Background: Ca2+ binding to the regulatory two-domain tandem (CBD12) activates ion transport in NCX.
Results: Ca2+ binding to high affinity sites of CBD12 results in a population shift, where more constraint conformational states become highly populated.
Conclusion: Ca2+-induced population shift governs NCX activation with no significant contribution of global conformational changes in CBD alignment.
Significance: Population shift may represent a general mechanism for regulating NCX.
Keywords: Allosteric Regulation, Calcium, Calcium-binding Proteins, Protein Conformation, Sodium-Calcium Exchange, Population Shift
Abstract
In eukaryotic Na+/Ca2+ exchangers (NCX) the Ca2+ binding CBD1 and CBD2 domains form a two-domain regulatory tandem (CBD12). An allosteric Ca2+ sensor (Ca3–Ca4 sites) is located on CBD1, whereas CBD2 contains a splice-variant segment. Recently, a Ca2+-driven interdomain switch has been described, albeit how it couples Ca2+ binding with signal propagation remains unclear. To resolve the dynamic features of Ca2+-induced conformational transitions we analyze here distinct splice variants and mutants of isolated CBD12 at varying temperatures by using small angle x-ray scattering (SAXS) and equilibrium 45Ca2+ binding assays. The ensemble optimization method SAXS analysis demonstrates that the apo and Mg2+-bound forms of CBD12 are highly flexible, whereas Ca2+ binding to the Ca3–Ca4 sites results in a population shift of conformational landscape to more rigidified states. Population shift occurs even under conditions in which no effect of Ca2+ is observed on the globally derived Dmax (maximal interatomic distance), although under comparable conditions a normal [Ca2+]-dependent allosteric regulation occurs. Low affinity sites (Ca1–Ca2) of CBD1 do not contribute to Ca2+-induced population shift, but the occupancy of these sites by 1 mm Mg2+ shifts the Ca2+ affinity (Kd) at the neighboring Ca3–Ca4 sites from ∼ 50 nm to ∼ 200 nm and thus, keeps the primary Ca2+ sensor (Ca3–Ca4 sites) within a physiological range. Thus, Ca2+ binding to the Ca3–Ca4 sites results in a population shift, where more constraint conformational states become highly populated at dynamic equilibrium in the absence of global conformational transitions in CBD alignment.
Introduction
Mammalian Na+/Ca2+ exchanger (NCX1–3)4 proteins extrude Ca2+, an essential signaling molecule, from the cell. Their activity is strongly and allosterically regulated by the interaction of Ca2+ with regulatory sites on the exchanger (1, 2). For example, in cardiomyocytes, NCX-mediated ion currents grow ∼25-fold when the [Ca2+]i rises from 0.1 to 2 μm during the action potential (3). The Ca2+ regulatory sites map to Ca2+ binding domains (CBD) CBD1 and CBD2, which form a tandem, CBD12. Four (Ca1–Ca4) and two (CaI and CaII) Ca2+ binding sites are located on CBD1 and CBD2, respectively (4–6). The two high affinity (Kd < 0.3 μm) sites of CBD1 (Ca3 and Ca4) serve as the primary Ca2+ sensor (7). A medium affinity site (CaI with a Kd of 2–10 μm) enables sustained NCX activation while also relieving Na+-dependent inactivation (5, 7, 8). The site CaII of CBD2 serves as the Mg2+ binding site (9, 10), whereas the role of low affinity (Kd > 10 μm) sites of CBD1 (Ca1 and Ca2) remains unclear (7). NCX proteins are expressed in a tissue-specific manner and contain an alternative splicing segment on CBD2, arising from a combination of six small exons, A, B, C, D, E, and F (1, 11). In isolated CBD12 and intact NCX, CBD2 interacts with CBD1 (8, 12, 13), thereby modifying the equilibrium binding and kinetic properties of the primary allosteric sensor (Ca3–Ca4) in a splice-variant-dependent manner (14). Thus, Ca2+, the exchanger substrate, allosterically regulates the exchanger in a feedback manner.
Structure-function relationships of allosteric regulation have been studied extensively in enzymes, although the underlying dynamic mechanisms of ligand-induced coupling remain unclear. The relevant mechanisms are even less obvious in ion pumps, channels, and transporters. The “induced fit” and “conformational selection” (population shift) hypotheses were proposed as principal mechanisms underlying ligand-induced dynamic coupling (15–17). However, induced fit is often mistakenly assigned simply because the conformational selection and population shift are not observed (15–17). This is because of time scales: induced fit is faster than conformational selection and population shift, which needs to overcome a barrier (a higher barrier leading to a slower population shift).
Recently, we discovered an interdomain Ca2+-driven “switch” in NCX proteins that couples the two regulatory domains upon Ca2+ binding to the allosteric sensor (18). To resolve the dynamic mechanisms of Ca2+-induced transitions in the two-domain regulatory tandem of NCX, we applied here EOM (ensemble optimization method) analyses to SAXS (small angle x-ray scattering) data (19, 20).
Different experimental approaches, used with isolated CBD12 and intact NCX, have shown that a short interdomain CBD1-CBD2 linker plays a critical role in Ca2+-driven coupling of CBDs (12, 13). The linker encodes specific information that governs CBD interactions (21). Recently, we identified a Ca2+-driven regulatory switch at the interface between the two CBDs. Ca2+ binding is associated with CBD tethering and slow dissociation of “occluded” Ca2+ from the Ca3–Ca4 sites (18). The structural properties of this interdomain module are very similar (if not identical) among diverse NCX orthologs and splice variants, exhibiting either positive, negative or no response to regulatory Ca2+ (18, 22). A slow dissociation of occluded Ca2+ from regulatory sites may represent a physiologically relevant slow inactivation of NCX variants (I2 state), observed in patch clamp experiments (12, 23).
Ca2+-induced conformational transitions in isolated CBD12 have been studied using NMR (24), FRET (23), and SAXS (8, 18) methods, but the results from the differing approaches do not agree. Whereas a large Ca2+-induced compaction (manifested as ∼25 Å decrease in Dmax) was observed in a standard SAXS analysis (8, 18), a small decrease in FRET (23) was observed upon Ca2+ binding, indicating a Ca2+-dependent increase of the interdomain distance. NMR studies reported a similarly extended shape for both the apo and bound states of CBD12, whereas Ca2+ binding resulted in rigidified motions of CBDs (24). Moreover, x-ray crystallography revealed a nearly identical interdomain angle in the Ca2+-bound CBD12-NCX1-AD (18) and CBD12-CALX1.1 (22), meaning that the phenotype differences in the Ca2+-dependent activation or inhibition, exhibited by these diverse orthologs, cannot be explained by interdomain angle or distance (18).
To this end, we sought to clarify the disparate results regarding the CBD12 conformational change. Subsequently, the next central question is which principal mechanism is responsible for ligand-induced regulatory coupling, i.e. conformational change in NCX, including understanding the role of low affinity Ca1–Ca2 sites and Mg2+-dependent modulation. One possibility is a Ca2+-induced shift in Dmax representing global conformational transitions. Alternatively, a Ca2+-driven population shift in conformational landscape manifested as rigidification may be an explanation. To address these questions, we quantitatively evaluated the equilibrium conformations of the brain (CBD12-AD) and kidney (CBD12-BD) splice variants, as well as the E454K mutant (CBD12-AD-E454K) in the presence and absence of Ca2+ by using EOM SAXS analysis and equilibrium 45Ca2+ binding assays. The only common sites among the constructs are the high affinity Ca3–Ca4 sites of CBD1 (8, 14, 18, 25). Collectively, our data indicate that Ca2+ binding to the Ca3–Ca4 sites results in domain rigidification through the population shift mechanism.
EXPERIMENTAL PROCEDURES
Expression, Mutagenesis, and Purification of CBD Proteins
The DNA constructs of CBD1-WT, CBD1-E454K, CBD12-AD, CBD12-AD-E454K, and CBD12-BD of canine NCX1 and CBD12–1.1 and CBD12–1.2 of CALX were expressed in Escherichia coli Rosetta2 (DE3) competent cells (Novagen), as described (10, 14, 18). Overexpressed proteins were purified on a Ni2+-nitrilotriacetic acid column followed by size exclusion chromatography on either Superdex-75 (CBD1 constructs) or Superdex-200 (CBD12 constructs) (>95% purity, judged by SDS-PAGE) (18).
Equilibrium 45Ca2+ Binding Assay
The equilibrium binding of 45Ca2+ to proteins was measured as the protein-bound radioactivity retained after ultrafiltration (10, 14). The assay medium (1.5 ml) containing 7–15 μm protein with 100 mm KCl, 10 mm Tris-HCl at pH 7.2 (TK buffer) was placed in the upper chamber of the Ultracel-3k concentrator, and the assay was performed as outlined previously (10, 14). The [Ca2+]free fraction was measured as a/b, where a represents the radioactivity of the ultrafiltate and b represents the radioactivity in the upper chamber; thus [Ca2+]free = [Ca2+]tot·(a/b). Bound Ca2+ is calculated as [Ca2+-CBD] = [Ca2+]tot − [Ca2+]free and binding stoichiometry as [Ca2+-CBD]/[CBD]. The 45Ca2+ titration curves of CBD12 were fit to Adair equations with a predefined number of binding sites according to the observed maximal binding capacity using Origin 7.0 software (OriginLab Corporation) (14). The 45Ca2+ titration curves of CBD1-WT and CBD1-E454K were fit to the Hill equation. [Ca2+]residual of decalcified buffer was measured by fluo-3 (10). All other procedures were described elsewhere (10).
Schild Plot Analysis
Log molar Mg2+ concentrations (x axis) were plotted against log (Kd ratio − 1) (y axis). Kd ratio represents the ratio between the apparent CBD1 Kd for Ca2+ in the absence and presence of each Mg2+ concentration. The Kd for Mg2+ (KMg) is obtained from the x axis intercept.
SAXS Data Collection and Analysis
SAXS data were measured at beamline X33 of the Deutsches Elektronen-Synchrotron, Hamburg, Germany, and at beamline BM29 of the European Synchrotron Radiation Facility, Grenoble, France. At beamline X33, data were collected with x-ray beam at wavelength λ = 1.5 Å, and the distance from the sample to detector (PILATUS 1M; Dectris Ltd.) was 2.7 m, covering a scattering vector range (q = 4πsinθ/λ) from 0.006 to 0.6 Å−1. Eight frames of two-dimensional images were recorded for each buffer or sample, with an exposure time of 15 s/frame. At beamline BM29, data were collected with x-ray beam at wavelength λ = 1.0 Å, and the distance from the sample to detector (PILATUS 1M) was 2.85 m, covering a scattering vector range (q = 4πsinθ/λ) from 0.004 to 0.45 Å−1. 10 frames of two-dimensional images were recorded for each buffer or sample, with an exposure time of 2 s/frame. The two-dimensional images were reduced to one-dimensional scattering profiles, and the scattering of the buffer was subtracted from the sample profile using the software on site.
The following samples were used for the experiments: Wild-type CBD12-AD, its mutant CBD12-AD-E454K, wild-type CBD12-BD, CALX-CBD12–1.1, and CALX-CBD12–1.2. The buffers contained 100 mm KCl, 20 mm β-mercaptoethanol, 20 mm Tris-HCl at pH 7.2 and either 10 mm CaCl2, 10 mm EDTA, or 1.1 mm MgCl2 + 1 mm EGTA. To account for possible interparticle effects, each sample was measured at three concentrations: ∼10, ∼5, and ∼2 mg/ml. In the presence of such effects, the lowest concentration curve was merged with a higher concentration curve at q ∼ 0.2 Å−1 to prevent distortion of the low angle data while preserving high signal-to-noise ratio at the higher angles, which are far less sensitive to interparticle effects (20). The experimental radius of gyration (Rg) and the forward scattering intensity I(0) were calculated from data at low q values in the range of qRg < 1.3, using the Guinier approximation: lnI(q) ≈ ln(I(0)) − Rg2q2/3.
EOM Analysis
Loops missing from the crystal structure of CBD12-E454K (PDB 3US9) were modeled using the I-TASSER server (26). The crystal structure of CALX-CBD12–1.1 (PDB 3RB5) was used for the CALX data. Unstructured residues at the N and C termini of CBD1 and CBD2 were truncated. The software RANCH (27) was used to generate a pool of 10,000 stereochemically feasible structures with a random CBD1-CBD2 linker and termini. These pools were input into GAJOE (27), which selects an ensemble with the best fit to the experimental data using a genetic algorithm: 50 ensembles of 20 orientations each were “crossed” and “mutated” for 1000 generations, and the process was repeated 50 times.
RESULTS
Global SAXS Parameters of NCX-CBD12 Constructs
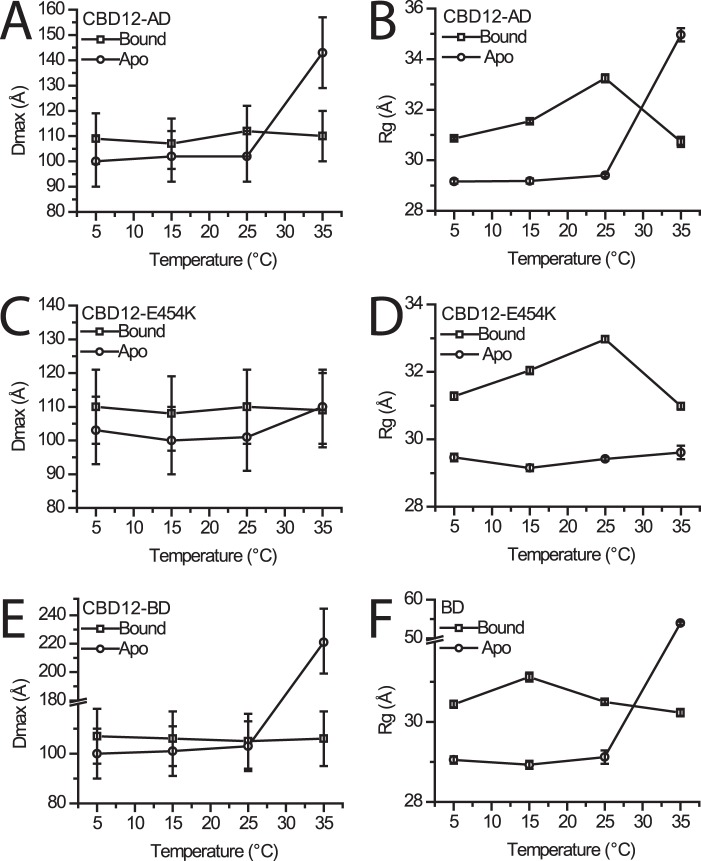
While measuring SAXS of previously characterized samples (8, 18), we observed varying results as a function of temperature. This observation led us to perform a systematic temperature study on the various protein constructs mentioned above. The results can be used to fully reconcile the disparate conclusions about CBD12 conformational changes, described above, and address the mechanism of allosteric regulation. Although the estimated errors overlap, a small but consistent increase in Dmax upon Ca2+ binding was detected between 5 and 25 °C in all constructs (Fig. 1, A, C, and E). The results for 5 °C were reproducible at two different beamlines, using different protein preparations, and are in line with previous FRET results indicating a slightly more compact apo conformation (23). We were able to recapitulate previous SAXS findings for CBD12-AD when measuring at 35 °C (Fig. 1A). In the case of CBD12-BD, higher order oligomers were formed at 35 °C as judged by dramatic increases in Dmax and Rg (Fig. 1, E and F). The mutant CBD12-E454K, whose crystal structure we reported (18), showed no notable temperature dependence, with nearly identical Dmax values also at 35 °C (Fig. 1, C and D). Further analysis was then pursued only for data collected at 5 °C, where oligomers are not formed. Also, these findings then largely represent the range between 5 and 25 °C. The FRET and NMR data, also measured in the range of 5–25 °C, are consistent with our findings. Moreover, allosteric regulation was studied under the same conditions (23, 24). Hence, our new SAXS data described here are physiologically relevant. We conclude that Ca2+ binding induces a slight extension of CBD12.
FIGURE 1.
Temperature dependence of SAXS-derived parameters. A, C, and E, observed Dmax values ± 10% (estimated range) for CBD12-AD, CBD12-AD-E454K, and CBD12-BD are shown, respectively, in the Ca2+-bound and -free forms. B, D, and F, observed Rg values ± S.E. for CBD12-AD, CBD12-AD-E454K, and CBD12-BD are shown, respectively, in the Ca2+-bound and -free forms.
The Rg and I(0) parameters were derived from the Guinier region (qRg <1.3) of each scattering profile using the PRIMUS software (28). The difference in Rg values between the Ca2+-bound and -free samples indicates that observed conformational transitions occur upon Ca2+ binding, despite the relatively small changes in Dmax at 5–25 °C. Porod volumes were derived from the paired-distance distribution functions and are consistent with that predicted for monomeric CBD12 for all samples, in the presence and absence of Ca2+ or Mg2+, eliminating the possibility that the observed differences among samples arise from oligomerization. The overall SAXS parameters at 5 °C are summarized in Table 1.
TABLE 1.
Data collection and scattering-derived parameters
1 Eight frames were measured for each sample.
2 ± S.E.
3 ± 10% (estimated range).
EOM Analysis of NCX-CBD12 SAXS Data
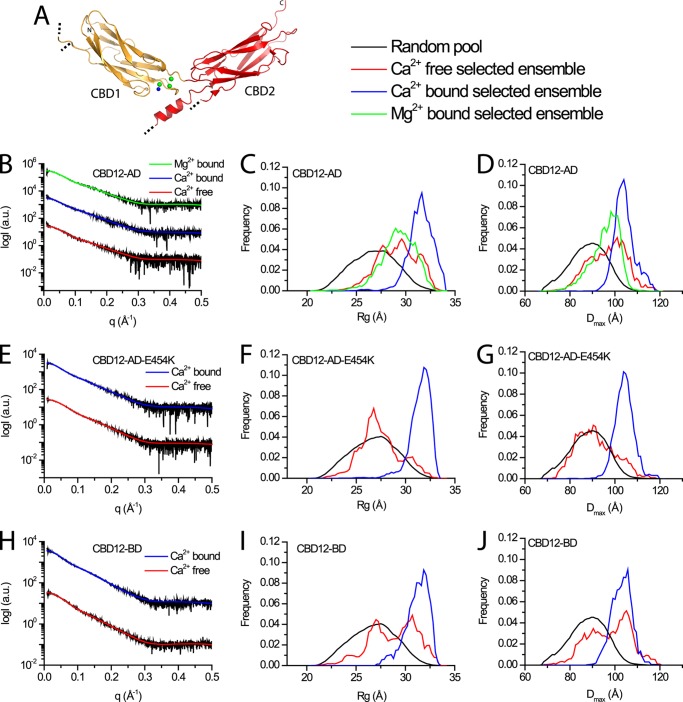
We then asked whether the principal mechanism of conformation change is induced or population shift. To test the possibility that the apo and bound forms are not distinct conformations but represent different conformational distributions, we used EOM to analyze the SAXS data. The selected ensembles fit the experimental data very well, as can be assessed from the low χ2 values (Table 1 and Fig. 2, B, E, and H). In all examined constructs, wide distributions of both Rg (Fig. 2, C, F, and I) and Dmax (Fig. 2, D, G, and J) are observed within the selected ensembles describing the apo form, implying a high degree of flexibility (27). Similar broad distributions are observed for CBD12-AD in the presence of Mg2+ (Fig. 2C). In fact, these wide distributions span almost the entire range of possible orientations in the random pool. However, the distributions are much narrower in the presence of Ca2+ (Fig. 2, C, D, F, G, I, and J) due to a bias toward a subset of conformations also present in the apo form distributions. These data support the notion that not only Ca2+ binding rigidifies the tandem CBD12 from a dynamic perspective (24), it also constrains the conformational fluctuation range, inducing a population shift.
FIGURE 2.
EOM analysis of NCX-CBD12 proteins. A, crystal structure of CBD12-E454K (PDB 3US9). CBD1 and CBD2 are colored orange and red, respectively. Green and blue spheres depict Ca2+ ions and water molecules, respectively. Dotted black lines denote chain breaks in the model due to poor electron density. B, E, and H, experimental SAXS curves and EOM fits for CBD12-AD, CBD12-AD-E454K, and CBD12-BD, respectively. C, F, and I, random Rg pools and EOM-selected ensemble distributions for CBD12-AD, CBD12-AD-E454K, and CBD12-BD, respectively. D, G, and J, random Dmax pools and EOM-selected ensemble distributions for CBD12-AD, CBD12-AD-E454K, and CBD12-BD, respectively.
EOM Analysis of CALX Splice Variants
CALX, the Drosophila NCX ortholog, has two splice variants. CALX1.1 is regulated negatively by Ca2+ whereas CALX1.2 is not Ca2+ regulated (22). We performed EOM analyses of the Ca2+-bound CBD12 variants of CALX to assess whether their conformations differ in solution (Fig. 3) in line with their physiological regulation. The analyses demonstrate that both variants span a relatively similar narrow conformation distribution compared with the random pool (Fig. 3, C and D). These distributions are also similar to those observed for NCX-CBD12 preparations (Figs. 2 and 3). However, a clear shoulder is observed for CALX-CBD12–1.2 (Fig. 3, C and D) in the narrower Rg and Dmax values, suggesting that it is not as rigid as the other proteins. In general, the CALX proteins show properties similar to those of NCX, meaning that the Ca2+-induced population shift, with more rigid conformational states, might be a common mechanism for both the NCX and CALX forms.
FIGURE 3.
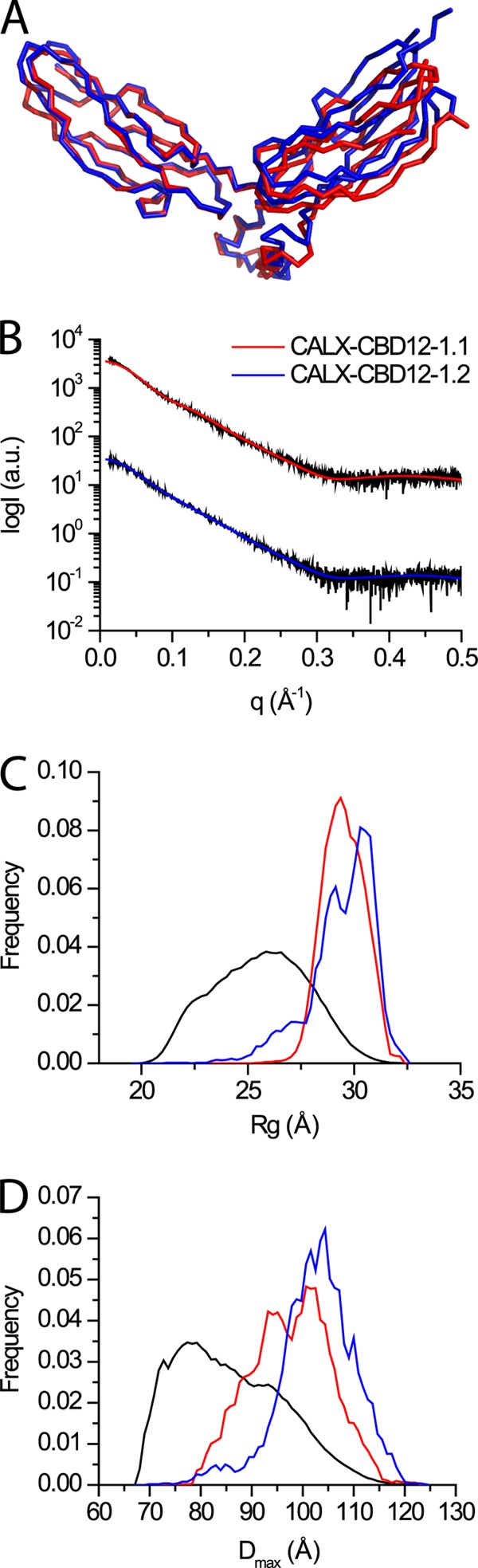
EOM analysis of CALX-CBD12 proteins. A, superposition of Cα representation of CBD12–1.1 (red, PDB 3RB5) and CBD12–1.2 (blue, PDB 3RB7) from CALX. B, respective experimental SAXS curves and EOM fits. C, respective random Rg pool and EOM-selected ensemble distributions. D, respective random Dmax pool and EOM-selected ensemble distributions.
Mg2+ Binds to Low Affinity Sites and Modulates High Affinity Sites
We have shown that Mg2+ did not induce conformational changes, in contrast to Ca2+ (Fig. 2C). However, Mg2+ at physiological concentrations is known to bind to both CBD1 and CBD2, modulating Ca2+ binding properties (9, 10). To map the Mg2+ effect, we measured the Mg2+ modulation of Ca2+ binding to WT and E454K mutants of CBD1 and CBD12 protein forms (Fig. 4). Measurement of the single versus tandem domains affords assignment of the effected binding sites. In addition, the E454K mutant has no Ca1 site and an altered Ca2 site but intact Ca3–Ca4 sites, further facilitating the assignment (18, 25).
FIGURE 4.
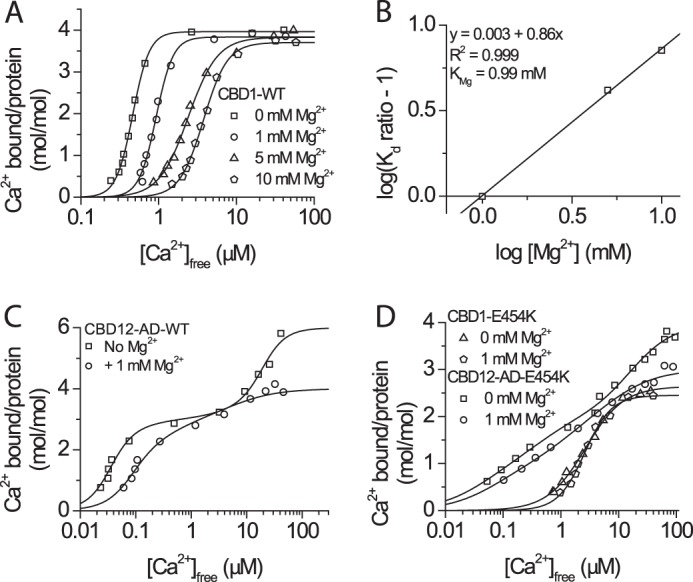
Effect of Mg2+ interaction with isolated CBD1 and CBD12 proteins. A, 45Ca2+ titration curves of CBD1 in the presence of 0, 1, 5, and 10 mm MgCl2, as indicated. B, Schild plot analysis of the CBD1 KMg (Kd for Mg2+). C, 45Ca2+ titration curves of CBD12-AD in the absence and presence of 1 mm MgCl2 showing agreement with the effects observed for CBD1. D, 45Ca2+ titration curves of CBD1-E454K and CBD12-E454K in the absence and presence of 1 mm MgCl2, as indicated.
In the presence of 1, 5, or 10 mm Mg2+, the maximal binding capacity of CBD1-WT is not altered, whereas the Kd for Ca2+ is increased with increasing [Mg2+], consistent with competitive inhibition (Fig. 4A). The Kd for Mg2+ (KMg), calculated using a Schild plot (Fig. 4B), is 0.99 mm, a physiological Mg2+ concentration. In the absence of Mg2+, CBD12-AD binds six Ca2+ ions, four in CBD1 and two in CBD2. In contrast, with Mg2+, the maximal binding capacity of CBD12-AD drops to four Ca2+ ions/protein (Fig. 4C) in addition to a ∼5-fold decrease in the affinity of Ca3–Ca4 sites. One Mg2+ ion binds to CBD2 (9), and one may bind to CBD1 even at saturating [Ca2+].
Because the reduced affinity may reflect competition of Mg2+ with Ca2+ for all Ca1–Ca4 sites, we tested the effect of Mg2+ on the E454K mutant (Fig. 4D). In contrast to CBD1-WT, the presence of 1 mm Mg2+ has no noticeable effect on CBD1-E454K (Fig. 4D). In contrast to CBD12-AD, Mg2+ does not alter the affinity of the Ca3–Ca4 sites in CBD12-AD-E454K, again demonstrating that Mg2+ and Ca2+ do not directly compete over the high affinity sites. The affinity of CBD12-AD-E454K in the absence of Mg2+ is ∼5-fold lower than that of CBD12-AD-WT, mimicking the effect of 1 mm Mg2+ upon WT. CBD12-AD-E454K binds a total of four Ca2+ ions; two bind to the CBD1 Ca3–Ca4 sites, and two bind to CBD2. In the presence of Mg2+, the mutant binds a total of three Ca2+ ions as Mg2+ replaces Ca2+ at CaII of CBD2 (9), again supporting the hypothesis that the second Mg2+ site in CBD12-AD-WT resides in CBD1.
DISCUSSION
Population Shift Underlies Ca2+-induced Regulatory Transitions in NCX Proteins
Previous NMR (24), FRET (23), and SAXS (8, 18) studies of isolated CBD12 tandem provided contradicting results, which were hard to reconcile. Here, after a careful temperature SAXS study, we used EOM, reconciling the different data. In agreement with the NMR (24) and FRET (23) data, large conformational changes are not detected under similar experimental conditions. The CBD12 tandem shows a Ca2+-modulated conformational flexibility, in agreement with NMR studies (24). Most importantly, our EOM analysis provides a detailed distribution model for the conformations of each state, revealing a prominent Ca2+-induced population shift (Figs. 2 and 3). Population shifts occur even under conditions in which no effect of Ca2+ is observed on the globally derived Dmax with either WT or E454K (Fig. 1, A, C, and E), under comparable conditions of normal [Ca2+]-dependent allosteric regulation as observed in patch clamp experiments (23).
We suggest that Ca2+-dependent alterations in Dmax, observed under certain conditions (8, 18), do not represent an obligate feature for coupling the Ca2+-driven allosteric regulation in NCX proteins. This study then suggests a model whereby Ca2+ binding to the primary allosteric sensor (Ca3–Ca4 sites) induces a population shift rather than global conformational changes of CBDs. The essence of a population shift mechanism supposes numerous conformational transitions over relatively small energetic barriers. The mechanism of population shift, described here, may have a physiological significance because induced fit can take place under one of two scenarios: high concentration of ligand or high affinity of protein to ligand (15–17). Neither of these regulatory conditions seems to take place in the cell (15, 16). Interestingly, induced fit may follow conformational selection to optimize the ligand-protein interactions (15–17). Molecular dynamics simulations may provide more details on the energetic landscape of Ca2+-induced dynamic transitions in the CBD12 tandem.
Recent crystal structures of CBD12, derived from NCX (18) and CALX (22), reveal three regions, two hydrophilic and one hydrophobic, at the interface of CBDs. The amino acids contributing to the interface are highly conserved among NCX orthologs and splice variants (18). Evidently, the interdomain linker governs CBD coupling both in intact NCX (13) or isolated CBD12 proteins (12, 21). Moreover, in Gly-503, its dihedral ϕ/ψ angles and its precise position in the linker are both essential for slow dissociation of occluded Ca2+ and CBD coupling (21). Thus, Gly-503 is indispensable for Ca2+-driven stabilization of interdomain salt bridges that limit the linker flexibility and thus, rigidifying CBD movements (21, 29). Therefore, the linker and perhaps additional components at the interface, e.g. Phe-450, may play a critical role in supporting the population shift mechanism.
An important feature of the interdomain Ca2+ switch is that Asp-499 and Asp-500 residues participate in coordinating the Ca3–Ca4 sites (on CBD1), while concomitantly stabilizing the two-domain interface through the salt bridge network via Arg-532, located on CBD2 (18). This network is the principal linchpin holding CBDs together upon Ca2+ binding to Ca3–Ca4 sites either in NCX (18) or CALX (22). Because CBD12-BD does not bind Ca2+ at CBD2 (8) and the low affinity sites of CBD1 are altered in CBD12-AD-E454K (18, 25), the population shift induced by Ca2+ can be attributed to the high affinity Ca3–Ca4 sites of CBD1 (Figs. 2 and 3). This is consistent with crystal structures of the CBD12 tandem, as occupation of these sites is involved in stabilizing the CBD1-CBD2 interface (18, 22). Importantly, only the Ca3–Ca4 sites are responsible for the brief “peak current” activation of NCX1, as shown in electrophysiological studies (7). This brief activation is observed for the respective NCX constructs of all the examined CBD12 constructs in this study (30). Thus, the population shift model, presented here, provides a detailed structural description of Ca2+ allosteric regulation of NCX orthologs and their splice variants.
Role of the Low Affinity Ca1–Ca2 Sites
Previous electrophysiological studies did not identify any specific role for the low affinity CBD1 Ca1–Ca2 sites (7), despite their strict conservation (8, 18). Our findings imply that these sites may function as Mg2+ sites while reducing the affinity of the Ca3–Ca4 sites from the Kd of ∼50 nm to the Kd of ∼200 nm (Fig. 4). Several independent findings support this hypothesis. First, CBD12-AD-E454K that mimics an occupied Ca1 site shows reduced Ca2+ affinity at the Ca3–Ca4 sites (Fig. 4D). Second, Mg2+ alone cannot induce the conformational change or population shift induced by Ca2+ binding to the high affinity Ca3–Ca4 sites (Fig. 2, C and D). Third, CBD12-AD-E454K, which has altered low affinity CBD1 sites, is not affected by Mg2+ (Fig. 4D). Fourth, the presence of Mg2+ reduces the total binding capacity of CBD12-AD and the binding affinity of the Ca3–Ca4 sites (Fig. 4C). Most importantly, the Ca3–Ca4 sites of CBD12-AD are titrated over the range of physiological [Ca2+] in the presence of Mg2+. Thus, we conclude that the low affinity Ca1 and/or Ca2 sites serve as Mg2+ sites, modulating the intrinsic affinity of the Ca3–Ca4 sites. This conclusion suggests a similar binding scheme for CBD1 compared with CBD2, where Mg2+ binding to the low affinity CaII site increases the affinity at the CaI site (9, 10). Therefore, collectively, the data suggest that occupation of Ca1–Ca2 sites by Mg2+ decreases the affinity of the primary Ca2+ sensor on CBD1 (Ca3–Ca4 sites), whereas the Mg2+ binding to CaII increases the affinity of the “secondary” Ca2+ sensor on CBD2 (CaI site). These dual effects of Mg2+ could be functionally relevant for keeping the sensitivity of CBD sensors within the physiological range of cytosolic [Ca2+] oscillations.
It was previously suggested that an electrostatic switch at CBD1 underlies Ca2+-dependent activation of NCX (8). However, our analysis suggests that this is not merely an electrostatic switch, but a Ca2+ switch situated at the interface of CBDs. Two findings support this notion. First, CBD12-AD-E454K undergoes the same Ca2+-induced population shift as CBD12-AD-WT, despite having less negative potential in the apo form and a constitutively “pseudo-bound” site (Ca1) (18, 25). Second, Mg2+ at physiologically relevant concentrations cannot induce the same effect as Ca2+, despite interacting with CBD1 (Fig. 2, C and D, and Fig. 4, A and B). Although the low affinity Ca1–Ca2 sites do not directly contribute to the population shift, the occupancy of these sites by Mg2+ decreases the affinity of the primary Ca3–Ca4 sensor, which is conditional for performing the population shift at increasing levels of cytosolic Ca2+ (>100 nm) during the action potential (e.g. in cardiomyocytes).
Alternative Splicing and NCX Regulation
Recently, the crystal structures of two CBD12 splice variants from CALX, a Drosophila melanogaster NCX ortholog, were determined (22). It was suggested that relatively small differences between their interdomain angles account for their differential regulatory responses. Considering the conservation of the domain interface and Ca2+ binding sites (18), our EOM analysis implies that in solution, both variants may have at least partially overlapping ranges of orientations. Taken together with the strikingly similar conformations of CBD12 from the differentially regulated AD and BD splice variants of NCX1 (Fig. 2), our results further support a common Ca2+ switch underlying eukaryotic NCX regulation (18). However, the differences between splice variants of NCX can be attributed to different binding properties of CBD2 (8), which is not the case for CALX (22). Interactions with other domains and differences in domains dynamics or population shifts may lead to specific regulation of the different exchangers and splice variants rather than different global conformations. Of course, this issue requires more dedicated experimentation in the future.
A Proposed Model for Allosteric Regulation of NCX
Fig. 5 presents a general scheme for transitions between possible states of CBD12. In the apo form, the domains are flexible, all six binding sites of CBD12 are available for binding, and Ca3–Ca4 sites possess the highest affinity. In the presence of physiological [Mg2+], Mg2+ occupies low affinity Ca2+ sites in CBD1 and CBD2 (Fig. 4). The presence of Mg2+, however, does not alter domain flexibility and conformation and hence, does not render NCX active (Fig. 2, C and D). In excitable tissues, the rise in the cytosolic [Ca2+] results in occupation of Ca3–Ca4 sites, rendering NCX active, manifest as peak current activation, via a population shift of CBD12 toward a narrowly distributed elongated shape (Fig. 2). Further rise in cytosolic Ca2+ may result in occupation of the CaI site at CBD2, alleviating Na+-dependent inactivation and thus, allowing sustained NCX activation (5, 8). The low affinity sites (Ca1–Ca2) at CBD1 may become Ca2+-occupied only in very restricted compartments of the cell, where [Ca2+]i may reach 200–300 μm. However, the most probable scenario is that under physiologically relevant conditions, the Ca1–Ca2 sites are constitutively occupied by Mg2+ to keep the affinity of the primary Ca2+ sensor (Ca3–Ca4 sites) within appropriate [Ca2+] ranges.
FIGURE 5.
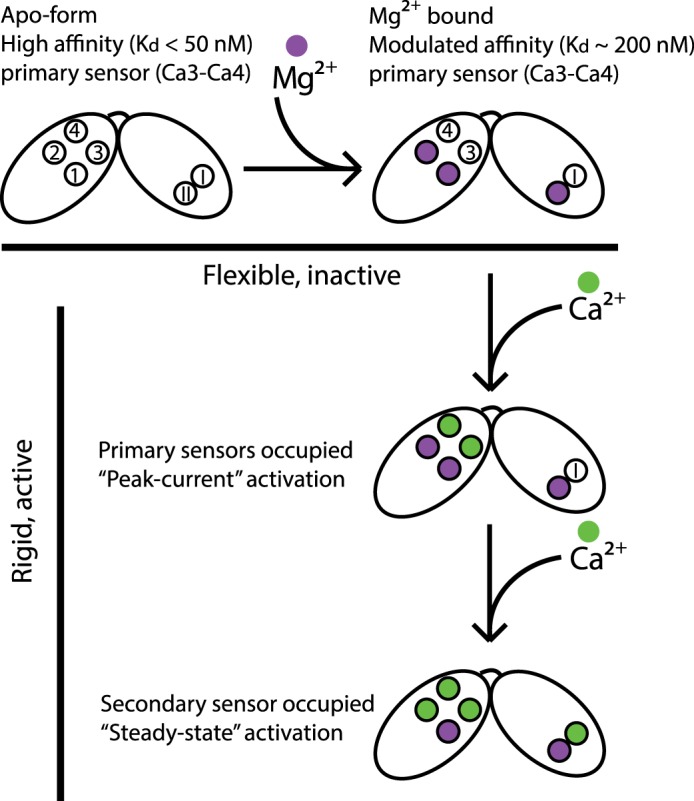
Model for NCX regulation. In the apo form, all six binding sites are not occupied (empty circles), and the Ca3–Ca4 sites possess high affinity. Under physiological conditions, Mg2+ occupies the low affinity sites (purple spheres) and modulates the remaining Ca2+ sites affinity. Rise in cytosolic [Ca2+] results in occupation of the Ca3–Ca4 sites, rendering CBD12 rigid and NCX active. Further rise in [Ca2+] alleviates Na+-dependent inactivation and underlies steady-state activation.
Acknowledgments
We thank the staffs of X33 at Deutsches Elektronen-Synchrotron and BM29 at European Synchrotron Radiation Facility for assistance with SAXS experimentation and Giancarlo Tria and Dr. Dmitry Svergun for fruitful discussions regarding SAXS data collection and analysis.
This work was partially funded by Israeli Ministry of Health Grant 2010-3-6266, United States-Israel Binational Foundation Research Grant 2009-334, Israel Science Foundation Grant 23/10 (to D. K.), a Deutsch-Israelische Projecktkooperation-Deutsche Forschungsgemeinschaft grant (to J. A. H.), and the Bernstein Foundation.
- NCX
- sodium-calcium exchanger
- CBD
- calcium binding domain
- Dmax
- maximal interatomic distance
- EOM
- ensemble optimization method
- fluo-3
- N-[2-[2[bis(carboxymethyl)amino]-5-(2,7dichloro-6-hydroxy-3-oxy-3Hxanthen-9-yl)henoxy]ethoxy]-4-methylphenyl]-N(carboxymethyl)glycine
- PDB
- Protein Data Bank
- Rg
- radius of gyration
- SAXS
- small angle x-ray scattering.
REFERENCES
- 1. Blaustein M. P., Lederer W. J. (1999) Sodium/calcium exchange: its physiological implications. Physiol. Rev. 79, 763–854 [DOI] [PubMed] [Google Scholar]
- 2. Philipson K. D., Nicoll D. A. (2000) Sodium-calcium exchange: a molecular perspective. Annu. Rev. Physiol. 62, 111–133 [DOI] [PubMed] [Google Scholar]
- 3. Boyman L., Hagen B. M., Giladi M., Hiller R., Lederer W. J., Khananshvili D. (2011) Proton-sensing Ca2+ binding domains regulate the cardiac Na+/Ca2+ exchanger. J. Biol. Chem. 286, 28811–28820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hilge M., Aelen J., Vuister G. W. (2006) Ca2+ regulation in the Na+/Ca2+ exchanger involves two markedly different Ca2+ sensors. Mol. Cell 22, 15–25 [DOI] [PubMed] [Google Scholar]
- 5. Besserer G. M., Ottolia M., Nicoll D. A., Chaptal V., Cascio D., Philipson K. D., Abramson J. (2007) The second Ca2+-binding domain of the Na+-Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis. Proc. Natl. Acad. Sci. U.S.A. 104, 18467–18472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nicoll D. A., Sawaya M. R., Kwon S., Cascio D., Philipson K. D., Abramson J. (2006) The crystal structure of the primary Ca2+ sensor of the Na+/Ca2+ exchanger reveals a novel Ca2+ binding motif. J. Biol. Chem. 281, 21577–21581 [DOI] [PubMed] [Google Scholar]
- 7. Ottolia M., Nicoll D. A., Philipson K. D. (2009) Roles of two Ca2+-binding domains in regulation of the cardiac Na+-Ca2+ exchanger. J. Biol. Chem. 284, 32735–32741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hilge M., Aelen J., Foarce A., Perrakis A., Vuister G. W. (2009) Ca2+ regulation in the Na+/Ca2+ exchanger features a dual electrostatic switch mechanism. Proc. Natl. Acad. Sci. U.S.A. 106, 14333–14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Breukels V., Konijnenberg A., Nabuurs S. M., Touw W. G., Vuister G. W. (2011) The second Ca2+-binding domain of NCX1 binds Mg2+ with high affinity. Biochemistry 50, 8804–8812 [DOI] [PubMed] [Google Scholar]
- 10. Boyman L., Mikhasenko H., Hiller R., Khananshvili D. (2009) Kinetic and equilibrium properties of regulatory calcium sensors of NCX1 protein. J. Biol. Chem. 284, 6185–6193 [DOI] [PubMed] [Google Scholar]
- 11. Kofuji P., Lederer W. J., Schulze D. H. (1994) Mutually exclusive and cassette exons underlie alternatively spliced isoforms of the Na/Ca exchanger. J. Biol. Chem. 269, 5145–5149 [PubMed] [Google Scholar]
- 12. Giladi M., Boyman L., Mikhasenko H., Hiller R., Khananshvili D. (2010) Essential role of the CBD1-CBD2 linker in slow dissociation of Ca2+ from the regulatory two-domain tandem of NCX1. J. Biol. Chem. 285, 28117–28125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ottolia M., Nicoll D. A., John S., Philipson K. D. (2010) Interactions between Ca2+ binding domains of the Na+-Ca2+ exchanger and secondary regulation. Channels 4, 159–162 [DOI] [PubMed] [Google Scholar]
- 14. Giladi M., Bohbot H., Buki T., Schulze D. H., Hiller R., Khananshvili D. (2012) Dynamic features of allosteric Ca2+ sensor in tissue-specific NCX variants. Cell Calcium 51, 478–485 [DOI] [PubMed] [Google Scholar]
- 15. Ma B., Nussinov R. (2010) Enzyme dynamics point to stepwise conformational selection in catalysis. Curr. Opin. Chem. Biol. 14, 652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boehr D. D., Nussinov R., Wright P. E. (2009) The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Okazaki K., Takada S. (2008) Dynamic energy landscape view of coupled binding and protein conformational change: induced-fit versus population-shift mechanisms. Proc. Natl. Acad. Sci. U.S.A. 105, 11182–11187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giladi M., Sasson Y., Fang X., Hiller R., Buki T., Wang Y. X., Hirsch J. A., Khananshvili D. (2012) A common Ca2+-driven interdomain module governs eukaryotic NCX regulation. PLoS One 7, e39985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bernadó P., Svergun D. I. (2012) Analysis of intrinsically disordered proteins by small-angle X-ray scattering. Methods Mol. Biol. 896, 107–122 [DOI] [PubMed] [Google Scholar]
- 20. Koch M. H., Vachette P., Svergun D. I. (2003) Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q. Rev. Biophys. 36, 147–227 [DOI] [PubMed] [Google Scholar]
- 21. Giladi M., Friedberg I., Fang X., Hiller R., Wang Y. X., Khananshvili D. (2012) G503 is obligatory for coupling of regulatory domains in NCX proteins. Biochemistry 51, 7313–7320 [DOI] [PubMed] [Google Scholar]
- 22. Wu M., Tong S., Gonzalez J., Jayaraman V., Spudich J. L., Zheng L. (2011) Structural basis of the Ca2+ inhibitory mechanism of Drosophila Na+/Ca2+ exchanger CALX and its modification by alternative splicing. Structure 19, 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. John S. A., Ribalet B., Weiss J. N., Philipson K. D., Ottolia M. (2011) Ca2+-dependent structural rearrangements within Na+-Ca2+ exchanger dimers. Proc. Natl. Acad. Sci. U.S.A. 108, 1699–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salinas R. K., Bruschweiler-Li L., Johnson E., Brüschweiler R. (2011) Ca2+ binding alters the interdomain flexibility between the two cytoplasmic calcium-binding domains in the Na+/Ca2+ exchanger. J. Biol. Chem. 286, 32123–32131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chaptal V., Ottolia M., Mercado-Besserer G., Nicoll D. A., Philipson K. D., Abramson J. (2009) Structure and functional analysis of a Ca2+ sensor mutant of the Na+/Ca2+ exchanger. J. Biol. Chem. 284, 14688–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bernadó P., Mylonas E., Petoukhov M. V., Blackledge M., Svergun D. I. (2007) Structural characterization of flexible proteins using small-angle x-ray scattering. J. Am. Chem. Soc. 129, 5656–5664 [DOI] [PubMed] [Google Scholar]
- 28. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 29. Matsuoka S., Nicoll D. A., Hryshko L. V., Levitsky D. O., Weiss J. N., Philipson K. D. (1995) Regulation of the cardiac Na+-Ca2+ exchanger by Ca2+: mutational analysis of the Ca2+-binding domain. J. Gen. Physiol. 105, 403–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dyck C., Omelchenko A., Elias C. L., Quednau B. D., Philipson K. D., Hnatowich M., Hryshko L. V. (1999) Ionic regulatory properties of brain and kidney splice variants of the NCX1 Na+-Ca2+ exchanger. J. Gen. Physiol. 114, 701–711 [DOI] [PMC free article] [PubMed] [Google Scholar]