Abstract
Bacterial type III protein export underlies flagellum assembly and delivery of virulence factors into eukaryotic cells. The sequence of protein interactions underlying the export pathway are poorly characterized; in particular, it is not known how chaperoned substrates in the cytosol are engaged by the membrane-localized export apparatus. We have identified a stalled intermediate export complex in the flagellar type III export pathway of Salmonella typhimurium by generating dominant-negative chaperone variants that are export-defective and arrest flagellar assembly in the wild-type bacterium. These chaperone variants bound their specific export substrates strongly and severely reduced their export. They also attenuated export of other flagellar proteins, indicating that inhibition occurs at a common step in the pathway. Unlike the cytosolic wild-type chaperone, the variants localized to the inner membrane, but not in the absence of the flagellar type III export apparatus. Membrane localization persisted in fliOPQR, flhB, flhA, fliJ, and fliH null mutants lacking specific flagellar export components but depended on the presence of the membrane-associated ATPase FliI. After expression of the variant chaperones in Salmonella, a stalled intermediate export complex, which contained chaperone, substrate, and the FliI ATPase with its regulator FliH, was isolated. Neither chaperone nor substrate alone was able to interact with liposome-associated FliI, but the chaperone-substrate-FliI(FliH) complex was assembled when chaperone was prebound to its substrate. Our data establish a key event in the type III protein export mechanism, docking of the cytosolic chaperone-substrate complex at the ATPase of the membrane-export apparatus.
Type III protein secretion systems are widely used by Gram-negative bacteria to assemble the cell-surface flagella required for motility and to deliver virulence effector proteins into eukaryotic host cells (1, 2). The flagellum-specific system of Salmonella typhimurium and Escherichia coli is genetically well characterized and is a model for the study of type III export, because components of the virulence and flagellar export machineries are homologous and their supramolecular structures are similar. The flagellum comprises a long filament that acts as a propeller anchored to the cell envelope by a flexible hook and basal body (1, 3). The flagellar “axial” proteins that polymerize to form the structure, including the hook-filament junction (FlgK and FlgL), filament (flagellin, FliC), and filament cap (FliD), are exported by the type III export apparatus to the growing flagellum (4, 5). Export of these structural components is strongly facilitated by cytosolic substrate-specific chaperones, which bind respectively to the C-terminal amphipathic domains of their substrates FlgK and FlgL (chaperone FlgN), FliD (FliT), and FliC (FliS) (6-8). Although primary sequence identity is low or inapparent between the chaperones of the flagellar and virulence type III export systems, most are small, homodimeric proteins with a predicted C-terminal amphipathic helix (6, 9, 10), and it is suggested that they act as “bodyguards” to maintain monomers in an export-competent form and prevent their interaction before export (6, 10-13). In the flagellar chaperones FlgN and FliT and the virulence chaperone CesT, the C-terminal region has been shown to mediate substrate binding, whereas the N terminus is responsible for homodimerization (7, 14).
Although the components of flagellar and virulence type III export systems are known, little detail is available describing the protein-protein interactions underlying the pathway, in particular the key transition of substrates from a cytosolic chaperone-bound state to their association with the export apparatus at the membrane. This is partly because of the difficulty of isolating intermediate export pathway complexes, because the substrate or chaperone-substrate complexes interact only transiently with downstream export components. We sought to identify the crucial docking event by generating dominant-negative export-defective chaperone variants that stall and accumulate intermediate complexes in the export pathway.
Materials and Methods
Bacterial Strains and Plasmids. Bacteria were cultured in Luria-Bertani broth at 37°C to late exponential phase (A600 ≈ 1.0), unless stated, containing (where appropriate) ampicillin (50 μg·ml-1) or chloramphenicol (20 μg·ml-1) and harvested by centrifugation (8,000 × g, 15 min). The wild-type S. typhimurium SJW1103 is motile (15). Derived mutants carry lesions in flagellar genes flgN (7); flhDC [SJW1368 (16)]; fliOPQR (SJW192), flhA (SJW1616), flhB (SJW1684), fliJ (SJW135), and fliI (SJW2702) (17); and fliH [MKM11 (18)]. Recombinant proteins were expressed in E. coli BL21 (DE3) (19) or its derivative C41 (20) from isopropyl β-d-thiogalactoside-inducible plasmids pET15b (21) or pACT7 (22) or in S. typhimurium from l-arabinose-inducible pBAD18 (23).
Generation of FlgN Chaperone Variants. In-frame internal deletions were created in FlgN by inverse PCR using pUC19flgN as a template and oligonucleotide primers engineered to introduce a SalI or XhoI site flanking the sequence to be deleted. After PCR amplification (Perkin-Elmer thermal cycler using Herculase Pfu DNA polymerase), DNA was digested with DpnI to remove methylated DNA template and purified by using a QIAquick PCR kit (Qiagen, Valencia, CA) followed by digestion with SalI or XhoI, as appropriate. After religation, plasmids were sequenced and digested with NdeI/BamHI or XbaI/HindIII before ligation of the deleted flgN inserts into vectors pET15b or pBAD18, respectively. C-terminal truncations of FlgN (NΔ120-140 and NΔ130-140) were created by introducing a stop codon by PCR at the appropriate position by using S. typhimurium genomic DNA as a template. The resulting PCR products were similarly digested NdeI/BamHI before ligation into pET15b and then XbaI/HindIII before ligation into pBAD18.
Protein Purification. FliI was purified from E. coli C41 (pGEX4T3-FliI) as described (24). FlgN and its deleted derivatives were purified from E. coli BL21 (pETN) as described (7). Chaperone-substrate complexes were purified on nickel nitrilotriacetic acid (Ni-NTA) agarose from E. coli BL21 (DE3) coexpressing pETN or pETNΔ130-140 and pACT7FlgK and dialyzed against 20 mM Tris·HCl/150 mM NaCl. Complexes were isolated from eluted fractions containing both proteins after passage through a Superdex-200 (Amersham Pharmacia) size-exclusion column.
Assay of Flagellar Protein Export. S. typhimurium culture supernatants were clarified by three rounds of centrifugation and passed through a 0.2-μm nitrocellulose filter (Millipore). Flagellar proteins were precipitated by 10% (wt/vol) trichloroacetic acid (TCA) on ice for 1 h, separated by SDS/PAGE, and visualized by Coomassie brilliant blue staining or immunoblotting with appropriate polyclonal antisera or a Ni-NTA-horse radish peroxidase conjugate (Qiagen) (7). Chemiluminescent detection was carried out by using Supersignal substrate (Pierce). Relative amounts of protein were estimated by densitometric analysis with nih image software.
Copurification Assays. Copurification (pull down) of flagellar protein complexes was achieved by Ni-NTA affinity chromatography (13) after coexpression in E. coli BL21 (DE3). The in vivo assay used S. typhimurium strains expressing (His)6-tagged FlgN or derivatives at wild-type levels as assessed by immunoblotting. Cells were resuspended in lysis buffer (50 mM NaH2PO4, pH 8.0/300 mM NaCl/10 mM imidazole) and passed through a French pressure cell (Aminco) three times at 82,800 kPa before the addition of 20 mM MgCl2. Membranes, unlysed cells, and insoluble proteins were removed by centrifugation (40,000 × g, 40 min). The resulting supernatant was incubated with Ni-NTA agarose, washed three times with lysis buffer, and eluted with the same buffer containing 500 mM imidazole. Eluted proteins were precipitated [10% (wt/vol) TCA] and visualized by Coomassie blue staining or immunoblotting.
Cell Fractionation. Cultures were fractionated essentially as described (25). To create spheroplasts, cell pellets were resuspended in 0.5 M sucrose/40 mM Tris·HCl, pH 7.4/5 mM EDTA before sequential addition of lysozyme (50 μg·ml-1), sterile distilled water, and 20 mM MgCl2. Spheroplasts were pelleted by centrifugation (16,000 × g, 15 min) and lysed in a hypotonic solution of 20 mM Tris·HCl, pH 7.4/5 mM EDTA/1 mM Pefabloc serine protease inhibitor (Roche Diagnostics). DNase I (50 μg·ml-1) was added, and the suspension was agitated until viscosity decreased. This was followed by a short spin (6,000 × g, 30 sec) to remove unlysed cells. Membranes then were pelleted (16,000 × g, 15 min) and resuspended in urea-SDS loading buffer. The supernatant was taken as the cytosolic fraction.
Preparation of S. typhimurium Membranes. Cell cultures were pelleted, washed, resuspended in 20 mM Tris·HCl, pH7.4/1 mM Pefabloc and passed through a French pressure cell (Aminco) three times at 82,800 kPa. After removal of intact cells (2,000 × g, 10 min), total membranes were separated from soluble proteins by high-speed centrifugation at 100,000 × g for 2 h (SW50.1 rotor, Beckman Coulter), and membranes were resuspended in 20 mM Tris·HCl, pH 7.4. Separation of inner and outer membranes was performed as described (26), and membrane fractions (in 5 mM EDTA) were layered onto sucrose gradients (16 ml) prepared in stepwise fashion with layers of 1.0, 1.2, 1.4, 1.6, 1.8, and 2.0 M sucrose containing 5 mM EDTA. Gradients were centrifuged for 16 h at 75,000 × g, after which 1-ml fractions were removed and proteins were precipitated with 10% (wt/vol) TCA.
Liposome-Protein Flotation Assay. E. coli total phospholipids (Avanti Polar Lipids) were prepared by sonication (27). Purified proteins (1-2 μg) were mixed with 40 μl of 10 mg·ml-1 lipid in Tris-buffered saline (20 mM Tris, pH 7.4/150 mM NaCl/TBS) (27) and incubated at room temperature for 15 min. Sucrose was added to a final concentration of 50% (wt/vol) in 1 ml of TBS (27), and samples were overlaid with 3.5 ml of 40% sucrose-TBS (wt/vol) and 0.5 ml of TBS. After centrifugation at 75,000 × g for 16 h at 16°C, 10 0.5-ml fractions were collected and pre-cipitated with 10% (wt/vol) TCA. Fractions 1-4, 5-7, and 8-10 were pooled to give top, middle, and bottom fractions, respectively.
Isothermal Titration Calorimetry. Isothermal titration calorimetry (ITC) measurements were performed by using a VP-ITC MicroCalorimeter (Microcal, Amherst, MA). Protein stock solutions were appropriately diluted in 20 mM Tris·HCl, pH 7.4/150 mM NaCl and degassed before each titration (ThermoVac, Microcal). Purified FlgK was placed in the calorimeter cell and mixed (300 rpm, 25°C). Typically, 5 μl of purified FlgN or NΔ130-140 was injected for 10 s at 4-min intervals until the titrant was in excess. Negative controls were performed by using injections with titrant buffer alone. Data were analyzed by using origin software.
Results
Export-Defective Chaperones That Block Flagellar Type III Export. FlgN chaperone variants were tested for a dominant-negative export defect, i.e., inability to restore FlgK-FlgL export to a nonmotile flgN null mutant while attenuating export of the same substrates in the wild type. FlgN function is retained when amino acids 1-67 of the 140-residue FlgN are replaced by dimeric glutathione S-transferase (7), and the putative substrate-binding amphipathic helix spans residues 74-114 (Fig. 1A). In-frame deletions were therefore made in the substrate-interacting C-terminal domain to generate (His)6-tagged variants NΔ80-100, NΔ90-110, NΔ100-120, NΔ120-140, and NΔ130-140, expressed from the arabinose-inducible pBAD plasmid (23). Similar to the native FlgN, full-length (His)6-FlgN complements the flgN null mutation (7) and moderately inhibits flagellum assembly when overexpressed in the motile wild type.
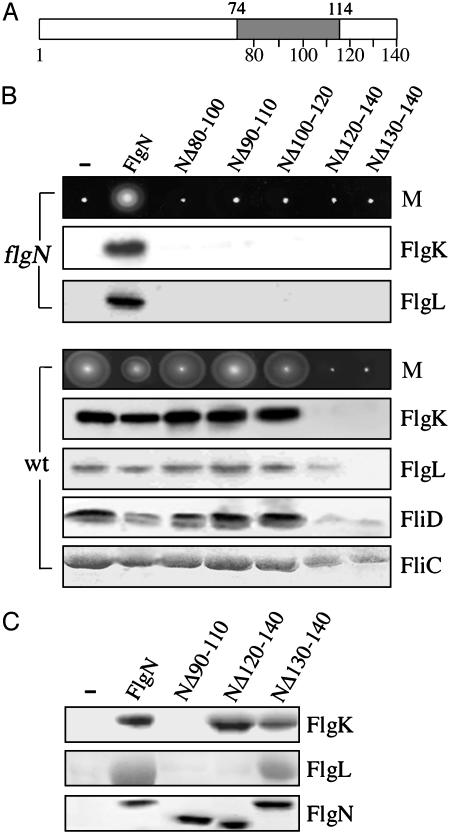
Fig. 1.
Identification of dominant-negative export-defective chaperone variants. (A) Representation of the 140-residue FlgN chaperone. In-frame deletions Δ80-100 to Δ130-140 were made in the substrate-binding C-terminal domain, in which the predicted amphipathic helix is shaded (residues 74-114). Vertical lines indicate intervals of 10 residues. The N-terminal domain determines homodimerization. (B) Motility (M) and flagellar protein export of the S. typhimurium flgN null mutant (flgN) and the wild-type (wt) carrying vector pBAD18 (-) or derivatives expressing the (His)6-tagged FlgN wild type or a deleted variant. Motility was assessed on 0.3% soft tryptone agar after point inoculation and incubation at 37°C for 6-8 h. Secreted proteins from late-exponential-phase cultures were separated by SDS (12%)/PAGE and immunoblotted with FlgK, FlgL, or FliD antisera or stained with Coomassie blue (FliC). In the flgN mutant, low-level expression was induced by 0.01% l-arabinose; in the wild type, high-level expression was induced by 0.1% l-arabinose. (C) Interaction of FlgN variants with export substrate. Lysates of E. coli BL21 (DE3)-coexpressing substrates FlgK or FlgL with (His)6-tagged wild-type or variant FlgN were incubated with Ni-NTA resin, and proteins were purified by affinity chromatography. Coeluted proteins were separated by SDS (15%)/PAGE and stained with Coomassie blue. Negative control (-) carries pBAD18.
The variant and wild-type FlgN proteins [(His)6-tagged throughout this study] were expressed in the nonmotile S. typhimurium flgN null mutant at approximately the wild-type (physiological) level by inducing with 0.01% (wt/vol) l-arabinose (as calibrated by immunoblotting; data not shown). In contrast to the (His)6-tagged wild type, none of the deleted FlgN variants were able to restore motility on soft agar, and immunoblotting of culture supernatants showed that this correlated with an inability to recover export of FlgK and FlgL (Fig. 1B, flgN). All of the variants therefore were export-defective. To ascertain whether any could block wild-type export, each was overexpressed in the motile S. typhimurium by induction with 0.1% (wt/vol) l-arabinose (generating an ≈10-fold higher level than induction with 0.01%). Full-length FlgN caused a slight inhibition of motility, whereas NΔ80-100, NΔ90-100, and NΔ100-120 had no effect (Fig. 1B, wt). In contrast, variants NΔ120-140 and NΔ130-140 caused a complete loss of motility. Immunoblotting of culture supernatants (Fig. 1B, wt) confirmed that overexpression of NΔ80-100, NΔ90-110, and NΔ100-120 had no effect on FlgK-FlgL export; the levels were comparable with those in the wild-type S. typhimurium carrying the expression vector (and the three defective nonattenuating variants), whereas by the same comparison NΔ130-140 and NΔ120-140 reduced export of FlgK at least 30-fold. NΔ130-140 caused a similar reduction in FlgL export, whereas NΔ120-140 reduced export of this substrate ≈5-fold. Wild-type FlgN caused a slight attenuation in the export of the filament-type type III export substrates bound by chaperones other than FlgN, namely, the filament subunit FliC and the cap subunit FliD. NΔ80-100, NΔ90-110, and NΔ100-120 had no effect, whereas NΔ130-140 and NΔ120-140 reduced their export ≈3-fold (FliC) and 5-fold (FliD).
To confirm that NΔ120-140 and NΔ130-140 bind their specific export substrates in the absence of any other flagellar proteins, the (His)6-tagged variants and wild-type FlgN each were coexpressed with untagged FlgK or FlgL in E. coli BL21 (DE3) and copurification-assayed after Ni-NTA affinity chromatography. Coeluted proteins were separated by SDS/PAGE and visualized by Coomassie blue staining (Fig. 1C). Neither FlgK nor FlgL copurified with any of the noninterfering export-defective variants NΔ80-100, NΔ90-110, and NΔ100-120 (Fig. 1C shows NΔ90-110 as an example), but FlgK copurified with both NΔ120-140 and NΔ130-140 (similar to the wild-type FlgN), and FlgL copurified only with NΔ130-140.
The data show that export-defective variants NΔ80-100, NΔ90-100, and NΔ100-120, disrupted in the potential amphipathic helix, are not dominant-negative. They do not bind substrate and presumably do not engage the export apparatus. In contrast, export-defective variants NΔ120-140 and NΔ130-140, deleted at the extreme C terminus outside the potential helix, bind substrate and exert a dominant-negative phenotype, strongly attenuating export of their substrates. They also inhibit export of the noncognate chaperoned substrates FliC and FliD, which would be compatible with the variants exerting their effect at a pivotal point in the export pathway.
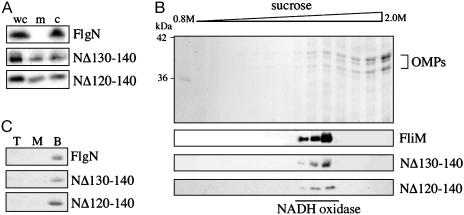
Dominant-Negative Export-Defective Chaperone Variants Localize to the Inner Membrane. Flagellar type III export chaperones are localized exclusively in the cytosol, in which they interact with their respective substrates (7, 8). Because the dominant-negative export-defective FlgN variants still bind substrate, it is possible that their interference with wild-type export arises from the formation of substrate complexes that stall at the membrane machinery. To assess this possibility, cell lysates of the S. typhimurium flgN null mutant expressing NΔ120-140 or NΔ130-140 were fractionated (25), separated by SDS/PAGE, and immunoblotted with anti-FlgN antiserum (Fig. 2A). In contrast to the exclusively cytosolic wild-type chaperone, both NΔ120-140 and NΔ130-140 were found in both cytosolic and membrane/insoluble fractions. No NADH oxidase activity was detected in cytosolic fractions, confirming that there was no membrane contamination (data not shown). To verify that the dominant-negative chaperones were not aggregating but rather localize to the inner membrane, membrane fractions were separated by density gradient centrifugation (26), TCA-precipitated, and separated by SDS/PAGE (Fig. 2B). Coomassie blue staining confirmed that outer membrane proteins located toward the bottom of the gradient, and immunoblotting revealed both the NΔ120-140 and NΔ130-140 in the inner-membrane fractions colocalizing with the NADH oxidase and flagellar C-ring component FliM (27, 28).
Fig. 2.
Membrane localization of dominant-negative chaperone variants. (A) Whole-cell lysates (wc) and membrane/insoluble (m) and cytoplasmic fractions of the S. typhimurium flgN mutant expressing either wild-type FlgN or variants NΔ120-140 or NΔ130-140 were separated by SDS (15%)/PAGE and immunoblotted with anti-FlgN antiserum. (B) Membrane fractions of S. typhimurium flgN expressing NΔ120-140 or NΔ130-140 were harvested (100,000 × g, 1 h) and centrifuged through a 0.8-2.0 M sucrose gradient (75,000 × g, 16 h) to separate inner and outer membranes. Precipitated proteins were separated by SDS (12%)/PAGE and stained with Coomassie blue (Top) or blotted with anti-FliM or anti-FlgN antisera. Positions of inner-membrane-associated NADH oxidase and outer membrane proteins (OMPs) are indicated. (C) Purified wild-type or variant FlgN was incubated with E. coli total phospholipid liposomes and placed at the bottom of a three-step sucrose density gradient. After centrifugation (75,000 × g, 16 h), fractions were collected, and proteins precipitated from the top (T), middle (M), and bottom (B) of the gradient were separated by SDS (15%)/PAGE and visualized with Coomassie blue.
This shift in chaperone localization was not caused by an intrinsic preference of the dominant-negative variants for lipid membranes, because when purified NΔ120-140 and NΔ130-140 were incubated with E. coli total phospholipid liposomes (Fig. 2C), neither colocalized (floated) with the phospholipid to the top of the sucrose density gradient, in contrast to inner-membrane-associated flagellar proteins (27).
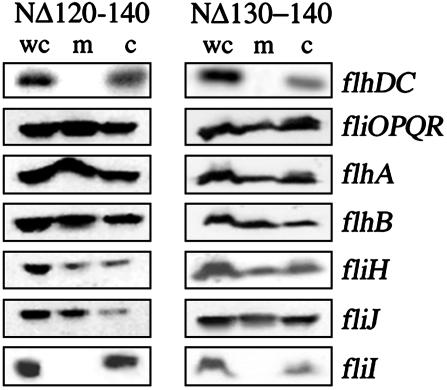
Membrane Localization Depends on the Type III Export Apparatus, Specifically the ATPase FliI. To establish whether localization to the membrane depends on the presence of the flagellar export apparatus, NΔ120-140, NΔ130-140, and wild-type FlgN were expressed in an flhDC null mutant, which lacks the flagellar master regulator required for expression of all flagellar proteins. Blotting of whole-cell, membrane, and cytosolic fractions with anti-FlgN antisera showed that, in the absence of flagellar proteins, the dominant-negative chaperones proteins localized exclusively to the cytosol (see Fig. 3). To establish whether this dependence could be ascribed to one or more components of the membrane apparatus, the same fractionation was repeated with specific flagellar null mutants, each expressing NΔ120-140 or NΔ130-140. Both chaperones localized to the membrane fraction in the fliOPQR, flhB, flhA, fliJ, and fliH null mutants (Fig. 3), but in the fliI mutant, they were exclusively cytosolic. These data suggest that variant chaperone localization to the inner membrane depends specifically on the membrane-associated flagellar ATPase FliI.
Fig. 3.
FliI-dependent membrane localization of dominant-negative variants. Whole-cell lysates (wc) and membrane/insoluble (m) and cytoplasmic (c) fractions of S. typhimurium null mutants (indicated on the right) expressing (His)6-tagged NΔ120-140 (Left) or NΔ130-140 (Right) were separated by SDS (15%)/PAGE, and FlgN proteins were probed with Ni-NTA-horseradish peroxidase conjugate (Qiagen).
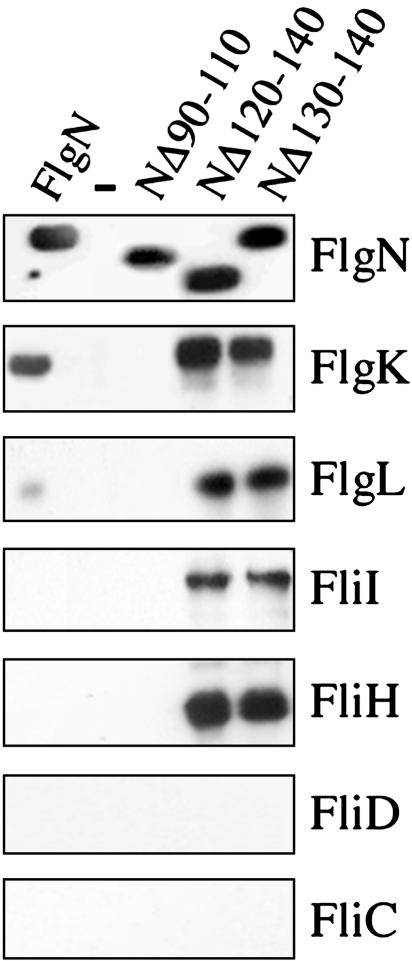
A Stalled Chaperone-Engaged Export Complex Containing the FliI ATPase and Its Regulator FliH. To isolate the putative “stalled” type III export complex(es) in vivo, cells were disrupted in a French press, which has been shown to release ≈30-50% of the inner-membrane-associated ATPase FliI, FliH inhibitor, and other peripheral membrane flagellar proteins into the soluble fraction (27, 29). (His)6-tagged dominant-negative variants NΔ120-140 and NΔ130-140 (and identically tagged wild-type FlgN and NΔ90-110 as positive and negative controls) were expressed at wild-type levels (0.01% l-arabinose) in the S. typhimurium flgN null mutant and affinity-purified. Coeluted proteins were separated and immunoblotted with anti-FlgK, anti-FlgL antisera, or Ni-NTA-horseradish peroxidase conjugate, which detects the (His)6 tag (Fig. 4). In vivo copurification allows identification of weaker interactions than the binding assay shown in Fig. 1. It detected an NΔ120-140 interaction with FlgL and revealed that more FlgK and FlgL are copurified (≈5- and 20-fold more, respectively) with both NΔ120-140 and NΔ130-140 than with wild-type FlgN. This seems not to reflect a higher affinity of the dominant-negative chaperone variants for substrate, because isothermal titration calorimetry (data not shown) indicates that FlgN and NΔ130-140 have the same KD for FlgK: ≈1.8 × 10-7 (±1.4 × 10-8) M. This may indicate that substrate complexes formed with the dominant-negative chaperones are more stable or long-lived than those with the wild-type FlgN.
Fig. 4.
Intermediate export complexes stalled by dominant-negative variants. Cultures of the S. typhimurium flgN null mutant expressing FlgN wild type or variants were lysed in a French pressure cell. After removal of cell debris, soluble fractions were incubated with Ni-NTA, and eluted proteins were separated by SDS (12%)/PAGE and immunoblotted with anti-FlgN, -FlgK, -FlgL, -FliI, -FliH, -FliD, or -FliC antisera. Negative control (-) carries the vector pBAD18.
The same samples were immunoblotted with anti-FliI, -FliH, FliD, and -FliC antisera. None of these proteins copurified with tagged wild-type FlgN, but both NΔ120-130 and NΔ130-140 copurified the FliI ATPase and its regulatory partner FliH (Fig. 4). These data demonstrate that the FliI-dependent membrane localization of the dominant-negative chaperones reflects an interaction with the export ATPase in vivo.
Export Substrate Is Required for Assembly of the Chaperone-Engaged ATPase Complex. FliI has been shown to colocalize (float) with liposomes of purified E. coli total phospholipid in sucrose density gradients (27), and this in vitro liposome flotation was applied to establish whether assembly of the stalled intermediate export complex is determined by FliI interaction with the chaperone, with the substrate, or with the chaperone-substrate complex.
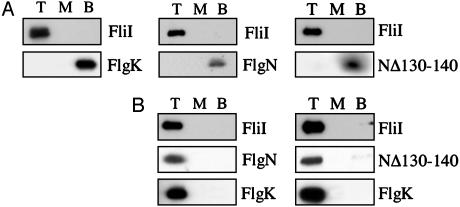
When purified FlgN, NΔ130-140, or FlgK were loaded individually at the bottom of a sucrose gradient with FliI and liposomes, each remained at the bottom after centrifugation, unlike FliI, which floated to the top (Fig. 5A). In contrast, preformed NΔ130-140-FlgK or FlgN-FlgK complexes cofloated with FliI (Fig. 5B). The same result was obtained when FliI was substituted by a preformed FliI-FliH complex (data not shown). These data indicate that only the substrate-bound form of the chaperone is able to interact with FliI either alone or in the FliI/H complex.
Fig. 5.
Interaction of chaperone-substrate complexes with the export ATPase FliI. (A) Purified FliI was preincubated with either FlgK substrate, FlgN wild type, or NΔ130-140 variant and added to E. coli total phospholipids liposomes. Sucrose was added to 55% (wt/vol) and overlaid with 40% (wt/vol) sucrose, and the protein-lipid vesicle mix was allowed to float through the density gradient during centrifugation (75,000 × g, 16 h). Proteins from the top (T), middle (M), and bottom (B) fractions of the gradient were precipitated and analyzed by SDS (15%)/PAGE before immunoblotting with anti-FliI or FlgK antisera. (B) Complex formation was assayed as described above after FliI was incubated with a preformed complex of FlgN-FlgK (Left) or NΔ130-140-FlgK (Right).
Discussion
Type III export of structural subunits to the nascent flagellum is facilitated by their interaction with specific cytosolic chaperones. We generated dominant-negative export-defective variants of the 140-aa FlgN, which chaperones the flagellum hook-junction proteins FlgK and FlgL, in an attempt to stall and accumulate transient complexes in the export pathway. The strategy has identified the key docking event in which chaperoned export substrates engage the membrane-export apparatus.
In contrast to export-defective variants that no longer bind their substrates, the dominant-negative chaperones NΔ130-140 and NΔ120-140 are C-terminally truncated, outside the predicted substrate-binding helix (residues 74-114). They were unable to restore FlgK-FlgL export and motility to an flgN null mutant when expressed at physiological levels, and they strongly attenuated motility and export of the normally motile wild type when overexpressed. They also caused a concomitant reduction in the export of other subunits not chaperoned by FlgN, i.e., FliC and FliD, which make up the filament and filament cap. This would be compatible with stalling of complexes at a common substrate-docking point. Although FlgK export was abolished by overexpression of both FlgN variants, FlgL export was similarly reduced only by NΔ130-140; when NΔ120-140 was overexpressed, it persisted at ≈5%. This correlates with an apparently weaker interaction of FlgL with NΔ120-140 and indicates that the substrate-binding sites for FlgK and FlgL on FlgN may not be identical. The dominant-negative chaperone variants localize to the inner membrane, in contrast to the exclusively cytosolic location of the wild-type FlgN. This shift is not caused by intrinsic lipid association, and the variants relocate to the cytosol in an aflagellate flhDC mutant, indicating dependence on other flagellar components. The shift in localization was not caused by any of the integral inner-membrane proteins FlhA, FlhB, FliO, FliP, FliQ, or FliR, components of the export machinery that are located at the base of the flagellum. It was, however, specifically dependent on the ATPase FliI, which is peripherally associated with the inner membrane (27). The ATPase is present in all type III export systems and is assumed to couple ATP hydrolysis to substrate translocation through the membrane-export machinery. The shift did not require the FliI-associated regulator FliH, which can localize to the membrane independently of FliI (27, 30).
The putative stalled chaperone-substrate complex with the ATPase was demonstrated after disruption of cells in a French press, which releases ≈50% of peripheral membrane protein from the membrane (27). A complex containing FliI, its regulator FliH, and export substrates FlgK and FlgL was isolated from Salmonella after affinity purification of each of the (His)6-tagged dominant-negative FlgN variants. The stalled multicomponent complex was not isolated by wild-type FlgN, which also copurified less FlgK and FlgL, suggesting that the in vivo variant chaperone-substrate complexes may be more stable and/or long-lived than those of the wild type. In vitro flotation assays showed that only the preformed chaperone-substrate complex interacts with FliI, not the chaperone or substrate alone. Because the substrate C-terminal domain is bound by chaperone, the export-essential N-terminal domain (31) would be free to interact with FliI. This would be compatible with the chaperone not being absolutely required for export but essential for efficient delivery of substrate to the machinery. Indeed, a basal level of exported FlgK and FlgL is observed in an flgN null mutant (≈50-fold less than wild type), and significant export in the absence of FlgN requires unphysiologically high concentrations of overexpressed substrate (7, 32).
Both wild-type and dominant-negative preformed chaperone substrate complexes interacted with liposome-associated FliI in vitro, but the unique accumulation of stalled export complexes in vivo by the dominant-negative chaperones suggests that they are defective in release of chaperone and/or substrate from the FliI-(FliH) export complex. This would be compatible with the involvement of other flagellar components in subsequent steps of the chaperone cycle of substrate delivery. Existing results suggest that a pool of chaperone is present in the cytosol, ready to bind the C-terminal domains of newly synthesized export substrates (6, 7). This binding prevents premature interactions between substrate monomers, because the chaperone binds to the disordered C-terminal regions required for substrate intersubunit interactions (13). Our results show that chaperone-substrate complexes then interact with FliI or a FliHI complex at the inner membrane. This interaction and/or subsequent interactions would preface dissociation of the negative regulator FliH, FliI ATP hydrolysis coupled to substrate export through the machinery, and release of the chaperone to additional cycles of substrate binding, FliI complex formation and release. It is possible also that the uncomplexed chaperone could exert a posttranscriptional regulatory effect during this cycle (33), and our data are compatible with both co- and posttranslational models for export of type III substrates (34).
There may be parallels between this improved view of the type III export pathway and the well characterized general secretory pathway (GSP). In the GSP, a subset of proteins to be exported across the inner membrane are bound by the GSP-specific cytosolic chaperone, SecB, which has at least two functions: keeping proteins in an unfolded, export-competent state, and facilitating targeting to the membrane-associated ATPase, SecA (35). The ATPase activity of SecA is proposed to supply the energy for transport through the SecYEG channel in the inner membrane (36). Regardless of whether such parallels prove to be of mechanistic significance, the work described here establishes a key docking event in the transition from the cytosolic to membrane stage of the pathway underlying type III export of flagellar components and, by extension, virulence effectors. This view is lent direct support by a parallel study of enteropathogenic E. coli virulence type III secretion, which indicated that the chaperone CesT and its export substrate Tir can interact independently or in complex with the export ATPase, EscN (37).
Acknowledgments
We thank Gillian Fraser and Richard Hayward for useful discussions and critical reading of the manuscript. This work was supported by a Wellcome Trust Program grant and a Biotechnology and Biological Sciences Research Council studentship.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ni-NTA, nickel nitrilotriacetic acid; TCA, trichloroacetic acid.
References
- 1.Macnab, R. M. (1996) in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), pp. 123-145.
- 2.Hueck, C. J. (1998) Microbiol. Mol. Biol. Rev. 62, 379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macnab, R. M. (2003) Annu. Rev. Microbiol. 57, 77-100. [DOI] [PubMed] [Google Scholar]
- 4.Yonekura, K., Maki, S., Morgan, D. G., DeRosier, D. J., Vonderviszt, F., Imada, K. & Namba, K. (2000) Science 290, 2148-2152. [DOI] [PubMed] [Google Scholar]
- 5.Yonekura, K., Maki-Yonekura, S. & Namba, K. (2003) Nature 424, 643-650. [DOI] [PubMed] [Google Scholar]
- 6.Fraser, G. M., Bennett, J. C. & Hughes, C. (1999) Mol. Microbiol. 32, 569-580. [DOI] [PubMed] [Google Scholar]
- 7.Bennett, J. C., Thomas, J., Fraser, G. M. & Hughes, C. (2001) Mol. Microbiol. 39, 781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auvray, F., Thomas, J., Fraser, G. M. & Hughes, C. (2001) J. Mol. Biol. 308, 221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wattiau, P., Woestyn, S. & Cornelis, G. R. (1996) Mol. Microbiol. 20, 255-262. [DOI] [PubMed] [Google Scholar]
- 10.Bennett, J. C. & Hughes, C. (2000) Trends Microbiol. 8, 202-204. [DOI] [PubMed] [Google Scholar]
- 11.Menard, R., Sansonetti, P., Parsot, C. & Vasselon, T. (1994) Cell 179, 515-525. [DOI] [PubMed] [Google Scholar]
- 12.Woestyn, S., Sory, M. P., Boland, A., Lequenne, O. & Cornelis, G. R. (1996) Mol. Microbiol. 20, 1261-1271. [DOI] [PubMed] [Google Scholar]
- 13.Ozin, A. J., Claret, L., Auvray, F. & Hughes, C. (2003) FEMS Microbiol. Lett. 219, 219-224. [DOI] [PubMed] [Google Scholar]
- 14.Delahay, R. M., Shaw, R. K., Elliot, S. J., Kaper, J. B., Knutton, S. & Frankel, G. (2002) Mol. Microbiol. 43, 61-73. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi, S., Fujita, H., Taira, T., Kutsukake, K., Homma, M. & Iino, T. (1984) J. Gen. Microbiol. 130, 3339-3342. [DOI] [PubMed] [Google Scholar]
- 16.Ohnishi, K., Ohto, Y., Aizawa, S., Macnab, R. M. & Iino, T. (1994) J. Bacteriol. 176, 2272-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubori, T., Shimamoto, N., Yamaguchi, S., Namba, K. & Aizawa, S. (1992) J. Mol. Biol. 226, 433-446. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Pedrajo, B., Fraser, G. M., Minamino, T. & Macnab, R. M. (2002) Mol. Microbiol. 45, 967-982. [DOI] [PubMed] [Google Scholar]
- 19.Studier, F. W., Rosenberg, A. H., Dunn, J. J. & Dubendorff, J. W. (1990) Methods Enzymol. 185, 60-89. [DOI] [PubMed] [Google Scholar]
- 20.Miroux, B. & Walker, J. E. (1996) J. Mol. Biol. 260, 289-298. [DOI] [PubMed] [Google Scholar]
- 21.Studier, F. W. & Moffatt, B. A. (1986) J. Mol. Biol. 189, 113-130. [DOI] [PubMed] [Google Scholar]
- 22.Thanabalu, T., Koronakis, E., Hughes, C. & Koronakis, V. (1998) EMBO J. 17, 6487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. (1995) J. Bacteriol. 177, 4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claret, L., Calder, S. R., Higgins, M. & Hughes, C. (2003) Mol. Microbiol. 48, 1349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koronakis, V., Hughes, C. & Koronakis, E. (1991) EMBO J. 10, 3263-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osborn, M. J. & Munson, R. (1974) Methods Enzymol. 31, 642-653. [DOI] [PubMed] [Google Scholar]
- 27.Auvray, F., Ozin, A. J., Claret, L. & Hughes, C. (2002) J. Mol. Biol. 318, 941-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubori, T., Yamaguchi, S. & Aizawa, S. (1997) Mol. Microbiol. 24, 399-410. [DOI] [PubMed] [Google Scholar]
- 29.Minamino, T. & Macnab, R. M. (1999) J. Bacteriol. 181, 1388-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minamino, T., Gonzalez-Pedrajo, B., Kihara, M., Namba, K. & Macnab, R. M. (2003) J. Bacteriol. 185, 3983-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuwajima, G., Kawagishi, I., Homma, M., Asaka, J., Kondo, E. & Macnab, R. M. (1989) Proc. Natl. Acad. Sci. USA 86, 4953-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aldridge, P., Karlinsey, J. & Hughes, K. T. (2003) Mol. Microbiol. 49, 1333-1345. [DOI] [PubMed] [Google Scholar]
- 33.Karlinsey, J. E., Lonner, J., Brown, K. L. & Hughes, K. T. (2000) Cell 102, 487-497. [DOI] [PubMed] [Google Scholar]
- 34.Aldridge, P. & Hughes, K. T. (2001) Trends Microbiol. 9, 209-214. [DOI] [PubMed] [Google Scholar]
- 35.Driessen, A. J. (2001) Trends Microbiol. 9, 193-196. [DOI] [PubMed] [Google Scholar]
- 36.Schiebel, E., Driessen, A. J., Hartl, F. U. & Wickner, W. (1991) Cell 64, 927-939. [DOI] [PubMed] [Google Scholar]
- 37.Gauthier, A. & Finlay, B. B. (2003) J. Bacteriol. 185, 6747-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]