Background: The conserved ATP-dependent chromatin remodeling enzyme Fun30 regulates heterochromatin silencing and DNA repair.
Results: Gene expression is up-regulated, and nucleosome positioning is altered in fun30Δ cells, and purified Fun30 slides nucleosome in vitro.
Conclusion: Fun30 represses gene expression by sliding promoter-proximal nucleosomes.
Significance: The function of Fun30 is expanded to regulation of transcription and chromatin remodeling by nucleosome sliding.
Keywords: Chromatin Regulation, Chromatin Remodeling, Gene Regulation, Nucleosome, Yeast Transcription, Fun30, Nucleosome Mapping, Nucleosome-free Region
Abstract
The evolutionarily conserved ATP-dependent chromatin remodeling enzyme Fun30 has recently been shown to play important roles in heterochromatin silencing and DNA repair. However, how Fun30 remodels nucleosomes is not clear. Here we report a nucleosome sliding activity of Fun30 and its role in transcriptional repression. We observed that Fun30 repressed the expression of genes involved in amino acid and carbohydrate metabolism, the stress response, and meiosis. In addition, Fun30 was localized at the 5′ and 3′ ends of genes and within the open reading frames of its targets. Consistent with its role in gene repression, we observed that Fun30 target genes lacked histone modifications often associated with gene activation and showed an increased level of ubiquitinated histone H2B. Furthermore, a genome-wide nucleosome mapping analysis revealed that the length of the nucleosome-free region at the 5′ end of a subset of genes was changed in Fun30-depleted cells. In addition, the positions of the −1, +2, and +3 nucleosomes at the 5′ end of target genes were shifted significantly, whereas the position of the +1 nucleosome remained largely unchanged in the fun30Δ mutant. Finally, we demonstrated that affinity-purified, single-component Fun30 exhibited a nucleosome sliding activity in an ATP-dependent manner. These results define a role for Fun30 in the regulation of transcription and indicate that Fun30 remodels chromatin at the 5′ end of genes by sliding promoter-proximal nucleosomes.
Introduction
Eukaryotic DNA is packaged with histone proteins to form chromatin, whose fundamental repeating unit is the nucleosome. Each canonical nucleosome contains ∼147 bp of DNA wrapped around a histone octamer, which comprises two each of the histones H2A, H2B, H3, and H4 (1). In addition, a fraction of the nucleosomes in chromatin contains nonallelic variant histones that have specific chromatin localization and tissue- or species-dependent expression patterns (2–4). The dynamics of the nucleosome, characterized by nucleosome position and occupancy, influence DNA accessibility during DNA metabolic processes, including transcription, replication, repair, and recombination (5–8).
Genome-wide high-resolution nucleosome mapping studies in yeast have revealed that ∼60–70% of nucleosomes are well positioned in an asynchronous cell population (9–11). Common primary chromatin features, including a relatively nucleosome-free region (NFR)2 and two flanking, well positioned nucleosomes (−1 and +1 nucleosomes), have been identified at the 5′ end of most genes (12–14). The NFR generally contains sequence-specific transcription factor binding sites and transcription start sites (TSS). The +1 nucleosome is highly enriched with H2AZ (a variant of histone H2A) containing nucleosomes (15–17), with flanking nucleosomes consisting of heterogeneous H2A, H2AZ, or H2A/H2AZ-containing nucleosome subtypes (17). While the −1 nucleosome resides in a region coinciding with the upstream activating sequence, the +1 nucleosome is located downstream of the TSS. As such, control of the dynamics of nucleosomes at the 5′ end of genes is expected to be important for transcription.
Both gene-specific and genome-wide studies indicate that nucleosome position and occupancy can be regulated by DNA sequence and trans-factors, including non-histone chromatin binding proteins, histone modification enzymes, and ATP-dependent chromatin remodeling enzymes (18–20). Of the trans-factors, the ATP-dependent chromatin remodeling enzymes belong to the Snf2 ATPase family, which can be classified into several subfamilies according to the ATPase domain and flanking sequences of their resident catalytic subunit (21). This class of enzymes uses the energy from ATP hydrolysis to break DNA-histone contacts transiently and locally. Consequently, the enzymes in this family could change nucleosome position and occupancy by either sliding or eviction of the histone octamer, eviction of the H2A-H2B dimer, or replacement of H2A-H2B by its variant (22, 23). However, how ATP-dependent chromatin remodeling enzymes alter nucleosome position and occupancy patterns in vivo is not well understood.
We searched the Saccharomyces Genome Database (SGD) for genes that are highly related to Drosophila ISWI, which encodes the ATPase subunit of the chromatin remodeling complex NURF (24). We found that one of the hits is an evolutionarily conserved Snf2 ATPase, Fun30 (Function unknown now 30). Fun30 was initially identified and named by yeast chromosome I cloning and sequencing projects (25, 26). Previous studies have implicated a role of Fun30 and its higher eukaryotic homolog Fft3 (pombe) or SMARCAD1 (mammalian) in promoting heterochromatin silencing (27–30). In addition, Fun30 and SMARCAD1 have been shown to facilitate DNA end resection in homologous recombination and regulate checkpoint deactivation (31–33). However, despite its emerging biological roles, how Fun30 remodels chromatin to regulate these processes in vivo is not well understood. Interestingly, a recent study has shown that Fun30 forms a homodimer and exhibits a histone dimer exchange activity in vitro (34).
Phylogenetic studies revealed that Fun30 is closely related to the Swr1 and Ino80 subfamily of ATP-dependent chromatin remodeling enzymes (21), both of which have important biological functions, including regulation of transcription (35). While Ino80 has nucleosome sliding, spacing, and displacement activities (36–38), the SWR1 enzyme complex is responsible for site-specific incorporation of H2AZ (39). Furthermore, a yeast synthetic genetic array analysis (40) revealed that Fun30 genetically interacts with four subunits of the SWR1 complex and H2AZ, suggesting a functional connection between the two activities. We hypothesize that Fun30 plays a role in regulation of gene expression through remodeling chromatin. To test this hypothesis, we sought to identify biological targets of Fun30 to define its role in regulation of transcription using cDNA microarray and chromatin immunoprecipitation (ChIP) assays. We mapped and analyzed genome-wide nucleosome patterns at the 5′ end of genes in wild-type and fun30Δ cells by Illumina mononucleosomal DNA sequencing technique. We purified Fun30 and characterized its nucleosome remodeling activity in vitro. Our results indicate that Fun30 represses transcription by sliding nucleosomes flanking the +1 H2AZ-containing nucleosome at the 5′ end of genes.
EXPERIMENTAL PROCEDURES
Yeast Strains, Medium, and Growth Conditions
Three copies of the FLAG epitope tag were fused to the C terminus of the FUN30 locus of W303 cells to generate strain YWH502, or the FUN30 locus of BY4741 cells to generate strain YWH505. A PCR-amplified kanMX4 dominant drug-resistance cassette, flanked by 40 nucleotides upstream and downstream of the FUN30 locus, was transformed into BY4741 cells to generate the fun30Δ strain (YWH15). Similarly, the htz1Δ strain (YWH367) was generated in BY4742 cells using the natMX4 cassette. The fun30Δhtz1Δ double mutant (YWH854) was generated by crossing strains YWH367 and YWH15, followed by tetrad dissection. All gene deletion or tagging strains were confirmed by PCR amplification of the targeting cassette that was used for single gene disruption or gene tagging.
The yeast rich medium YPD (1% yeast extract, 2% peptone, 2% dextrose) was prepared according to standard recipes. Yeast was grown at 30 °C in YPD medium until A600−1 if not stated otherwise in the text or figure legends.
RNA Isolation, cDNA Microarray, and Data Analysis
The wild type and mutants were harvested at ∼A600 in YPD medium at 30 °C. Total RNA was isolated using the standard hot acid phenol extraction method, followed by purification with an RNeasy kit (Qiagen). RNA reverse transcription, cDNA labeling, and microarray hybridization were performed as described previously (39). For each strain, three biological replicates and experimental dye swap duplicates of each replicate were used. Microarray raw data sets were analyzed using the LIMMA package in R. Briefly, for data normalization, we used functions for background correction, normalization within and between arrays, and correlation between duplicates. For differential expression gene analysis, we used a linear fitting model and eBayes statistics to select differentially expressed genes with false discovery rates of less than 0.05. The output from LIMMA is the average of changes (in log2 value) of three replicates of the mutant. A gene ontology analysis was performed using Gene Ontology Slim Mapper in SGD and a functional hierarchy of the Saccharomyces cerevisiae transcriptional system (41). When comparing differential expression gene generated from different mutants using a Venn diagram, the significance of overlap was calculated by hypergeometric distribution. An examination of the individual up-regulated gene by quantitative PCR was performed using the CFX96 Touch real-time PCR detection system (Bio-Rad) following reverse transcription. ACT1 was used as a control for normalization.
Fun30 ChIP
The Fun30-3FLAG strain was grown at 30 °C until A600. Formaldehyde cross-linking (1%) was performed at room temperature for 20 min with gentle shaking. Cell breaking, DNA shearing, and FLAG immunoprecipitation were performed essentially as described previously (42). Primers for quantitative PCR were designed every 500–800 base pairs using the primer design function in SGD. A NFR at the HSC82 locus was used as control for signal normalization. No-tag ChIP negative control was performed once, and no signal was detected.
Mononucleosome Isolation, Illumina Sequencing, and Data Analysis
The wild type and mutant were grown at ∼A600 in YPD at 30 °C. Cells were fixed with 1% paraformaldehyde at room temperature for 30 min. Crude yeast nuclei were extracted as described (43). Micrococcal nuclease (MNase) titrations were performed at 37 °C for 10 min as described (12). Mononucleosomal DNA was gel-purified and subjected to Illumina sequencing as described previously. Sequence reads (∼25 base pairs) were mapped to the reference yeast genome (2003) in Bowtie 0.12.7. Reads that were mapped to a unique location and contained up to two mismatches were selected. The FASTQ files for the wild type and mutant were then analyzed by a template filtering algorithm to extract nucleosome features including nucleosome center position, nucleosome left and right boundary, and nucleosome occupancy (44). Nucleosome occupancy for each nucleosome was first normalized with the total uniquely mapped reads of the corresponding strain. Quantile normalization was then performed for normalized nucleosome occupancy between the wild-type and mutant strains before further analysis.
When a TSS was located within 200 bp upstream of the ATG start codon, the +1 nucleosome was defined as the nucleosome whose center was closest to ATG between the TSS and ATG. In all other cases, the +1 nucleosome was defined as the nucleosome whose center was closest to ATG in a 350-bp window flanking the ATG start codon (-200 bp < ATG < 150 bp).
For NFR length analysis, the NFR was defined as the nucleosome-free region immediately adjacent to the left boundary of the +1 nucleosome. Genes were divided into five groups according to NFR length by k-means clustering. The frequency for every NFR length was counted in each group. A smoothed distribution of relative frequency (the frequency for each NFR length normalized by total NFR frequencies) was then plotted as a function of NFR length (base pairs). A two-sample Kolmogorov-Smirnov test for the distributions of the wild type and mutant in each group was performed.
For nucleosome center shift analysis, the −1 nucleosome was defined as the last nucleosome whose 3′ end was located within 287 bp upstream of the 5′ end of the +1 nucleosome. The +2 nucleosome was the first nucleosome whose 5′ end was located within 165 bp downstream of the 3′ end position of the +1 nucleosome, while the +3 nucleosome was the first nucleosome whose 5′ end was located within 165 bp from the 3′ end of the +2 nucleosome. In the absence of the +2 nucleosome, the +3 nucleosome was defined as the first nucleosome whose 5′ end was located within 330 bp from the 3′ end of the +1 nucleosome. Nucleosome center difference of each nucleosome (i.e. the −1 nucleosome) between the wild type and mutant (nucleosome center coodinatemutant − nucleosome center coodinatewild type for genes on the Watson strand and nucleosome center coodinatewild type − nucleosome center coodinatemutant for genes on the Crick strand) in all annotated genes was calculated. Kernel density estimation (KDE) of the nucleosome center differences (i.e. the −1 nucleosome) of 83 Fun30-target genes was plotted as a function of base pair difference. The random 83 genes were sampled 100 times. The KDE plot for the random 83 genes is an averaged density estimation of 100 random 83-gene lists. A randomization test in the range of one standard deviation (± 72 bp) was performed as described previously (45). Briefly, the relative frequency (scores) of the center differences between the 83 Fun30-target genes and each of the100 random 83 genes were calculated within one standard deviation, respectively. The p values were obtained by finding the rank of Fun30-target genes among the 100 scores.
For nucleosome occupancy analysis, occupancy ratios of normalized nucleosome occupancy (> 0) of the corresponding nucleosomes in wild type and mutant were compared (ratio = normalized nucleosome occupancymutant nucleosome/normalized nucleosome occupancywild type nucleosome). The corresponding mutant nucleosome was defined as the nucleosome whose center was closest to the center of the wild type nucleosome in the range of ± 80 bp from the wild-type nucleosome center. When the nucleosome at the boundary of the 1000-bp window was absent in either the wild type or mutant, that nucleosome would be excluded for comparison. For 0.6-fold change, genes in the mutant that have a maximum ratio < 1.2 and a minimum ratio < 0.6 were selected. For 1.8-fold change, genes in the mutant that have a minimum ratio of > 0.8 and a maximum ratio of > 1.8 were selected. A paired Student's t test for corresponding nucleosome occupancy in the wild type and mutant was performed. Genes with a p value < 0.05 were selected. The corresponding nucleosome data from the wild type and mutant were plotted.
Protein Purification
Yeast wild-type and ATPase mutant Fun30 proteins were purified using one-step affinity purification with anti-FLAG M2-agarose beads (Sigma). Fun30 was purified using the 0.5 m KCl wash condition and eluted with FLAG peptide as described (46).
ATPase Assay
Each standard reaction mix, containing 2 nm purified wild-type or ATPase mutant Fun30 enzyme, 30 μm ATP, 5 μCi [α-32P]ATP (PerkinElmer), was incubated with 50 ng of DNA or nucleosome (50 ng of DNA equivalents) at 30 °C for 30 min. One microliter of reaction mixture was separated on a TLC plate and later visualized by autoradiography. Signals were quantified on a Typhoon 9400 (GE Healthcare).
Mononucleosome Sliding Assay
A standard nucleosome mobilization assay was performed as described previously (47). In brief, mononucleosomes were assembled using a 359-bp Drosophila hsp70 promoter and reconstituted Drosophila histone octamers. Each 10-μl reaction contained 10 mm Tris-HCl (pH7.5), 50 mm NaCl, 3 mm MgCl2, 1 mm β-mercaptoethanol, 100 μg/ml BSA, 1 mm ATP, 5.8 nm mononucleosome, and ∼1 nm or 2 nm of wild-type Fun30 or ∼4.8 nm of ATPase mutant Fun30 (K603A). The reactions were incubated at 37 °C for 30 min and analyzed using a 4% PAGE gel. Signals visualized by SYBR Green I staining were quantified on a Typhoon 9400 (GE Healthcare).
RESULTS
Derepression of Stress Response and Metabolism Genes in fun30Δ
A synthetic lethal genetic screen found that Fun30 genetically interacts with subunits of the SWR1 complex and histone H2AZ (40). Given that synthetic lethality or sickness can occur between genes acting in the same biochemical pathway or in distinct but compensatory pathways (48, 49), the synthetic sickness phenotype of the double deletion mutant fun30Δhtz1Δ (HTZ1 is the gene name for H2AZ in yeast) suggests that Fun30 and H2AZ are functionally connected. Because H2AZ nucleosomes have been implicated in regulation of gene activation or repression (50), we examined the role of Fun30 in the regulation of gene expression. To identify gene targets of Fun30 under physiological conditions, we synthesized cDNA using total RNA isolated from log phase cells grown under YPD medium conditions, followed by two-color gene expression microarray hybridization and data analysis.
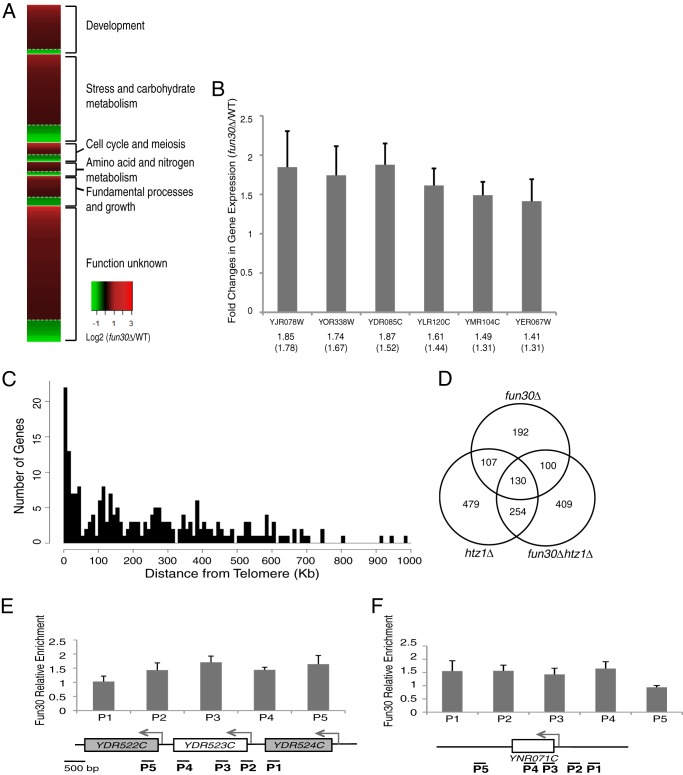
We identified a total of 275 genes as Fun30-dependent genes whose RNA levels in fun30Δ were at least 1.3-fold higher or 0.65-fold lower than those in the wild type (Fig. 1A). A gene ontology analysis revealed that 60% of these genes are annotated and required for various biological processes. Among them, ∼50% are required for the stress response, meiosis, and carbohydrate, amino acid, and nitrogen metabolism (Fig. 1A). In addition, of the 275 Fun30-dependent genes, the expression levels of 223 (88%) were up-regulated. This up-regulation of gene expression was confirmed by RT-PCR of individual affected genes (Fig. 1B). It is possible that many of these Fun30-repressed genes are located at or near heterochromatin because Fun30 has been shown recently to promote heterochromatin silencing (27, 30). We then plotted the number of Fun30-dependent genes as a function of distance from telomere to centromere. While there are ∼15% of the genes locating in the telomere and subtelomere regions, the majority of the genes locate widespread along chromosomes (Fig. 1C). Taken together, these results suggest that Fun30 is required for transcriptional repression of a subset of genes.
FIGURE 1.
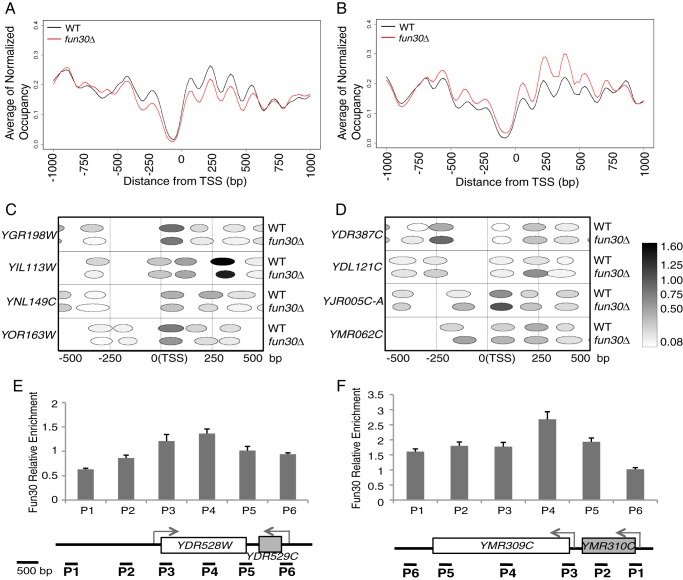
Fun30-dependent genes. A, Fun30 represses the transcription of genes required for various biological functions. Fun30-dependent genes were defined as genes whose expression is more than 1.3-fold or less than 0.65-fold in the mutant compared with the wild type with a false discovery rate value of p < 0.05. B, gene expression measurement of individual up-regulated genes by quantitative RT-PCR. Values indicate relative fold changes in gene expression detected by quantitative RT-PCR or microarray (in parentheses), respectively. C, chromosomal locations of Fun30-dependent genes relative to the telomere. The diagram shows the number of Fun30-dependent genes (y axis) plotted as a function of distance from telomeres (x axis). D, Venn diagram showing the overlap of affected genes among the three mutants (fun30Δ, 1.2-fold up and 0.65-fold down; htz1Δ, 1.3-fold up and 0.6-fold down; and fun30Δhtz1Δ, 1.3-fold up and 0.6-fold down). E and F, Fun30 is enriched at its target genes. The bar diagrams show in vivo binding of Fun30-FLAG as measured by real-time PCR quantification of Fun30-FLAG ChIP-enriched DNA at the indicated loci (from P1 through P5). Error bars represent the S.D. of three independent experiments of a total of six ChIP reactions.
Fun30 and H2AZ Regulate the Expression of a Common Subset of Genes
We compared the transcription profile of the fun30Δ strain to the profiles of htz1Δ and fun30Δhtz1Δ strains and found that the expression of 130 genes was affected in all three strains (Fig. 1D). These 130 overlapping genes can be further divided into three groups: 12 genes belong to both Fun30 and H2AZ-activated genes, 72 genes belong to both Fun30 and H2AZ-repressed genes, and 46 genes are Fun30-repressed but H2AZ-activated genes. These results suggest that although Fun30 and H2AZ each have different functions, as shown by the non-overlapping affected genes, they also regulate the expression of a common set of genes. Moreover, the transcription profile of the fun30Δhtz1Δ mutant revealed that many affected genes on chromosome III increased about 2-fold and were located next to each other (data not shown), suggesting regional chromosomal duplications on chromosome III in the double mutant. This may partially explain the high degree of non-overlapping genes between the double mutant and the fun30Δ or htz1Δ single mutant.
To determine whether these 130 genes are direct targets of Fun30, we examined Fun30 localization by ChIP. We found that Fun30 is uniformly distributed along the genes, including the 5′ and 3′ ends and within open reading frames (Fig. 1, E and F). Here we refer to the 130 overlapping genes among the three mutant strains as Fun30-target genes, and we proceeded to analyze the nucleosome positions in these genes (see later).
Local but Not Global Nucleosome Organization at the 5′ End of Genes Is Affected in fun30Δ
Genome-wide nucleosome distributions have been mapped in yeast by high-resolution tilling array or next-generation sequencing. Nucleosomes at the 5′ end of the genes are better phased than those in the coding regions or 3′ end. To examine nucleosomes across the genome in wild-type and mutant cells, we isolated mononucleosome DNA from MNase-digested chromatin for Illumina sequencing. Following sequence read alignments against the yeast reference genome (2003 Saccharomyces Genome Database genome build) in Bowtie, we determined the position and occupancy of nucleosomes using a template filtering algorithm (44).
To compare nucleosome features at the 5′ end of genes, we aligned nucleosomes using a TSS of each gene (51). We found that the average genome-wide nucleosome organization in a 1000-bp window aligned by TSS in fun30Δ cells was similar to that of the wild type (data not shown), which suggests that Fun30 is not required for maintaining global chromatin structure under physiological conditions, unlike the Isw1 and Chd1 chromatin remodeling enzymes (52).
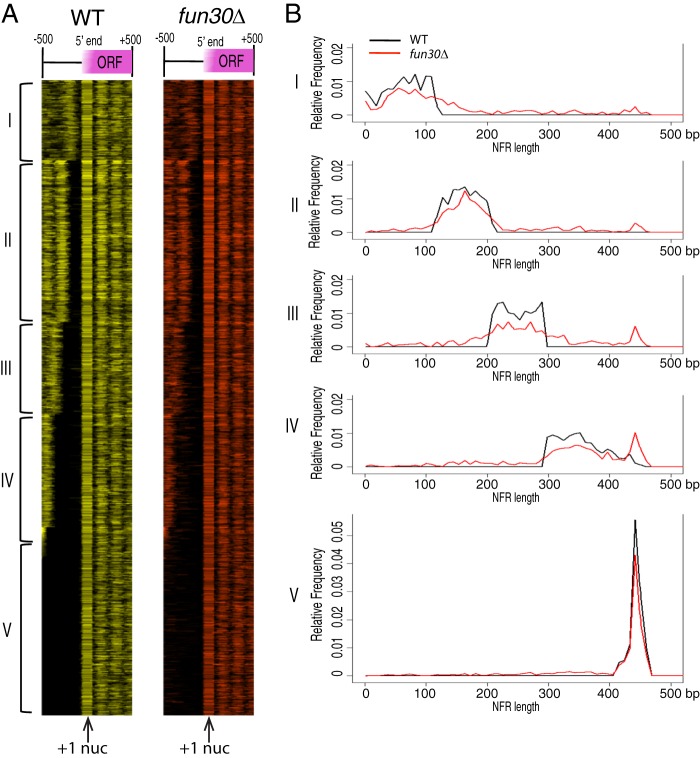
The length of the NFR at the 5′ end of genes can influence transcription (53, 54). To examine the NFR in fun30Δ cells, we aligned the nucleosomes of 5062 genes by the +1 nucleosome center (Fig. 2A). We noticed that many genes in our data, in addition to the highly transcribed genes such as ribosome protein genes, lacked the −1 and −2 nucleosomes. This is likely to be caused by MNase overdigestion because the pattern is similar to the nucleosome pattern from overdigested chromatin by MNase, as described by Weiner et al. (44). We separated the genes into five groups by k-means clustering, with each group containing genes that have similar nucleosome patterns flanking the NFR (Fig. 2A). For each group of genes, we plotted the smoothed relative NFR frequency as a function of the NFR length. Comparing with the wild type, we found that the NFR length distribution was significantly (p < 2.2e-16) altered in all five groups in Fun30-depleted cells (Fig. 2B). These results suggest that Fun30 remodels nucleosomes at the 5′ end of genes.
FIGURE 2.
Nucleosome organization and NFR length distribution at the 5′ end of genes in the wild type and fun30Δ. A, nucleosomes (nuc) at the 5′ ends of 5062 genes were aligned according to the +1 nucleosome center. Genes were clustered into five groups by k-means clustering on the basis of NFR length. B, relative frequency distribution of NFR length in the wild type and fun30Δ. A two-sample Kolmogorov-Smirnov test (p < 2.2e-16) was performed to show the significant difference in distribution between the wild type and mutant in each group.
Nucleosome Centers at the 5′ End of Fun30 Target Genes Are Shifted in fun30Δ
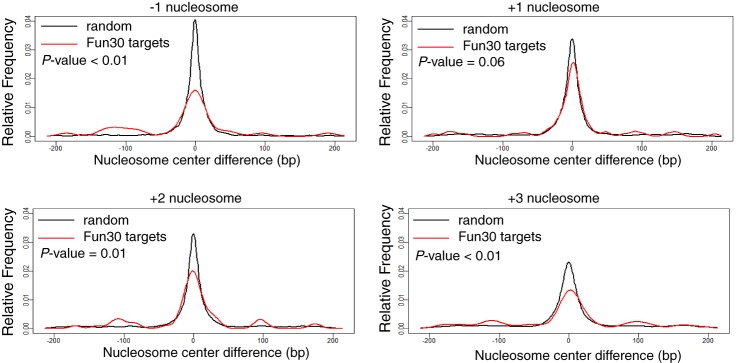
Altered NFR length could result from changes in nucleosome positioning. We next examined the center position of individual nucleosomes (−1, +1, +2, or +3 nucleosome) at the 5′ end of Fun30-target genes. Of the 130 Fun30 target genes identified by microarray data analysis, 83 genes have nucleosome sequencing data. For each nucleosome position of the 83 genes, we calculated the KDE using nucleosome center differences between fun30Δ and the wild type. We then compared the KDE plot of Fun30-target genes to the plot of the randomly selected 83 genes. We found that, while there are minor changes found at the +1 nucleosomes, the peak of KDE is changed from 0.04 in the wild type to 0.016 in the fun30Δ mutant at the −1 nucleosome, from 0.023 to 0.013 at the +3 nucleosome and, to a lesser extent, from 0.033 to 0.020 at the +2 nucleosome (Fig. 3). These results indicate that Fun30 changes nucleosome positioning at the 5′ end of its target genes, preferentially at −1, +2, and +3 nucleosomes, nucleosomes that are around the +1 H2AZ-containing nucleosome.
FIGURE 3.
Fun30 slides nucleosomes at the 5′ end of genes in vivo. The KDE of center difference at each nucleosome (−1, +1, +2, and +3 nucleosomes) of 83 Fun30 target genes was compared with that of 83 randomly selected genes. The p values for significant difference in the nucleosome center shift between Fun30 targets and random genes are shown.
Nucleosomes at Promoters of Fun30-repressed Genes Show an Increased Level of Ubiquitinated Histone H2B and a Reduced Level of Histone H3 Lysine 4 Trimethylation, Lysine14 Acetylation, and Histone H4 Acetylations
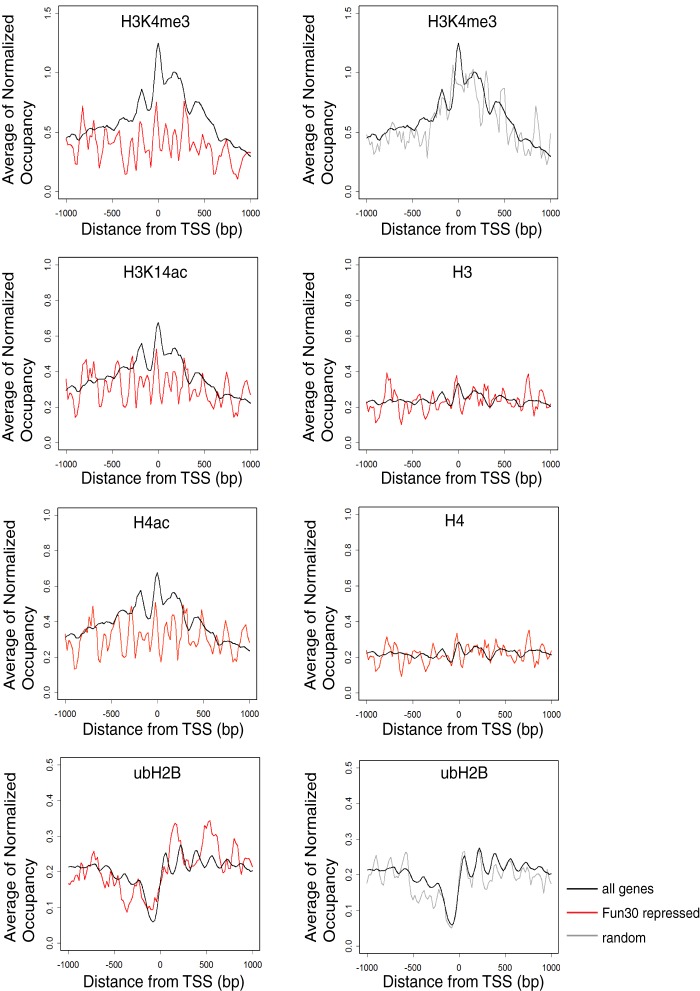
To further characterize the nucleosomes at Fun30 target genes, we examined their histone modification patterns using published data sets (55, 56). We compared histone acetylation, methylation, and ubiquitination patterns at the 5′ end of 73 Fun30-repressed genes and total genes in wild-type cells. While histone H3K4me1, H3K4me2, H3K36me3, H3K79me3, and H3K9ac modifications of Fun30-target genes are slightly reduced or indistinguishable from the total genes (data not shown), there is a substantially reduced level of H3K4me3, H3K14ac, and H4Kac at the promoters of these genes (Fig. 4). Interestingly, we also observed an increase of ubH2B (H2BK123 ubiquitination) in the open reading frames of Fun30 target genes (Fig. 4). These results corroborate our finding that Fun30 plays a role in transcriptional repression.
FIGURE 4.
Histone modifications at the 5′ end of Fun30-repressed genes. Average occupancies of histone H3; H3 lysine 4 trimethylation; H3 lysine 14 acetylation; H4; H4 acetylation; and ubiquitinated histone H2B were compared between 73 Fun30-repressed genes and all genes or 73 randomly selected genes.
Chromatin Remodeling by Fun30 Is Independent of Transcription
The chromatin remodeling activity of Fun30 could be independent of transcription. To address this possibility, we next analyzed nucleosome data generated from 5062 genes by comparing nucleosome occupancy. In a 1000-bp window centered by a TSS, we selected genes that have at least one nucleosome whose occupancy is lower (< 0.6-fold) or higher (> 1.8-fold) in fun30Δ than in wild-type cells. We then plotted the average nucleosome features (nucleosome occupancy, center position, and width) of the selected genes in a 2000-bp window. We found a total of 181 genes that showed an altered average nucleosome profile at the 5′ end of genes in Fun30-depleted cells (Fig. 5, A and B). When we plotted the nucleosomes of individual genes in the 1000-bp window flanking the TSS, we observed that the position or width of many “occupancy”-changed nucleosomes was altered as well (Fig. 5, C and D). Interestingly, we also observed an occupancy change in the +1 H2AZ-containing nucleosomes as exemplified in YJR005C-A and YMR062C, suggesting that remodeling of flanking nucleosomes by Fun30 may regulate SWR1-mediated H2AZ exchange. Furthermore, these changes are likely caused directly by Fun30, as confirmed by Fun30 ChIP at these regions (Fig. 5, E and F). Given that the expression of many of these genes is not affected in fun30Δ, we conclude that, in addition to transcription, chromatin remodeling by Fun30 may be important for other biological processes.
FIGURE 5.
Chromatin remodeling by Fun30 at the 5′ end of a subset of genes is independent of transcription. A and B, pairwise comparison of averages of nucleosome features (occupancy, center position, width) of 86 or 95 genes whose nucleosome occupancies were lower or higher in fun30Δ. The y axis is an average of normalized occupancy. C and D, plots of nucleosome organization at the 5′ end of individual genes. The color key indicates the range of normalized nucleosome occupancy of depicted nucleosomes. E and F, Fun30 localization at occupancy-changed genes. Bar diagrams show in vivo binding of Fun30-FLAG as measured by real-time PCR quantification of Fun30-FLAG ChIP-enriched DNA at the indicated loci (from P1 to P6). Error bars represent the S.D. of three independent experiments of a total of six ChIP reactions.
Fun30 Slides Nucleosomes in Vitro in an ATP-dependent Manner
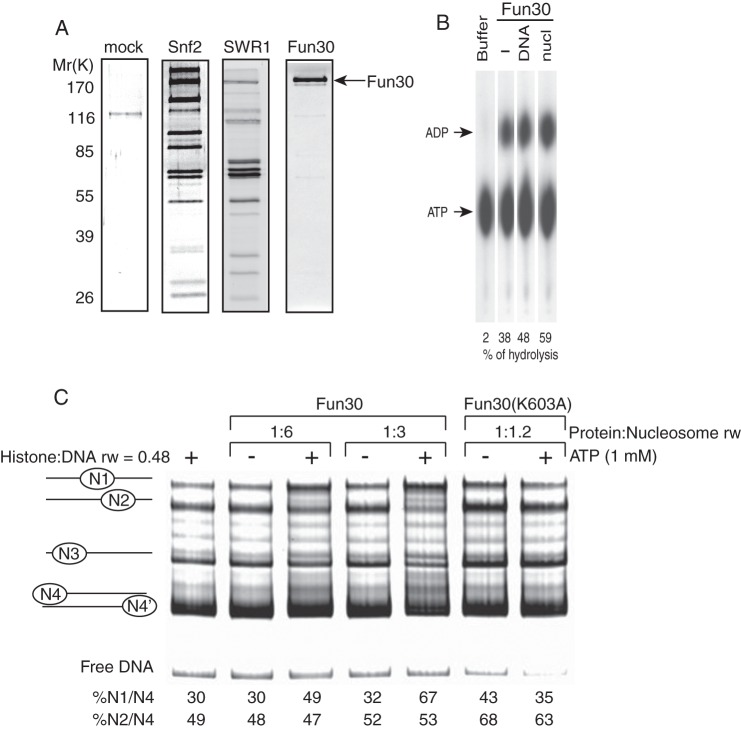
Previous studies have demonstrated that chromatin remodeling enzymes, including NURF and Ino80, remodel chromatin by moving nucleosomes along the DNA in vitro using a mononucleosome sliding assay (37, 47). Our observations that Fun30 changes nucleosome positioning in vivo prompted us to examine whether purified Fun30 by itself has nucleosome sliding activity using the same assay. We first purified the protein by affinity purification from a strain in which a triple FLAG epitope tag was inserted at the C terminus of the FUN30 locus. Fun30-FLAG appears to function as the wild-type Fun30 because the constructed strain does not display mutant phenotypes. Unlike the Snf2 or SWR1 chromatin-remodeling enzyme, which contains 11 or 14 identified subunits (39, 57), anti-FLAG beads pulled down only Fun30 under the same purification and high salt washing conditions (Fig. 6A), which is consistent with a previous study that Fun30 exists as a single component protein (34). Our purified Fun30 also has both DNA and nucleosome-stimulated ATPase activity (Fig. 6B), as measured by the yield of ADP. While DNA stimulated 48% of ATP hydrolysis, nucleosomes stimulated 59% of ATP hydrolysis by Fun30. We observed that Fun30 has a high level of intrinsic ATPase activity (38% of ATP hydrolysis). This could be contributed by the endogenous DNA or substoichiometric nucleosomes that copurified with Fun30, as was shown previously for the intrinsic ATPase activity of the Ino80 complex (37).
FIGURE 6.
Purified Fun30 slides nucleosomes in an ATP-dependent manner. A, Fun30 is a single-component enzyme. Shown are SDS-PAGE (10% gel) and silver staining analysis of anti-FLAG affinity-purified Snf2, SWR1 complexes, and Fun30 under the same purification and wash conditions. The mock purification shows one major contaminant. B, thin-layer chromatography analysis shows ATPase activity of Fun30. nucl denotes nucleosome. C, Fun30 slides nucleosomes in an ATP-dependent manner. The native PAGE gel shows the positions of mononucleosomes from N1 to N4. Mononucleosome (5.8 nm) mobilization by Fun30 in the presence of ATP is indicated by reduction of the N2 and N4 band intensity and increase of the N1 band intensity. rw denotes ratio of molar mass weight.
The mononucleosomes in our sliding assay can be resolved as four major nucleosome species (N1-N4) on a native PAGE. These N1-N4 species represent thermodynamically stable positions of nucleosome core particles on the 359-bp DNA, with N1 at the center and N4 or N4′ at either end of the DNA. In the presence of ATP and substoichiometric amounts of Fun30, the relative band intensity enrichment of N1 over N4-N4′ was increased substantially, from 30 to 49% or from 32 to 67% (Fig. 6C). In contrast, the band intensities of N2, N3, and N4-N4′ were reduced and displayed a similar degree of reduction because the ratios of band intensities of N2 over N4-N4′ were similar with or without ATP (Fig. 6C). In addition, the nucleosome sliding activity of Fun30 requires the energy from ATP hydrolysis because we failed to observe changes in nucleosome band intensities when the purified ATPase mutant Fun30 (K603A) was used in the reaction (Fig. 6C). These results corroborate our in vivo findings by showing a direct nucleosome sliding activity of Fun30.
DISCUSSION
In this study, we identified a role of Fun30 in the regulation of transcriptional repression by altering nucleosome positioning. We found that Fun30 represses gene expression, including genes involved in amino acid and carbohydrate metabolism, the stress response, and meiosis. Consistent with its role in transcriptional repression, we found that the Fun30 target genes are mildly enriched with ubH2B, a histone modification associated with gene repression (58), while lacking the gene activation-associated histone modifications, including H3K4me3 and H3K16ac. We also observed that the length of the NFR at the promoters of many genes is altered in the absence of Fun30. Furthermore, consistent with its in vitro nucleosome sliding activity, we found that Fun30 slides the −1 and +2 nucleosomes flanking the +1 nucleosome, as well as the +3 nucleosome, at the 5′ end of its target genes in vivo. Finally, we showed that Fun30 also changes the chromatin structure at genes whose expression is not affected, suggesting that the chromatin remodeling activity of Fun30 is independent of transcription.
Under physiological conditions, unlike Isw1 and Chd1, which are required for global nucleosome organization (52), we found that Fun30 is required for nucleosome structure at specific loci. We and others (59) have shown that Fun30 is enriched at the gene coding regions, including the promoter, open reading frame, and 3′ end. In addition, Fun30 is also enriched at the boundary of the telomeres and euchromatin, centromeres, double strand breaks, non-coding RNAs, Ty element, and autonomously replicating sequence (33, 59), suggesting that Fun30 plays a role in various biological processes. Indeed, recent studies have linked Fun30 to promoting heterochromatin silencing, maintaining chromatin structure at the centromere, and DNA repair (27, 30–33, 59). Our study extended these findings by linking Fun30 to repression of transcription. In addition, our observation that Fun30 remodels nucleosomes at loci where gene expression was not changed suggest that first, the function of Fun30 at these loci might be important for transcriptional regulation under non-physiological growth conditions; second, Fun30 plays a role in other important biological processes because chromosome features, including non-coding RNA and autonomously replicating sequences, often coincide at the 5′ end of genes.
Both sequence-specific DNA-binding transcription factors and chromatin remodeling around the NFR have been shown to regulate NFR length at promoters, which could influence gene activation and repression. Earlier studies showed that the DNA binding factors Reb1 and Abf1 are highly enriched in the NFRs of a subset of promoters (16). Consistent with this observation, it has been reported that NFR-bound Reb1 recruits the RSC chromatin remodeling enzyme, which would increase the NFR length by moving NFR flanking nucleosomes away from the NFR (60). In contrast, chromatin remodeling enzyme Isw2 has been shown to reduce NFR length by moving flanking nucleosomes toward the NFR, presumably through the recruitment of Ume6 or the Tup1-Ssn6 complex (53, 61, 62). Interestingly, Tup1-Ssn6 was shown recently to be required for stable occupancy of the promoter nucleosome (P nucleosome), a nucleosome occupying the NFR that is frequently lost in Ssn6- or Tup1-depleted cells (54). Our studies extend these findings by showing that position of −1 nucleosomes was shifted in the absence of Fun30, which led to the changes in NFR length to up-regulate transcription. In addition, unlike other known chromatin remodeling enzymes that exhibit directionality in nucleosome remodeling (60, 61, 63), of the gene promoters we examined, nucleosome sliding by Fun30 in vivo appears to lack directionality. Therefore, additional factors, including histone modifications or other chromatin binding proteins, could also contribute to Fun30-regulated transcriptional repression. Nonetheless, our results indicate that Fun30 is required for proper NFR length in the regulation of transcription.
A recent study has shown that Fun30 exhibits histone dimer exchange activity in vitro (34). During the course of nucleosome data analysis, in addition to nucleosome position shift, we also observed nucleosome loss or gain at the 5′ end of a subset of genes in the fun30Δ mutant (Fig. 2A, data not shown). Strikingly, the position, occupancy, or width of most, but not all, flanking nucleosomes was changed as well. We speculate several possibilities that cause nucleosome “loss” or “gain” in the mutant. First, Fun30 could be directly involved in the displacement or deposition of nucleosomes, as it has been shown that Fun30 exhibits histone dimer displacement and deposition activity in vitro. Second, Fun30 could slide nucleosomes to specific loci to render the nucleosomes in wild-type cells either more sensitive (gain in fun30Δ) or resistant (loss in fun30Δ) to MNase digestion. Third, Fun30 could slide nucleosomes to the neighboring predicted nucleosome position (gain in mutant) or to an NFR (loss in mutant) as NFRs have been found throughout the genome in addition to gene promoters (53). Fourth, Fun30 could slide flanking nucleosomes to facilitate nucleosome gain or loss by another chromatin remodeler or nucleosome chaperone. This latter possibility could explain recent findings that H2AZ nucleosomes mislocalize in Fun30-depleted cells (29, 59). It will be interesting to examine the fate of H2AZ nucleosomes at our identified Fun30 target genes. Depending on the local chromatin context, these possibilities are not mutually exclusive.
Chromatin remodeling enzymes can be recruited to chromatin through gene-specific transcription factors, general transcription factors, or recognition of histone modifications. Although Fun30 has been shown to have a higher affinity for unphosphorylated H2A-containing nucleosomes than for γ-H2AX-containing nucleosomes in vitro (33), it is not clear how Fun30 is recruited to chromatin. Fun30 and its homologs have a putative ubiquitin interaction domain, the CUE motif. Although deletion of the CUE motif exhibits a reduced level of silencing at the HMR locus, overexpression of CUE-deleted or mutated FUN30 results in slow growth in galactose medium (27). A possible role of the CUE motif could be recognizing ubiquitinylated histones. Our observation that Fun30 targets had an increased level of ubH2B suggests a functional interaction between the CUE motif of Fun30 and ubH2B. Extensive efforts are needed to uncover specific histone modifications, protein factors such as transcription factors, and Fun30 modifications that are critical for Fun30 recruitment and its functions.
Acknowledgments
We thank Iouri Chepelev, Oliver Rando, and Assaf Weiner for advice on DNA reads alignment, normalization, and analysis; members of the Carl Wu laboratory for helpful advice and discussions; Xuetong Shen, Ju-Gyeong Kang, Paul Badenhorst, Michael Lichten, Yikang Rong, Ed Luk, Toshio Tsukiyama, and James Haber for helpful advice and discussions. We also thank Keith Robertson and Darrell Brann for critical reading of the manuscript.
This work was supported, in whole or in part, by NHLBI National Institutes of Health intramural research support (to K. Z.), NCI National Institutes of Health intramural research support (to C. W.), and NCI National Institutes of Health Grant 1K22CA122453 (to W.-H. W.).
The microarray data and the mononucleosomal DNA sequencing data sets reported in this paper have been submitted to the Gene Expression Omnibus Repository. The accession number is GSE48571.
- NFR
- nucleosome-free region
- TSS
- transcription start site
- MNase
- micrococcal nuclease
- KDE
- kernel density estimation.
REFERENCES
- 1. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2. Banaszynski L. A., Allis C. D., Lewis P. W. (2010) Histone variants in metazoan development. Dev. Cell 19, 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ray-Gallet D., Almouzni G. (2010) Nucleosome dynamics and histone variants. Essays Biochem. 48, 75–87 [DOI] [PubMed] [Google Scholar]
- 4. Talbert P. B., Henikoff S. (2010) Histone variants. Ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11, 264–275 [DOI] [PubMed] [Google Scholar]
- 5. Clapier C. R., Cairns B. R. (2009) The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 [DOI] [PubMed] [Google Scholar]
- 6. Jiang C., Pugh B. F. (2009) Nucleosome positioning and gene regulation. Advances through genomics. Nat. Rev. Genet. 10, 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ransom M., Dennehey B. K., Tyler J. K. (2010) Chaperoning histones during DNA replication and repair. Cell 140, 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seeber A., Hauer M., Gasser S. M. (2013) Nucleosome remodelers in double-strand break repair. Curr. Opin. Genet. Dev. 23, 174–184 [DOI] [PubMed] [Google Scholar]
- 9. Rando O. J., Winston F. (2012) Chromatin and transcription in yeast. Genetics 190, 351–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiang C., Pugh B. F. (2009) A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 10, R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brogaard K., Xi L., Wang J. P., Widom J. (2012) A map of nucleosome positions in yeast at base-pair resolution. Nature 486, 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yuan G. C., Liu Y. J., Dion M. F., Slack M. D., Wu L. F., Altschuler S. J., Rando O. J. (2005) Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309, 626–630 [DOI] [PubMed] [Google Scholar]
- 13. Schones D. E., Zhao K. (2008) Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 9, 179–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cairns B. R. (2009) The logic of chromatin architecture and remodelling at promoters. Nature 461, 193–198 [DOI] [PubMed] [Google Scholar]
- 15. Albert I., Mavrich T. N., Tomsho L. P., Qi J., Zanton S. J., Schuster S. C., Pugh B. F. (2007) Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446, 572–576 [DOI] [PubMed] [Google Scholar]
- 16. Raisner R. M., Hartley P. D., Meneghini M. D., Bao M. Z., Liu C. L., Schreiber S. L., Rando O. J., Madhani H. D. (2005) Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123, 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luk E., Ranjan A., Fitzgerald P. C., Mizuguchi G., Huang Y., Wei D., Wu C. (2010) Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell 143, 725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Segal E., Widom J. (2009) What controls nucleosome positions? Trends Genet. 25, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jansen A., Verstrepen K. J. (2011) Nucleosome positioning in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 75, 301–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sadeh R., Allis C. D. (2011) Genome-wide “re”-modeling of nucleosome positions. Cell 147, 263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dürr H., Flaus A., Owen-Hughes T., Hopfner K. P. (2006) Snf2 family ATPases and DExx box helicases. Differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 34, 4160–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Narlikar G. J. (2010) A proposal for kinetic proof reading by ISWI family chromatin remodeling motors. Curr. Opin. Chem. Biol. 14, 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hota S. K., Bartholomew B. (2011) Diversity of operation in ATP-dependent chromatin remodelers. Biochim. Biophys. Acta 1809, 476–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsukiyama T., Daniel C., Tamkun J., Wu C. (1995) ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 83, 1021–1026 [DOI] [PubMed] [Google Scholar]
- 25. Clark M. W., Zhong W. W., Keng T., Storms R. K., Barton A., Kaback D. B., Bussey H. (1992) Identification of a Saccharomyces cerevisiae homolog of the SNF2 transcriptional regulator in the DNA sequence of an 8.6 kb region in the LTE1-CYS1 interval on the left arm of chromosome I. Yeast 8, 133–145 [DOI] [PubMed] [Google Scholar]
- 26. Barton A. B., Kaback D. B. (1994) Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae. Analysis of the genes in the FUN38-MAK16-SPO7 region. J. Bacteriol. 176, 1872–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Neves-Costa A., Will W. R., Vetter A. T., Miller J. R., Varga-Weisz P. (2009) The SNF2-family member Fun30 promotes gene silencing in heterochromatic loci. PLoS ONE 4, e8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rowbotham S. P., Barki L., Neves-Costa A., Santos F., Dean W., Hawkes N., Choudhary P., Will W. R., Webster J., Oxley D., Green C. M., Varga-Weisz P., Mermoud J. E. (2011) Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol. Cell 42, 285–296 [DOI] [PubMed] [Google Scholar]
- 29. Strålfors A., Walfridsson J., Bhuiyan H., Ekwall K. (2011) The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLoS Genet. 7, e1001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu Q., Zhang X., Bi X. (2011) Roles of chromatin remodeling factors in the formation and maintenance of heterochromatin structure. J. Biol. Chem. 286, 14659–14669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Costelloe T., Louge R., Tomimatsu N., Mukherjee B., Martini E., Khadaroo B., Dubois K., Wiegant W. W., Thierry A., Burma S., van Attikum H., Llorente B. (2012) The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 489, 581–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen X., Cui D., Papusha A., Zhang X., Chu C. D., Tang J., Chen K., Pan X., Ira G. (2012) The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489, 576–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eapen V. V., Sugawara N., Tsabar M., Wu W. H., Haber J. E. (2012) The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol. Cell Biol. 32, 4727–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Awad S., Ryan D., Prochasson P., Owen-Hughes T., Hassan A. H. (2010) The Snf2 homolog Fun30 acts as a homodimeric ATP-dependent chromatin-remodeling enzyme. J. Biol. Chem. 285, 9477–9484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morrison A. J., Shen X. (2009) Chromatin remodelling beyond transcription. The INO80 and SWR1 complexes. Nat. Rev. Mol. Cell Biol. 10, 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Udugama M., Sabri A., Bartholomew B. (2011) The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol. Cell Biol. 31, 662–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shen X., Mizuguchi G., Hamiche A., Wu C. (2000) A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544 [DOI] [PubMed] [Google Scholar]
- 38. Papamichos-Chronakis M., Watanabe S., Rando O. J., Peterson C. L. (2011) Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144, 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mizuguchi G., Shen X., Landry J., Wu W. H., Sen S., Wu C. (2004) ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 [DOI] [PubMed] [Google Scholar]
- 40. Krogan N. J., Keogh M. C., Datta N., Sawa C., Ryan O. W., Ding H., Haw R. A., Pootoolal J., Tong A., Canadien V., Richards D. P., Wu X., Emili A., Hughes T. R., Buratowski S., Greenblatt J. F. (2003) A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12, 1565–1576 [DOI] [PubMed] [Google Scholar]
- 41. Wapinski I., Pfeffer A., Friedman N., Regev A. (2007) Natural history and evolutionary principles of gene duplication in fungi. Nature 449, 54–61 [DOI] [PubMed] [Google Scholar]
- 42. Fazzio T. G., Gelbart M. E., Tsukiyama T. (2005) Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo. Mol. Cell Biol. 25, 9165–9174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thoma F. (1996) Mapping of nucleosome positions. Methods Enzymol. 274, 197–214 [DOI] [PubMed] [Google Scholar]
- 44. Weiner A., Hughes A., Yassour M., Rando O. J., Friedman N. (2010) High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 20, 90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cristianini N., Hahn M. W. (2007) Introduction to Computational Genomics. A case Studies Approach, Chapter 2, pp. 34–35, Cambridge University Press, Cambridge, UK [Google Scholar]
- 46. Mizuguchi G., Wu W. H., Alami S., Luk E. (2012) Biochemical assay for histone H2A.Z replacement by the yeast SWR1 chromatin remodeling complex. Methods Enzymol. 512, 275–291 [DOI] [PubMed] [Google Scholar]
- 47. Hamiche A., Sandaltzopoulos R., Gdula D. A., Wu C. (1999) ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97, 833–842 [DOI] [PubMed] [Google Scholar]
- 48. Hartman J. L., 4th, Garvik B., Hartwell L. (2001) Principles for the buffering of genetic variation. Science 291, 1001–1004 [DOI] [PubMed] [Google Scholar]
- 49. Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., Chu C. S., Schuldiner M., Gebbia M., Recht J., Shales M., Ding H., Xu H., Han J., Ingvarsdottir K., Cheng B., Andrews B., Boone C., Berger S. L., Hieter P., Zhang Z., Brown G. W., Ingles C. J., Emili A., Allis C. D., Toczyski D. P., Weissman J. S., Greenblatt J. F., Krogan N. J. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–810 [DOI] [PubMed] [Google Scholar]
- 50. Marques M., Laflamme L., Gervais A. L., Gaudreau L. (2010) Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics 5, 267–272 [DOI] [PubMed] [Google Scholar]
- 51. Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Münster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L. M. (2009) Bidirectional promoters generate pervasive transcription in yeast. Nature 457, 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gkikopoulos T., Schofield P., Singh V., Pinskaya M., Mellor J., Smolle M., Workman J. L., Barton G. J., Owen-Hughes T. (2011) A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 333, 1758–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yadon A. N., Van de Mark D., Basom R., Delrow J., Whitehouse I., Tsukiyama T. (2010) Chromatin remodeling around nucleosome-free regions leads to repression of noncoding RNA transcription. Mol. Cell Biol. 30, 5110–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen K., Wilson M. A., Hirsch C., Watson A., Liang S., Lu Y., Li W., Dent S. Y. (2013) Stabilization of the promoter nucleosomes in nucleosome-free regions by the yeast Cyc8-Tup1 corepressor. Genome Res. 23, 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., Lee T. I., Bell G. W., Walker K., Rolfe P. A., Herbolsheimer E., Zeitlinger J., Lewitter F., Gifford D. K., Young R. A. (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527 [DOI] [PubMed] [Google Scholar]
- 56. Batta K., Zhang Z., Yen K., Goffman D. B., Pugh B. F. (2011) Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 25, 2254–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cairns B. R., Kim Y. J., Sayre M. H., Laurent B. C., Kornberg R. D. (1994) A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl. Acad. Sci. U.S.A. 91, 1950–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chandrasekharan M. B., Huang F., Sun Z. W. (2010) Histone H2B ubiquitination and beyond. Regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics 5, 460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Durand-Dubief M., Will W. R., Petrini E., Theodorou D., Harris R. R., Crawford M. R., Paszkiewicz K., Krueger F., Correra R. M., Vetter A. T., Miller J. R., Kent N. A., Varga-Weisz P. (2012) SWI/SNF-like chromatin remodeling factor Fun30 supports point centromere function in S. cerevisiae. PLoS Genet. 8, e1002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hartley P. D., Madhani H. D. (2009) Mechanisms that specify promoter nucleosome location and identity. Cell 137, 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Whitehouse I., Rando O. J., Delrow J., Tsukiyama T. (2007) Chromatin remodelling at promoters suppresses antisense transcription. Nature 450, 1031–1035 [DOI] [PubMed] [Google Scholar]
- 62. Goldmark J. P., Fazzio T. G., Estep P. W., Church G. M., Tsukiyama T. (2000) The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103, 423–433 [DOI] [PubMed] [Google Scholar]
- 63. Yen K., Vinayachandran V., Batta K., Koerber R. T., Pugh B. F. (2012) Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 149, 1461–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]