Background: Sea anemone peptides are promising tools for understanding physiological functions of ion channels.
Results: A new peptide, Ugr 9-1, was isolated from the sea anemone venom and was shown to inhibit the acid-sensing ion channel 3 (ASIC3) channel.
Conclusion: Ugr 9-1 affects the ASIC3 channel, produces analgesic effects, and has a unique spatial structure and mechanism of action.
Significance: Ugr 9-1 represents a novel structural fold of natural short peptides modulating neuronal channels.
Keywords: Acid Sensing Ion Channels (ASIC), Gene Structure, Nuclear Magnetic Resonance, Peptides, Protein Chemistry, Protein Purification, Protein Sequence, Protein Structure
Abstract
Three novel peptides were isolated from the venom of the sea anemone Urticina grebelnyi. All of them are 29 amino acid peptides cross-linked by two disulfide bridges, with a primary structure similar to other sea anemone peptides belonging to structural group 9a. The structure of the gene encoding the shared precursor protein of the identified peptides was determined. One peptide, π-AnmTX Ugr 9a-1 (short name Ugr 9-1), produced a reversible inhibition effect on both the transient and the sustained current of human ASIC3 channels expressed in Xenopus laevis oocytes. It completely blocked the transient component (IC50 10 ± 0.6 μm) and partially (48 ± 2%) inhibited the amplitude of the sustained component (IC50 1.44 ± 0.19 μm). Using in vivo tests in mice, Ugr 9-1 significantly reversed inflammatory and acid-induced pain. The other two novel peptides, AnmTX Ugr 9a-2 (Ugr 9-2) and AnmTX Ugr 9a-3 (Ugr 9-3), did not inhibit the ASIC3 current. NMR spectroscopy revealed that Ugr 9-1 has an uncommon spatial structure, stabilized by two S-S bridges, with three classical β-turns and twisted β-hairpin without interstrand disulfide bonds. This is a novel peptide spatial structure that we propose to name boundless β-hairpin.
Introduction
The phylum Cnidaria species have a rich variety of biologically active compounds, which are enclosed in specialized, stinging organelles (nematocysts) and serve as chemical weapons to capture prey and as defensive mechanisms against numerous predators. The nematocysts in an anemone body are distributed unequally. In particular, the ectoderm of the column (the outer surface of the anemone body) contains relatively few nematocysts, nematocysts in the tentacles are more numerous, and some internal structures (mesenteric filaments, the ectodermal lining of the throat) contain large amounts of different types of nematocysts (1).
In humans, the stinging cells can cause itching and burning sensations as a reaction to the injected venom, and the site of contact may suffer tissue necrosis from the action of the abundant cytotoxic venom components on the membrane of mammalian cells (2, 3). In addition to these membrane-disrupting components, sea anemone venom contains a variety of peptides with different biological activity. Among these are many protease inhibitors and peptide toxins acting on ion channels. The much researched site-3 sodium channel peptide toxins, which slow the kinetics of the Na+-channel inactivation, and potassium channel inhibitory toxins have been identified in various species of sea anemones (4–6). Promising peptides affecting the receptors involved in nociception have been identified in the venom of sea anemones. An analgesic effect on animal models was observed after the introduction of sea anemone peptides, such as APETx2, a selective blocker of the proton-sensitive channel acid-sensing ion channel 3 (ASIC3)2 (7), and APHC1, the modulator of vanilloid receptor TRPV1 activity (8, 9).
The ASIC3 channel belongs to a group of sodium-selective acid-sensing ion channels (ASIC) that could be activated by extracellular acidosis (7, 10) and considered to be a perspective target for treatment of inflammation and pain. The ASIC3 channel is predominantly expressed in peripheral sensory neurons (11). ASIC3 channels in response to a drop of extracellular pH generate a transient inactivating current followed by a sustained component (12). ASIC3 have been shown to contribute to acidic and inflammatory pain during pathological conditions such as inflammation, ischemia, hematomas, etc. (13, 14).
Here, we report the identification from the sea anemone Urticina grebelnyi of a peptidic molecule affecting the ASIC3 channel. The structure and the mechanism of action of this peptide differ from the well known sea anemone toxin APETx2.
EXPERIMENTAL PROCEDURES
Venom Collection
The U. grebelnyi specimens were collected in the vicinity of Petropavlovsk-Kamchatsky using SCUBA equipment at depths from 5 to ∼20 m. The living specimens were kept in a cold water aquarium (a year and more) to make them available at demand for experiments. To collect venom, a living specimen was briefly placed on a clean dish and the surface of the ectoderm scraped, allowing nematocysts to release the crude venom. The obtained venom was immediately washed with 10 mm EDTA and 1 mm PMSF solution, collected in tubes, and frozen until separation.
Purification Procedure
In the first stage of separation, crude venom was subjected to size-exclusion chromatography on a TSK-2000SW column (7.5 × 600 mm), using 10% acetonitrile solution with 0.1% trifluoroacetic acid (TFA) as a mobile phase to remove any large cytotoxins (Fig. 1A). The next stage of separation was done on a reverse-phase column Jupiter C5 (4.6 × 150 mm), using a 60-min linear gradient from 0 to 60% of acetonitrile concentration in the presence of 0.1% TFA and a constant flow rate 1 ml/min (Fig. 1B). Finally, the active peptide and its homologues were isolated on a Vydac C18 column (250 × 4.6 mm), using a linear gradient of acetonitrile concentration (0–15% in 5 min, 15–55% in 80 min) in 0.1% TFA and a constant flow rate 1 ml/min (Fig. 1C).
FIGURE 1.
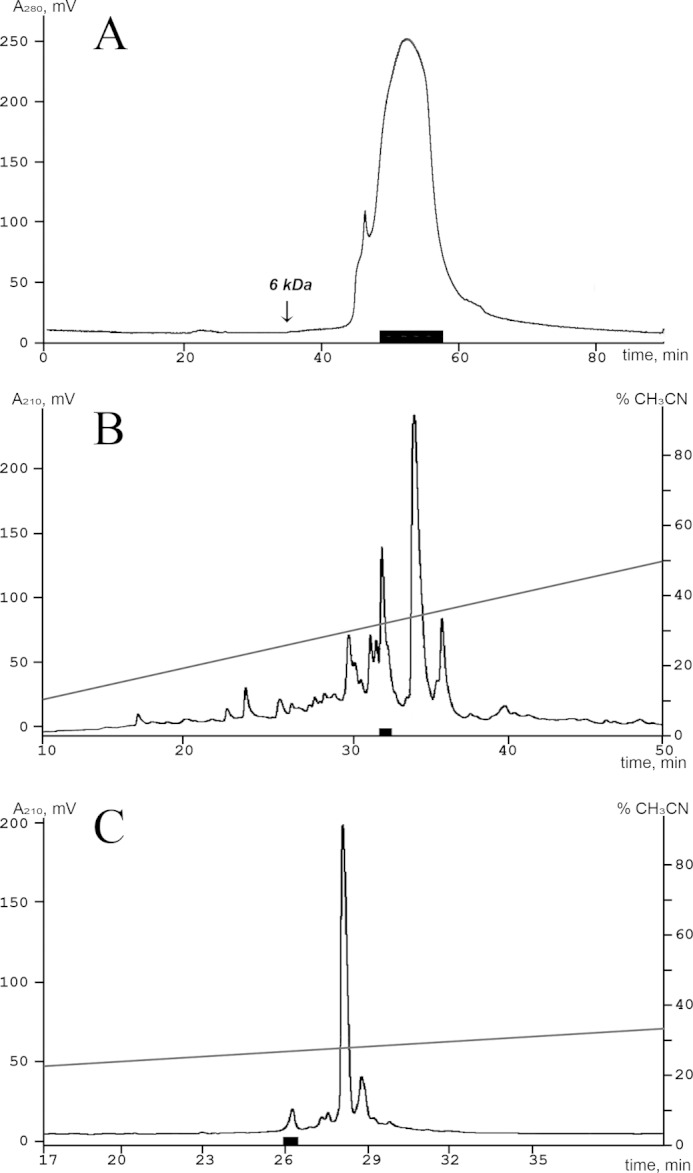
Isolation of the active peptide from the venom of U. grebelnyi. Fractions marked with a black box exercised an inhibitory activity on the ASIC3 channel in all separation steps. A, the first separation stage of the crude peptide fraction by size exclusion chromatography on a TSK-2000SW (7.5 × 600 mm) column in 10% acetonitrile solution with 0.1% trifluoroacetic acid and a flow rate of 0.5 ml/min. B, second separation step on a reverse-phase column Jupiter C5 column (4.6 × 150 mm) in 0.1% trifluoroacetic acid with a flow rate of 1 ml/min using a linear gradient of acetonitrile concentration. C, final separation step on a Vydac C18 column (250 × 4.6 mm) using a linear gradient of acetonitrile concentration in 0.1% trifluoroacetic acid with a flow rate 1 ml/min.
Mass Spectrometry
Molecular weight measurement was carried out by matrix-assisted laser desorption ionization (MALDI), time-of-flight mass spectrometry (Micromass, UK), and Ultraflex TOF-TOF (Bruker Daltonik, Germany) instruments. Calibration was performed using either a ProteoMass peptide and protein MALDI-MS calibration kit, with a mass range of 700 to 66,000 Da, or a ProteoMass peptide MALDI-MS calibration kit, with a mass range of 700 to 3,500 Da (both from Sigma). The molecular mass was determined in linear or reflector positive ion mode, using samples prepared by the dried-droplet method with 2.5-dihydroxybenzoic acid (10 mg/ml in 70% acetonitrile with 0.1% TFA) or α-cyano-4-hydroxycinnamic acid (10 mg/ml in 50% acetonitrile with 0.1% TFA) matrices.
Reduction of Disulfide Bonds and Modification of Thiol Groups
The dried sample was dissolved in 40 μl of a solution containing 6 m guanidine hydrochloride, 3 mm EDTA, and 0.1 m Tris-HCl (pH 8.5) and then incubated overnight at 45 °C. Next, the denaturated protein was incubated for 4 h at 40 °C with 2 μl of 1.4 m 1,4-dithiothreitol solution in 0.1 m Tris-HCl (pH 8.5). Free cysteines were alkylated for 15 to 20 min at room temperature in the dark by addition of 2 μl of 50% 4-vinylpyridine in methanol. The modified peptides were immediately separated from the excess reagents by HPLC on a reverse-phase Luna C18 column (150 × 3 mm), using a 60-min linear gradient (15–55%) of acetonitrile concentration in 0.1% TFA at a flow rate of 0.3 ml/min.
Amino Acid Sequence Analysis
N-terminal sequencing of alkylated peptide was carried out by automated stepwise Edman degradation, using a Procise model 492 protein sequencer (Applied Biosystems) according to the manufacturer's protocol.
Precursor Determination
Total RNA was purified from the tentacles of U. grebelnyi, using Trisol® Reagent (Ambion, Canada) as directed by the manufacturer's protocols. cDNA was synthesized from 5 μg of total RNA, using the MINT kit (Evrogen, Russia) following the manufacturer's recommendation. Rapid amplification of cDNA ends (RACE) was carried out, using the universal primer T7cap (GTAATACGACTCACTATAGGGCAAGCAGTGGTAACAACGCAGAGT) and degenerated primers Ug1 (ATCTGTATCGATCCNCCNTGYMG) and Ug2 (CCACCCTGTAGNTTYTGYTAYCA) for 3′ terminus determination (3′-RACE) and Ug3 (CGGTCATCACAGTTATATTATGAG) and Ug4 (ATTATGAGTTTTTGACAGGATTAC) for 5′ terminus determination (5′-RACE). DNA sequencing was carried out on Applied Biosystems 3730 DNA Analyzer.
Gene Synthesis
The DNA encoding of the π-AnmTX Ugr 9a-1 mature peptide sequence was constructed from three synthetic oligonucleotides using the PCR technique. The target PCR fragment was amplified using a forward primer containing a site for restriction enzyme BglII and the Met codon for BrCN cleavage (5′-GAATTAGATCTCATGATTTCCATTGATCCGCCGTGCCGTTTTTGCTATCAT-3′), reverse primer 1 (5′-CGCATCATACACGCAATTGCCGGAGCCATCACGATGATAGCAAAAACGGCA-3′), reverse primer 2 containing the site for restriction enzyme XhoI and the stop codon (5′-GGATTCCTCGAGCTACACCGCGCCGCAGCCATACGCATCATACACGCAATT-3′). The target PCR fragment encoding the peptide gene was gel-purified, digested by BglII/XhoI, and cloned into the expression vector pET32b+ (Novagen). The resulting construct was confirmed by sequencing.
Recombinant Peptide Production
Recombinant peptide was produced through fusion with a thioredoxin domain. Escherichia coli BL21(DE3) cells transformed with the expression vector were cultured at 37 °C in an LB medium containing 100 μg/ml of ampicillin up to the culture density of A600 ∼ 0.6–0.8. To induce expression, up to 0.2 mm isopropyl 1-thio-β-d-galactopyranoside was added. The cells were cultured at 25 °C for 18 h, harvested, re-suspended in the start buffer for affinity chromatography (400 mm NaCl, 20 mm Tris-HCl buffer, pH 7.5), and ultrasonicated and centrifuged for 15 min at 14,000 × g to remove all insoluble particles. The supernatant was applied to a TALON Superflow metal affinity resin (Clontech), and the fusion protein purified according to the manufacturer's instructions. Protein cleavage was performed overnight at room temperature in the dark as described previously (15). HCl to a final concentration of 0.2 m and CNBr with a molar ratio CNBr to fusion protein of 600:1 were added. Recombinant peptide was purified from the reaction mixture on a reverse-phase Jupiter C5 column (250 × 10 mm). The purity of the target peptide was checked by MALDI-TOF mass spectrometry and N-terminal sequencing.
NMR Spectroscopy and Spatial Structure Calculation
The sample for NMR spectroscopy was prepared by dissolving 5 mg of recombinant peptide in 350 ml of buffer containing 5% D2O, 30 mm Na2HPO4, 15 mm citric acid, and 1 mm sodium azide (pH 5.0). All spectra were recorded at 30 °C on the Bruker Avance spectrometer with the working frequency on the proton channel equal to 700 Mhz. Proton and 13C resonance assignment were obtained through the standard procedure (16), based on spectra MLEV-TOCSY (80 ms mixing time), NOESY (40 and 80 ms mixing times), 1H,13C-HSQC, and DQF-COSY. After assignment, the sample was lyophilized and dissolved in pure D2O buffer with the same contents to measure the rate of exchange of amide protons to deuterium and additional NOESY (80 ms mixing time) and DQF-COSY were recorded.
Spatial structure calculation was performed using the simulated annealing/molecular dynamics protocol as implemented in CYANA software package version 3.0 (17). Upper interproton distance constraints were derived from NOESY (τm = 80 ms) cross-peaks via a 1/r6 calibration. The torsion angle restraints and stereo-specific assignments were obtained from J couplings and NOE intensities. 3JHNHα couplings were measured from the splitting of cross-peaks in NOESY spectra and 3JHαHβ couplings were obtained using ACME software (18) in the COSY spectrum of the peptide in D2O solution (relaxation delay 3 s). Hydrogen bonds were introduced as distance restraints on the final stage of structure calculation based on the temperature coefficients of the chemical shifts and deuterium exchange rates of HN protons (protons with gradients less than 4.5 ppb/K and with a H-D exchange rate slower than 0.03 min−1 at 10 °C were supposed to participate in hydrogen bonding, hydrogen bond acceptors were determined based on the preliminary NMR structure). Chemical shifts, NMR constraints, and derived atomic coordinates (20 models) of π-AnmTX Ugr 9a-1 were deposited into the Protein Data Bank with accession code PDB code 2LZO.
Electrophysiology
Xenopus laevis oocytes were removed surgically, defolliculated, and injected with 2.5 to 10 ng of human ASIC3 cRNA (AF057711.1). cRNA transcripts were synthesized from NaeI-linearized ASIC3 cDNA template (pcDNA3.1 + human ASIC3 subcloned from clone EX-Q0260-B02 (GeneCopoeia, Inc.)), using a RiboMAXTM large-scale RNA production system T7 (Promega) according to the manufacturer's protocol for capped transcripts. After injection, oocytes were kept for 2 to 3 days at 19 °C and then up to 7 days at 15 to 19 °C in a ND-96 medium containing (in mm) 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, and 5 HEPES titrated to pH 7.4 with NaOH supplemented with gentamycin (50 μg/ml). Two-electrode voltage clamp recordings were performed using a GeneClamp 500 amplifier (Axon Instruments), and the data were filtered at 20 Hz and digitized at 100 Hz by an AD converter L780 (LCard, Russia) using in-house software. To induce transient and/or sustained currents, three different protocols with different sample time applications, pH declines, and activation times were used. Microelectrodes were filled with a 3 m KCl solution. The working buffer solution was ND-96 titrated by NaOH to pH 7.8 or 7.3. The solution for the pH shift was constructed based on the ND-96 solution in which 5 mm HEPES was replaced with 10 mm acetic acid (pH 4.0) or 5 mm MES (pH 5.5) in a supplementary 0.1% BSA solution. The duration of the activation pulses varied from 1 s to measure transient current to 1.5 s to measure both current components and to 3 s to measure the sustained current. The four parameter logistic equation was used for curve-fitting analysis: F(x) = ((a1 − a2)/(1 + (x/x0)n)) + a2, where x is the concentration of peptide; F(x) is the response value at given peptide concentration; a1 is the control response value (fixed at 100%); x0 is the IC50 value; n = Hill coefficient (slope factor); and a2 the response value at maximal inhibition (% of control).
Animal Models
Adult male CD-1 mice (Animal Breeding Facility Branch of Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Pushchino, Russia) weighing 20 to 25 g were used. The animals were housed at a room temperature of 23 ± 2 °C and subjected to a 12-h light-dark cycle, with food and water available ad libitum. Approval for all the experiments was given by the Animal Care and Use Committee of the Branch of the IBCh RAS (Pushchino, Russia Federation). Samples or saline were administered intravenously 30 min before testing.
Complete Freund's adjuvant (CFA) induced thermal hyperalgesia was measured in adult mice weighing about 20 g. CFA suspended in an oil/saline (1:1) emulsion was injected into the dorsal surface of the left hind-paw of the mice (20 μl/paw). The control mice received 20 μl of saline. Paw withdrawal latencies to thermal stimulation (53 °C) were measured 24 h after CFA injection.
The abdominal constriction test of visceral pain is based on the acetic acid-induced writhing count. Separate groups of mice were injected with 0.6% acetic acid in saline (10 ml/kg intraperitoneally). The mice were immediately placed inside transparent glass cylinders, and the number of writhes recorded for 15 min.
Rota-rod test was performed using a Dual Species Economex Rota-Rod system (Columbus Instruments). The full test consisted of two parts: pre-training and 3 test trials. For pre-training the mice were trained to stay and walk on the rod for 30 s at 5, 10, and 20 rounds per min (rpm). Mice that were not able to do this were excluded from the testing. Peptides Ugr 9-1, Ugr 9-2, and Ugr 9-3 at a dose of 0.5 mg/kg or saline were administrated intravenously 15 min before the first trial. During the trial mice were placed on the rota-rod and speed was accelerated from 5 to 22 rpm in 30 s. The latency to fall from the rod was recorded. Each mouse was tested in 3 trials with a 5-min resting time between trials. The average time spent on the rota-rod across the three trials was used in the analyses.
The significance of the data were determined by performing an analysis of variance followed by a Tukey's test. All data are presented as mean ± S.E.
Assay of Crayfish Toxicity
Crayfish toxicity was tested using noble crayfish (Astacus astacus) weighing about 35 g. Peptides as well as negative control and U. grebelnyi crude venom were administered into each crayfish at the junction between the body and the leg at a dose of 1 mg/kg in 100 μl of water. The negative control group was injected with 100 μl of water. Each group consisted of five crayfishes. Crayfish were observed for any symptoms for 2 h after injection.
Computation
The similarity of the mature chain sequences was determined by both a BLAST search and SRDA (19–21). The sequence and spatial structure data for analyses were retrieved from the UniProt Data Bank and the Information Portal to Biological Macromolecular Structures. The sequence alignment was built with the Megalign module from Dnastar Inc. The visual analysis of structures and the figure drawings were performed using the MOLMOL and PyMOL software. The elements of the secondary structure were analyzed with the STRIDE package (22).
RESULTS
Peptides Isolation and Primary Structure Determination
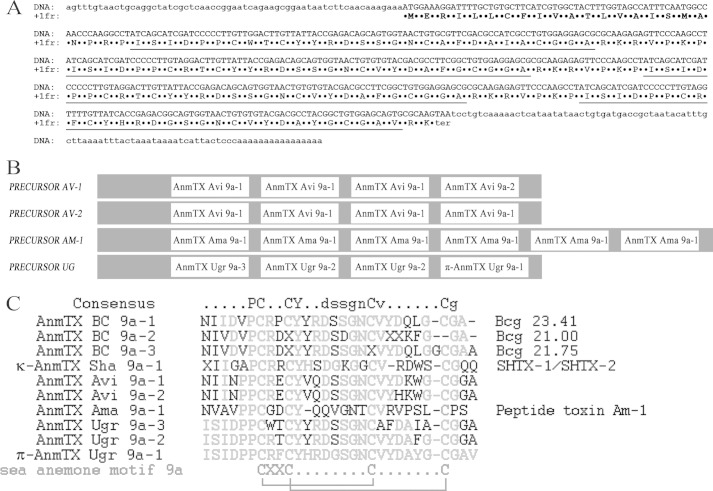
Activity testing was performed using the two-electrode voltage clamp technique on X. laevis oocytes that express human ASIC3 channels. It demonstrated that the venom of sea anemone U. grebelnyi produced a significant inhibitory effect. A combination of gel filtration and reversed-phase chromatography resulted in isolation of one active compound (Fig. 1). This peptide had an average molecular mass 3135.5 Da, and its N-terminal sequence of 25 amino acid residues was established by Edman degradation. To complete structure elucidation, the cDNA encoding the precursor protein named UG (GenBankTM number HF562346) was obtained using 3′- and 5′-RACE approaches.
The precursor protein UG consists of a signal peptide and sequences of four short peptides separated by spacer regions (Fig. 2A). The active peptide is in the C terminus of the precursor, whereas two copies of the peptide with an estimated molecular mass of 3087.4 Da are in the middle, and one copy of the peptide with an estimated molecular mass of 3053.4 Da is located on the N terminus (Fig. 2B). Each mature peptide sequence in the precursor was established using mature chains prediction (23). The spacer regions consist of proteolysis signals removed by various enzymes during maturation (Fig. 2A).
FIGURE 2.
Structural peculiarity of precursor protein organization. A, nucleotide sequence of peptide UG accompanied with deduced amino acids. The signal peptide sequence is marked in bold, and the determined mature peptides sequences are underlined. B, schematic structural organization of structural class 9a toxin precursors. The gray sections are removed by maturation processes. C, primary structure of mature peptides. The multiple sequence alignment of known toxins from structural class 9a included three novel peptides from U. grebelnyi; toxin SHTX-I/SHTX-2 (P0C7W7) from S. haddoni; Bcg toxins (P86466, P86465, and P86467) from B. cangicum; peptide toxin Am-1 (P69929) from A. maculate; and two peptide toxins from precursors AV-1 and AV-2 from A. viridis (20). Residues different from the π-AnmTX Ugr 9a-1 sequence are highlighted. The structural class 9a distribution pattern for cysteines is shown in the bottom, along with the disulfide bond connections determined in this paper by NMR spectroscopy for π-AnmTX Ugr 9a-1. The names in the left side of the figure are shown an improved rational nomenclature for sea anemone toxins (also shown as part of precursor sequences on panel B), accompanied by the original names on the right.
The primary structure of the mature peptide consists of 29 amino acid residues cross-linked by two disulfide bridges (Fig. 2C). It does not share any sequence homology to another ASIC3 inhibitor APETx2 previously isolated from sea anemone Anthopleura elegantissima (24). The calculated molecular weight accords well with the measured weight, indicating an absence of post-translational modifications.
To date, all cysteine-containing peptides from sea anemone venoms have been categorized into 10 major structural classes by a typical distribution pattern for Cys residues (25). All three peptides derived from the precursor protein UG belong to structural class 9a, which includes small peptide molecules stabilized by 2 disulfide bonds. A few members of structural class 9a members have been identified, as displayed in Fig. 2C. According to the improved nomenclature for toxins from sea anemone (25), the active peptide was named π-AnmTX Ugr 9a-1 (short name Ugr 9-1), the peptide with a molecular mass of 3087 Da was named AnmTX Ugr 9a-2 (Ugr 9-2), and the peptide with a molecular mass of 3053 was named AnmTX Ugr 9a-3 (Ugr 9-3).
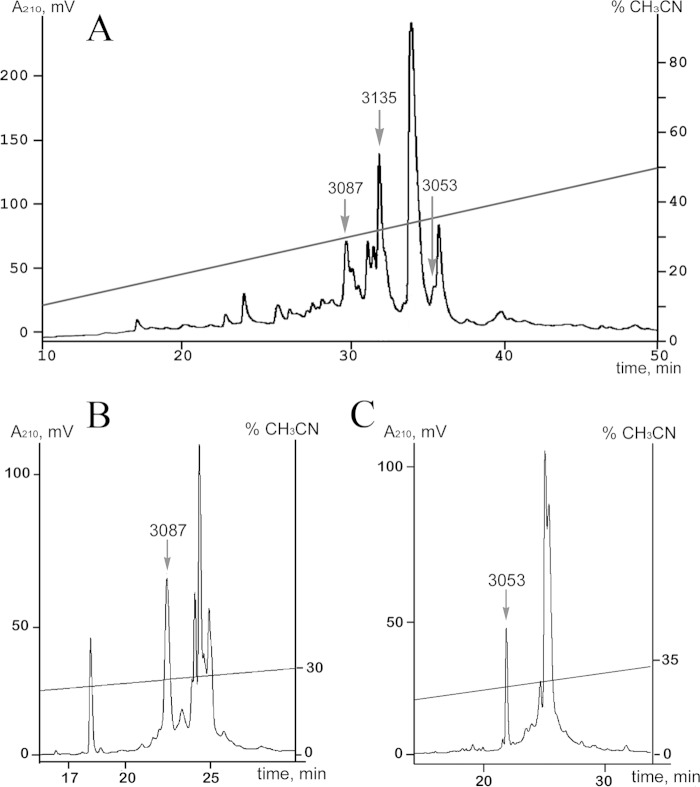
Mass spectrometry analysis of the venom separation profile revealed two peptides with a molecular weight similar to that predicted for both peptides Ugr 9-2 and 9-3 (Fig. 3A). These peptides were isolated (Fig. 3, B and C), and their identity to the predicted sequences was confirmed by MALDI mass spectrometry and partial N-terminal Edman sequencing. None of them inhibited or activated the ASIC3 current in concentrations up to 40 μm.
FIGURE 3.
Isolation of Ugr 9-2 and Ugr 9-3. A, the separation profile of active peptides fraction on a reverse-phase Jupiter C5 column (4.6 × 150 mm). All conditions are equal to those described in the legend to Fig. 1. The purification procedure step for Ugr 9-2 (B) and Ugr 9-3 (C) on a Vydac C18 column (250 × 4.6 mm) in a linear gradient of acetonitrile concentration with 0.1% trifluoroacetic acid and a flow rate 1 ml/min. The elution times for peptides are marked by arrows with the appropriate molecular weight.
The molecular targets of these peptides are different from previously described homologous peptides SHTX-1/SHTX-2 that are potassium channel blockers and cause paralysis in crabs with ED50 of 430 μg/kg (26). None of the Ugr 9-1, Ugr 9-2, and Ugr 9-3 caused lethality or paralysis of noble crayfish (A. astacus) at a dose of 1 mg/kg.
Recombinant Peptide Production
To provide a sufficient amount of peptide for functional characterization and spatial structure determination, recombinant Ugr 9-1 was produced through the prokaryotic expression system. Thioredoxin was chosen as the fusion partner for expression, because it is ensures high yields of cysteine-containing peptides with native conformation. A synthetic gene coding for the mature peptide was constructed and cloned into a pET-32b(+) expression vector, and the resulting plasmid was used to transform E. coli BL21(DE3) cells. Thioredoxin-Ugr 9-1 fusion protein production and purification were followed by CNBr cleavage to release the target peptide.
The recombinant peptide was purified by reverse-phase high-performance liquid chromatography. The final yield of the purified recombinant peptide was estimated to be ∼8 mg/liter of cell culture. The molecular weight of the recombinant product was equal to that of the native molecule, and the amino acid sequence of 5 N-terminal residues was also determined. The recombinant Ugr 9-1 and the natural peptide had the same retention time when they were co-injected in a reverse-phase column that confirms the proper recombinant peptide folding. In the electrophysiological study, the recombinant peptide showed the same ASIC3 receptor inhibitory activity as the native peptide.
Spatial Structure Determination
To determine the tertiary organization of the short peptide from structural group 9a, the Ugr 9-1 structure was determined by NMR spectroscopy. The NMR data on the recombinant peptide is summarized in Table 1 and Fig. 4. The set of structures was calculated in CYANA from 100 random start points using following experimental data: upper and lower NOE-based distance restraints, J coupling-based torsion angle restraints, and hydrogen bonds restraints. The resultant structures are characterized by low CYANA target function and residual restraint violations and quite a low root mean square deviation value for backbone atoms, indicating that the structure of the peptide is accurately and precisely defined by the experimental data in the 6–27 region.
TABLE 1.
Statistics for the best peptide structures
| Distance and angle restraints | |
| Total NOEs | 281 |
| Intraresidual | 140 |
| Interresidual | 141 |
| Sequential (|i-j| = 1) | 80 |
| Medium range (1<|i-j|≤4) | 26 |
| Long range (|i-j|>4) | 35 |
| Hydrogen bond restraints (upper/lower) | 16/16 |
| S-S bond restraints (upper/lower) | 6/6 |
| J couplings | 54 |
| JHNCαH | 19 |
| JHCαCβH | 35 |
| Total restraints/per residue | 379/13 |
| Statistics of calculated set of structures | |
| CYANA target function (Å2) | 0.42 ± 0.02 |
| Restraint violations | |
| Distance (>0.2 Å) | 0 |
| Angle (>5°) | 0 |
| Root mean square deviations on the region with defined structure (6–27) (Å) | |
| Backbone | 0.18 ± 0.14 |
| All heavy atoms | 0.96 ± 0.18 |
FIGURE 4.
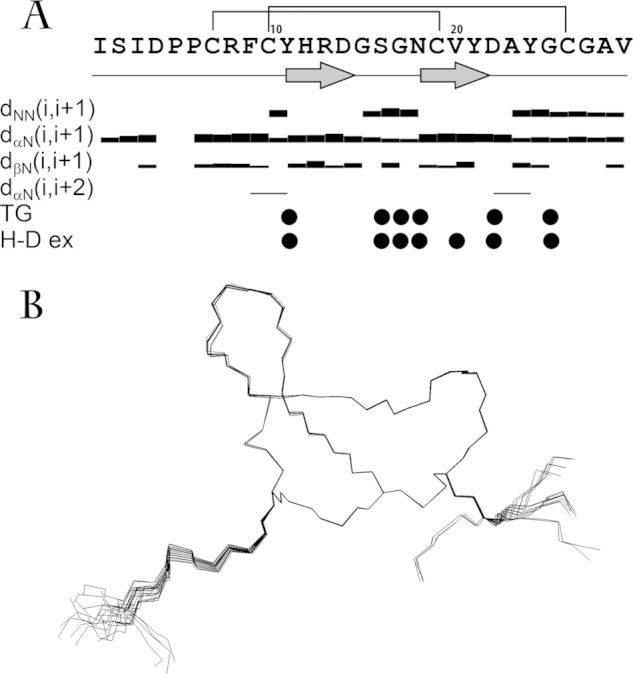
A, overview of the NMR data, defining the secondary structure of Ugr 9-1 peptide. NOE connectivities, exchange rate of HN protons with solvent (W-ex), and temperature gradients of HN chemical shifts (TempGrad) are shown versus the sequence. The widths of the bars represent the relative intensity of the cross-peak in 80-ms NOESY spectrum. Filled circles denote HN protons, which signal either a temperature gradient higher than −4.5 ppb/K or an exchange with the solvent with characteristic times longer than 15 m. The elements of the secondary structure are shown on the separate line, and β-strands are denoted by arrows. B, backbones and disulfide bridges of 20 NMR-derived structures with the best target function of the set of 100 structures, calculated from random start points. The backbones are overlaid by superimposing atoms of residues 6 to 27.
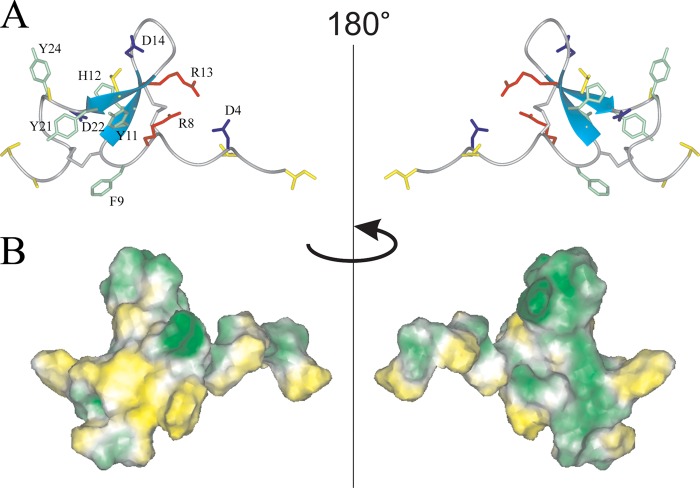
The ribbon representation of the spatial structure of Ugr 9-1 is displayed in Fig. 5A. The peptide itself presents a twisted β-hairpin in the Tyr11-Tyr21 region with 3 classical β-turns in regions Arg8–Tyr11 (type 2), Asp14–Gly17 (type 1), and Asn22–Gly25 (type 1). Two disulfide bonds stabilize the fold: Cys10–Cys26 and Cys7–Cys19, along with 6 backbone-backbone and 2 side chain-backbone hydrogen bonds. Alternative disulfide linkages were tested during the structure calculation, but none yielded the spatial structure of Ugr 9-1 with a reasonably low target function. In addition, a salt bridge between the charged Asp4 and Arg13 side changes might occur according to the determined set of spatial structure; however, no direct NMR data supporting this interaction was found.
FIGURE 5.
Two-sided view of the determined spatial structure of Ugr 9-1. A, ribbon representation. Positively and negatively charged side chains are shown in red and blue, respectively. The aromatic and hydrophobic (Leu, Ile, Val, Thr, and Ala) side chains are shown in green and yellow, and cysteine side chains in gray. B, hydrophobicity of the peptide surface. The peptide surface is colored from yellow (hydrophobic) to green (hydrophilic), according to the molecular hydrophobic potential (49).
The analysis of the electrostatic properties of Ugr 9-1 did not reveal any peculiarities. Both the positively and negatively charged side chains are scattered homogeneously on the peptide surface. Hydrophobic residues are clustered on the C-terminal region of the peptide (Fig. 5B), which may be important for peptide activity.
Comparison of Structural Class 9a Peptides from Sea Anemones
A BLAST search found the homology of the primary structures of Ugr 9-1, -2, and -3 to be similar to previously known peptides from the Bcg III group (Bcg 21.00, Bcg 21.75, and Bcg 23.41) and to the potassium channel inhibitors SHTX-1/SHTX-2 (Fig. 2C).
Peptides SHTX-1/SHTX-2 from the sea anemone Stichodactyla haddoni, which are characterized as potassium channel inhibitors differ in one post-translational modification of Pro6. In the peptide SHTX-1, Pro is a hydroxyproline, whereas in SHTX-2, Pro is unmodified (26).
Peptides of the Bcg III group were discovered during the proteomic study of the venom of the sea anemone Bunodosoma cangicum. Their structures were partially determined through both Edman degradation and mass spectrometry (27). For one peptide, the Bcg 23.41 toxin was assumed to have an inhibitory activity on K+-channels, but this was not confirmed by direct experimentation. The structures of three Bcg toxins were detected in the crude venom, suggesting the possible presence of a common complex protein precursor. This precursor may be constructed by the same principles as other precursor proteins of this group.
Other experimental approaches, including the detailed analysis of sea anemone sequences derived from nucleotide sequences using the algorithm SRDA (19), found structural similarities to more peptides: peptide toxin Am-1 and peptide toxins AV-1 and AV-2 (Fig. 2C), deduced from the nucleotide sequences of the extended peptide precursor protein, shown in Fig. 2B. Peptide toxin Am-1 was isolated from the venom of Antheopsis maculata as the toxin possessing weak lethal activity toward crabs (LD50 = 830 μg/kg). Further cDNA cloning based on 3′-RACE and 5′-RACE established the structure of its precursor, which contained six copies of Am I (28). Two other precursors, peptide toxins AV-1 and AV-2, were deduced from the dbEST of sea anemone Anemonia viridis (20); therefore, no biological activity was reported.
Thus, so far, structural class 9a of sea anemone toxins has been determined to consist of 10 related structures and common features, such as a cysteine pattern with a 1–3, 2–4 pairing and conservative residues Pro6, Tyr11, Asp14, Asn18, Val20, and Gly27. Additionally, a typical region of 15–17 corresponds to the central β-turn containing mainly Ser and Gly residues that are important for structure folding.
Electrophysiology
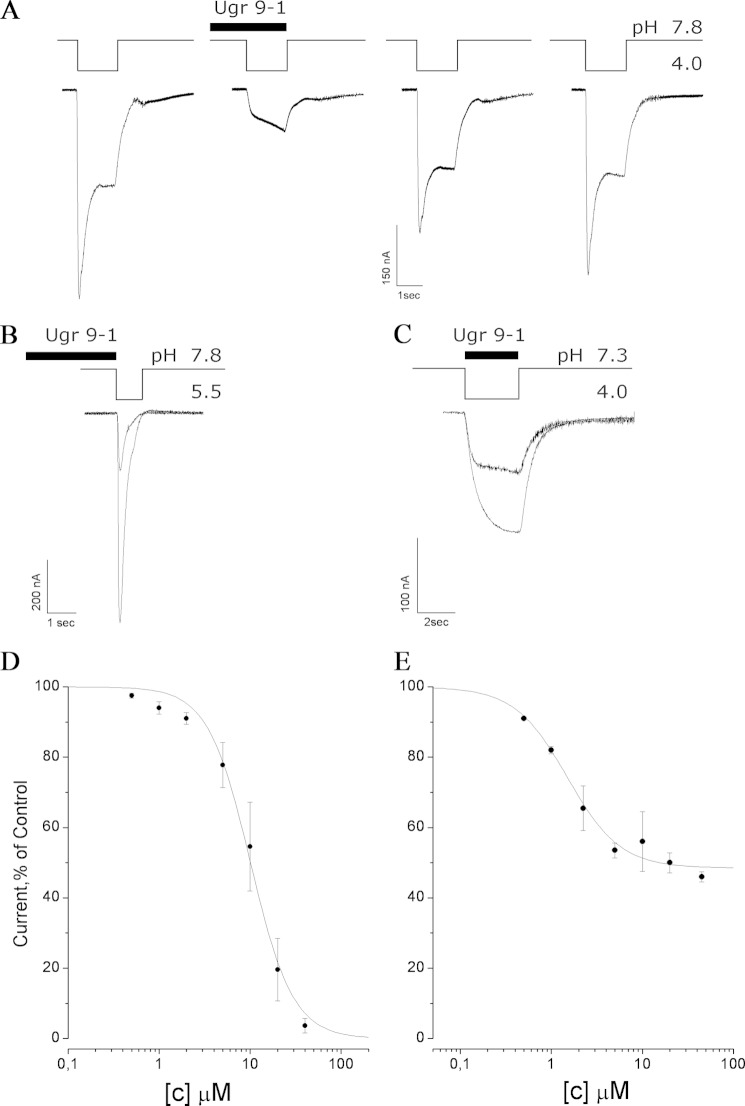
The standard two-electrode voltage clamp technique on X. laevis oocytes expressing human ASIC3 receptors was used to analyze venom, fractions, and recombinant peptide. Human ASIC3 currents (both the transient and sustained components of integral current) were evoked by a 1.5-s pH drop from the base value of 7.8 to 4.0 in the external oocyte solution. Measured compounds were applied for up to 10 s before and during the activation impulse. The measured effects of the application of crude U. grebelnyi venom, pure native peptide, and recombinant Ugr 9-1 did not differ significantly. Experimental traces for recombinant Ugr 9-1 testing are displayed in Fig. 6A. Ugr 9-1 produced a significant inhibition of the sustained and transient components of ASIC3 currents. The inhibition was shown to be reversible, because after the next two pH 4.0 drops, the response parameters recovered completely.
FIGURE 6.
Electrophysiological characterization of Ugr 9-1 action on human ASIC3 expressed in the X. laevis oocyte. Acid-induced currents were evoked by pH drop; the time intervals between the recordings were about 1 to 2 min, and the holding potential was −50 mV. A, effect on both current components by application of Ugr 9-1 at a concentration of 40 μm; B, effect on the transient current by application of Ugr 9-1 at a concentration of 20 μm; C, effect on the sustained current by application of Ugr 9-1 at a concentration of 10 μm; D and E, dose-response curves for the Ugr 9-1 inhibitory activity separately calculated for the transient (D) and sustained (E) components. Each point is the mean ± S.D. of 3–7 measurements. Data were fitted by the logistic equation.
Two Different Protocols Were Used to Independently Measure the Effect on Each Current Component
To analyze the inhibition of the transient components of ASIC3 currents, the channels were activated by a short 1-s pH drop from 7.8 to 5.5 (Fig. 6B). Ugr 9-1 was applied 10 s before activation. It completely inhibited the transient component of the ASIC3 currents in concentrations of more than 40 μm. The inhibitory effect was concentration dependent and fitted well the logistic equation with the half-maximal inhibitory concentration (IC50) of 10 ± 0.6 μm and Hill coefficient (nH) of 1.86 ± 0.19 (Fig. 6D).
Protocol, including dropping the pH from 7.3 to 4.0, was used to elicit the sustained component of the current with none or a minor transient component (Fig. 6C). The Ugr 9-1 application was combined with a 3-s pH activation. The amplitude of the sustained component of the ASIC3 currents in the control and sample experiments was calculated as a quasi-stationary value of the current on the third second. Dose-response analysis of the inhibitory effect of Ugr 9-1 on the sustained component of the ASIC3 current estimates a maximal inhibitory effect to be 48 ± 2%, IC50 = 1.44 ± 0.19 μm, and nH = 1.48 ± 0.27 (Fig. 6E). The peptide Ugr 9-1 was tested on several ion channels for inhibitory/activation effects and showed neither agonistic nor antagonistic activity on ASIC1a, ASIC1b, and ASIC2a at concentrations up to 50 μm as well as on the human KV1.3 channel at concentrations up to 1 μm in oocyte electrophysiology experiments.
Analgesic Effects of Ugr 9-1
Molecules capable of inhibiting ASIC3 in vitro were shown to produce an analgesic effect in animal models of pain (29). We checked the two animal models most closely associated with the acid-sensing channels and detected analgesic activity comparable with known pharmacological agents.
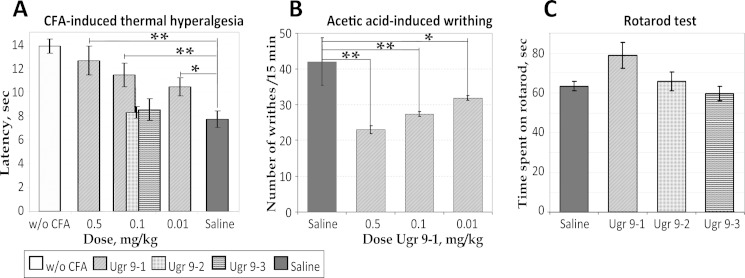
To characterize the ability of Ugr 9-1 to block an inflammation, we used a CFA-induced thermal hyperalgesia model (Fig. 7A). Groups of mice treated with saline within 24 h of CFA injection showed a significantly lower inflamed hind-paw withdrawal latency (7.7 ± 0.7 s) on a hot plate than the control group treated with saline twice (13.8 ± 0.6 s). Intravenous administration of Ugr 9-1 (30 min before testing) significantly reversed thermal hyperalgesia in a dose-dependent manner. Latency in response to the thermal stimulus increased from 7.7 ± 0.7 s (CFA/saline group) to 12.6 ± 1.2, 11.4 ± 1, and 10.4 ± 0.75 s following 0.5, 0.1, and 0.01 mg/kg treatment, respectively. The maximal effect, calculated as a percentage of the reduction of hyperalgesia, was 80 ± 19% for a 0.5 mg/kg dose. Peptides Ugr 9-2 and Ugr 9-3 at 0.1 mg/kg failed to reverse thermal hyperalgesia in this test (Fig. 7A).
FIGURE 7.
Efficacy of peptides in animal models. A, effect of peptides Ugr 9-1, Ugr 9-2, and Ugr 9-3 on the mice withdrawal latency on a hot plate in the thermal hyperalgesia test after CFA injection (n = 7). B, the attenuated response of Ugr 9-1-treated mice to intraperitoneal administration of acetic acid estimated by the number of writhes in 15-min intervals (n = 7). C, Ugr 9-1, Ugr9-2, and Ugr 9-3 at 0.5 mg/kg (n = 10) did not significantly alter time spent on the rota-rod. Results presented as mean ± S.E.; **, p < 0.005; *, p < 0.05.
The common pain test for response to low pH is an acetic acid writhing test. Intraperitoneal administration of acetic acid provoked a stereotyped behavior in mice characterized by abdominal contractions (acetic acid-induced writhes). The number of writhes in the control group treated by saline was 42.0 ± 6.6, whereas the number of writhes decreased dramatically in the groups pretreated with Ugr 9-1 by up to 23 ± 1.1 for a 0.5 mg/kg dose, 27.3 ± 0.7 for 0.1 mg/kg, and 31.8 for 0.01 mg/kg (Fig. 7B).
We also evaluated peptides Ugr 9-1, Ugr 9-2, and Ugr 9-3 in rota-rod tests as compounds capable of producing motor deficit could misrepresent results of other behavioral tests (particularly in pain models). Peptides Ugr 9-1, Ugr 9-2, and Ugr 9-3 at 0.5 mg/kg did not decrease the time spent on the rod compared with saline (Fig. 7C). Moreover Ugr 9-1 even tended to prolong the time spent on the rod but this effect was not statistically significant in our experiments. Therefore the efficacy of Ugr 9-1 in pain models did not result from a toxic effect of the peptide, locomotor impairment, or sedation.
DISCUSSION
Sea anemone peptide toxins are promising tools to characterize diverse ion channels. Studying the spatial structure of the active peptides is important to identify their structural and functional properties and to establish the molecular mechanisms of the interaction of the peptide ligand with the target. Therefore, the spatial structure of the 18 cysteine-containing sea anemone peptide toxins with 4 different folds have been dissolved over the last 25 years (25).
We have isolated and characterized peptide Ugr 9-1 in reference to class 9a and its fold, which to our knowledge does not match any known peptide folds. The spatial structure of Ugr 9-1 is centered on the flattened β-hairpin attached to both termini, so the molecule has a flat nonglobular shape (25 × 20 × 9 Å) with an overhang of the 6 N-terminal residues. Some similarity is observed in the backbone fold of class 5a short toxins, which have overlapped β-turns. Notably, both toxin classes have similar molecular weights but different numbers of disulfide bonds due to a dissimilar shapes.
Short venom peptides are an example of a rational natural design. They are designed to function for a long time by being folded into stable structures, usually fixed by several disulfide bonds. We examined all resolved structures of short venom peptides to find possible similarities. We searched the PDB for molecules with folds composed of only β-hairpins and found a small number of structures: 3 peptides from spiders, 1 peptide from cones (30–32), 3 antimicrobial peptides, and 1 hormone-like peptide that suppresses the growth of insects (33–36).
The structure of spider toxins is more distinct from that of Ugr 9-1. The β-structure element is close to the C terminus, has a longer turn region, and is stabilized by disulfide bond. In addition, a long N terminus with loops is connected to the β-structure through disulfide bonds (for example, the structure of purotoxin (PDB 2KGU)). The conotoxin MrIA structure has a β-hairpin, but in contrast to the Ugr 9a-1 structure, two disulfide bonds firmly connect the antiparallel β-strands on both sides (PDB 2J15). Antimicrobial peptides have a similar elongated shape (for example, the structure of bovine lactoferricin (PDB 1LFC)), and their β-hairpin element is also stabilized by at least 1 disulfide bond.
The β-hairpin structure of the Ugr 9-1 is not stabilized by disulfide bonds (Fig. 5A). One bond attaches a prolonged N terminus to the β-core, whereas the other bond attaches the C terminus to a β-turn. Thus, this β-hairpin without interstrand disulfide bonds is the novel spatial fold of short peptides. We propose naming this new structural fold the boundless β-hairpin.
We assume that peptides of structure class 9a have a unique non-globular structure that may be a result of directed evolution to create molecules with higher permeability and stability. Similar to many other secreted peptides from animal venoms, peptides Ugr 9-1, 9-2, and 9-3 are encoded in a single precursor protein, UG in this case (37–40). The complex precursors can be obviously more effective both for short peptides and intensive expression in animals and for production of recombinant peptides in prokaryotic expression systems (41).
Two peptides, Ugr 9-2 and 9-3, do not modulate ASIC3 activity but have significant homology with the active Ugr 9-1 peptide and evidently have the same fold type. All together, the three novel peptides are a visual confirmation of the combinatorial biochemistry model, according to which several sequence alterations of peptides with similar folds result in selectivity to different cellular receptors.
The Ugr 9-1 molecule can be divided into two surfaces: the inner face, which is the surface that includes the N terminal overhang, and the outer face, which is the opposite side of the molecule. Peptides Ugr 9-1, 9-2, and 9-3 were found to differ in 10 positions. Two C terminal substitutions could be considered homologous. The ASIC3-active peptide sequence differs from the Ugr 9-2 sequence only at 6 positions; therefore, we can presume that certain residues are important for the biological activity: Phe9, His12, Gly14, and Tyr24, located on the border between the inner and outer surfaces. In our opinion, it is the most reasonable to assume that the key residues are Phe, Tyr, and His, which, in combination with any other residues, can be involved in the recognition of ASIC3 receptors. Further experiments by site-directed mutagenesis could confirm this speculation.
The restricted expression of ASIC3 in sensory neurons and their contribution to acid sensing, development, and maintenance of inflammatory pain led to the exploration of this receptor as a pharmacological target for pain treatment. Several ligands are known to inhibit ASIC3 currents with different degrees of efficacy. Amiloride, peptides APETx2 and π-AnmTX Hcr lb-1 (a structural homologue of APETx2) inhibit transient ASIC3 currents but have no effect on the sustained component (24, 42, 43), whereas salicylic acid (IC50 260 μm) and diclofenac (IC50 92 μm) inhibit the sustained, but not the transient, component of ASIC3 current (44). Sevanol, an analgesic compound from thyme, inhibits both components of ASIC3 current but also ASIC1a (45).
Similar to sevanol, Ugr 9-1 reversibly and completely blocks the ASIC3 transient component with an IC50 = 9 μm and, with an IC50 = 1.5 μm, can reversibly inhibit ∼50% of the amplitude of the ASIC3 sustained component. Because the effective concentrations and Hill coefficients for inhibition of the transient and sustained components are different, we can suggest that distinct mechanisms of inhibition are involved for each component. Therefore, the mode of action of Ugr 9-1 on the ASIC3 receptor is absolutely new.
Ugr 9-1 produced an inhibition effect in vitro with IC50 less than that of small organic molecules but higher than that of APETx2. In vivo studies have shown that the administration of Ugr 9-1 reverses acid-induced and CFA-induced inflammatory pain as much as APETx2, which attenuates acid-induced and inflammatory pain at doses of 0.01 to 0.5 mg/kg in rats (7, 46).
Doubtless, an advantage of Ugr 9-1 over APETx2 for pain relief is its short length, which reduces the resources that must be consumed for production by chemical or biotechnological synthesis. The folding problem for recombinant APETx2 is also quite acute (47, 48). In our experiments, we did not have any problems with recombinant peptide production and folding, most probably because of small size and low number of disulfide bonds.
We should conclude that π-AnmTX Ugr 9a-1 is a unique, potentially natural compound possessing a new spatial structure fold, exercising an inhibitory activity on the ASIC3 channel, and producing an analgesic effect in animal models of pain. Moreover, π-AnmTX Ugr 9a-1 can serve as a research tool to determine the physiological involvement of ASIC channels in neuronal excitability and pain perception and transmission.
Acknowledgments
We are grateful to Ts. A. Egorov and A. Kh. Musolyamov (Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, the Russian Academy of Sciences) for their help in amino acid sequencing, Sylvie Diochot (Institutde Pharmacologie Moleculaire et Cellulaire, Valbonne, France) for PCi plasmids containing cDNA of rat ASIC1a, ASIC1b, and ASIC2a, and Antonina Berkut (Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, the Russian Academy of Sciences), Steve Peigneur, and Jan Tytgat (University of Leuven, Belgium) for electrophysiological experiments on KV1.3.
This work was supported by the Programs of the Russian Academy of Sciences “Molecular and Cell Biology” and “Fundamental Sciences for Medicine,” the Federal Program of The Ministry of Education and Science of the Russian Federation (Agreement 8063), and Russian Foundation for Basic Research Grants 12-04-33151, 12-04-90442, and 12-04-01068.
The atomic coordinates and structure factors (code 2LZO) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The nucleotide sequence reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number HF562346.
- ASIC3
- acid-sensing ion channel type 3
- CFA
- complete Freund's adjuvant
- COSY
- correlation spectroscopy
- MALDI
- matrix-assisted laser desorption/ionization
- RACE
- rapid amplification of cDNA ends
- TOCSY
- total correlation spectroscopy
- TRPV1
- transient receptor potential vanilloid type 1
- PDB
- Protein Data Bank.
REFERENCES
- 1. Sanamyan N., Sanamyan K. (2006) The genera Urticina and Cribrinopsis (Anthozoa: Actiniaria) from the northwestern Pacific. J. Nat. Hist. 40, 359–393 [Google Scholar]
- 2. Carli A., Bussotti S., Mariottini G. L., Robbiano L. (1996) Toxicity of jellyfish and sea-anemone venoms on cultured V79 cells. Toxicon. 34, 496–500 [DOI] [PubMed] [Google Scholar]
- 3. Marino A., Valveri V., Muià C., Crupi R., Rizzo G., Musci G., La Spada G. (2004) Cytotoxicity of the nematocyst venom from the sea anemone Aiptasia mutabilis. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 139, 295–301 [DOI] [PubMed] [Google Scholar]
- 4. Shiomi K. (2009) Novel peptide toxins recently isolated from sea anemones. Toxicon. 54, 1112–1118 [DOI] [PubMed] [Google Scholar]
- 5. Castañeda O., Harvey A. L. (2009) Discovery and characterization of cnidarian peptide toxins that affect neuronal potassium ion channels. Toxicon 54, 1119–1124 [DOI] [PubMed] [Google Scholar]
- 6. Bosmans F., Tytgat J. (2007) Sea anemone venom as a source of insecticidal peptides acting on voltage-gated Na+ channels. Toxicon 49, 550–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deval E., Noël J., Lay N., Alloui A., Diochot S., Friend V., Jodar M., Lazdunski M., Lingueglia E. (2008) ASIC3, a sensor of acidic and primary inflammatory pain. EMBO J. 27, 3047–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andreev Y. A., Kozlov S. A., Kozlovskaya E. P., Grishin E. V. (2009) Analgesic effect of a polypeptide inhibitor of the TRPV1 receptor in noxious heat pain models. Dokl. Biochem. Biophys. 424, 46–48 [DOI] [PubMed] [Google Scholar]
- 9. Andreev Y. A., Kozlov S. A., Koshelev S. G., Ivanova E. A., Monastyrnaya M. M., Kozlovskaya E. P., Grishin E. V. (2008) Analgesic compound from sea anemone Heteractis crispa is the first polypeptide inhibitor of vanilloid receptor 1 (TRPV1). J. Biol. Chem. 283, 23914–23921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kellenberger S., Schild L. (2002) Epithelial sodium channel/degenerin family of ion channels. A variety of functions for a shared structure. Physiol. Rev. 82, 735–767 [DOI] [PubMed] [Google Scholar]
- 11. Molliver D. C., Immke D. C., Fierro L., Paré M., Rice F. L., McCleskey E. W. (2005) ASIC3, an acid-sensing ion channel, is expressed in metaboreceptive sensory neurons. Mol. Pain 1, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Salinas M., Lazdunski M., Lingueglia E. (2009) Structural elements for the generation of sustained currents by the acid pain sensor ASIC3. J. Biol. Chem. 284, 31851–31859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yen Y. T., Tu P. H., Chen C. J., Lin Y. W., Hsieh S. T., Chen C. C. (2009) Role of acid-sensing ion channel 3 in sub-acute-phase inflammation. Mol. Pain 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deval E., Gasull X., Noël J., Salinas M., Baron A., Diochot S., Lingueglia E. (2010) Acid-sensing ion channels (ASICs). Pharmacology and implication in pain. Pharmacol. Ther. 128, 549–558 [DOI] [PubMed] [Google Scholar]
- 15. Andreev Y. A., Kozlov S. A., Vassilevski A. A., Grishin E. V. (2010) Cyanogen bromide cleavage of proteins in salt and buffer solutions. Anal. Biochem. 407, 144–146 [DOI] [PubMed] [Google Scholar]
- 16. Wuthrich K. (1986) NMR of Proteins and Nucleic acids, John Wiley and Sons, New York [Google Scholar]
- 17. Kozlov S., Malyavka A., McCutchen B., Lu A., Schepers E., Herrmann R., Grishin E. (2005) A novel strategy for the identification of toxinlike structures in spider venom. Proteins 59, 131–140 [DOI] [PubMed] [Google Scholar]
- 18. Delaglio F., Wu Z., Bax A. (2001) Measurement of homonuclear proton couplings from regular 2D COSY spectra. J. Magn. Reson. 149, 276–281 [DOI] [PubMed] [Google Scholar]
- 19. Kozlov S., Grishin E. (2005) Classification of spider neurotoxins using structural motifs by primary structure features. Single residue distribution analysis and pattern analysis techniques. Toxicon 46, 672–686 [DOI] [PubMed] [Google Scholar]
- 20. Kozlov S., Grishin E. (2011) The mining of toxin-like polypeptides from EST database by single residue distribution analysis. BMC Genomics 12, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST. A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frishman D., Argos P. (1995) Knowledge-based protein secondary structure assignment. Proteins 23, 566–579 [DOI] [PubMed] [Google Scholar]
- 23. Kozlov S. A., Grishin E. V. (2007) The universal algorithm of maturation for secretory and excretory protein precursors. Toxicon 49, 721–726 [DOI] [PubMed] [Google Scholar]
- 24. Diochot S., Baron A., Rash L. D., Deval E., Escoubas P., Scarzello S., Salinas M., Lazdunski M. (2004) A new sea anemone peptide, APETx2, inhibits ASIC3, a major acid-sensitive channel in sensory neurons. EMBO J. 23, 1516–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kozlov S., Grishin E. (2012) Convenient nomenclature of cysteine-rich polypeptide toxins from sea anemones. Peptides 33, 240–244 [DOI] [PubMed] [Google Scholar]
- 26. Honma T., Kawahata S., Ishida M., Nagai H., Nagashima Y., Shiomi K. (2008) Novel peptide toxins from the sea anemone Stichodactyla haddoni. Peptides 29, 536–544 [DOI] [PubMed] [Google Scholar]
- 27. Zaharenko A. J., Ferreira W. A., Jr., Oliveira J. S., Richardson M., Pimenta D. C., Konno K., Portaro F. C., de Freitas J. C. (2008) Proteomics of the neurotoxic fraction from the sea anemone Bunodosoma cangicum venom. Novel peptides belonging to new classes of toxins. Comp. Biochem. Physiol. Part D Genomics Proteomics 3, 219–225 [DOI] [PubMed] [Google Scholar]
- 28. Honma T., Hasegawa Y., Ishida M., Nagai H., Nagashima Y., Shiomi K. (2005) Isolation and molecular cloning of novel peptide toxins from the sea anemone Antheopsis maculata. Toxicon 45, 33–41 [DOI] [PubMed] [Google Scholar]
- 29. Andreev Y. A., Vassilevski A. A., Kozlov S. A. (2012) Molecules to selectively target receptors for treatment of pain and neurogenic inflammation. Recent Pat. Inflamm Allergy Drug Discov. 6, 35–45 [DOI] [PubMed] [Google Scholar]
- 30. Corzo G., Bernard C., Clement H., Villegas E., Bosmans F., Tytgat J., Possani L. D., Darbon H., Alagón A. (2009) Insecticidal peptides from the theraposid spider Brachypelma albiceps. An NMR-based model of Ba2. Biochim. Biophys. Acta 1794, 1190–1196 [DOI] [PubMed] [Google Scholar]
- 31. Grishin E. V., Savchenko G. A., Vassilevski A. A., Korolkova Y. V., Boychuk Y. A., Viatchenko-Karpinski V. Y., Nadezhdin K. D., Arseniev A. S., Pluzhnikov K. A., Kulyk V. B., Voitenko N. V., Krishtal O. O. (2010) Novel peptide from spider venom inhibits P2X3 receptors and inflammatory pain. Ann. Neurol. 67, 680–683 [DOI] [PubMed] [Google Scholar]
- 32. Lovelace E. S., Armishaw C. J., Colgrave M. L., Wahlstrom M. E., Alewood P. F., Daly N. L., Craik D. J. (2006) Cyclic MrIA. A stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter. J. Med. Chem. 49, 6561–6568 [DOI] [PubMed] [Google Scholar]
- 33. Fahrner R. L., Dieckmann T., Harwig S. S., Lehrer R. I., Eisenberg D., Feigon J. (1996) Solution structure of protegrin-1, a broad-spectrum antimicrobial peptide from porcine leukocytes. Chem. Biol. 3, 543–550 [DOI] [PubMed] [Google Scholar]
- 34. Hwang P. M., Zhou N., Shan X., Arrowsmith C. H., Vogel H. J. (1998) Three-dimensional solution structure of lactoferricin B, an antimicrobial peptide derived from bovine lactoferrin. Biochemistry 37, 4288–4298 [DOI] [PubMed] [Google Scholar]
- 35. Umetsu Y., Aizawa T., Muto K., Yamamoto H., Kamiya M., Kumaki Y., Mizuguchi M., Demura M., Hayakawa Y., Kawano K. (2009) C-terminal elongation of growth-blocking peptide enhances its biological activity and micelle binding affinity. J. Biol. Chem. 284, 29625–29634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shenkarev Z. O., Balandin S. V., Trunov K. I., Paramonov A. S., Sukhanov S. V., Barsukov L. I., Arseniev A. S., Ovchinnikova T. V. (2011) Molecular mechanism of action of β-hairpin antimicrobial peptide arenicin. Oligomeric structure in dodecylphosphocholine micelles and pore formation in planar lipid bilayers. Biochemistry 50, 6255–6265 [DOI] [PubMed] [Google Scholar]
- 37. Vassilevski A. A., Kozlov S. A., Grishin E. V. (2008) Antimicrobial peptide precursor structures suggest effective production strategies. Recent Pat. Inflamm. Allergy Drug Discov. 2, 58–63 [DOI] [PubMed] [Google Scholar]
- 38. Lai R., Zheng Y. T., Shen J. H., Liu G. J., Liu H., Lee W. H., Tang S. Z., Zhang Y. (2002) Antimicrobial peptides from skin secretions of Chinese red belly toad Bombina maxima. Peptides 23, 427–435 [DOI] [PubMed] [Google Scholar]
- 39. Kozlov S. A., Vassilevski A. A., Feofanov A. V., Surovoy A. Y., Karpunin D. V., Grishin E. V. (2006) Latarcins, antimicrobial and cytolytic peptides from the venom of the spider Lachesana tarabaevi (Zodariidae) that exemplify biomolecular diversity. J. Biol. Chem. 281, 20983–20992 [DOI] [PubMed] [Google Scholar]
- 40. Terry A. S., Poulter L., Williams D. H., Nutkins J. C., Giovannini M. G., Moore C. H., Gibson B. W. (1988) The cDNA sequence coding for prepro-PGS (prepro-magainins) and aspects of the processing of this prepro-polypeptide. J. Biol. Chem. 263, 5745–5751 [PubMed] [Google Scholar]
- 41. Shlyapnikov Y. M., Andreev Y. A., Kozlov S. A., Vassilevski A. A., Grishin E. V. (2008) Bacterial production of latarcin 2a, a potent antimicrobial peptide from spider venom. Protein Expr. Purif. 60, 89–95 [DOI] [PubMed] [Google Scholar]
- 42. Escoubas P., De Weille J. R., Lecoq A., Diochot S., Waldmann R., Champigny G., Moinier D., Ménez A., Lazdunski M. (2000) Isolation of a tarantula toxin specific for a class of proton-gated Na+ channels. J. Biol. Chem. 275, 25116–25121 [DOI] [PubMed] [Google Scholar]
- 43. Kozlov S. A., Osmakov D. I., Andreev Y. A., Koshelev S. G., Gladkikh I. N., Monastyrnaya M. M., Kozlovskaya E. P., Grishin E. V. (2012) A sea anemone polypeptide toxin inhibiting the ASIC3 acid-sensitive channel. Bioorg. Khim. 38, 578–583 [DOI] [PubMed] [Google Scholar]
- 44. Voilley N., de Weille J., Mamet J., Lazdunski M. (2001) Nonsteroid anti-inflammatory drugs inhibit both the activity and the inflammation-induced expression of acid-sensing ion channels in nociceptors. J. Neurosci. 21, 8026–8033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dubinnyi M. A., Osmakov D. I., Koshelev S. G., Kozlov S. A., Andreev Y. A., Zakaryan N. A., Dyachenko I. A., Bondarenko D. A., Arseniev A. S., Grishin E. V. (2012) Lignan from thyme possesses inhibitory effect on ASIC3 channel current. J. Biol. Chem. 287, 32993–33000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karczewski J., Spencer R. H., Garsky V. M., Liang A., Leitl M. D., Cato M. J., Cook S. P., Kane S., Urban M. O. (2010) Reversal of acid-induced and inflammatory pain by the selective ASIC3 inhibitor, APETx2. Br. J. Pharmacol. 161, 950–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Anangi R., Rash L. D., Mobli M., King G. F. (2012) Functional expression in Escherichia coli of the disulfide-rich sea anemone peptide APETx2, a potent blocker of acid-sensing ion channel 3. Mar. Drugs 10, 1605–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vetter I., Davis J. L., Rash L. D., Anangi R., Mobli M., Alewood P. F., Lewis R. J., King G. F. (2011) Venomics. A new paradigm for natural products-based drug discovery. Amino Acids 40, 15–28 [DOI] [PubMed] [Google Scholar]
- 49. Efremov R. G., Vergoten G. (1995) Hydrophobic nature of membrane-spanning α-helical peptides as revealed by Monte Carlo simulations and molecular hydrophobicity potential analysis. J. Phys. Chem. 99, 10658–10666 [Google Scholar]