Background: Within the skeletal muscle Ca2+ release channel RyR1, S-oxidation and S-nitrosylation of allosterically linked Cys residues are coupled to oxygen tension (pO2).
Results: Mass spectrometry identifies multiple Cys residues as oxidized at high versus low pO2.
Conclusion: Endogenous H2O2 catalyzes pO2-coupled disulfide formation within an allosteric Cys network that gates S-nitrosylation.
Significance: Dynamic disulfide formation subserves physiological redox regulation of RyR1.
Keywords: Disulfide, Reactive Oxygen Species (ROS), Redox Regulation, Redox Signaling, Ryanodine Receptor, Skeletal Muscle, S-Oxidation
Abstract
In mammalian skeletal muscle, Ca2+ release from the sarcoplasmic reticulum (SR) through the ryanodine receptor/Ca2+-release channel RyR1 can be enhanced by S-oxidation or S-nitrosylation of separate Cys residues, which are allosterically linked. S-Oxidation of RyR1 is coupled to muscle oxygen tension (pO2) through O2-dependent production of hydrogen peroxide by SR-resident NADPH oxidase 4. In isolated SR (SR vesicles), an average of six to eight Cys thiols/RyR1 monomer are reversibly oxidized at high (21% O2) versus low pO2 (1% O2), but their identity among the 100 Cys residues/RyR1 monomer is unknown. Here we use isotope-coded affinity tag labeling and mass spectrometry (yielding 93% coverage of RyR1 Cys residues) to identify 13 Cys residues subject to pO2-coupled S-oxidation in SR vesicles. Eight additional Cys residues are oxidized at high versus low pO2 only when NADPH levels are supplemented to enhance NADPH oxidase 4 activity. pO2-sensitive Cys residues were largely non-overlapping with those identified previously as hyperreactive by administration of exogenous reagents (three of 21) or as S-nitrosylated. Cys residues subject to pO2-coupled oxidation are distributed widely within the cytoplasmic domain of RyR1 in multiple functional domains implicated in RyR1 activity-regulating interactions with the L-type Ca2+ channel (dihydropyridine receptor) and FK506-binding protein 12 as well as in “hot spot” regions containing sites of mutation implicated in malignant hyperthermia and central core disease. pO2-coupled disulfide formation was identified, whereas neither S-glutathionylated nor sulfenamide-modified Cys residues were observed. Thus, physiological redox regulation of RyR1 by endogenously generated hydrogen peroxide is exerted through dynamic disulfide formation involving multiple Cys residues.
Introduction
Redox-based post-translational regulation of protein function is exerted principally through modification of Cys thiol side chains. S-Nitrosylation provides the best characterized example with a large number of endogenous substrates identified in the context of cellular signal transduction along myriad pathways (1, 2). Accumulating evidence also points to a potentially broad regulatory influence of S-oxidation, catalyzed by reactive oxygen species (effectively, hydrogen peroxide (H2O2)), which could in principle be exerted through a number of independent or coupled oxidative modifications: the reversible and dynamic formation of disulfide bonds, sulfenic (and possibly sulfinic) acid, and sulfenamide as well as S-glutathionylation and S-sulfhydration (2–4). A broad role for physiological S-oxidation would be consistent with the ubiquitous expression across multicellular organismal phylogeny and cell types of the NADPH oxidases (Noxs),3 which function to generate reactive oxygen species (5). However, although there have been significant recent advances (6, 7), there are relatively few examples in which mediation or modulation of physiological signal transduction by S-oxidation has been characterized fully, that is with respect to the (enzymatic) source of endogenous H2O2 and the generative stimulus, the protein targets subject to S-oxidation and the specific Cys residues modified within those targets, the chemical nature of the modification, the effect of modification on protein function, and the consequences of altered protein function in the cellular milieu.
The ryanodine receptor/Ca2+ release channel (RyR), which serves as the essential source of Ca2+ release from the sarcoplasmic reticulum (SR) that mediates excitation-contraction coupling in skeletal and cardiac striated muscle, has emerged as a paradigmatic example of redox regulation of protein function through Cys-directed post-translational modification (8–16). RyRs, which function as tetramers, are the largest channel proteins described. Monomers of RyR1, the predominant form of RyR in skeletal muscle, comprise about 5000 amino acids (∼565 kDa). One hundred of those residues are Cys, and half of those Cys residues on average are in the reduced form (sulfhydryl bearing a free thiol) under basal conditions (8, 16). Studies using application in vitro of exogenous redox agents (reduced glutathione, oxidized glutathione, S-nitrosoglutathione, nitric oxide (NO), and NO donors (NOC and NOR)) have reported modification of multiple Cys residues within RyR1 by S-glutathionylation, S-nitrosylation, and unspecified S-oxidation (8–11, 13, 14, 17, 18). It has also been reported that tetanic stimulation of skeletal muscle elicits Nox-dependent production of H2O2 (19) and that RyR1 can be modified by S-glutathionylation via Nox2 localized to transverse tubule membranes (20). Increasing evidence thus supports the possibility that Nox-derived reactive oxygen species may play a physiological role in skeletal muscle excitation-contraction coupling.
Physiological S-oxidation of RyR1 is coupled to endogenous S-nitrosylation of the channel, providing a paradigmatic example of allosteric cross-talk by redox-based modifications. More specifically, under physiological conditions in situ, it has been shown that RyR1 is modified by S-nitrosylation that reflects the production of NO by neuronal nitric-oxide synthase (8, 10, 12) and by S-oxidation that is dependent upon the production of H2O2 by Nox4 (16). Physiological S-oxidation is governed by a redox cycle intrinsic to the SR that is coupled to muscle pO2: in isolated SR (SR vesicles) at relatively high pO2 (21% O2), an average of about six to eight free Cys thiols are oxidized consequent upon H2O2 production by Nox4, and the transition to relatively low pO2 (1% O2) is accompanied by reduction that reflects suppressed H2O2 production and the operation of an as yet unspecified reductive mechanism (8, 16). Oxygen-coupled S-oxidation activates RyR1, enhances Ca2+ release, and gates S-nitrosylation, which is blocked at high versus low pO2. S-Nitrosylation at low pO2, which also activates RyR1, targets a single Cys residue (Cys3635) that is not a member of the set of Cys residues oxidized at high versus low pO2 (10, 13). Thus, S-oxidation allosterically regulates RyR1 activity, and S-oxidation and S-nitrosylation operate together to modulate Ca2+ release through RyR1 over a range of pO2 that extends from the low pO2 that characterizes working muscle to oxidative stress.
The analysis in skeletal muscle SR provides an example of physiological S-oxidation in which the source of H2O2 (Nox4) and the generative stimulus (pO2), the specific protein target (RyR1), and the effects of modification on protein function (activation of RyR1 and enhanced Ca2+ release from the SR) have been described. However, although it has been shown that only a relatively small subset of Cys residues on average are subject to pO2-coupled S-oxidation, neither the identity of the targeted Cys residues nor the nature of the oxidative modification(s) are known. We developed a mass spectrometry-based scheme for analysis of regulated protein S-oxidation that we applied to identify the sites of physiological S-oxidation within RyR1 and the form of modification. Here we identify 13 Cys residues subject to oxidation at high versus low pO2 in isolated SR and eight additional Cys residues subject to oxidation at high versus low pO2 but only when NADPH levels are supplemented to enhance Nox4 activity. pO2-coupled S-oxidation targets Cys residues that are distributed widely within the cytoplasmic domain of RyR1 in multiple functional domains. Furthermore, we show that disulfide formation is the likely form of S-oxidation.
EXPERIMENTAL PROCEDURES
Preparation of SR Vesicles and Purification of RyR1
All preparations here and below utilized hind limb muscle from rabbit. SR vesicles were prepared essentially as described (21). Briefly, muscle was homogenized in buffer containing 20 mm Hepes, pH 7.4, 2 mm EDTA, 0.2 mm EGTA, 0.3 m sucrose, and protease inhibitors (100 nm aprotinin, 20 μm leupeptin, 1 μm pepstatin, 0.2 mm phenylmethylsulfonyl fluoride, 1 mm benzamidine). Homogenates were centrifuged at 9,200 × g for 20 min, and the resultant supernatant was centrifuged at 100,000 × g for 1 h. The resultant pellet (membrane fraction) was resuspended and fractionated on a continuous 20–45% (w/v) sucrose gradient by centrifugation at 100,000 × g for 14 h. Fractions containing SR vesicles were eluted, collected by centrifugation at 120,000 × g, resuspended, aliquoted, and stored in liquid nitrogen. RyR1 was purified from SR vesicles solubilized with CHAPS by sucrose density gradient centrifugation as described (22). Protein concentrations were determined with a bicinchoninic acid-based assay.
Quantification of Total RyR1 Free Thiols (Sulfhydryls)
The free thiol content of RyR1 was quantified by monobromobimane (MBB) fluorescence. As described (8, 16), SR vesicles held at 21% or 1% O2 (glove box) were exposed to MBB in the presence of 10 μm Ca2+ prior to isolation of RyR1 as above and quantification of fluorescence. When used, NADPH (1 mm) was added 30 min before MBB.
Isotope-coded Affinity Tag (ICAT) Labeling
SR vesicles were incubated in 50 mm phosphate buffer containing 10 μm CaCl2, pH 7.5 at 1 or 21% O2 for 30 min. When used, 10 units/ml polyethylene glycol (PEG)-coupled catalase, 20 μm diphenyleneiodonium (DPI), or 1 mm NADPH was added at the beginning of the incubation. Samples were then incubated with 10× excess (over calculated Cys content) ICAT reagent for 4 h in the presence of 2% SDS. Light ICAT (12C) and heavy ICAT (13C) were used to label samples incubated at low pO2 (1% O2) or high pO2 (21% O2), respectively. Samples (1 and 21% O2) were then mixed 1:1, and excess ICAT was removed by size exclusion filtration through a P6 gel column. Proteins were separated by reducing SDS-PAGE, and the band containing RyR1 was excised and incubated with DTT followed by iodoacetamide (IA) to alkylate previously oxidized Cys residues (23). In-gel digestion by trypsin or chymotrypsin was carried out overnight in 100 mm ammonium bicarbonate at 37 °C. ICAT-labeled peptides were purified on avidin affinity columns, and the biotin moiety was cleaved (AB Sciex). Both ICAT-labeled peptides (avidin-bound) and unlabeled peptides (flow-through) were collected, desalted with a C18 UltraMicro Tip Column, and resuspended in 0.1% (v/v) formic acid for analysis by liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS). The ICAT-based analytic scheme is illustrated in Fig. 1A.
FIGURE 1.
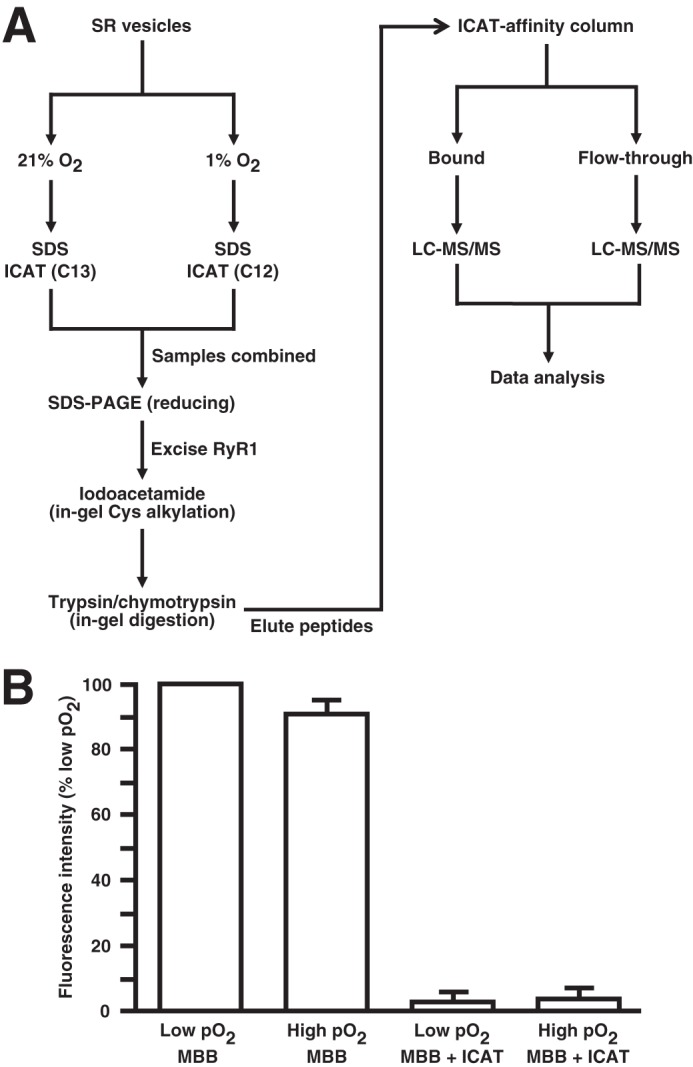
The MS-based analytic scheme utilized in the identification of sites of pO2-coupled redox regulation of RyR1. A, to identify pO2-sensitive Cys residues within RyR1, SR vesicles maintained at 21% O2 or 1% O2 are solubilized in SDS, and free thiols are labeled with heavy or light ICAT, respectively. Samples are then combined, and RyR1 is isolated by SDS-PAGE under reducing conditions. The band containing RyR1 is excised, and free thiols are alkylated with iodoacetamide followed by in-gel digestion with either trypsin or chymotrypsin. Eluted peptides are passed over an ICAT affinity column, and both bound peptides and the flow-through are analyzed by LC-MS/MS. Thus, for a given peptide, an increase in the light:heavy ratio at 1% O2 versus 21% O2 indicates Cys residues that are oxidized at high versus low pO2. B, ICAT fully labels RyR1 free Cys thiols. SR vesicles maintained at 1% O2 (low pO2) or 21% O2 (high pO2) were solubilized in SDS, and free thiols were labeled with the fluorescent reporter MBB. Prior incubation with ICAT reagent eliminates MBB fluorescence, demonstrating that free thiols are quantitatively labeled by ICAT reagent. Note that MBB fluorescence is decreased at high versus low pO2, indicating a loss of free thiols to S-oxidation (8, 16). Error bars represent S.E.
Dimedone Labeling
SR vesicles were incubated with 10 mm dimedone in 50 mm phosphate buffer containing 10 μm CaCl2, pH 7.5 at room temperature for 1 h at 21% O2. Proteins were separated by reducing SDS-PAGE, and the band containing RyR1 was excised and incubated with DTT followed by IA. Following in-gel digestion by trypsin or chymotrypsin, eluted peptides were desalted and analyzed by LC-MS/MS. Alternatively, muscle homogenates were incubated with 10 mm dimedone at 21% O2 for 1 h prior to isolation of SR vesicles and analysis as above.
Preparation of RyR1 for Native Identification of Disulfide and Other Oxidative Modifications
SR vesicles were incubated with 50 mm phosphate buffer containing 10 μm CaCl2, pH 7.5 at 1 or 21% O2 for 20 min. Samples were then held at 1 or 21% O2 and incubated with a 50-fold excess of IA or N-ethylmaleimide for 20 min at room temperature in the dark followed by the addition of 2% (w/v) SDS and incubation for an additional hour. Proteins were separated by non-reducing SDS-PAGE followed by excision of the band containing RyR1, in-gel digestion with trypsin or chymotrypsin, and analysis by LC-MS/MS.
Mass Spectrometric Analysis
LC-MS/MS was performed using an LTQ Orbitrap XL linear ion trap mass spectrometer (Thermo Fisher Scientific). Reverse-phase high performance liquid chromatography (HPLC) was conducted with an Ultimate 3000 HPLC system (Dionex) equipped with a Dionex C18 Acclaim PepMap 100 column (0.075 × 150 mm) in a linear gradient of acetonitrile from 2 to 60% (v/v) for a period of 60 min at a flow rate of 0.3 μl/min. Spectra were acquired in the positive ionization mode by data-dependent methods consisting of a full MS scan in high mass accuracy Fourier transform MS mode at 60,000 resolution and MS/MS on the six most abundant precursor ions in collision-induced dissociation mode with a normalized collision energy of 35%. A dynamic exclusion function was applied with repeat count of 2, repeat duration of 45 s, exclusion duration of 15 s, and exclusion size list of 350. Peptides were identified with Mascot Daemon (version 2.3.0, Matrix Science), and the data were searched against the RyR1 primary sequence. The mass tolerance was set at 10 ppm for precursor ions and 0.8 Da for product ions. Carbamidomethylation or N-ethylmaleimide labeling of Cys residues, dimedone labeling of Cys residues and sulfenic acids (when used), and oxidation of Met residues were set as variable modifications. Other possible modifications of Cys, including sulfenation, sulfination, sulfonation, glutathionylation, and sulfenamide formation, were also tested. The identification of disulfide bonds was performed using the on-line version of MassMatrix software (24). The modification(s), including disulfide bond formation, suggested by the search engine were verified by manual examination of each tandem mass spectrum and compared with corresponding unmodified peptides. For ICAT labeling analysis, both light and heavy ICAT-labeled peptides were first identified using Mascot Daemon and then confirmed by manual examination, especially in the case of peptides containing multiple Cys residues and mixed labeling with IA and ICAT.
MS Data Analysis
Quantification of ICAT labeling was conducted using Mascot Distiller (Matrix Science). All reported peptides labeled with ICAT were identified with standard error <0.1 and correlation coefficient >0.95. The two peaks from heavy and light ICAT labeling were identified as separated by 9 Da. The ratio of light ICAT (labeled at 1% oxygen) to heavy ICAT (labeled at 21% oxygen) was calculated and averaged on the basis of peptide peak heights for each individual scan. The ratios for the same peptide but with different charge and methionine oxidation state were then averaged manually. For peptides with multiple Cys residues and mixed labeling by IA and ICAT, the ratio was manually calculated from the peptide peak heights extracted from the selected ion chromatogram. Standard errors were derived from replicated biological samples, different charge and methionine oxidation states, or multiple scanning. Quantification of disulfide was conducted manually. Two native reference peptides, A (LSLPVQFHQHFR) and B (GDGGEGEGEVQFLR), that appeared unmodified in each of several repeated experiments were chosen (25). Peaks from the same disulfide-containing peptide with different methionine oxidation states were added together. After normalization with respect to the two reference peptides at the different pO2 values, relative disulfide abundance at high versus low pO2 was calculated as an average of three biological replicates.
RESULTS
Identification of Cys Residues within RyR1 Subject to pO2-dependent Oxidative Modification
RyR1 is an unusually large protein (∼5,000 amino acids) that contains 100 Cys residues. We developed an approach that allowed us to assess by mass spectrometric analysis the redox state of 93 of 100 Cys residues. In this approach (Fig. 1A), an RyR1-enriched subcellular fraction (SR vesicles) is solubilized in SDS under non-reducing conditions, and free thiols are labeled with ICAT reagent. The efficacy of ICAT labeling under the conditions used is demonstrated by the finding that incubation with ICAT reagent at either low or high pO2 eliminates subsequent labeling by the thiol-specific agent MBB (Fig. 1B). RyR1 is then enriched by SDS-PAGE under reducing conditions followed by in-gel alkylation with IA of thiols freed by reduction and in-gel digestion with either trypsin or chymotrypsin. Digests are subjected to avidin affinity purification, and both bound and unbound peptides are analyzed by LC-MS/MS. The dual enrichment provided by selective ICAT labeling of Cys-containing peptides and gel purification resulted in coverage of 93% of RyR1 Cys residues when the results of trypsin and chymotrypsin digestion were combined.
After labeling SR vesicles held at low versus high pO2 (1% O2 versus 21% O2) with light (12C) versus heavy (13C) ICAT reagent, respectively, followed by reduction and labeling with IA, RyR1 Cys residues fell into one of four principal groups. Seven Cys residues were not identified in our analysis (Cys146, Cys1489, Cys1491, Cys2651, Cys4657, Cys4876, and Cys4882) (including additional experiments using digestion with Asp-N), although the predicted masses of the peptides containing those Cys residues were within the detectable limit (∼800–3,000 Da). Fifteen Cys residues were labeled with IA only and were therefore identified as fully oxidized (although reducible by DTT) at both low and high pO2 (Fig. 2), consistent with their participation in “structural” disulfides. Remarkably, 78 of 93 identified Cys residues were detected with ICAT labeling and were thus present in some proportion in reduced form. Twenty-two of those were labeled with ICAT reagent but not IA (Fig. 2) and displayed a light:heavy ratio of ∼1:1, consistent with their presence in fully reduced form at both low and high pO2 (Table 1). An additional two Cys residues detected with ICAT only (Cys566 and Cys762) were pO2-sensitive (increased light:heavy ratio) (Table 2), although no IA-labeled form was detected presumably because the IA-labeled peptide was of low abundance (consistent with the existence of those Cys residues in predominantly reduced form at low pO2).
FIGURE 2.
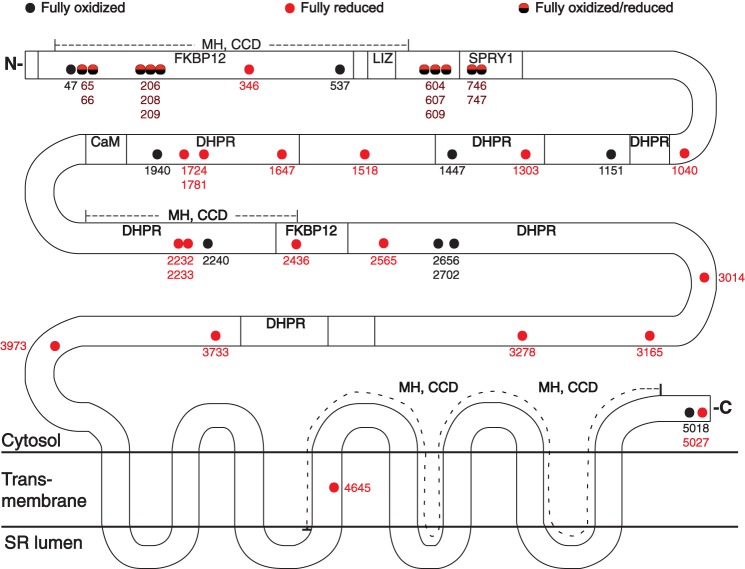
RyR1 Cys residues insensitive to pO2. Following analysis according to the scheme illustrated in Fig. 1A, 72 Cys residues were identified as pO2-insensitive, including a set of 37 Cys residues labeled with IA or ICAT only and therefore members of a fully oxidized or a fully reduced population, respectively. Fully oxidized Cys residues are likely to participate in structural disulfides with the exception of Cys537, which could be labeled with dimedone (see Table 5). Note that the analysis could not discriminate between multiple Cys residues located within a single peptide. In the four cases in which multiple Cys residues were present in a single peptide (Cys65/66, Cys206/208/209, Cys604/607/609, and Cys746/747), one Cys was ICAT-labeled, and the remainder were IA-labeled in a pO2-insensitive fashion. The locations within RyR1 of fully oxidized or reduced Cys residues are indicated. Transmembrane domains; regions implicated in the interaction of RyR1 with the L-type Ca2+ channel (dihydropyridine receptor (DHPR)), FK506-binding protein 12 (FKBP12), and calmodulin (CaM); leucine/isoleucine zipper (LIZ) motifs; and the SPRY1 domain as well as hot spot regions implicated in malignant hypothermia/central core disease (MH, CCD) are delineated on the basis of the human RyR1 sequence (for a recent review, see Ref. 33). Quantitative data are provided in Table 1. Black circles, fully oxidized; red circles, fully reduced; half red-half black circles, fully oxidized or reduced.
TABLE 1.
pO2-insensitive RyR1 Cys residues
Cys residues labeled with IA alone (fully oxidized at low and high pO2) are not shown (see Fig. 2). Cys residues labeled with ICAT alone (fully reduced at high and low pO2) are underlined. For all remaining pO2-independent Cys residues, some proportions were detected with IA and with ICAT labeling, and L:H ICAT ratios were ∼1:1 at low or high pO2 and at high pO2 with NADPH supplementation. Note anomalous values for Cys3193 and Cys3635 (see Fig. 3 and text). Standard error was derived from replicated biological samples (n = 2–5), multiple peptides with different charge and/or methionine oxidation, or multiple scanning. PEG-CAT, polyethylene glycol-coupled catalase; DPI, diphenyleneiodonium; —, data not available.
| Cys | Control | DPI | PEG-CAT | NADPH | Cys | Control | DPI | PEG-CAT | NADPH |
|---|---|---|---|---|---|---|---|---|---|
| 24 | 0.94 ± 0.06 | 1.10 ± 0.10 | 0.99 ± 0.04 | 1.12 ± 0.06 | 2436 | 1.16 ± 0.01 | 1.02 ± 0.01 | 0.97 ± 0.02 | 1.13 ± 0.17 |
| 230 | 1.15 ± 0.01 | 0.98 ± 0.02 | 1.05 ± 0.03 | 1.19 ± 0.02 | 2565 | 1.20 ± 0.12 | 1.00 ± 0.01 | 0.92 ± 0.02 | 1.14 ± 0.16 |
| 315 | 1.11 ± 0.05 | 0.90 ± 0.02 | 0.98 ± 0.01 | 0.98 ± 0.02 | 3014 | 1.07 ± 0.07 | 0.94 ± 0.01 | 0.93 ± 0.01 | 1.15 ± 0.07 |
| 346 | 1.00 ± 0.06 | 1.03 ± 0.02 | 0.94 ± 0.05 | 1.06 ± 0.04 | 3044 | 1.09 ± 0.04 | 0.99 ± 0.05 | 0.94 ± 0.01 | 1.12 ± 0.09 |
| 393 | 0.96 ± 0.05 | 0.98 ± 0.01 | 1.00 ± 0.01 | 1.14 ± 0.09 | 3067 | 1.09 ± 0.03 | 1.04 ± 0.01 | 0.91 ± 0.02 | 1.20 ± 0.33 |
| 811 | 1.01 ± 0.07 | 0.98 ± 0.01 | 0.98 ± 0.03 | 1.18 ± 0.08 | 3165 | 1.01 ± 0.01 | 0.98 ± 0.03 | 0.95 ± 0.03 | 1.05 ± 0.01 |
| 906 | 0.96 ± 0.02 | 0.97 ± 0.03 | 0.94 ± 0.01 | 1.23 ± 0.12 | 3170 | 1.12 ± 0.04 | 0.96 ± 0.02 | 0.92 ± 0.02 | 1.16 ± 0.11 |
| 937 | 1.07 ± 0.05 | 1.01 ± 0.03 | 0.97 ± 0.02 | 1.16 ± 0.16 | 3193 | 0.75 ± 0.01 | 0.80 ± 0.01 | 0.83 ± 0.01 | 1.09 ± 0.06 |
| 1040 | 1.07 ± 0.11 | 0.84 ± 0.01 | 0.89 ± 0.04 | 1.11 ± 0.04 | 3216 | 1.13 ± 0.12 | 0.99 ± 0.02 | 0.97 ± 0.02 | 1.14 ± 0.10 |
| 1192 | 0.98 ± 0.02 | 1.01 ± 0.01 | 0.99 ± 0.01 | 1.17 ± 0.10 | 3240 | 1.07 ± 0.08 | 0.95 ± 0.03 | 0.94 ± 0.05 | 1.09 ± 0.07 |
| 1217 | 1.17 ± 0.03 | 0.96 ± 0.04 | 0.95 ± 0.05 | 1.19 ± 0.20 | 3278 | 1.09 ± 0.19 | 0.98 ± 0.02 | 0.92 ± 0.08 | 1.12 ± 0.17 |
| 1269 | 1.11 ± 0.18 | 0.96 ± 0.01 | 0.95 ± 0.01 | 1.12 ± 0.06 | 3304 | 1.03 ± 0.13 | 1.02 ± 0.01 | 0.99 ± 0.02 | 1.08 ± 0.01 |
| 1303 | 1.05 ± 0.13 | 0.97 ± 0.01 | 0.97 ± 0.01 | 1.12 ± 0.02 | 3402 | 1.11 ± 0.05 | 1.01 ± 0.02 | 0.97 ± 0.02 | 1.04 ± 0.05 |
| 1518 | 1.21 ± 0.02 | 1.03 ± 0.02 | 0.96 ± 0.06 | — | 3525 | 1.14 ± 0.04 | 0.97 ± 0.01 | 0.96 ± 0.03 | 0.99 ± 0.09 |
| 1591 | 1.04 ± 0.04 | 0.99 ± 0.01 | 0.98 ± 0.01 | 1.13 ± 0.10 | 3635 | 1.10 ± 0.21 | 0.79 ± 0.02 | 0.77 ± 0.01 | 1.11 ± 0.01 |
| 1630 | 1.21 ± 0.02 | — | — | — | 3650 | 1.08 ± 0.04 | 0.98 ± 0.05 | 0.96 ± 0.01 | 1.05 ± 0.10 |
| 1647 | 1.06 ± 0.05 | 1.05 ± 0.03 | 0.94 ± 0.04 | 1.12 ± 0.12 | 3733 | 1.03 ± 0.06 | 1.00 ± 0.06 | 0.94 ± 0.03 | 1.13 ± 0.05 |
| 1724 | 0.98 ± 0.02 | — | 1.02 ± 0.04 | — | 3786 | 1.16 ± 0.05 | 1.03 ± 0.01 | 1.01 ± 0.02 | 0.98 ± 0.10 |
| 1781 | 0.94 ± 0.11 | 0.98 ± 0.02 | 0.95 ± 0.04 | 1.08 ± 0.05 | 3839 | — | 0.98 ± 0.05 | 0.91 ± 0.06 | 1.15 ± 0.06 |
| 1947 | 1.13 ± 0.02 | 0.99 ± 0.04 | 0.94 ± 0.04 | 1.09 ± 0.03 | 3918 | — | 1.16 ± 0.05 | 1.07 ± 0.02 | 1.07 ± 0.18 |
| 2042 | 1.17 ± 0.02 | 1.02 ± 0.02 | 0.96 ± 0.02 | 1.22 ± 0.14 | 3973 | 1.09 ± 0.11 | 1.05 ± 0.08 | 1.00 ± 0.04 | 1.05 ± 0.04 |
| 2158 | 1.15 ± 0.27 | 1.02 ± 0.04 | 0.83 ± 0.01 | 1.21 ± 0.21 | 4114 | 0.98 ± 0.02 | 1.01 ± 0.03 | 1.02 ± 0.04 | 1.12 ± 0.13 |
| 2232/2233 | 1.19 ± 0.02 | 0.95 ± 0.04 | 0.97 ± 0.04 | 0.96 ± 0.07 | 4645 | 1.07 ± 0.03 | 1.01 ± 0.02 | — | — |
| 2237 | 0.94 ± 0.14 | — | — | — | 4958 | 1.06 ± 0.02 | 1.00 ± 0.02 | 1.07 ± 0.01 | — |
| 2326 | 1.11 ± 0.12 | 0.94 ± 0.01 | 0.95 ± 0.03 | 1.08 ± 0.15 | 4961 | 1.21 ± 0.03 | 0.91 ± 0.07 | 1.02 ± 0.01 | — |
| 2363 | 1.09 ± 0.04 | 0.98 ± 0.01 | 0.98 ± 0.01 | 1.06 ± 0.09 | 5027 | 0.87 ± 0.01 | 1.02 ± 0.01 | 0.97 ± 0.01 | 1.08 ± 0.01 |
TABLE 2.
pO2-sensitive RyR1 Cys residues
For most pO2-dependent Cys residues, some proportions were detected with IA and with ICAT labeling, and ICAT L:H ratios ≥1:1.24 (L represents light ICAT, low pO2; H represents heavy ICAT, high pO2) indicate enhanced oxidation at high versus low pO2. In two cases (indicated by underlining), only ICAT labeling was detected. In all cases, the increase in L:H ratio at high versus low pO2 was eliminated by inhibiting NADPH oxidase activity with DPI or scavenging H2O2 with polyethylene glycol-coupled catalase (PEG-CAT) and was enhanced by supplementation with NADPH. Standard error was derived from replicated biological samples (n = 2–5), multiple peptides with different charge and/or methionine oxidation, or multiple scanning. —, data not available.
| Cys | Control | DPI | PEG-CAT | NADPH |
|---|---|---|---|---|
| 36 | 1.33 ± 0.02 | 1.04 ± 0.02 | 0.98 ± 0.03 | 1.42 ± 0.22 |
| 566 | 1.25 ± 0.04 | 1.04 ± 0.02 | 1.00 ± 0.03 | 1.47 ± 0.08 |
| 762 | 1.27 ± 0.05 | — | 1.03 ± 0.06 | 1.28 ± 0.23 |
| 845/854 | 1.28 ± 0.06 | 1.14 ± 0.02 | 1.06 ± 0.01 | 1.53 ± 0.28 |
| 1674 | 1.27 ± 0.08 | 1.03 ± 0.01 | 0.94 ± 0.08 | 1.56 ± 0.03 |
| 2305/2310 | 1.47 ± 0.05 | 0.84 ± 0.01 | 0.93 ± 0.08 | 1.30 ± 0.11 |
| 2555 | 1.27 ± 0.08 | 1.08 ± 0.03 | 0.94 ± 0.03 | 1.32 ± 0.17 |
| 2606 | 1.30 ± 0.08 | 1.22 ± 0.01 | 1.05 ± 0.02 | 1.33 ± 0.01 |
| 2611 | 1.39 ± 0.35 | 1.10 ± 0.01 | 0.86 ± 0.01 | 1.35 ± 0.01 |
| 2704 | 1.25 ± 0.03 | — | 0.98 ± 0.03 | — |
| 4238 | 1.24 ± 0.03 | 1.08 ± 0.01 | 0.98 ± 0.02 | — |
Each of the remaining 54 Cys residues were labeled with IA or ICAT reagent and identified as members of a population that contained both reduced and oxidized Cys residues at both low and high pO2. The observed light ICAT labeling versus heavy ICAT labeling (L:H) ratios ranged from 0.75 to 1.47 with the large majority distributed from 0.94 to 1.21 (Fig. 3A). Greater relative abundance of light ICAT labeling versus heavy ICAT labeling identified Cys residues subject to oxidation at high versus low pO2. Based upon an arbitrary threshold L:H of 1.24 (which in terms of tetrameric RyR1 would represent at the extremes a population of ∼25% of RyR where all subunits are modified or 100% of RyR where a single monomer is modified), the redox states of 13 Cys residues (L:H of 1.24–1.47) were coupled to pO2 (Fig. 4 and Table 2). Importantly, as illustrated in Table 2, in every case examined, the increase in L:H ratio at high versus low pO2 was eliminated when SR vesicles were exposed to 20% O2 in the presence of either PEG-coupled catalase to remove H2O2 or the Nox inhibitor DPI, confirming the essential role of endogenous H2O2 production by SR-resident Nox4 (16).
FIGURE 3.
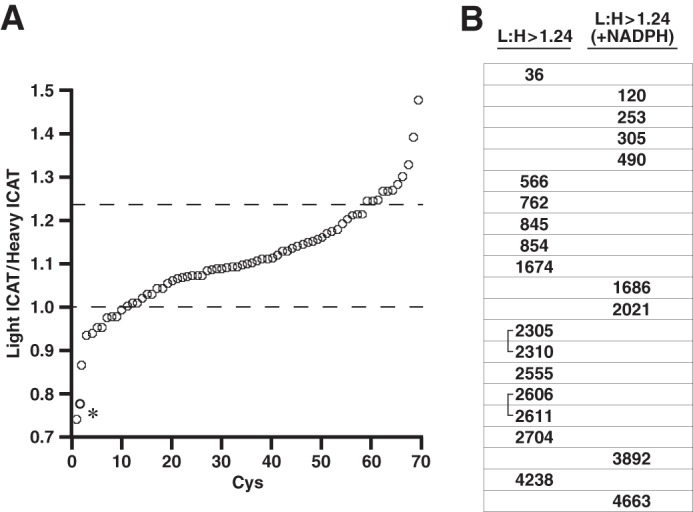
Effect of pO2 on redox state of Cys residues within RyR1. A, SR vesicles held at low pO2 (1% O2) or high pO2 (21% O2) were labeled with light or heavy ICAT reagent, respectively, before isolation of RyR1, trypsin digest, and analysis by LC-MS/MS (see Fig. 1A). The graph includes the 71 Cys residues that were identified as ICAT-labeled under control conditions (low and high pO2 with no exogenous additives) in more than one biological replicate and that were contained in peptides that included only a single Cys (allowing unambiguous identification). Light:heavy ratios >1 indicate Cys oxidation (loss of free thiol) at high versus low pO2. A light:heavy cutoff value of 1.24:1 (indicated by the upper dotted line) yielded a set of 13 Cys residues. An asterisk indicates Cys3193 and Cys3635, which exhibited anomalous behavior (see Table 1). B, Cys residues identified as subject to oxidation at high versus low pO2 are listed with reference to their location within RyR1. Note that the L:H ratio was enhanced for all listed Cys residues by supplemental NADPH (see Table 2), but that the L:H ratio for Cys residues listed in the right-hand column was ∼1:1 in the absence of supplemental NADPH; that is, pO2-dependent oxidation required supplemental NADPH (see Table 3). Identified disulfides (2305/2310 and 2606/2611) are indicated by brackets (see Figs. 5 and 6).
FIGURE 4.
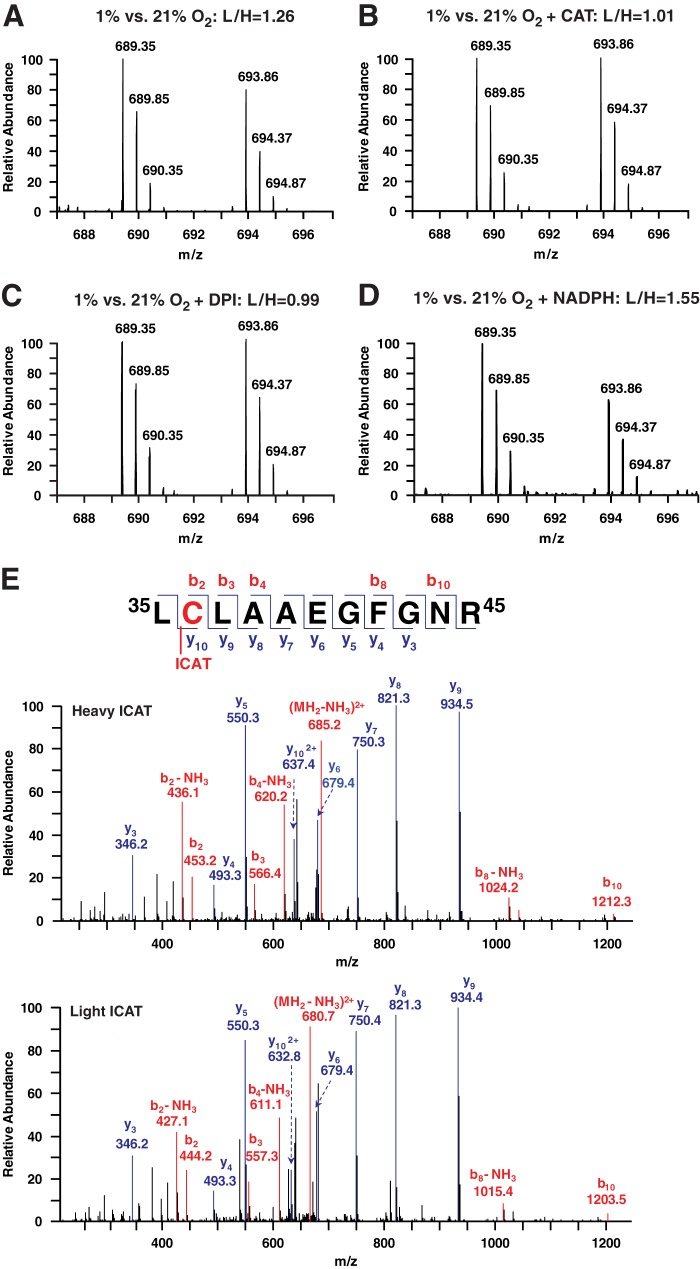
An illustrative example of ICAT-based identification and quantification of pO2-coupled oxidation of RyR1 Cys (Cys36). Mass spectra of RyR1 tryptic peptide 35LCLAAEGFGNR45 (M2+) derived from SR vesicles incubated at 1% O2 or 20% O2 (A–D) with the addition of PEG-coupled catalase (CAT; 10 units/ml) (B), DPI (100 μm) (C), or NADPH (1 mm) (D). In A–D, the left-hand and right-hand peaks represent labeling by light (at 1% O2) or heavy (at 20% O2) ICAT (difference of 4.5 m/z), respectively. The L:H ratio of 1.26 at 1% O2 versus 20% O2 (A) indicates a 26% increase in the yield of peptides containing oxidized Cys36 at high versus low pO2, and this difference is eliminated by diminishing H2O2 with catalase (B) or NADPH oxidase activity with DPI (C). The NADPH-coupled increase in L:H ratio to 1.55 (D) indicates increased oxidation when pO2-dependent NADPH oxidase activity is enhanced. E, peptide MS/MS spectra identify the site of ICAT labeling as Cys36.
It is of note that the two Cys residues exhibiting anomalous behavior, Cys3193 and Cys3635, include the identified site of endogenous S-nitrosylation (Cys3635) (Fig. 3A and Table 1). Cys3635 is pO2-insensitive (L:H ∼ 1 under control conditions), consistent with the previous finding that Cys3635 is not subject to pO2-coupled S-oxidation (13). However, the L:H ratio drops to ∼0.78 when H2O2 is removed at high pO2 (Table 1). One possible explanation for this finding is that, inasmuch as some proportion of Cys3635 is present in isolated SR vesicles in the S-nitrosylated form (not shown), enhanced denitrosylation (26) at high pO2 in the absence of H2O2 (as in the hypothetical case of a denitrosylase that is inhibited by reactive oxygen species) would increase ICAT labeling at high versus low pO2 and thereby decrease the L:H ratio.
We reported previously that both generation of H2O2 and RyR1 activity in SR vesicles were enhanced at 21 versus 1% oxygen and that these effects were associated with pO2-coupled oxidation of a set of thiols within RyR1 (16). In addition, we reported that supplementation with NADPH (to enhance Nox4 activity) enhanced H2O2 production (16). Here we confirmed directly that the loss of free thiols within RyR1 in SR vesicles exposed to high versus low pO2 was enhanced by addition of NADPH (Table 3). When SR vesicles were incubated with NADPH (1 mm), the L:H ratios were also enhanced for 6 of 11 tested Cys residues of the 13 Cys residues that were oxidized at high versus low pO2 (Fig. 4 and Table 2). Furthermore, addition of NADPH resulted in an increase in L:H ratio from ∼1:1 to >1.24:1 in a population of eight additional Cys residues (Fig. 3B; Table 4). Thus, 21 Cys residues within RyR1 are subject to pO2-coupled redox regulation.
TABLE 3.
Oxidation of RyR1 Cys thiols at high pO2 is enhanced by NADPH
In SR vesicles incubated at 1% O2 or 20% O2 and in the presence or absence of supplementary NADPH (1 mm), RyR1 free thiols were labeled with MBB prior to purification by density gradient centrifugation and fluorescence quantification. Values are mol of free thiol/mol of RyR1. Note that about five free thiols on average are oxidized at high versus low pO2 and that oxidation is enhanced by NADPH at high but not at low pO2. p values are from paired t tests (n = 5).
| High pO2 (21% O2) | Low pO2 (1% O2) | |
|---|---|---|
| Control | 35.30 ± 1.56 | 40.31 ± 1.59 |
| +NADPH | 32.98 ± 1.42 | 39.42 ± 2.07 |
| p value vs. control | <0.05 | 0.41 |
TABLE 4.
NADPH-dependent oxidation of Cys thiols in RyR1 at high pO2
In SR vesicles incubated at 1% O2 or 20% O2 and in the presence or absence of supplementary NADPH (1 mm), a set of Cys residues was identified whose members were oxidized at high versus low pO2 only in the presence of supplemental NADPH (L:H ratios ≥1:1.24) (L represents light ICAT, low pO2; H represents heavy ICAT, high pO2). All of these Cys residues were detected with both IA and ICAT labeling. Standard errors are derived from replicated biological samples (n = 2–5), peptides with different charge and methionine oxidation, or multiple scanning. PEG-CAT, polyethylene glycol-coupled catalase. —, data not available.
| Cys | Control | DPI | PEG-CAT | NADPH |
|---|---|---|---|---|
| 120 | 1.14 ± 0.08 | 0.86 ± 0.02 | 0.98 ± 0.01 | 1.24 ± 0.21 |
| 253 | 1.07 ± 0.11 | 0.97 ± 0.01 | 0.99 ± 0.01 | 1.26 ± 0.35 |
| 305 | 1.13 ± 0.23 | 0.89 ± 0.04 | 0.90 ± 0.03 | 1.27 ± 0.09 |
| 490 | 1.10 ± 0.05 | 0.96 ± 0.05 | 0.90 ± 0.05 | 1.55 ± 0.07 |
| 1686 | 1.10 ± 0.02 | 1.04 ± 0.03 | 0.97 ± 0.01 | 1.35 ± 0.19 |
| 2021 | 1.02 ± 0.04 | 0.97 ± 0.01 | 0.94 ± 0.01 | 1.27 ± 0.08 |
| 3892 | 1.18 ± 0.01 | 1.03 ± 0.04 | 0.99 ± 0.01 | 1.25 ± 0.16 |
| 4663 | — | 1.03 ± 0.04 | — | 1.31 ± 0.06 |
Identification of the Form of pO2-coupled S-Oxidation
To investigate the form of pO2-coupled S-oxidation, we incubated SR at low or high pO2, blocked free thiols with IA or N-ethylmaleimide, and then purified RyR1 by non-reducing SDS-PAGE prior to trypsin or chymotrypsin digest and analysis by LC-MS/MS. We did not detect S-glutathionylation at either low or high pO2. Western blot analysis of intact RyR1 using anti-glutathione antibodies also failed to detect S-glutathionylation, although S-glutathionylation of RyR1 was detected by Western blot when SR vesicles were incubated with oxidized glutathione as a positive control (data not shown). We also did not detect sulfenamide (27), and in addition, neither sulfenic (SOH) nor sulfinic acid (SO2H) was detected. We identified the sulfonic acid form (SO3H) of Cys1192 and Cys3067, but quantification revealed that sulfonation was not pO2-coupled (not shown).
Intrapeptide disulfide linkages were identified between Cys2305 and Cys2310 and between Cys2606 and Cys2611 (Figs. 3B, 5, and 6). Both disulfide pairs were identified in each of three separate biological samples. In both cases, Cys2305/2310 and Cys2606/2611 could also be alkylated by N-ethylmaleimide or IA at both low and high pO2 (Fig. 5). Quantification of disulfide formation with a native peptide reference method (see “Experimental Procedures”) (25) yielded H:L ratios of 1.23 ± 0.16 and 1.45 ± 0.04 (n = 3 experiments) for Cys2305/2310 and Cys2602/2611, respectively. We did not detect disulfide formation between Cys residues located in separate peptides, but theoretical analysis of the results of trypsin/chymotrypsin digest indicates that very few composite peptides resulting from disulfide formation would possess a mass low enough to allow their detection by MS/MS.
FIGURE 5.
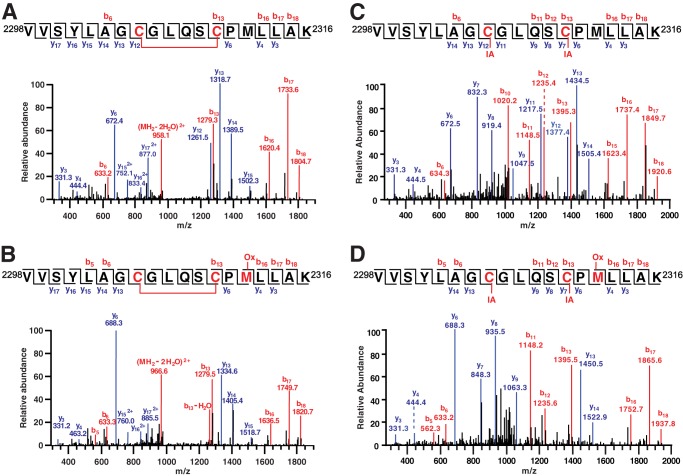
pO2-coupled disulfide formation within RyR1 (Cys2305/2310). A–D, MS/MS spectra illustrate the four forms of peptide 2298VVSYLAGCGLQSCPMLLAK2316 observed. An intrapeptide disulfide between Cys2305 and Cys2310 (A) was also seen in conjunction with oxidation (Ox) of Met2312 (B). IA alkylation of Cys2305 and Cys2310 (C) also seen in conjunction with oxidation (Ox) of Met2312 (D) indicates that both Cys residues were in the reduced state prior to initial blocking by alkylation. Note that all four forms are detected in samples prepared from SR vesicles held at high or low pO2 but that the disulfide form is more abundant (1.23-fold) in samples prepared at high pO2 (see “Results”).
FIGURE 6.
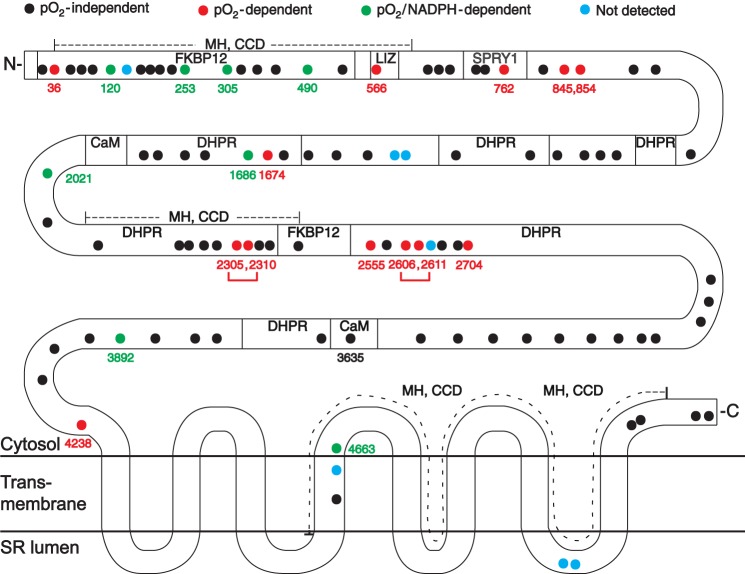
Schematic illustration of the distribution within RyR1 of Cys residues subject to pO2-coupled redox modification. Among the 100 Cys residues of rabbit RyR1, color-coding identifies Cys residues unaffected by pO2 (n = 72; black circles), oxidized at high versus low pO2 (enhanced by exogenous NADPH) (n = 13; red circles), and oxidized at high pO2 only with addition of exogenous NADPH (n = 8; green circles). Cys residues not detected in our analysis are also indicated (n = 7; blue circles). Identified disulfides formed more abundantly at high versus low pO2 are indicated by brackets (Cys2305-Cys2310 and Cys2606-Cys2611), and for reference, the previously identified site of S-nitrosylation is indicated (Cys3635). The following functional domains within RyR1 are delineated: transmembrane domains; regions implicated in the interaction of RyR1 with the L-type Ca2+ channel (dihydropyridine receptor (DHPR)), FK506-binding protein 12 (FKBP12), and calmodulin (CaM); leucine/isoleucine zipper (LIZ) motifs; and the SPRY1 domain as well as hot spot regions implicated in malignant hypothermia/central core disease (MH, CCD).
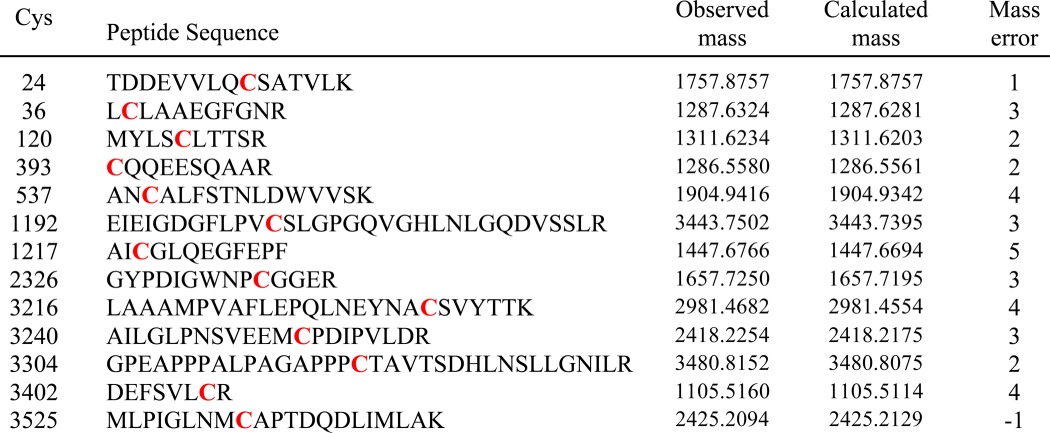
Although we did not observe sulfenic acid at either low or high pO2, we considered the possibility that sulfenic acid might represent a transient intermediate en route to disulfide formation. We did not identify sulfenic acid by dimedone labeling (28) following incubation of SR vesicles at low or high pO2. However, if muscle homogenates were incubated with dimedone (in room air; 21% O2) prior to the isolation of SR vesicles and purification of RyR1, then 13 dimedone-modified Cys residues were detected (Table 5). Eleven of those 13 Cys residues were not among the set of pO2-sensitive Cys residues (listed in Tables 2 and 4). However, Cys36 and Cys120 were identified as both dimedone-labeled and pO2-sensitive, consistent with a role for sulfenic acid as a transient intermediate to higher oxidation, presumably disulfide formation.
TABLE 5.
Dimedone labeling of Cys thiols in RyR1
Muscle homogenates were incubated with dimedone prior to preparation of SR vesicles and analysis. Dimedone labeling (red) was identified by MS/MS. Mass is given as Daltons.
DISCUSSION
It has been established that the redox state of pO2-sensitive RyR1 Cys residues is regulated by a cycle of dynamic S-oxidation and reduction that operates within the SR, but the identity of pO2-sensitive Cys residues has remained unknown (16, 17). We show here that pO2-coupled redox regulation of RyR1 is exerted through S-oxidation of 21 Cys residues that are distributed widely within RyR1. Previous reports based upon thiol labeling have reported pO2-coupled oxidation of a set of approximately six to eight thiols on average (8). The analytic scheme used here based upon ICAT labeling and mass spectrometric analysis revealed that this set of thiols represents substoichiometric oxidation of a larger set. (Indeed, only two of the Cys residues we identified as pO2-sensitive appeared to be fully reduced at low pO2, and more generally, 54 of the 93 Cys residues we identified comprise mixed populations of reduced and oxidized Cys residues at both low and high pO2). In the best characterized cases of redox regulation of protein function by S-oxidation, including most prominently protein-tyrosine phosphatases, modification preferentially targets a single, active site Cys within a minor population of protein (27). Thus, our results provide an apparently novel example of multisite redox regulation by physiological S-oxidation.
Physiological S-Oxidation of Cys Residues in RyR1
There are few analyses of oxidative modification of protein function in situ that have identified the source and nature of oxidizing equivalents, the protein targets and Cys residues modified, the nature of the modification, and the consequences of modification for protein function. Skeletal muscle RyR1, which contains 100 Cys residues, has long provided a model of Cys-based redox regulation. In particular, it has been reported that Nox2 is localized to transverse tubule membranes in skeletal muscle and that addition of NADPH to isolated triads (containing transverse tubule as well as SR membranes) results in the glutathionylation of RyR1 (20). However, we did not identify S-glutathionylated Cys residues within RyR1 following pO2-coupled activation of RyR1, consistent with previous work showing that Nox2 may play a role in tetanic contraction but is not a source of pO2-coupled reactive oxygen species production (16). In contrast, Nox4 was identified previously as the source of pO2-coupled S-oxidation of RyR1 that activates RyR1 to enhance Ca2+ release from the SR (16). Here, in addition to identifying pO2-sensitive Cys residues within RyR1, we identified disulfide as a stable product of S-oxidation but not other potential, reversible oxidative modifications, including S-glutathionylation, sulfenic acid, sulfinic acid, or sulfenamide (a form of modification within protein-tyrosine phosphatases (29, 30)). Thus, glutathionylation and disulfide modification may represent signatures of contraction- and oxygen-regulated RyR1 activity, respectively.
Previous studies using mass spectrometric analysis but relying primarily upon the administration of exogenous oxidants reported modification of multiple Cys residues within RyR1 by glutathionylation or unidentified S-oxidation as well as (by inference) disulfide formation (14). Although it is not possible to deduce the coverage of RyR1 Cys residues in previous analyses, remarkably, none of the Cys residues reported previously to be oxidatively modified are among the set of Cys residues shown here to be subject to physiological, pO2-coupled S-oxidation with the exception of Cys36, which was identified previously as a possible participant in disulfide formation catalyzed by the addition of high concentrations of H2O2 (5 mm) (14). Conversely, with the exception of Cys36, Cys residues identified here as pO2-sensitive do not appear among the Cys residues reported to be oxidatively modified by exogenous agents (14), although Cys2606/2611, which we identified as participating in a pO2-sensitive disulfide, were identified as hyperreactive by selective labeling with a thiol-reactive coumarin (31). Interestingly, a recent crystal structure of the N-terminal domain of RyR2 in which Cys36 is conserved indicated a conformation in which Cys36 could form a vicinal pair with Cys65 (32). However, we did not identify Cys65 as pO2-sensitive.
Generally, studies of oxidative modification of Cys residues within proteins that are involved in cellular signal transduction have relied on the in vitro application of oxidants, principally H2O2, to address the form of modification. In the much studied case of protein-tyrosine phosphatases, in vitro analysis has identified multiple possible oxidative modifications, including intramolecular disulfides and sulfenamide (27). However, there is little evidence bearing on the nature of oxidative modification of Cys residues induced by endogenously generated H2O2, and more generally, the nature of oxidative modification that may subserve physiological signal transduction remains largely unexplored (2). Our results indicate that disulfide formation serves as a principal dynamic modification of Cys residues within RyR1 that is governed by endogenous oxidative and reductive mechanisms to regulate RyR1 function allosterically.
Our finding that ∼21 Cys residues contribute to a total of 6–8 mol of thiol/mol of RyR monomer that are subject to pO2-coupled oxidation is in keeping with the evolving understanding that regulatory post-translational modifications of proteins are typically present in substoichiometric amounts. Subpopulations of proteins may be subject to differential regulation, and sub-stoichiometric oxidative modification of RyR1 may be viewed as providing a selective gain of function. However, we cannot determine whether a subpopulation of RyR1 is subject to privileged modification (e.g. by Nox4 that may be associated with a fraction of RyR1) or whether redox modifications are in fact evenly distributed across the entire population of RyR1 in which case oxidation of single subunits of the RyR1 tetramer may cooperatively influence the activity of other subunits to subserve pO2-coupled regulation of calcium release.
The Distribution of Physiologically S-Oxidized Cys residues in RyR1
The localization of identified Cys residues within RyR1 is illustrated in Fig. 6. All pO2-sensitive Cys residues are located in cytoplasmic portions of RyR1 and with a single exception (Cys4663) within the N-terminal domain. Cys residues subject to pO2-dependent S-oxidation are located within multiple functional domains (for a recent review, see Ref. 33), including domains implicated in activity-regulating interactions of RyR1 with FK506-binding protein 12 (33, 34) and the dihydropyridine receptor (33, 35). In addition, pO2-sensitive Cys residues are located in domains implicated in malignant hyperthermia and central core disease (33). It is not possible to deduce which identified Cys residues play a role in pO2-dependent enhancement of RyR1 activity or may be subject to dysregulated S-oxidation in disease, and concerted effects involving multiple Cys residues seem likely. However, it should be noted that mutation of Cys36 (Cys35 in the human sequence) has been implicated directly in malignant hyperthermia and central core disease (36) and that oxidative stress associated with aging and in a model of muscular dystrophy results in the displacement of FK506-binding protein 12 from RyR1 and consequently Ca2+ leakage through RyR1 that contributes to loss of muscle function (37, 38). It is also of interest that Cys3635, which is not a target of pO2-coupled S-oxidation but which serves as the principal target of endogenous S-nitrosylation that is gated allosterically by RyR1 S-oxidation of alternative Cys residues (10, 13), is located distantly in the primary sequence from identified pO2-sensitive Cys residues within RyR1 (Fig. 6).
Ninety-eight of 100 RyR1 Cys residues are conserved in mammals, including all pO2-sensitive Cys residues identified here with the exception of Cys305 (oxidized at high pO2 only in the presence of NADPH). Furthermore, the majority of RyR1 Cys residues that we identified as subject to physiological redox regulation are also conserved in RyR2 and RyR3, consistent with the finding that the activity of RyR2 is enhanced at high versus low pO2 (15). Thus, our findings suggest that S-oxidation of Cys residues within the RyRs is likely to serve as a mechanism for physiological redox regulation of RyR function across many mammalian cell types and tissues.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 HL0591130 (to J. S. S.).
- Nox
- NADPH oxidase
- DPI
- diphenyleneiodonium
- IA
- iodoacetamide
- ICAT
- isotope-coded affinity tag
- MBB
- monobromobimane
- RyR
- ryanodine receptor/Ca2+ release channel
- SR
- sarcoplasmic reticulum
- L
- light ICAT labeling
- H
- heavy ICAT labeling.
REFERENCES
- 1. Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. (2005) Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 6, 150–166 [DOI] [PubMed] [Google Scholar]
- 2. Janssen-Heininger Y. M., Mossman B. T., Heintz N. H., Forman H. J., Kalyanaraman B., Finkel T., Stamler J. S., Rhee S. G., van der Vliet A. (2008) Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic. Biol. Med. 45, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chung H. S., Wang S. B., Venkatraman V., Murray C. I., Van Eyk J. E. (2013) Cysteine oxidative posttranslational modifications: emerging regulation in the cardiovascular system. Circ. Res. 112, 382–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Paul B. D., Snyder S. H. (2012) H2S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 13, 499–507 [DOI] [PubMed] [Google Scholar]
- 5. Bedard K., Krause K. H. (2007) The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 87, 245–313 [DOI] [PubMed] [Google Scholar]
- 6. Paulsen C. E., Truong T. H., Garcia F. J., Homann A., Gupta V., Leonard S. E., Carroll K. S. (2012) Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat. Chem. Biol. 8, 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Svoboda L. K., Reddie K. G., Zhang L., Vesely E. D., Williams E. S., Schumacher S. M., O'Connell R. P., Shaw R., Day S. M., Anumonwo J. M., Carroll K. S., Martens J. R. (2012) Redox-sensitive sulfenic acid modification regulates surface expression of the cardiovascular voltage-gated potassium channel Kv1.5. Circ. Res. 111, 842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eu J. P., Sun J., Xu L., Stamler J. S., Meissner G. (2000) The skeletal muscle calcium release channel: coupled O2 sensor and NO signaling functions. Cell 102, 499–509 [DOI] [PubMed] [Google Scholar]
- 9. Pessah I. N., Feng W. (2000) Functional role of hyperreactive sulfhydryl moieties within the ryanodine receptor complex. Antioxid. Redox Signal. 2, 17–25 [DOI] [PubMed] [Google Scholar]
- 10. Sun J., Xin C., Eu J. P., Stamler J. S., Meissner G. (2001) Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc. Natl. Acad. Sci. U.S.A. 98, 11158–11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aracena P., Sánchez G., Donoso P., Hamilton S. L., Hidalgo C. (2003) S-Glutathionylation decreases Mg2+ inhibition and S-nitrosylation enhances Ca2+ activation of RyR1 channels. J. Biol. Chem. 278, 42927–42935 [DOI] [PubMed] [Google Scholar]
- 12. Eu J. P., Hare J. M., Hess D. T., Skaf M., Sun J., Cardenas-Navina I., Sun Q. A., Dewhirst M., Meissner G., Stamler J. S. (2003) Concerted regulation of skeletal muscle contractility by oxygen tension and endogenous nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 100, 15229–15234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun J., Xu L., Eu J. P., Stamler J. S., Meissner G. (2003) Nitric oxide, NOC-12, and S-nitrosoglutathione modulate the skeletal muscle calcium release channel/ryanodine receptor by different mechanisms. An allosteric function for O2 in S-nitrosylation of the channel. J. Biol. Chem. 278, 8184–8189 [DOI] [PubMed] [Google Scholar]
- 14. Aracena-Parks P., Goonasekera S. A., Gilman C. P., Dirksen R. T., Hidalgo C., Hamilton S. L. (2006) Identification of cysteines involved in S-nitrosylation, S-glutathionylation, and oxidation to disulfides in ryanodine receptor type 1. J. Biol. Chem. 281, 40354–40368 [DOI] [PubMed] [Google Scholar]
- 15. Sun J., Yamaguchi N., Xu L., Eu J. P., Stamler J. S., Meissner G. (2008) Regulation of the cardiac muscle ryanodine receptor by O2 tension and S-nitrosoglutathione. Biochemistry 47, 13985–13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun Q. A., Hess D. T., Nogueira L., Yong S., Bowles D. E., Eu J., Laurita K. R., Meissner G., Stamler J. S. (2011) Oxygen-coupled redox regulation of the skeletal muscle ryanodine receptor-Ca2+ release channel by NADPH oxidase 4. Proc. Natl. Acad. Sci. U.S.A. 108, 16098–16103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun J., Xu L., Eu J. P., Stamler J. S., Meissner G. (2001) Classes of thiols that influence the activity of the skeletal muscle calcium release channel. J. Biol. Chem. 276, 15625–15630 [DOI] [PubMed] [Google Scholar]
- 18. Aracena P., Tang W., Hamilton S. L., Hidalgo C. (2005) Effects of S-glutathionylation and S-nitrosylation on calmodulin binding to triads and FKBP12 binding to type 1 calcium release channels. Antioxid. Redox Signal. 7, 870–881 [DOI] [PubMed] [Google Scholar]
- 19. Espinosa A., Leiva A., Peña M., Müller M., Debandi A., Hidalgo C., Carrasco M. A., Jaimovich E. (2006) Myotube depolarization generates reactive oxygen species through NAD(P)H oxidase; ROS-elicited Ca2+ stimulates ERK, CREB, early genes. J. Cell. Physiol. 209, 379–388 [DOI] [PubMed] [Google Scholar]
- 20. Hidalgo C., Sánchez G., Barrientos G., Aracena-Parks P. (2006) A transverse tubule NADPH oxidase activity stimulates calcium release from isolated triads via ryanodine receptor type 1 S-glutathionylation. J. Biol. Chem. 281, 26473–26482 [DOI] [PubMed] [Google Scholar]
- 21. Anderson K., Cohn A. H., Meissner G. (1994) High-affinity [3H]PN200–110 and [3H]ryanodine binding to rabbit and frog skeletal muscle. Am. J. Physiol. Cell Physiol. 266, C462–C466 [DOI] [PubMed] [Google Scholar]
- 22. Lai F. A., Erickson H. P., Rousseau E., Liu Q. Y., Meissner G. (1988) Purification and reconstitution of the calcium release channel from skeletal muscle. Nature 331, 315–319 [DOI] [PubMed] [Google Scholar]
- 23. Hellman U., Wernstedt C., Góñez J., Heldin C. H. (1995) Improvement of an “In-Gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 224, 451–455 [DOI] [PubMed] [Google Scholar]
- 24. Xu H., Zhang L., Freitas M. A. (2008) Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine. J. Proteome Res. 7, 138–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruse C. I., Willard B., Jin J. P., Haas T., Kinter M., Bond M. (2002) Quantitative dynamics of site-specific protein phosphorylation determined using liquid chromatography electrospray ionization mass spectrometry. Anal. Chem. 74, 1658–1664 [DOI] [PubMed] [Google Scholar]
- 26. Beigi F., Gonzalez D. R., Minhas K. M., Sun Q. A., Foster M. W., Khan S. A., Treuer A. V., Dulce R. A., Harrison R. W., Saraiva R. M., Premer C., Schulman I. H., Stamler J. S., Hare J. M. (2012) Dynamic denitrosylation via S-nitrosoglutathione reductase regulates cardiovascular function. Proc. Natl. Acad. Sci. U.S.A. 109, 4314–4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ostman A., Frijhoff J., Sandin A., Böhmer F. D. (2011) Regulation of protein tyrosine phosphatases by reversible oxidation. J. Biochem. 150, 345–356 [DOI] [PubMed] [Google Scholar]
- 28. Burgoyne J. R., Eaton P. (2011) Contemporary techniques for detecting and identifying proteins susceptible to reversible thiol oxidation. Biochem. Soc. Trans. 39, 1260–1267 [DOI] [PubMed] [Google Scholar]
- 29. Salmeen A., Andersen J. N., Myers M. P., Meng T. C., Hinks J. A., Tonks N. K., Barford D. (2003) Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 423, 769–773 [DOI] [PubMed] [Google Scholar]
- 30. van Montfort R. L., Congreve M., Tisi D., Carr R., Jhoti H. (2003) Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature 423, 773–777 [DOI] [PubMed] [Google Scholar]
- 31. Voss A. A., Lango J., Ernst-Russell M., Morin D., Pessah I. N. (2004) Identification of hyperreactive cysteines within ryanodine receptor type 1 by mass spectrometry. J. Biol. Chem. 279, 34514–34520 [DOI] [PubMed] [Google Scholar]
- 32. Lobo P. A., Van Petegem F. (2009) Crystal structures of the N-terminal domains of cardiac and skeletal muscle ryanodine receptors: insights into disease mutations. Structure 17, 1505–1514 [DOI] [PubMed] [Google Scholar]
- 33. Hwang J. H., Zorzato F., Clarke N. F., Treves S. (2012) Mapping domains and mutations on the skeletal muscle ryanodine receptor channel. Trends Mol. Med. 18, 644–657 [DOI] [PubMed] [Google Scholar]
- 34. Gaburjakova M., Gaburjakova J., Reiken S., Huang F., Marx S. O., Rosemblit N., Marks A. R. (2001) FKBP12 binding modulates ryanodine receptor channel gating. J. Biol. Chem. 276, 16931–16935 [DOI] [PubMed] [Google Scholar]
- 35. Sheridan D. C., Takekura H., Franzini-Armstrong C., Beam K. G., Allen P. D., Perez C. F. (2006) Bidirectional signaling between calcium channels of skeletal muscle requires multiple direct and indirect interactions. Proc. Natl. Acad. Sci. U.S.A. 103, 19760–19765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lynch P. J., Krivosic-Horber R., Reyford H., Monnier N., Quane K., Adnet P., Haudecoeur G., Krivosic I., McCarthy T., Lunardi J. (1997) Identification of heterozygous and homozygous individuals with the novel RYR1 mutation Cys35Arg in a large kindred. Anesthesiology 86, 620–626 [DOI] [PubMed] [Google Scholar]
- 37. Andersson D. C., Betzenhauser M. J., Reiken S., Meli A. C., Umanskaya A., Xie W., Shiomi T., Zalk R., Lacampagne A., Marks A. R. (2011) Ryanodine receptor oxidation causes intracellular calcium leak and muscle weakness in aging. Cell Metab. 14, 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Andersson D. C., Meli A. C., Reiken S., Betzenhauser M. J., Umanskaya A., Shiomi T., D'Armiento J., Marks A. R. (2012) Leaky ryanodine receptors in beta-sarcoglycan deficient mice: a potential common defect in muscular dystrophy. Skelet. Muscle 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]