR product was ligated into the corresponding sites of pcDNA3.1/myc-His, resulting in the formation of pNRP2b(0)-Myc. Isoform b(5) was amplified by reverse transcription (RT)-PCR from HEK-293 mRNA using the SuperScript First-Strand Synthesis System (Invitrogen) and primers MR114 and MR25. The obtained PCR-product was digested with NotI/XbaI and ligated into the corresponding sites of pNRP2b(0)-Myc. The identity of all constructs was confirmed by sequencing. The sequences of the indicated primers are given in supplemental Table 1.
Generation of NRP2 Glycosylation Variants
Individual glycosylation sites were destroyed by site-directed mutagenesis using a double-joint PCR strategy. All mutagenesis PCR reactions were performed with Phusion DNA polymerase (New England Biolabs) and pNRP2b(5)-Myc as template. O-glycosylation mutants were generated by a subsequent exchange of residues Thr-607, -613, -614, -615, -619, and -624 against alanine resulting in a hexa mutant. Based on this mutant, penta mutants were obtained by individual back mutation of single alanine residues to threonine. Briefly, two overlapping gene fragments were amplified using a mutagenesis primer in combination with a vector primer (antisense and sense mutagenesis primer in combination with primer T7 and BGH-reverse, respectively). PCR products were joined by PCR and amplified by nested PCR using the primers NRP2C and MR25. Nested PCR products were ligated into Bsu36I/XhoI sites of pNRP2b(5)-Myc. N-Glycosylation mutants were generated using the same strategy, except for the nested PCR, which was performed using the primer pair MR23 and NRP2B. Nested PCR products were ligated into HindIII/BamHI sites of pNRP2b(5)-Myc. The identity of all constructs was confirmed by sequencing. Sequences of all mutagenesis primers are given in supplemental Table 2.
Cell Culture and Transfection of COS-7 Cells
COS-7 cells were maintained in DMEM/Ham's F-12 1:1 (Seromed) supplemented with 10% FCS and 1 mm sodium pyruvate in a 37 °C, 5% CO2 incubator. Transient transfections were performed with FuGENE HD (Roche Applied Science) according to the manufacturer's recommendations. Inhibition of O-glycosylation was achieved by feeding cells with 2 mm α-benzyl-GalNAc (Merck) before start of the transfection.
Generation of Murine Bone Marrow-derived Dendritic Cells
Bone marrow-derived dendritic cells (BM-DCs) were generated from 8-week-old wild-type, St8sia2−/− (40), and St8sia4−/− (41) mice. Knock-out animals were backcrossed to C57BL/6J mice for six generations, and genotyping was performed as described (24). Mice were sacrificed by cervical dislocation, hind limbs were collected, and bone marrow was flushed out from the femur and tibia using a 27-Gauge needle syringe. Red blood cells were removed by ammonium chloride treatment. Cells were filtered through a nylon mesh, suspended in RPMI 1640 medium supplemented with 20 ng/ml recombinant, murine granulocyte-macrophage colony-stimulating factor, 10% FCS, 100 units/ml penicillin, 100 units/ml streptomycin, and plated at a concentration of 1 × 106 cells/ml. Medium was changed on days 3, 5, and 7. On day 8, DCs were matured using 5 nm CpG-ODN 1668 (TIB Molbiol Syntheselabor GmbH, Berlin, Germany), and cells were harvested on day 9. Differentiation into DCs was analyzed on day 9 using allophycocyanin-labeled anti-CD11c and phycoerythrin-labeled anti-MHC class II mouse monoclonal antibodies using a BD LSR II Flow Cytometer (BD Biosciences).
RT-PCR Analysis
Total RNA was extracted from BM-DCs using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. 2.5 μg of total RNA was reverse-transcribed with random hexamer primers using RevertAid reverse transcriptase (Fermentas) according to the manufacturer's recommendations. Gene-specific cDNA was amplified by PCR with the following primer pairs: 5′-GGCTGTGGCCAGGAGATTG-3′ and 5′-GGCATACTCCTGAACTGGAGCC-3′ (st8sia2); 5′-GGTCCAGTTGGCTTGGCCTG-3′ and 5′-CAGTCGTTTGGGTGACAGGTG-3′ (st8sia3); 5′-GCACCAAGAGACGCAACTCATC-3′ and 5′-CAGAGCTGTTGACAAGTGATCTGC-3′ (st8sia4); 5′-CCTCAGCTGCACTGAAGACAC-3′ and 5′-GGTTGTCTTCTCATCCAGATATG-3′ (st8sia6); and 5′-TTCCTCATGGACTGATTATGGACA-3′ and 5′-AGAGGGCCACAATGTGATGG-3′ (hprt).
PCR was performed with a denaturation step at 98 °C for 30 s followed by 30 cycles of denaturation at 98 °C for 10 s, primer annealing at 60 °C for 30 s, and primer extension at 72 °C for 20 s. Upon completion of the cycling steps, a final extension at 72 °C for 2.5 min was done. PCR products were separated on a 10% acrylamide gel and stained with ethidium bromide.
Protein Extraction, Immunoprecipitation, and Western Blot Analysis
Transfected cells of one well of a 6-well plate were lysed in 200 μl of lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5 mm EDTA) containing 1% Triton X-100, 200 units/ml aprotinin, 2 mm phenylmethylsulfonyl fluoride, and 10 μg/ml leupeptin. For detection of polysialylated protein, one-half of the lysate was treated with 1 μg of endoN for 30 min on ice. BM-DCs were lysed with lysis buffer containing 2% Triton-X-100, 50 μl/ml protease inhibitor mixture (Sigma), 20 μg/ml calpain inhibitor II, 1 mm benzamidine, and 2 mm phenylmethylsulfonyl fluoride. For immunoprecipitation of polySia-NRP2, mAb 735 was covalently coupled to M-280 tosylactivated Dynabeads (Invitrogen) according to the manufacturer's instructions. One part of the isolated polysialylated protein was treated with 600 ng of endoN for 30 min at 37 °C. Immunoprecipitation of Myc-tagged NRP2 was performed with anti-Myc mAb 9E10 coupled to Protein G-Sepharose (GE Healthcare). Lysates and immunoprecipitated proteins were separated by SDS-PAGE under reducing conditions and transferred to polyvinylidene difluoride membrane (Immobilon-P; Millipore). Primary antibodies were used at a concentration of 5 μg/ml and detected by peroxidase-conjugated anti-mouse IgG or anti-rabbit IgG antibodies followed by enhanced chemiluminescence (ECL) detection.
Enzymatic Desialylation of NRP2
Immunoprecipitated NRP2 was washed twice with 1 ml of sialidase buffer (50 mm sodium acetate, pH 5.5) and incubated for 3 h at 37 °C in 50 μl of sialidase buffer containing 12.5 milliunits of sialidase from Arthrobacter ureafaciens (EY Laboratories).
Lectin Staining
Western blot analysis with digoxigenin-labeled peanut agglutinin (PNA) was performed with the DIG glycan differentiation kit (Roche Applied Science) according to the manufacturer's recommendations. Bound lectin was detected by peroxidase-conjugated anti-digoxigenin Fab fragments (Roche Applied Science) followed by ECL detection. To allow parallel visualization of Myc-tagged NRP2, blots were re-probed with anti-Myc mAb 9E10 followed by alkaline phosphatase-conjugated anti-mouse IgG antibodies (Southern Biotech) and colorimetric detection with 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium.
Gel-based O-Glycomics
Coomassie-stained protein bands in SDS-gels were treated with Pronase to generate Pronase-stable glycopeptides for the off-gel liberation of O-linked glycans by reductive β-elimination (42). The permethylated glycan alditols were analyzed by matrix-assisted laser desorption ionization (MALDI)-TOF-TOF mass spectrometry.
MALDI-TOF-TOF Mass Spectrometry
MALDI mass spectrometry was performed on an UltrafleXtreme instrument (Bruker Daltonics). The methylated glycan alditols contained in methanol were applied to the stainless steel target by mixing a 0.75-μl aliquot of sample with the same volume of matrix (saturated solution of 4-hydroxy-α-cyanocinnamic acid in acetonitrile, 0.1% TFA). Analyses were performed by positive ion detection in the reflectron mode. Ionization of co-crystallized analytes was induced with a pulsed Smart-beam laser (accumulation of about 5000 shots), and the ions were accelerated in a field of 20 kV and reflected at 23 kV. The structural assignments of major glycan components in the peptide mixture were confirmed by MS/MS analysis of post-source decay fragments in the laser-induced dissociation mode.
Immunofluorescence Analysis
One day before transfection, 1.8 × 105 COS-7 cells were seeded per well of a 6-well plate containing four glass coverslips. 24 h after transfection cells were fixed in 4% paraformaldehyde and stained with anti-NRP2 polyclonal antibody H-300 (4 μg/ml in 20% horse serum) followed by incubation with Alexa568-conjugated anti-rabbit IgG secondary antibody. Coverslips were mounted in Vectashield mounting medium containing DAPI (Vector Laboratories) and analyzed under a Zeiss Axiovert 200M microscope equipped with ApoTome module, AxioCam MRm digital camera, and Axio Vision software (Zeiss).
RESULTS
Polysialylation of NRP2 on Murine Bone Marrow-derived DCs Strictly Depends on ST8SiaIV
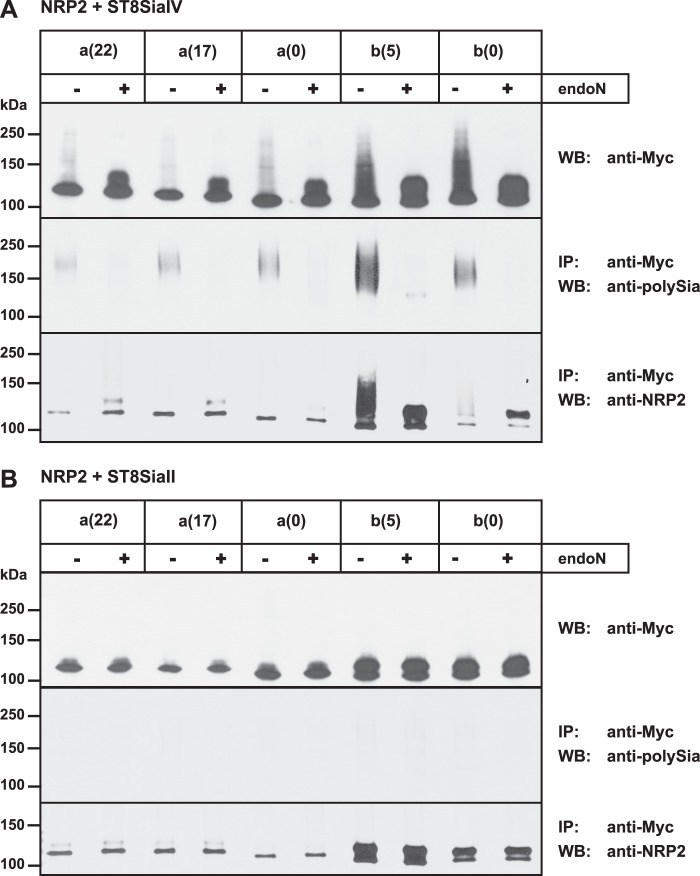
To unambiguously define the impact of the two polysialyltransferases in vivo, we made use of the two existing mouse models lacking either ST8SiaII (40) or ST8SiaIV (41). Because polySia-NRP2 has so far only been described in the human system, we first analyzed the expression and polysialylation status of NRP2 on mature BM-DCs from wild-type mice. Western blot analysis of whole cell lysate with an anti-polySia mAb revealed a diffuse signal that was completely abolished after treatment by endosialidase (endoN), an enzyme that specifically removes α2,8-linked polySia (43) (Fig. 1A, top panel). Immunoprecipitation of the polysialylated material and staining with an anti-NRP2 mAb revealed a single band at ∼125 kDa that was shifted to ∼110 kDa after enzymatic removal of polySia by endoN (Fig. 1A, bottom panel), demonstrating that polySia-NRP2 is expressed on murine BM-DCs. Similar to what was found for human monocyte-derived DCs (38), the total polySia signal was much broader than the signal obtained for polySia-NRP2. Moreover, like human monocyte-derived DCs, we found no polySia-NCAM on murine BM-DCs, and BM-DCs derived from Ncam−/− mice showed the same polySia and polySia-NRP2 signals as the corresponding wild-type cells (data not shown).
FIGURE 1.
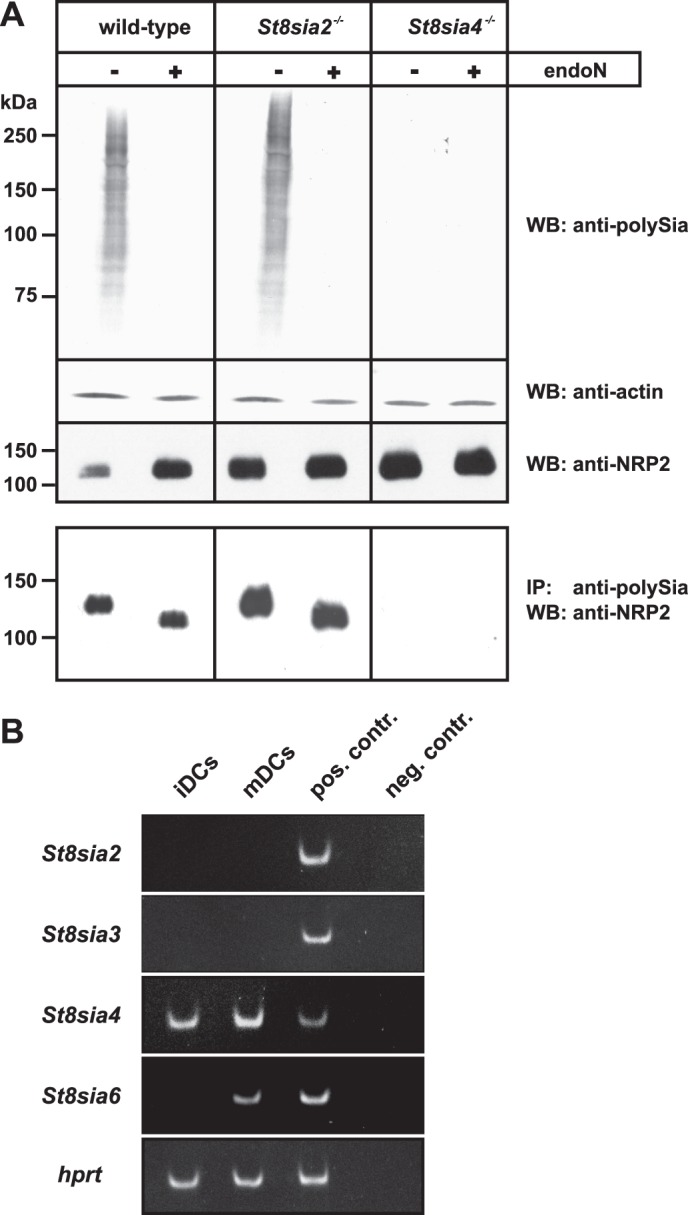
PolySia-NRP2 in murine BM-DCs. A, shown is a Western blot analysis of mature murine BM-DCs generated from wild-type, St8sia2−/−, and St8sia4−/− mice. Before and after removal of polySia by endoN, whole cell lysates were separated by SDS-PAGE and analyzed by Western blotting with anti-polySia mAb 735 (top panel), anti-actin mAb (second panel), and anti-NRP2 mAb (third panel). The bottom panel shows the Western blot analysis of immunoprecipitated polySia-NRP2. Using mAb 735, polysialylated proteins were immunoprecipitated from whole cell lysates of murine BM-DCs generated from wild-type, St8sia2−/−, and St8sia4−/− mice, and the isolated material was then analyzed before and after endoN treatment with anti-NRP2 mAb D39A5. WB, Western blot; IP, immunoprecipitation. B, shown is RT-PCR of protein-specific α2,8-sialyltransferase mRNA expression in immature and mature BM-DCs (iDCs and mDCs, respectively) from wild-type mice. Ethidium bromide-stained PCR products of mouse St8sia2 (72 bp), St8sia3 (82 bp), St8sia4 (68 bp), and St8sia6 (86 bp) are shown, whereas RT-PCR products of the house keeping gene hypoxanthine-guanine phosphoribosyltransferase (hprt, 86 bp) were used as controls for quality and equal loading of the cDNAs. cDNA from postnatal day 1 mouse brain was used as the positive control (pos. contr.). Water instead of cDNA was used in the negative control (neg. contr.) reactions for each PCR.
While in BM-DCs derived from St8sia2−/− mice normal polySia-NRP2 level were detected (Fig. 1A, bottom panel), the same cell type derived from St8sia4−/− mice was completely devoid of polySia (Fig. 1A, upper panel). Although NRP2 expression was not affected as demonstrated by direct analysis of cell lysate with an anti-NRP2 antibody (see the last row of the upper panel in Fig. 1A), no polySia-NRP2 was found in BM-DCs of St8sia4−/− mice (Fig. 1A, bottom panel). This finding demonstrated that in murine BM-DCs, polysialylation of NRP2 is strictly dependent on the presence of ST8SiaIV.
Using RT-PCR, we analyzed the expression pattern of the polysialyltransferases in BM-DCs of wild-type mice (Fig. 1B). ST8SiaIV transcripts were already detectable in immature DCs, and expression was up-regulated after maturation induced by the toll-like receptor 9 agonist CpG. By contrast, no ST8SiaII transcripts were detected, highlighting that ST8SiaIV is the only polysialyltransferase expressed in murine BM-DCs. We also analyzed the expression of ST8SiaIII and ST8SiaVI, two other members of the α2,8-sialyltransferase family that use glycoproteins as acceptors. While ST8SiaVI is a monosialyltransferase (44, 45), ST8SiaIII has been shown to form oligosialic acid in vitro (46, 47). During maturation of BM-DCs, the expression of ST8SiaVI was up-regulated, and transcripts were clearly detectable in mature DCs (Fig. 1B). In contrast, neither immature DCs nor mature DCs expressed ST8SiaIII transcripts, excluding a contribution of this enzyme to the polysialylation status of NRP2.
Polysialylation of NRP2 Is Restricted to a Particular Glyco- but Not Isoform
To address the question of whether ST8SiaII per se is able to polysialylate NRP2, we analyzed polysialylation of NRP2 in a cell culture system that allowed co-expression of NRP2 with either ST8SiaII or ST8SiaIV. To provide optimal conditions for polysialylation of NRP2, we first evaluated whether all isoforms of NRP2 serve as targets for polysialylation. Alternative splicing of NRP2 transcripts results in the formation of a soluble and several transmembrane isoforms (48). Based on differences in the primary sequence of transmembrane domain (TMD) and cytosolic domain, the membrane-bound isoforms fall into two classes, termed NRP2a and NRP2b (see Fig. 2A). In humans, two NRP2a forms have been described that are characterized by insertion of 17 and 22 (17 plus 5) amino acids and were termed NRP2a(17) and NRP2a(22), respectively. By database mining, an additional NRP2a isoform was found (GenBankTM accession number AF022859.1) that lacks any short variable insertions and was termed NRP2a(0) (Fig. 2A). In the case of NRP2b, two isoforms, termed b(5) and b(0), are known that differ only by insertion of a five-amino acid peptide of the sequence GENFK that is also part of isoform NRP2a(22) (48). Rey-Gallardo et al. (16) showed polysialylation for both one NRP2a and one NRP2b form but did not further discriminate between NRP2a and NRP2b subforms. To study a possible contribution of the variable peptide sequences on NRP2 polysialylation, we analyzed all five transmembrane isoforms for their capacity to serve as a target for polysialylation. Therefore, cDNAs encoding each human isoform with a C-terminal Myc tag were generated and expressed in COS-7 cells. Western blot analysis with an anti-Myc antibody revealed for all variants a double band (Fig. 2B). Although the signal for the lower band was in all cases of almost equal intensity, the signal of the upper band was much more prominent for the NRP2b forms. Mass spectrometric analysis of the upper and lower band of NRP2b(5) confirmed that both bands contained protein that was N-glycosylated at positions Asn-152 and Asn-157 located in the a2 domain (supplemental Fig. S1). To investigate potential differences in O-glycosylation, immunoprecipitated NRP2 was analyzed by PNA (Fig. 2C), a plant lectin that recognizes O-GalNAc glycans of the structure Galβ1,3GalNAc-Thr/Ser (49). Because sialylation of this glycotope blocks PNA binding, NRP2 was analyzed before and after sialidase treatment. Only after de-sialylation, a single band was stained by PNA that was most prominent for the NRP2b isoforms b(5) and b(0). Re-staining of the PNA blots with anti-Myc mAb demonstrated that de-sialylation increased the mobility of the upper band of NRP2 and that the signal obtained by PNA staining corresponded to the upper band. Thus, in COS-7 cells, NRP2 was expressed as two glycoforms that differed in the presence or absence of sialylated O-glycans.
FIGURE 2.
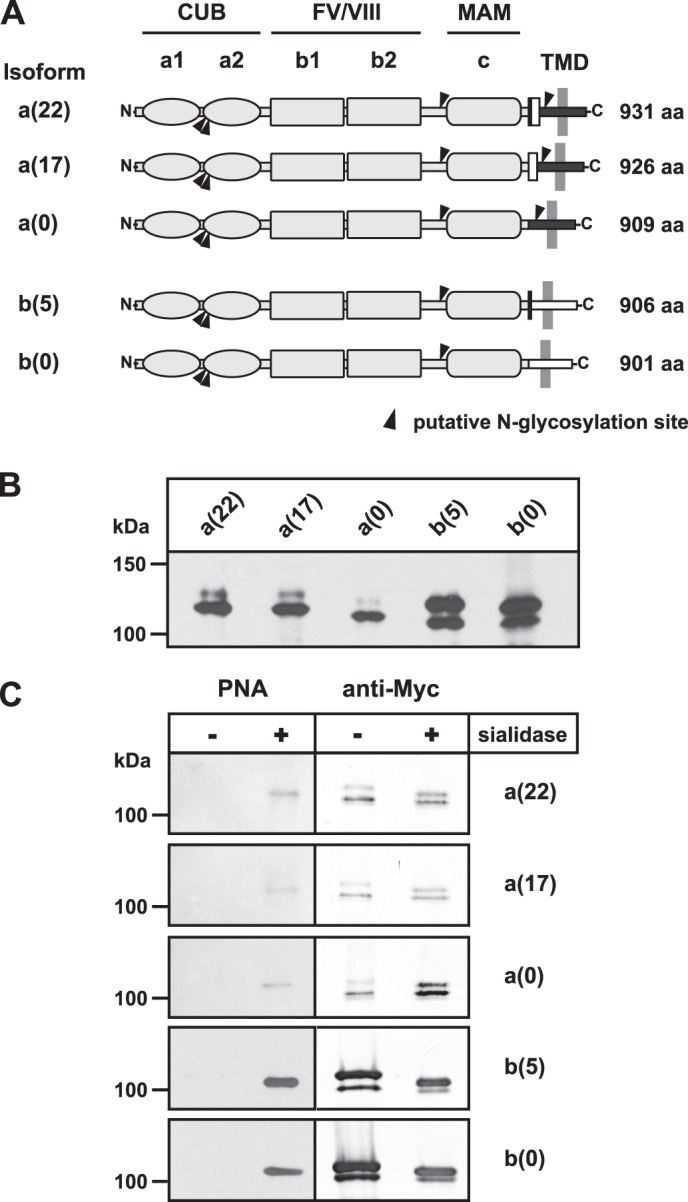
NRP2 isoforms are expressed as two glycoforms. A, shown is a schematic representation of human NRP2. The N-terminal extracellular part (amino acids (aa) 1–808) is shared by all isoforms and consists of two CUB domains (a1 and a2), two coagulation factors V/VIII (FV/VIII) homologous domains (b1 and b2), and one MAM or c domain. This common part is followed by a short stem region, a single transmembrane domain (TMD), and a cytosolic part. Based on two alternative C-terminal transmembrane domain/cytoplasmic regions (represented by black and white horizontal boxes), the membrane-bound isoforms are classified as NRP2a or NRP2b forms. Variable insertion of two short peptides comprising 5 and 17 amino acids (depicted as black and white vertical rectangles, respectively) gives rise to three NRP2a isoforms. They contain 22 (17 + 5), 17, or 0 additional amino acids and are denoted as a(22), a(17), or a(0), respectively. The two known NRP2b isoforms b(5) and b(0) differ by the insertion of the same variable five amino acid peptide that is found in NRP2a(22). The number of amino acids of each isoform is indicated in the right margin. Black triangles indicate the position of potential N-glycosylation sites. B, shown is a Western blot analysis of NRP2 isoforms. COS-7 cells were transiently transfected with cDNAs of full-length NRP2 isoforms carrying a C-terminal Myc-epitope. Twenty-four hours after transfection, cells were harvested, and whole cell lysates were separated by 7% SDS-PAGE. Western blot analysis was performed with the anti-Myc mAb 9E10 followed by ECL detection. C, shown is lectin analysis of NRP2 isoforms. NRP2 isoforms were expressed in COS-7 cells as described in B. After immunoprecipitation with anti-Myc mAb 9E10, proteins were separated by 7% SDS-PAGE before and after treatment with sialidase from A. ureafaciens. Western blot analysis was performed with digoxigenin-labeled PNA followed by peroxidase-conjugated anti-digoxigenin antibodies and ECL detection (left panel). Thereafter, the same blot was re-probed with anti-Myc mAb 9E10 followed by alkaline phosphatase-conjugated anti-mouse IgG antibodies and colorimetric detection (right panel).
To investigate whether all isoforms of NRP2 can serve as polySia acceptor, ST8SiaIV was expressed together with all of the five transmembrane isoforms of human NRP2, each tagged with a C-terminal Myc-epitope. Cell lysates were analyzed either directly (Fig. 3A, top panel) or after immunoprecipitation with an anti-Myc antibody (Fig. 3A, middle and bottom panel). Direct analysis of lysates was performed before and after treatment with endoN, and the subsequent Western blot analysis with anti-Myc antibody revealed a diffuse endoN-sensitive signal that was much more intense in the case of NRP2b compared with NRP2a variants (Fig. 3A, top panel). However, in all cases endoN treatment resulted in the collapse of this diffuse signal and appearance of a focused band, clearly demonstrating that all isoforms had been polysialylated by ST8SiaIV. Notably, the lower NRP2 band at ∼105–120 kDa was not affected by endoN treatment, indicating that the PNA-negative glycoform did not serve as a target for polysialylation (Fig. 3A, upper panel). Polysialylation of all NRP2 isoforms was further confirmed by analyzing immunoprecipitated NRP2 with an anti-polySia mAb (Fig. 3A, middle panel). For each isoform, an endoN-sensitive signal was observed. These signals were most prominent for the NRP2b isoforms with the highest intensity seen for NRP2b(5). In parallel, the immunoprecipitated material was analyzed with an anti-NRP2 antibody (Fig. 3A, bottom panel). Although low amounts of polySia-NRP2 were difficult to stain with anti-NRP2 antibody, the material was well seen after endoN treatment when it had collapsed into a focused band. Comparison of the band pattern before and after endoN treatment again showed that the upper band, representing the PNA-positive glycoform, served as polySia acceptor. Thus, ST8SiaIV was able to modify all transmembrane isoforms of NRP2 but acted exclusively on the PNA-positive glycoform. Because this glycoform was predominantly found for NRP2b variants (Fig. 2, B and C), this might explain why these variants showed enhanced polysialylation compared with NRP2a variants.
FIGURE 3.
Polysialylation of NRP2 isoforms is mediated by ST8SiaIV but not ST8SiaII. In COS-7 cells the indicated Myc-tagged NRP2 isoforms were transiently co-expressed with either ST8SiaIV (A) or ST8SiaII (B). Cells were harvested 24 h after transfection, and cell lysates were analyzed either directly (top panels) or after immunoprecipitation (IP) with anti-Myc mAb 9E10 (middle and bottom panels). For direct analysis, one aliquot of each lysate was treated with endoN to remove polySia. After separation by 7% SDS-PAGE, samples were analyzed by Western blotting (WB) using anti-Myc mAb 9E10 (top panels). Immunoprecipitated NRP2 was separated by 7% SDS-PAGE before and after endoN treatment and analyzed by Western blotting using anti-polySia mAb 735 (middle panels) or anti-NRP2 antibody H-300 (bottom panels). Because endoN cannot remove the most proximal α2,8-linked sialic acid residues of a protein-bound polySia chain (26), the endoN-treated polySia-NRP2 shown in A migrates slightly slower than the non-polysialylated NRP2 shown in B.
Differences in the Acceptor Specificity of the Polysialyltransferases ST8SiaII and ST8SiaIV
To study the ability of ST8SiaII to use NRP2 as an acceptor substrate, each NRP2 isoform was individually expressed with ST8SiaII, and cell lysates were analyzed as described above. However, a high molecular weight smear was not detected for either of the isoforms (Fig. 3B, top panel). Moreover, no polySia was detected if immunoprecipitated NRP2 was stained with an anti-polySia mAb (Fig. 3B, middle panel), and analysis of the immunoprecipitated material with an anti-NRP2 antibody showed no difference in the band pattern before and after endoN treatment. Taken together, these data demonstrated that none of the NRP2 isoforms had been polysialylated by ST8SiaII.
PolySia on NRP2 Is Selectively Attached to O-Linked Glycans
The finding that in human DCs most of the polySia could be removed from NRP2 by alkali treatment led to the conclusion that polySia is predominantly attached to O-glycans (38). The residual amount of polySia that was still detectable after alkali treatment could be either attributed to incomplete removal of O-glycans or to polysialylated N-glycans. Enzymatic removal of N-glycans by peptide N-glycosidase F did not affect polysialylation of NRP2 (38). However, because peptide N-glycosidase F treatment did not lead to a decrease in molecular mass of NRP2 as expected, removal of N-glycans could not be verified (38). To address the impact of N-glycans for NRP2 polysialylation, we mutated the Asn residue in the Asn-XXX-(Ser/Thr) sequon of the three potential N-glycosylation sites of NRP2b (Asn-152, Asn-157, and Asn-629). Because polysialylation of NRP2 was most efficient for NRP2b(5) (Fig. 3A), this isoform was used for all subsequent analyses. Based on a C-terminal-Myc-tagged variant of NRP2b(5), single (ΔAsn-629), double (ΔAsn-152, ΔAsn-157) and triple mutants (ΔAsn-152, ΔAsn-157, and ΔAsn-629) were generated and expressed in COS-7 cells either alone (Fig. 4A) or in combination with ST8SiaIV (Fig. 4B). Like wild-type NRP2b(5), all mutant proteins migrated as double bands in the SDS-PAGE (Fig. 4A), ruling out that loss of N-glycosylation abolished O-glycosylation. Deletion of Asn-152 and Asn-157 but not Asn-629 resulted in increased mobility of NRP2 (Fig. 4A), suggesting that only Asn-152 and Asn-157 are glycosylated. As shown in Fig. 4B, even combined deletion of all three N-glycosylation sites did not abolish polysialylation by ST8SiaIV, highlighting that N-glycans are not essential for the polysialylation of NRP2.
FIGURE 4.
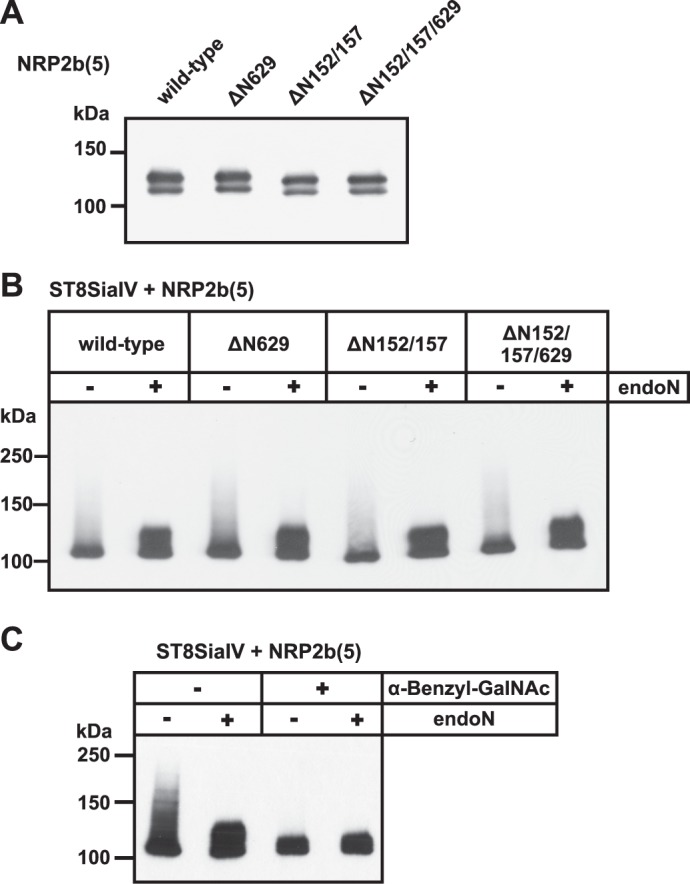
Impact of N- and O-glycosylation on NRP2 polysialylation. A, analysis of N-glycosylation-deficient NRP2 variants is shown. C-terminal-Myc-tagged NRP2b(5) variants lacking N-glycosylation site Asn-629 (ΔN629), Asn-152 and Asn-157 (ΔN152/157), or all three sites (ΔN152/157/629) were expressed in COS-7 cells. Cell lysates were analyzed by Western blotting with anti-Myc mAb 9E10. B, polysialylation of N-glycosylation-deficient NRP2 is shown. Wild-type and the N-glycosylation-deficient NRP2b(5) variants shown in A were co-expressed with ST8SiaIV. Cells lysates were analyzed before and after endoN treatment by Western blotting using anti-Myc mAb 9E10. C, analysis of O-glycosylation-deficient NRP2 is shown. C-terminal Myc-tagged NRP2b(5) and ST8SiaIV were co-expressed in COS-7 cells in the absence or presence of the O-glycosylation inhibitor α-benzyl-GalNAc. Cell lysates were analyzed by Western blotting with anti-Myc mAb 9E10.
To investigate whether polySia is exclusively attached to O-glycans, we blocked the synthesis of mucin-type O-glycans by treatment of COS-7 cells with α-benzyl-GalNAc. This structural analog of GalNAc-Ser/Thr acts as a competitive inhibitor of O-glycan extension by serving as a decoy acceptor for enzymes involved in these extensions (50, 51). NRP2b(5) and ST8SiaIV were co-expressed in the presence or absence of α-benzyl-GalNAc, and polysialylation was analyzed by Western blotting. In the presence of inhibitor, NRP2 was completely devoid of polySia (Fig. 4C), demonstrating that polysialylation of NRP2 occurs exclusively on O-glycans. Moreover, NRP2b(5) expressed in the presence of α-benzyl-GalNAc appeared as a single band at ∼105 kDa, confirming the finding that the additional band at ∼125 kDa observed in the absence of inhibitor (see Figs. 2B and 4A) represents NRP2 carrying mucin-type O-glycans.
Mass Spectrometric Characterization of NRP2-derived O-Glycans
So far, nothing is known on the O-glycosylation pattern of NRP2. To obtain structural information of the O-glycans attached to NRP2, a mass spectrometry approach was performed with NRP2b(5) expressed in COS-7 cells.
Consistent with the PNA-staining, no O-glycans were detected for NRP2b(5) obtained from NRP2 migrating in the lower band. By contrast, three major signals at m/z 1256, 1344, and 1705 were detected for NRP2b(5) extracted from the upper band, corresponding to di-sialylated core 1, mono-sialylated core 2, and di-sialylated core 2 O-glycans, respectively (Fig. 5 and supplemental Table 3). No indications were found for the presence of fucosylated species.
FIGURE 5.
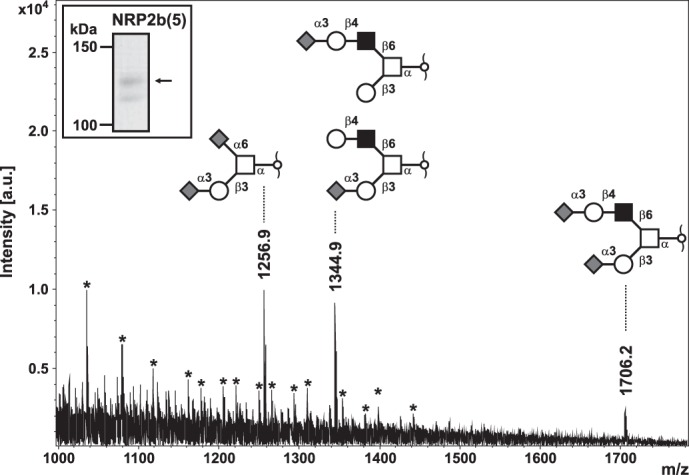
Gel-based O-glycomics of NRP2. C-terminal Myc-tagged NRP2b(5) expressed in COS-7 cells was immunoprecipitated by anti-Myc mAb 9E10. After separation by SDS-PAGE, Coomassie-stained upper and lower band of NRP2b(5) (see the inset) were cut out individually and digested in-gel with Pronase E to generate Pronase-stable glycopeptides (42). Glycopeptides eluted from gel slices were off-gel-treated with 50 mm NaOH in the presence of sodium borohydride to liberate O-linked glycans by reductive β-elimination. The permethylated glycan alditols were analyzed by MALDI-TOF mass spectrometry for O-glycoprofiling. Major sialyl-oligosaccharides corresponding to core 1 and core 2 mucin-type glycans (refer to inset structural models) were identified in the upper gel band on the basis of their molecular masses and their calculated monosaccharide compositions in concert with MS/MS analyses by post-source decay fragmentation of precursor ions under laser-induced dissociation conditions (supplemental Table 3). The mass spectrum shown revealed strong contamination with detergent, and the corresponding signal series is indicated by asterisks. For NRP2b(5) of the lower gel-band, no evidence for O-glycosylation was obtained in parallel analyses (data not shown). a.u., arbitrary units.
Core 1 O-Glycans Are Sufficient for Polysialylation of NRP2
To investigate whether ST8SiaIV is selective for particular O-glycan structures such as β6-branched core 2 glycans, we analyzed polysialylation of NRP2 in CHO cells. These cells produce exclusively core 1 structures due to a lack of core 2 β1,6-N-acetylglucosaminyltransferase (C2GnT), essential for the β6-branching (52, 53). NRP2b(5) expressed in CHO cells showed the same double band pattern as in COS-7 cells (Fig. 6). Moreover, co-expression with ST8SiaIV resulted in polysialylation of NRP2, a process that was again restricted to NRP2 migrating in the upper band. Although the extent of NRP2 polysialylation was lower in CHO compared with COS-7 cells, these data demonstrated that core 1 glycans were sufficient for polysialylation of NRP2.
FIGURE 6.
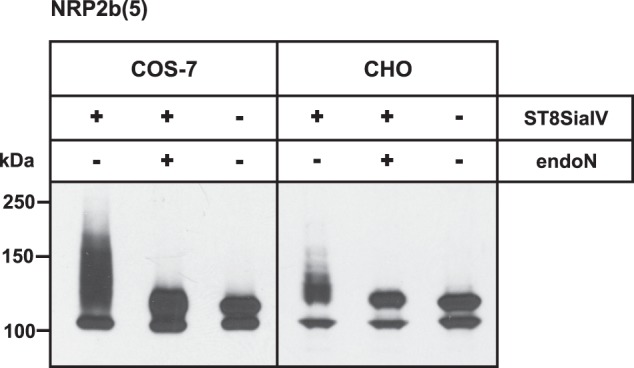
Core 1 O-glycans are sufficient for NRP2 polysialylation. C-terminal Myc-tagged NRP2b(5) was transiently expressed with or without ST8SiaIV in COS-7 cells (left panel) and CHO cells (right panel). Cell lysates were separated before or after treatment with endoN by 7% SDS-PAGE followed by Western blot analysis using anti-Myc mAb 9E10.
Determination of the O-Glycosylation Sites Involved in NRP2 Polysialylation
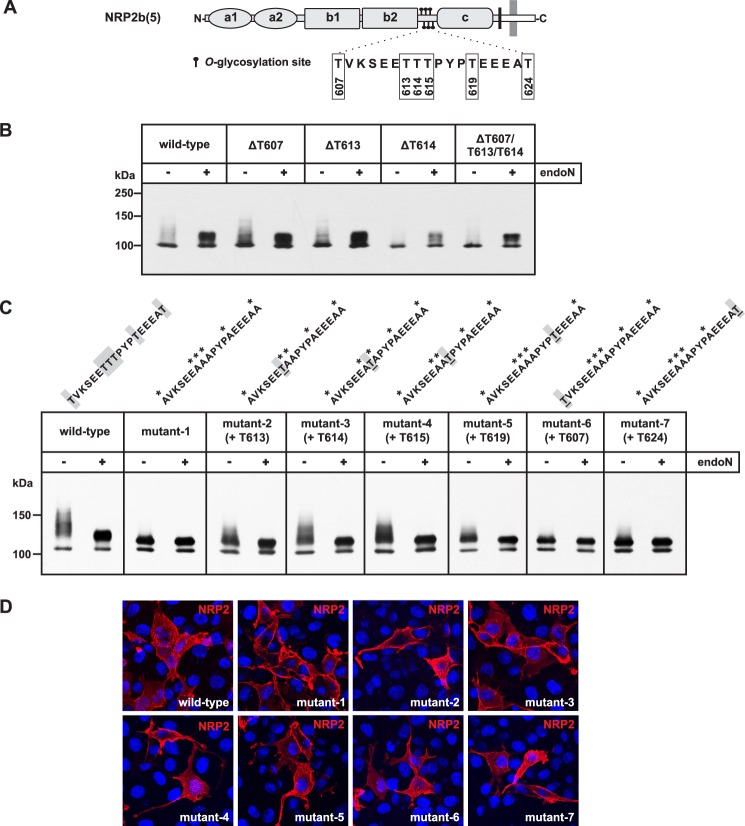
To identify the O-glycan attachment sites required for polysialylation of NRP2, we performed a comprehensive site-directed mutagenesis approach. Because no consensus recognition sequence for O-glycosyltransferases is known, putative sites for mucin-type O-GalNAc-glycosylation were predicted by NetOGlyc 3.1 (54). Although almost 150 sites stretching over the whole protein were identified, only a few of them were predicted with high probability (supplemental Fig. S2). The highest scores were obtained for residues Thr-607, Thr-613, and Thr-614 followed by Thr-615, Thr-619, and Thr-624. As shown schematically in Fig. 7A, these residues are all localized in the linker region between b2 and c domain.
FIGURE 7.
Identification of particular O-glycosylation sites involved in NRP2 polysialylation. A, a schematic representation of NRP2(b) shows the location of the six O-glycosylation sites targeted in this study. The peptide sequence carrying these sites is shown below with the amino acid numbers of the corresponding threonine residues. B, shown is analysis of single and triple mutants. In COS-7 cells, ST8SiaIV was co-expressed with C-terminal Myc-tagged NRP2b(5) either as the wild-type form or as a mutant variant lacking the indicated O-glycosylation sites due to a corresponding threonine to alanine exchange. Cell lysates were separated before and after treatment with endoN by 7% SDS-PAGE followed by Western blot analysis using anti-Myc mAb 9E10. C, shown is analysis of hexa and penta mutants. ST8SiaIV was co-expressed with Myc-tagged NRP2b(5) lacking either all six O-glycosylation sites highlighted in A (mutant-1) or a combination of five sites (mutant-2 to -7). As in B, cell lysates were separated by 7% SDS-PAGE before and after treatment with endoN, and Western blot analysis was performed with anti-Myc mAb 9E10. Above each panel, the peptide sequence of the linker region is given with alanine exchanges marked by an asterisk. In the case of the penta mutants, the retained threonine residue that can still serve as O-glycosylation sites is underlined. D, shown is surface expression of wild-type and mutant forms of NRP2b(5). COS-7 cells were transfected with either wild-type NRP2b(5) or the mutant forms of NRP2b(5) analyzed in C (mutant-1 to -7). 24 h after transfection, cells were fixed with 4% paraformaldehyde and stained with anti-NRP2 antibody followed by Alexa568-conjugated secondary antibody (red). Nuclei were stained with DAPI (blue).
Based on isoform NRP2b(5), we first analyzed the impact of Thr-607, Thr-613, and Thr-614 for NRP2 polysialylation. Each site was deleted by single or combined exchange to alanine. The ability of the mutants to serve as an acceptor for polysialylation was analyzed in COS-7 cells by co-expressing the mutants with ST8SiaIV. In each case, polysialylation of NRP2 was observed by the appearance of an endoN-sensitive smear and even the NRP2 variant lacking all three sites was still polysialylated (Fig. 7B). However, polysialylation of NRP2 was lost by combined deletion of all six O-glycosylation sites shown in Fig. 7A (see mutant-1 in Fig. 7C). Notably, the obtained hexa-mutant still migrated as a double band, indicating the presence of additional O-glycans that are located outside of the targeted amino acid stretch.
To study the impact of individual threonine residues on the polysialylation of NRP2, penta mutants were generated that lacked all but one of the O-glycosylation sites deleted in mutant-1. Restoration of one of the centrally located threonine residues, i.e. Thr-613, -614, -615, or -619, resulted in decreased but clearly visible polysialylation, evident by comparison of the upper band before and after endoN digest (see mutant-2, -3, -4, and -5 in Fig. 7C). The presence of a single threonine in one of the flanking positions, i.e. Thr-607 or Thr-624, allowed, if at all, the synthesis of only minute amounts of polySia (see mutant-6 and -7 in Fig. 7C), suggesting that the O-glycans attached to Thr-613, Thr-614, Thr-615, and Thr-619 serve as the major polySia attachment sites.
To exclude that the high number of introduced amino acid exchanges led to impaired folding and trafficking, we analyzed the cell surface localization of hexa- and penta-mutants by immunofluorescence analysis with an anti-NRP2 antibody directed against the extracellular part of the protein. For both wild-type and mutant forms of NRP2, the same cell surface staining was observed (Fig. 7D), excluding the possibility that the mutant forms were retained in the endoplasmic reticulum and were thus not accessible to the Golgi-localized polysialyltransferase ST8SiaIV.
Consistent with the finding that polySia is attached to O-glycans in the linker region between b2 and c domain, mass spectrometric analyses of NRP2(5) revealed a clear difference in the peptide pattern of this region between the two glycoforms separated as upper and lower band by SDS-PAGE. For the lower band glycoform, which did not serve as polySia acceptor, non-glycosylated peptides covering the linker region were observed, whereas no corresponding peptides were found for the upper band glycoform (supplemental Fig. S3). This indicates that the whole pool of the latter glycoform has an O-glycosylated linker region. In line with this, the upper band glycoform was fully converted to polySia-NRP2 in the presence of ST8SiaIV as indicated by disappearance of the band and full transformation into a diffuse microheterogeneous signal (see wild-type NRP2 in Fig. 7, B and C).
DISCUSSION
PolySia is a unique glycotope that largely impacts the function of the underlying protein (21, 22). Despite the restricted number of polySia-carriers, two enzymes have been evolved in vertebrates that catalyze polysialylation. In the nervous system, both enzymes, ST8SiaII and ST8SiaIV, contribute to polysialylation of the major polySia-carrier NCAM, and genetic ablation of both enzymes was necessary to fully abrogate polysialylation during brain development (24, 25, 40, 41, 55). Moreover, the fact that only minor differences were found between the polysialylated N-glycans of NCAM isolated from perinatal brain of wild-type, St8sia2−/−, and St8sia4−/− mice highlighted very similar catalytic functions of the two polysialyltransferases (28). Beyond NCAM polysialylation, however, the two polysialyltransferases may exert very different functions by targeting distinct acceptor proteins. Recently, we showed that polysialylation of SynCAM 1 strictly depends on the presence of ST8SiaII (56). In the developing brain, the modification of SynCAM 1 by polySia is cell type-specific and restricted to NG2 cells, a class of multifunctional precursor cells (33, 57). Polysialylation of SynCAM 1 was completely lost in St8sia2−/− but not in St8sia4−/− mice, and in cell culture experiments, SynCAM 1 was efficiently polysialylated by ST8SiaII but served only as a poor substrate for ST8SiaIV (56).
Here, we now showed that in contrast to SynCAM 1, NRP2 is a substrate for ST8SiaIV but not for ST8SiaII. In COS-7 cells, co-expression of NRP2 with ST8SiaIV but not ST8SiaII resulted in the formation of polySia-NRP2. Consistently, polysialylation of NRP2 was completely abrogated in BM-DCs of St8sia4−/− mice, whereas modification of NRP2 by polySia was not affected in BM-DCs of St8sia2−/− mice. Like in human monocyte-derived DCs (38), the total polySia signal of murine BM-DCs was stretched over a much broader molecular mass region than the polySia-NRP2 signal. As discussed by Curreli et al., (38) this might be either due to impaired binding of anti-NRP2 antibodies to highly polysialylated NRP2 or to the presence of additional, not yet identified polySia-carriers. However, even if other carriers contributed to the total polySia signal, these carriers were exclusively polysialylated by ST8SiaIV because the polySia was completely lost in St8sia4−/−-derived BM-DCs.
Through co-expression experiments, we demonstrated that ST8SiaIV can modify all transmembrane isoforms of NRP2, excluding the possibility that particular variable peptide sequences serve as recognition sites for ST8SiaIV. However, modification of NRP2 by polySia was strictly dependent on the presence of O-glycans, and combined deletion of all three N-glycosylation sites did not affect NRP2 polysialylation. This is in contrast to both NCAM and SynCAM 1 polysialylation, which occur selectively on N-glycans (23, 33, 58). In COS-7 cells, we blocked mucin-type O-glycosylation at the level of the Tn antigen (GalNAc-Thr/Ser) by α-benzyl-GalNAc. This sugar analog mimics the structure of the Tn antigen and acts as competitive decoy acceptor for enzymes involved in subsequent O-glycan extension (50, 51). The observed block of NRP2 polysialylation in α-benzyl-GalNAc-treated COS-7 cells indicated that the Tn antigen is not sufficient for polysialylation of NRP2 and suggested a crucial requirement for O-glycan extensions. Previously, we showed that terminal α2,3- or α2,6-sialylation of acceptor glycans is a prerequisite for the polysialylation of NCAM and SynCAM 1 (26, 33). Accordingly, polysialylation of NRP2 might crucially depend on O-glycan capping by terminal sialic acids. Consistent with this, structural analysis of the O-glycans of NRP2 expressed in COS-7 cells demonstrated the presence of di-sialylated core 1 as well as mono- and di-sialylated core 2 glycans. NRP2 expressed in CHO cells, which synthesize exclusively core 1 glycans due to a lack of the core 2 β1,6-N-acetylglucosaminyltransferase (52, 53), was still polysialylated by ST8SiaIV. Thus, a simple core 1 structure capped by terminal sialylation may represent the minimal glycan structure required for NRP2 polysialylation. The fact that polysialylation of NRP2 was more pronounced in COS-7 than in CHO cells (Fig. 6) suggests that core 2 glycans were more efficiently polysialylated than core 1 glycans. However, it has to be considered that in contrast to COS-7 cells, CHO cells express ST8SiaIV and NCAM (59). Thus, in CHO cells, NRP2 has to compete with NCAM for polysialylation. This fact is highlighted by the observation that despite endogenous ST8SiaIV expression in CHO cells, polysialylation of NRP2 was only achieved after the ST8SiaIV level was increased by co-expression of exogenous ST8SiaIV (Fig. 6).
During maturation of murine BM-DCs, we also observed an up-regulation of ST8SiaVI (Fig. 1B). This enzyme transfers single sialic acid residues preferentially to α2,3-sialylated O-glycans, forming an α2,8-linked di-Sia motif (44, 45). Thus, in theory, capping of NRP2 O-glycans by a di-Sia motif could occur in BM-DCs and could provide a recognition signal for the polysialyltransferase. However, by demonstrating polysiaylation of NRP2 in COS-7 cells, we can rule out that this motif is prerequisite for NRP2 polysialyation, as ST8SiaVI is not expressed in COS-7 cells (data not shown) and we found no indications for the presence of a di-Sia motif on the O-glycans of COS-7 cell-expressed NRP2 (Fig. 5 and supplemental Table 3).
A comprehensive site-directed mutagenesis approach finally allowed us to localize the O-glycans that serve as polySia acceptor sites in NRP2. Notably, the corresponding O-glycosylation sites were all clustered in a short stretch of 17 amino acids located between b2 and c domain. Combined alanine exchange of all six threonine residues located in this stretch completely abolished polysialylation of NRP2. Reintroduction of single threonine residues resulted only in a partial rescue of NRP2 polysialylation, indicating that in the native protein, polySia is attached to more than one O-glycosylation site. Because the rescue was most pronounced after reintroduction of single threonines at the centrally located positions Thr-613, -614, -615, and -619, these sites might represent the major polySia attachment sites. Future work will be necessary to precisely define whether all six or only particular sites are polysialylated in vivo. However, due to the close proximity of mucin-type O-glycan sites, O-glyopeptide analysis is in general still a challenge (60).
NRP1, the closely related homologue of NRP2, lacks the cluster of threonines identified as polySia attachment site of NRP2 (Fig. 8), which might explain why it does not serve as a target for polysialylation (38). However, NRP1 can be posttranslationally modified by a glycosaminoglycan (GAG) chain at a single serine residue at position 612 located in precisely the same linker region that encompasses the polySia attachment sites of NRP2 (61–63). NRP1 exists as heparan sulfate or chondroitin sulfate proteoglycan and the predominant GAG type as well as the degree and length of modification depends on the cell type. In vascular smooth muscles cells, chondroitin sulfate-modified NRP1 was shown to enhance platelet-derived growth factor (PDGF)-driven chemotaxis and cell migration (63). Initiation of GAGylation depends on a serine residue located in a Ser-Gly/Ala dipeptide that has one or more acidic amino acids in close proximity (64). As highlighted in Fig. 8, NRP2 lacks the Ser-612—Gly-613 dipeptide used for GAGylation in NRP1 and is, therefore, not modified by GAGs (61, 63). However, its posttranslational modification by polySia provides similar functions. As highly negatively charged polymers, both GAGs and polySia are known to serve as scavenger molecules for various positively charged soluble factors such as chemokines and growth factors. For example, polySia on NCAM of oligodendrocyte precursor cells is needed for directional cell migration in response to concentration gradients of PDGF (65). On hippocampal and hypothalamic neurons, polySia-NCAM enhances the sensitivity to brain-derived neurotrophic factor (BDNF), for which direct binding to polySia has been shown (66, 67). Besides the well studied interaction of GAGs with fibroblast growth factors, GAGs were reported to bind chemokines, increase their local concentration, and present them to their G-protein-coupled receptors (68–70). The chemokine CCL21 binds to both polySia and GAGs via its C-terminal basic region (15, 71). Similar to the function of GAGs on NRP1, polySia on NRP2 was suggested to trap CCL21, increase the local concentration of this chemokine, and facilitate the interaction with its receptor CCR7 (cysteine-cysteine chemokine receptor 7) (15, 16).
FIGURE 8.
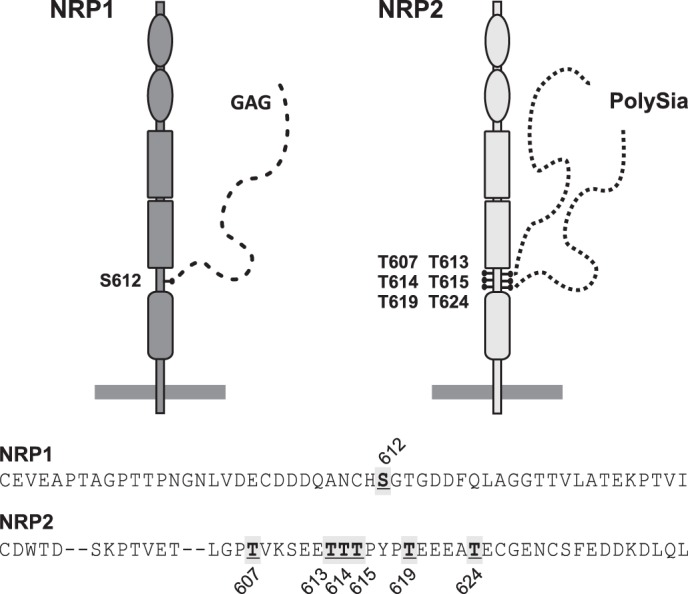
Glycosaminoglycan and polySia attachment sites in NRP1 and NRP2, respectively. Shown is a schematic representation of NRP1 and NRP2 modified by GAGylation and polySia, respectively. The amino acid sequence of the linker region between b2 and c domain is given for human NRP1 and NRP2, and the amino acids used as attachment sites for GAGylation or polysialylation are indicated by gray boxes.
The finding that almost the same position is used for GAGylation of NRP1 and polysialylation of NRP2 suggests that for both types of carbohydrate polymers, attachment to the linker region between b2 and c domain provides optimal positioning to fulfill their function in presenting or sequestering adsorbed chemokines to their cell surface receptors. However, as highlighted for FGF2 bound to heparan sulfate and polySia, the type of carbohydrate chain will determine how a soluble factor is displayed (72).
Supplementary Material
Acknowledgment
We thank Maike Hartmann for excellent technical assistance during the revision of this manuscript.
This work was supported by Deutsche Forschungsgemeinschaft Grant MU 1774/3 (to M. M.).

This article contains supplemental Tables 1–3 and Figs. 1–3.
- NRP2
- neuropilin 2
- NRP1
- neuropilin 1
- CCL19
- cysteine-cysteine motif chemokine 19
- CCL21
- cystein-cystein motif chemokine 21
- polySia
- polysialic acid
- DC
- dendritic cell
- BM-DC
- bone marrow-derived DC
- GAG
- glycosaminoglycan
- NCAM
- neural cell adhesion molecule
- endoN
- endoneuraminidase
- α-benzyl-GalNAc
- α-benzyl-N-acetylgalactosamine.
REFERENCES
- 1. Kitsukawa T., Shimizu M., Sanbo M., Hirata T., Taniguchi M., Bekku Y., Yagi T., Fujisawa H. (1997) Neuropilin-semaphorin III/D-mediated chemorepulsive signals play a crucial role in peripheral nerve projection in mice. Neuron 19, 995–1005 [DOI] [PubMed] [Google Scholar]
- 2. Kawasaki T., Kitsukawa T., Bekku Y., Matsuda Y., Sanbo M., Yagi T., Fujisawa H. (1999) A requirement for neuropilin-1 in embryonic vessel formation. Development 126, 4895–4902 [DOI] [PubMed] [Google Scholar]
- 3. Chen H., Bagri A., Zupicich J. A., Zou Y., Stoeckli E., Pleasure S. J., Lowenstein D. H., Skarnes W. C., Chédotal A., Tessier-Lavigne M. (2000) Neuropilin-2 regulates the development of selective cranial and sensory nerves and hippocampal mossy fiber projections. Neuron 25, 43–56 [DOI] [PubMed] [Google Scholar]
- 4. Giger R. J., Cloutier J. F., Sahay A., Prinjha R. K., Levengood D. V., Moore S. E., Pickering S., Simmons D., Rastan S., Walsh F. S., Kolodkin A. L., Ginty D. D., Geppert M. (2000) Neuropilin-2 is required in vivo for selective axon guidance responses to secreted semaphorins. Neuron 25, 29–41 [DOI] [PubMed] [Google Scholar]
- 5. Yuan L., Moyon D., Pardanaud L., Bréant C., Karkkainen M. J., Alitalo K., Eichmann A. (2002) Abnormal lymphatic vessel development in neuropilin 2 mutant mice. Development 129, 4797–4806 [DOI] [PubMed] [Google Scholar]
- 6. Chen H., Chédotal A., He Z., Goodman C. S., Tessier-Lavigne M. (1997) Neuropilin-2, a novel member of the neuropilin family, is a high affinity receptor for the semaphorins Sema E and Sema IV but not Sema III. Neuron 19, 547–559 [DOI] [PubMed] [Google Scholar]
- 7. He Z., Tessier-Lavigne M. (1997) Neuropilin is a receptor for the axonal chemorepellent Semaphorin III. Cell 90, 739–751 [DOI] [PubMed] [Google Scholar]
- 8. Kolodkin A. L., Levengood D. V., Rowe E. G., Tai Y. T., Giger R. J., Ginty D. D. (1997) Neuropilin is a semaphorin III receptor. Cell 90, 753–762 [DOI] [PubMed] [Google Scholar]
- 9. Pellet-Many C., Frankel P., Jia H., Zachary I. (2008) Neuropilins. Structure, function and role in disease. Biochem. J. 411, 211–226 [DOI] [PubMed] [Google Scholar]
- 10. Schwarz Q., Ruhrberg C. (2010) Neuropilin, you gotta let me know. Should I stay or should I go? Cell Adh. Migr. 4, 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tordjman R., Lepelletier Y., Lemarchandel V., Cambot M., Gaulard P., Hermine O., Roméo P. H. (2002) A neuronal receptor, neuropilin-1, is essential for the initiation of the primary immune response. Nat. Immunol. 3, 477–482 [DOI] [PubMed] [Google Scholar]
- 12. Sarris M., Andersen K. G., Randow F., Mayr L., Betz A. G. (2008) Neuropilin-1 expression on regulatory T cells enhances their interactions with dendritic cells during antigen recognition. Immunity 28, 402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takamatsu H., Takegahara N., Nakagawa Y., Tomura M., Taniguchi M., Friedel R. H., Rayburn H., Tessier-Lavigne M., Yoshida Y., Okuno T., Mizui M., Kang S., Nojima S., Tsujimura T., Nakatsuji Y., Katayama I., Toyofuku T., Kikutani H., Kumanogoh A. (2010) Semaphorins guide the entry of dendritic cells into the lymphatics by activating myosin II. Nat. Immunol. 11, 594–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bax M., van Vliet S. J., Litjens M., García-Vallejo J. J., van Kooyk Y. (2009) Interaction of polysialic acid with CCL21 regulates the migratory capacity of human dendritic cells. PLoS ONE 4, e6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rey-Gallardo A., Escribano C., Delgado-Martín C., Rodriguez-Fernández J. L., Gerardy-Schahn R., Rutishauser U., Corbi A. L., Vega M. A. (2010) Polysialylated neuropilin-2 enhances human dendritic cell migration through the basic C-terminal region of CCL21. Glycobiology 20, 1139–1146 [DOI] [PubMed] [Google Scholar]
- 16. Rey-Gallardo A., Delgado-Martín C., Gerardy-Schahn R., Rodríguez-Fernández J. L., Vega M. A. (2011) Polysialic acid is required for neuropilin-2a/b-mediated control of CCL21-driven chemotaxis of mature dendritic cells and for their migration in vivo. Glycobiology 21, 655–662 [DOI] [PubMed] [Google Scholar]
- 17. Sozzani S., Allavena P., Vecchi A., Mantovani A. (2000) Chemokines and dendritic cell traffic. J. Clin. Immunol. 20, 151–160 [DOI] [PubMed] [Google Scholar]
- 18. Förster R., Davalos-Misslitz A. C., Rot A. (2008) CCR7 and its ligands. Balancing immunity and tolerance. Nat. Rev. Immunol. 8, 362–371 [DOI] [PubMed] [Google Scholar]
- 19. Randolph G. J., Ochando J., Partida-Sánchez S. (2008) Migration of dendritic cell subsets and their precursors. Annu. Rev. Immunol. 26, 293–316 [DOI] [PubMed] [Google Scholar]
- 20. Sánchez-Sánchez N., Riol-Blanco L., Rodríguez-Fernández J. L. (2006) The multiple personalities of the chemokine receptor CCR7 in dendritic cells. J. Immunol. 176, 5153–5159 [DOI] [PubMed] [Google Scholar]
- 21. Rutishauser U. (2008) Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat. Rev. Neurosci. 9, 26–35 [DOI] [PubMed] [Google Scholar]
- 22. Mühlenhoff M., Oltmann-Norden I., Weinhold B., Hildebrandt H., Gerardy-Schahn R. (2009) Brain development needs sugar. The role of polysialic acid in controlling NCAM functions. Biol. Chem. 390, 567–574 [DOI] [PubMed] [Google Scholar]
- 23. Liedtke S., Geyer H., Wuhrer M., Geyer R., Frank G., Gerardy-Schahn R., Zähringer U., Schachner M. (2001) Characterization of N-glycans from mouse brain neural cell adhesion molecule. Glycobiology 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 24. Weinhold B., Seidenfaden R., Röckle I., Mühlenhoff M., Schertzinger F., Conzelmann S., Marth J. D., Gerardy-Schahn R., Hildebrandt H. (2005) Genetic ablation of polysialic acid causes severe neurodevelopmental defects rescued by deletion of the neural cell adhesion molecule. J. Biol. Chem. 280, 42971–42977 [DOI] [PubMed] [Google Scholar]
- 25. Galuska S. P., Oltmann-Norden I., Geyer H., Weinhold B., Kuchelmeister K., Hildebrandt H., Gerardy-Schahn R., Geyer R., Mühlenhoff M. (2006) Polysialic acid profiles of mice expressing variant allelic combinations of the polysialyltransferases ST8SiaII and ST8SiaIV. J. Biol. Chem. 281, 31605–31615 [DOI] [PubMed] [Google Scholar]
- 26. Mühlenhoff M., Eckhardt M., Bethe A., Frosch M., Gerardy-Schahn R. (1996) Polysialylation of NCAM by a single enzyme. Curr. Biol. 6, 1188–1191 [DOI] [PubMed] [Google Scholar]
- 27. Angata K., Suzuki M., Fukuda M. (1998) Differential and cooperative polysialylation of the neural cell adhesion molecule by two polysialyltransferases, PST and STX. J. Biol. Chem. 273, 28524–28532 [DOI] [PubMed] [Google Scholar]
- 28. Galuska S. P., Geyer R., Gerardy-Schahn R., Mühlenhoff M., Geyer H. (2008) Enzyme-dependent variations in the polysialylation of the neural cell adhesion molecule (NCAM) in vivo. J. Biol. Chem. 283, 17–28 [DOI] [PubMed] [Google Scholar]
- 29. Fujimoto I., Bruses J. L., Rutishauser U. (2001) Regulation of cell adhesion by polysialic acid. Effects on cadherin, immunoglobulin cell adhesion molecule, and integrin function and independence from neural cell adhesion molecule binding or signaling activity. J. Biol. Chem. 276, 31745–31751 [DOI] [PubMed] [Google Scholar]
- 30. Johnson C. P., Fragneto G., Konovalov O., Dubosclard V., Legrand J. F., Leckband D. E. (2005) Structural studies of the neural-cell-adhesion molecule by X-ray and neutron reflectivity. Biochemistry 44, 546–554 [DOI] [PubMed] [Google Scholar]
- 31. Johnson C. P., Fujimoto I., Rutishauser U., Leckband D. E. (2005) Direct evidence that neural cell adhesion molecule (NCAM) polysialylation increases intermembrane repulsion and abrogates adhesion. J. Biol. Chem. 280, 137–145 [DOI] [PubMed] [Google Scholar]
- 32. Gascon E., Vutskits L., Kiss J. Z. (2007) Polysialic acid-neural cell adhesion molecule in brain plasticity. From synapses to integration of new neurons. Brain Res. Rev. 56, 101–118 [DOI] [PubMed] [Google Scholar]
- 33. Galuska S. P., Rollenhagen M., Kaup M., Eggers K., Oltmann-Norden I., Schiff M., Hartmann M., Weinhold B., Hildebrandt H., Geyer R., Mühlenhoff M., Geyer H. (2010) Synaptic cell adhesion molecule SynCAM 1 is a target for polysialylation in postnatal mouse brain. Proc. Natl. Acad. Sci. U.S.A. 107, 10250–10255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zuber C., Lackie P. M., Catterall W. A., Roth J. (1992) Polysialic acid is associated with sodium channels and the neural cell adhesion molecule N-CAM in adult rat brain. J. Biol. Chem. 267, 9965–9971 [PubMed] [Google Scholar]
- 35. Yabe U., Sato C., Matsuda T., Kitajima K. (2003) Polysialic acid in human milk. CD36 is a new member of mammalian polysialic acid-containing glycoprotein. J. Biol. Chem. 278, 13875–13880 [DOI] [PubMed] [Google Scholar]
- 36. Mühlenhoff M., Eckhardt M., Bethe A., Frosch M., Gerardy-Schahn R. (1996) Autocatalytic polysialylation of polysialyltransferase-1. EMBO J. 15, 6943–6950 [PMC free article] [PubMed] [Google Scholar]
- 37. Close B. E., Colley K. J. (1998) In vivo autopolysialylation and localization of the polysialyltransferases PST and STX. J. Biol. Chem. 273, 34586–34593 [DOI] [PubMed] [Google Scholar]
- 38. Curreli S., Arany Z., Gerardy-Schahn R., Mann D., Stamatos N. M. (2007) Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J. Biol. Chem. 282, 30346–30356 [DOI] [PubMed] [Google Scholar]
- 39. Frosch M., Görgen I., Boulnois G. J., Timmis K. N., Bitter-Suermann D. (1985) NZB mouse system for production of monoclonal antibodies to weak bacterial antigens. Isolation of an IgG antibody to the polysaccharide capsules of Escherichia coli K1 and group B meningococci. Proc. Natl. Acad. Sci. U.S.A. 82, 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Angata K., Long J. M., Bukalo O., Lee W., Dityatev A., Wynshaw-Boris A., Schachner M., Fukuda M., Marth J. D. (2004) Sialyltransferase ST8Sia-II assembles a subset of polysialic acid that directs hippocampal axonal targeting and promotes fear behavior. J. Biol. Chem. 279, 32603–32613 [DOI] [PubMed] [Google Scholar]
- 41. Eckhardt M., Bukalo O., Chazal G., Wang L., Goridis C., Schachner M., Gerardy-Schahn R., Cremer H., Dityatev A. (2000) Mice deficient in the polysialyltransferase ST8SiaIV/PST-1 allow discrimination of the roles of neural cell adhesion molecule protein and polysialic acid in neural development and synaptic plasticity. J. Neurosci. 20, 5234–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Breloy I., Pacharra S., Ottis P., Bonar D., Grahn A., Hanisch F. G. (2012) O-Linked N,N′-diacetyllactosamine (LacdiNAc)-modified glycans in extracellular matrix glycoproteins are specifically phosphorylated at subterminal N-acetylglucosamine. J. Biol. Chem. 287, 18275–18286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stummeyer K., Dickmanns A., Mühlenhoff M., Gerardy-Schahn R., Ficner R. (2005) Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat. Struct. Mol. Biol. 12, 90–96 [DOI] [PubMed] [Google Scholar]
- 44. Takashima S., Ishida H. K., Inazu T., Ando T., Ishida H., Kiso M., Tsuji S., Tsujimoto M. (2002) Molecular cloning and expression of a sixth type of α2,8-sialyltransferase (ST8Sia VI) that sialylates O-glycans. J. Biol. Chem. 277, 24030–24038 [DOI] [PubMed] [Google Scholar]
- 45. Teintenier-Lelièvre M., Julien S., Juliant S., Guerardel Y., Duonor-Cérutti M., Delannoy P., Harduin-Lepers A. (2005) Molecular cloning and expression of a human hST8Sia VI (α2,8-sialyltransferase) responsible for the synthesis of the diSia motif on O-glycosylproteins. Biochem. J. 392, 665–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yoshida Y., Kojima N., Kurosawa N., Hamamoto T., Tsuji S. (1995) Molecular cloning of Sia α2,3Gal β1,4GlcNAc α2,8-sialyltransferase from mouse brain. J. Biol. Chem. 270, 14628–14633 [DOI] [PubMed] [Google Scholar]
- 47. Angata K., Suzuki M., McAuliffe J., Ding Y., Hindsgaul O., Fukuda M. (2000) Differential biosynthesis of polysialic acid on neural cell adhesion molecule (NCAM) and oligosaccharide acceptors by three distinct α2,8-sialyltransferases, ST8Sia IV (PST), ST8Sia II (STX), and ST8Sia III. J. Biol. Chem. 275, 18594–18601 [DOI] [PubMed] [Google Scholar]
- 48. Rossignol M., Gagnon M. L., Klagsbrun M. (2000) Genomic organization of human neuropilin-1 and neuropilin-2 genes. Identification and distribution of splice variants and soluble isoforms. Genomics 70, 211–222 [DOI] [PubMed] [Google Scholar]
- 49. Gillespie W., Paulson J. C., Kelm S., Pang M., Baum L. G. (1993) Regulation of α2,3-sialyltransferase expression correlates with conversion of peanut agglutinin (PNA)+ to PNA− phenotype in developing thymocytes. J. Biol. Chem. 268, 3801–3804 [PubMed] [Google Scholar]
- 50. Huang J., Byrd J. C., Yoon W. H., Kim Y. S. (1992) Effect of benzyl-α-GalNAc, an inhibitor of mucin glycosylation, on cancer-associated antigens in human colon cancer cells. Oncol. Res. 4, 507–515 [PubMed] [Google Scholar]
- 51. Huet G., Kim I., de Bolos C., Lo-Guidice J. M., Moreau O., Hemon B., Richet C., Delannoy P., Real F. X., Degand P. (1995) Characterization of mucins and proteoglycans synthesized by a mucin-secreting HT-29 cell subpopulation. J. Cell Sci. 108, 1275–1285 [DOI] [PubMed] [Google Scholar]
- 52. Bierhuizen M. F., Fukuda M. (1992) Expression cloning of a cDNA encoding UDP-GlcNAc:Gal β1–3-GalNAc-R (GlcNAc to GalNAc) β1–6GlcNAc transferase by gene transfer into CHO cells expressing polyoma large tumor antigen. Proc. Natl. Acad. Sci. U.S.A. 89, 9326–9330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dennis J. W. (1993) Core 2 GlcNAc-transferase and polylactosamine expression in O-glycans. Glycobiology 3, 91–93 [DOI] [PubMed] [Google Scholar]
- 54. Julenius K., Mølgaard A., Gupta R., Brunak S. (2005) Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15, 153–164 [DOI] [PubMed] [Google Scholar]
- 55. Oltmann-Norden I., Galuska S. P., Hildebrandt H., Geyer R., Gerardy-Schahn R., Geyer H., Mühlenhoff M. (2008) Impact of the polysialyltransferases ST8SiaII and ST8SiaIV on polysialic acid synthesis during postnatal mouse brain development. J. Biol. Chem. 283, 1463–1471 [DOI] [PubMed] [Google Scholar]
- 56. Rollenhagen M., Kuckuck S., Ulm C., Hartmann M., Galuska S. P., Geyer R., Geyer H., Mühlenhoff M. (2012) Polysialylation of the synaptic cell adhesion molecule 1 (SynCAM 1) depends exclusively on the polysialyltransferase ST8SiaII in vivo. J. Biol. Chem. 287, 35170–35180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nishiyama A., Komitova M., Suzuki R., Zhu X. (2009) Polydendrocytes (NG2 cells). Multifunctional cells with lineage plasticity. Nat. Rev. Neurosci. 10, 9–22 [DOI] [PubMed] [Google Scholar]
- 58. von der Ohe M., Wheeler S. F., Wuhrer M., Harvey D. J., Liedtke S., Mühlenhoff M., Gerardy-Schahn R., Geyer H., Dwek R. A., Geyer R., Wing D. R., Schachner M. (2002) Localization and characterization of polysialic acid-containing N-linked glycans from bovine NCAM. Glycobiology 12, 47–63 [DOI] [PubMed] [Google Scholar]
- 59. Eckhardt M., Mühlenhoff M., Bethe A., Koopman J., Frosch M., Gerardy-Schahn R. (1995) Molecular characterization of eukaryotic polysialyltransferase-1. Nature 373, 715–718 [DOI] [PubMed] [Google Scholar]
- 60. Jensen P. H., Kolarich D., Packer N. H. (2010) Mucin-type O-glycosylation. Putting the pieces together. FEBS J. 277, 81–94 [DOI] [PubMed] [Google Scholar]
- 61. Shintani Y., Takashima S., Asano Y., Kato H., Liao Y., Yamazaki S., Tsukamoto O., Seguchi O., Yamamoto H., Fukushima T., Sugahara K., Kitakaze M., Hori M. (2006) Glycosaminoglycan modification of neuropilin-1 modulates VEGFR2 signaling. EMBO J. 25, 3045–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Frankel P., Pellet-Many C., Lehtolainen P., D'Abaco G. M., Tickner M. L., Cheng L., Zachary I. C. (2008) Chondroitin sulphate-modified neuropilin 1 is expressed in human tumour cells and modulates 3D invasion in the U87MG human glioblastoma cell line through a p130Cas-mediated pathway. EMBO Rep. 9, 983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pellet-Many C., Frankel P., Evans I. M., Herzog B., Jünemann-Ramírez M., Zachary I. C. (2011) Neuropilin-1 mediates PDGF stimulation of vascular smooth muscle cell migration and signalling via p130Cas. Biochem. J. 435, 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Esko J. D., Zhang L. (1996) Influence of core protein sequence on glycosaminoglycan assembly. Curr. Opin. Struct. Biol. 6, 663–670 [DOI] [PubMed] [Google Scholar]
- 65. Zhang H., Vutskits L., Calaora V., Durbec P., Kiss J. Z. (2004) A role for the polysialic acid-neural cell adhesion molecule in PDGF-induced chemotaxis of oligodendrocyte precursor cells. J. Cell Sci. 117, 93–103 [DOI] [PubMed] [Google Scholar]
- 66. Vutskits L., Djebbara-Hannas Z., Zhang H., Paccaud J. P., Durbec P., Rougon G., Muller D., Kiss J. Z. (2001) PSA-NCAM modulates BDNF-dependent survival and differentiation of cortical neurons. Eur. J. Neurosci. 13, 1391–1402 [DOI] [PubMed] [Google Scholar]
- 67. Kanato Y., Kitajima K., Sato C. (2008) Direct binding of polysialic acid to a brain-derived neurotrophic factor depends on the degree of polymerization. Glycobiology 18, 1044–1053 [DOI] [PubMed] [Google Scholar]
- 68. Hoogewerf A. J., Kuschert G. S., Proudfoot A. E., Borlat F., Clark-Lewis I., Power C. A., Wells T. N. (1997) Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry 36, 13570–13578 [DOI] [PubMed] [Google Scholar]
- 69. Kuschert G. S., Coulin F., Power C. A., Proudfoot A. E., Hubbard R. E., Hoogewerf A. J., Wells T. N. (1999) Glycosaminoglycans interact selectively with chemokines and modulate receptor binding and cellular responses. Biochemistry 38, 12959–12968 [DOI] [PubMed] [Google Scholar]
- 70. Handel T. M., Johnson Z., Crown S. E., Lau E. K., Proudfoot A. E. (2005) Regulation of protein function by glycosaminoglycans. As exemplified by chemokines. Annu. Rev. Biochem. 74, 385–410 [DOI] [PubMed] [Google Scholar]
- 71. Hirose J., Kawashima H., Swope Willis M., Springer T. A., Hasegawa H., Yoshie O., Miyasaka M. (2002) Chondroitin sulfate B exerts its inhibitory effect on secondary lymphoid tissue chemokine (SLC) by binding to the C terminus of SLC. Biochim. Biophys. Acta 1571, 219–224 [DOI] [PubMed] [Google Scholar]
- 72. Ono S., Hane M., Kitajima K., Sato C. (2012) Novel regulation of fibroblast growth factor 2 (FGF2)-mediated cell growth by polysialic acid. J. Biol. Chem. 287, 3710–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.