Background: Control of thioredoxin-interacting protein (TXNIP) expression is critical for pancreatic beta cell survival.
Results: FOXO1 binds to the TXNIP promoter; blocks ChREBP occupancy, and inhibits glucose-induced beta cell TXNIP transcription.
Conclusion: FOXO1 controls glucose-induced gene expression by competing with ChREBP at target promoters, e.g. TXNIP and L-PK.
Significance: This represents a novel gene regulatory mechanism and is the first demonstration of FOXO1-ChREBP cross-talk.
Keywords: Beta Cell, Diabetes, FOXO, Thioredoxin, Transcription Regulation, ChREBP, TXNIP
Abstract
Thioredoxin-interacting protein (TXNIP) has emerged as an important factor in pancreatic beta cell biology, and tight regulation of TXNIP levels is necessary for beta cell survival. However, the mechanisms regulating TXNIP expression have only started to be elucidated. The forkhead boxO1 transcription factor (FOXO1) has been reported to up-regulate TXNIP expression in neurons and endothelial cells but to down-regulate TXNIP in liver, and the effects on beta cells have remained unknown. We now have found that FOXO1 binds to the TXNIP promoter in vivo in human islets and INS-1 beta cells and significantly decreases TXNIP expression. TXNIP promoter deletion analyses revealed that an E-box motif conferring carbohydrate response element-binding protein (ChREBP)-mediated, glucose-induced TXNIP expression is necessary and sufficient for this effect, and electromobility shift assays confirmed FOXO1 binding to this site. Moreover, FOXO1 blocked glucose-induced TXNIP expression and reduced glucose-induced ChREBP binding at the TXNIP promoter without affecting ChREBP expression or nuclear localization, suggesting that FOXO1 may compete with ChREBP for binding to the TXNIP promoter. In fact, a FOXO1 DNA-binding mutant (FOXO1-H215R) failed to inhibit TXNIP transcription, and the effects were not restricted to TXNIP as FOXO1 also inhibited transcription of other ChREBP target genes such as liver pyruvate kinase. Together, these results demonstrate that FOXO1 inhibits beta cell TXNIP transcription and suggest that FOXO1 confers this inhibition by interfering with ChREBP DNA binding at target gene promoters. Our findings thereby reveal a novel gene regulatory mechanism and a previously unappreciated cross-talk between FOXO1 and ChREBP, two major metabolic signaling pathways.
Introduction
We identified thioredoxin-interacting protein (TXNIP)2 as the most highly up-regulated gene in human pancreatic islet cells exposed to high levels of glucose using microarray analysis (1). TXNIP, also called vitamin D3-up-regulated protein 1 (VDUP1) (2), binds and inhibits the antioxidant protein thioredoxin (3–6). Further studies revealed that TXNIP overexpression leads to beta cell apoptosis (7–9) and that TXNIP is overexpressed in islets of insulin-resistant and diabetic mice (8). Importantly, we discovered that TXNIP deficiency protects against type 1 and type 2 diabetes in mice (10, 11). These results suggest that TXNIP plays an integral role in beta cell biology, but the mechanisms and factors regulating beta cell TXNIP expression have only started to be elucidated.
We recently established that glucose-induced TXNIP expression is mediated by the carbohydrate response element-binding protein (ChREBP) (12). At this point, ChREBP is the only major nutrient- and glucose-responsive transcription factor known to be capable of regulating a number of target genes involved in glucose and lipid metabolism (13, 14). Most prominently, ChREBP has been shown to regulate glucose-induced transcription of liver pyruvate kinase (L-PK) in liver (15) and in beta cells (12, 16). Upon high glucose exposure, ChREBP enters the nucleus and binds two E-box motifs that make up the carbohydrate response element (ChoRE) in the promoter of target genes such as TXNIP and L-PK, leading to transcription (15, 17, 18).
The transcription factor forkhead boxO1 (FOXO1) is known to regulate many cellular processes, including cell cycle progression, cell death, differentiation, stress resistance, and metabolism (19–21). FOXO1 has been shown to be involved in a number of metabolic pathways in many different cells types. The cellular localization and activity of FOXO1 are controlled by post-translational modifications such as phosphorylation. Insulin signaling, through the activation of the phosphoinositide 3-kinase (PI3K) pathway, leads to the phosphorylation and nuclear exclusion of FOXO1 (19–21). Conversely, when FOXO1 is phosphorylated via oxidative stress pathways, such as the c-Jun N-terminal kinase (JNK) pathway, it is shuttled into the nucleus (19). In beta cells, FOXO1 has been shown to play a role in both beta cell function and proliferation. Nuclear localization of FOXO1 is required for the induction of MafA and NeuroD, two critical transcription factors necessary for proper beta cell function as they regulate insulin gene expression (22, 23). In contrast, nuclear localization of FOXO1 is mutually exclusive with that of Pdx1, a transcription factor required for beta cell differentiation and proliferation (24–26), and FOXO1 has been shown to compete with FOXA2 for binding to the Pdx1 promoter (27). These findings suggest that FOXO1 promotes normal beta cell function but inhibits beta cell proliferation. Interestingly, in liver cells, FOXO1 has been shown to down-regulate TXNIP (28); however, in neurons (29) and glucose-treated endothelial cells (30), FOXO1 has been demonstrated to up-regulate TXNIP expression. These studies suggest that FOXO1 may regulate TXNIP in a tissue-specific manner. However, no data are available on how FOXO1 may regulate TXNIP expression in the beta cell. The present studies were therefore aimed at determining the effects of FOXO1 on beta cell TXNIP expression and elucidating the molecular mechanisms involved. Surprisingly, they revealed a novel cross-talk between the FOXO1 and ChREBP signaling pathways, both of which control gene expression of a variety of important metabolic factors.
EXPERIMENTAL PROCEDURES
Cell Culture
Rat insulinoma (INS-1) beta cells were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, 1 mm sodium pyruvate, 2 mm l-glutamine, 10 mm HEPES, and 0.05 mm 2-mercaptoethanol. Cells were passaged using 0.25% trypsin and kept in a 37 °C incubator at 5% CO2. Unless otherwise noted, cells were maintained at normal growth conditions with 11.1 mm glucose. Human islets were obtained from the University of Alabama at Birmingham Islet Resource Facility and incubated overnight at 5 mm glucose prior to being used in the studies described. Islets from the same donor were always used as a control.
Transient Transfection Assays
For luciferase reporter assays, INS-1 cells were grown in 12-well plates and transfected with TXNIP promoter-driven luciferase reporter plasmid or SV40-driven pGL3 control plasmid (0.2 μg/well) and FOXO1 (Addgene; plasmid 13507), FOXO1-H215R (Addgene; plasmid 13509), or LacZ expression plasmid (0.4 μg/well) using DharmaFECT Duo transfection reagent (Dharmacon) (2 μl/well). Cells were harvested 48 h after transfection, and firefly luciferase activity was determined using the Dual Luciferase Assay kit (Promega). For RNA and protein samples, INS-1 cells were grown in 6-well plates and transfected with FOXO1, FOXO1-H215R, or LacZ expression plasmid (1 μg/well) using DharmaFECT Duo transfection reagent (4 μl/well). Cells were harvested 48 h after transfection for RNA and 72 h after transfection for protein.
Quantitative RT-PCR
Total RNA was extracted using an RNeasy kit (Qiagen) according to the manufacturer's instructions. 1.25 μg of RNA was reverse-transcribed to cDNA using SuperScript III First-Strand kit (Invitrogen). Quantitative real-time PCR was performed on a PRISM 7000 sequence detection system using SYBR Green (Applied Biosystems). Primers used are listed in supplemental Table S1. All samples were corrected for the 18 S ribosomal subunit (Applied Biosystems) run as an internal standard.
Western Blotting
Whole cell lysates and nuclear fractions were prepared as described previously (7). Protein extracts were diluted in 2× Tris-glycine SDS sample buffer and boiled 3 min. Samples were analyzed on a 10–20% Tris-glycine gel. Immunoblotting was performed using the following primary antibodies: anti-TXNIP JY2 (1:1000; MBL), anti-FOXO1 (1:1000; Abcam), anti-ChREBP (1:1000; Santa Cruz Biotechnology), and anti-β-actin (1:1000; Abcam). The secondary antibodies used were anti-mouse IgG (1:5000; GE Healthcare) and anti-rabbit IgG (1:5000; GE Healthcare). Bands were visualized by ECL Plus detection reagent (Pierce) and quantified by ImageJ.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as described previously (12). In brief, human islets or INS-1 cells were cross-linked by adding 1% formaldehyde in PBS to the cells for 15 min. Glycine was added at a final concentration of 0.125 m to terminate the cross-linking reaction. Sonication was performed with a Branson Sonifier 250 for 10 30-s pulses at 17.5% output. Protein concentrations were determined by BCA assay (Pierce), and 500 μg of precleared lysates was incubated with 4 μg of ChREBP (Santa Cruz), FOXO1 (Abcam), or rabbit IgG overnight at 4 °C. Immune complexes were captured with 50 μl of a 50% protein A-Sepharose slurry for 3 h at 4 °C. DNA fragments were purified using a Qiagen PCR purification kit and quantified by real-time PCR with the primers listed in supplemental Table S1.
Electromobility Shift Assay (EMSA)
Probes containing the ChoRE or FOXO1 consensus binding site sequences of the TXNIP promoter were generated using the oligonucleotides 5′-gaactgtgcacgagggatgcacgagcctccggg-3′, and 3′-cccggaggctcgtgcatccctcgtgcacagttc-5′ or 5′-gttagaggcctggtaaacaaggaccaagtagccaatgggagaactgtg-3′ and 3′-cagttctcccattggctacttggtccttgtttaccaggcctctaacc, respectively. Oligonucleotide labeling and EMSA procedures were performed using DIG Gel Shift Kit second Generation (Roche Applied Science). Briefly, 8 ng of labeled oligonucleotides was mixed with 15 μg of whole cell protein extract prepared from INS-1 cells overexpressing FOXO1, ChREBP, or with 180 ng of BSA in the binding buffer provided, and the binding reaction was incubated at room temperature for 15 min. For the competition assay, 125× molar excess unlabeled oligonucleotides was added. For the antibody inhibition assay, 2 μg of anti-FOXO1 (Abcam) or anti-ChREBP antibody (Santa Cruz Biotechnology) was added to the binding reaction, and the reaction was extended for another 20 min. Protein-DNA complexes were then separated on 6% DNA retardation gels (Invitrogen), transferred onto nylon transfer membranes, cross-linked by UV cross-linker (Stratagene), and detected by digoxigenin antibody.
Statistical Analysis
p values were calculated by Student's t test or by one-way analysis of variance for data sets of more than two groups.
RESULTS
FOXO1 Binds to the TXNIP Promoter and Inhibits TXNIP Transcription in Pancreatic Beta Cells
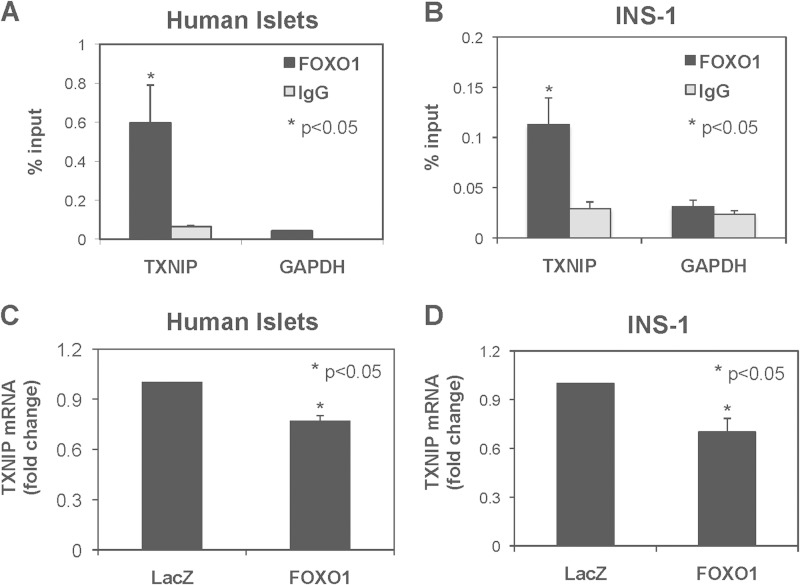
The transcription factor FOXO1 has previously been shown to down-regulate TXNIP in liver (28) and to up-regulate TXNIP in neurons (29) and glucose-treated endothelial cells (30), suggesting that the effects might be tissue-dependent. However, it remained unknown whether FOXO1 had any effects on TXNIP in pancreatic beta cells. To establish whether FOXO1 could affect TXNIP transcription in pancreatic beta cells, we first used ChIP assays to determine whether FOXO1 occupies the TXNIP promoter in human islets and rat INS-1 beta cells. We saw a significant enrichment of endogenous FOXO1 at the TXNIP promoter in both human islets (Fig. 1A) and INS-1 cells (Fig. 1B), whereas there was no enrichment with the IgG negative control or at the GAPDH internal control. These results suggest that in pancreatic beta cells, FOXO1 indeed binds to the TXNIP promoter in vivo and may regulate TXNIP transcription. Next, to study the effect of FOXO1 on beta cell TXNIP expression we transfected human islets and INS-1 cells with a FOXO1 expression plasmid. In both, human islets (Fig. 1C) and INS-1 cells (Fig. 1D) endogenous TXNIP mRNA levels were significantly decreased in response to FOXO1 compared with the LacZ control, suggesting that in beta cells FOXO1 inhibits TXNIP expression.
FIGURE 1.
FOXO1 effects on TXNIP transcription. A and B, ChIP assays were performed using human islets (A) and INS-1 cells (B) and FOXO1 antibodies or rabbit IgG. The TXNIP promoter region and GAPDH coding region were amplified by quantitative real-time PCR, and the percentage of bound promoter was calculated. C and D, human islets (C) or INS-1 cells (D) were transiently transfected with FOXO1 expression plasmid or LacZ control plasmid. Cells were harvested 48 h after transfection, and TXNIP mRNA levels were measured by quantitative RT-PCR. Bars represent means ± S.E. (error bars); n = 3–4.
An E-box Repeat in the Proximal TXNIP Promoter Is Necessary and Sufficient for the Inhibitory Effects of FOXO1 on TXNIP Transcription
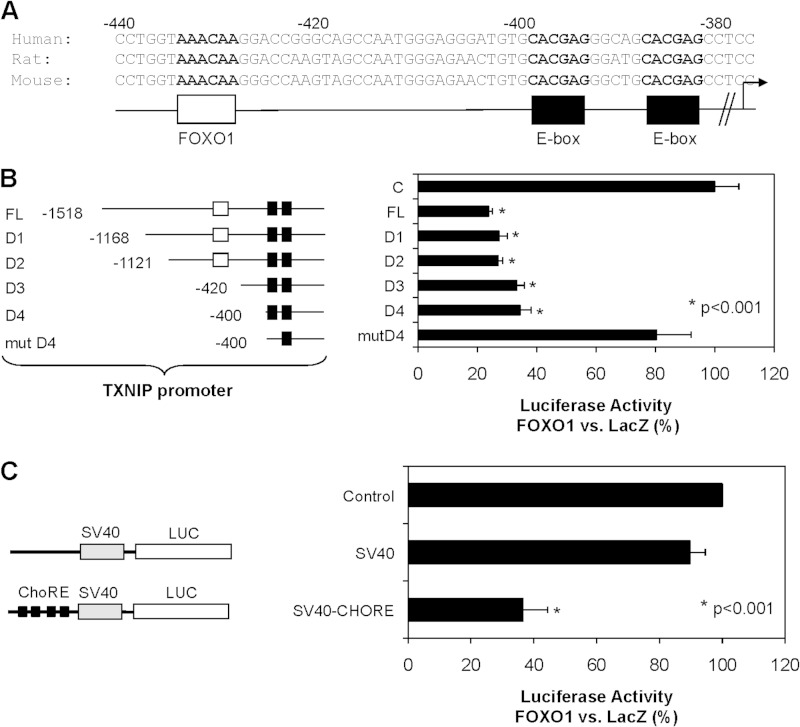
To assess the mechanisms by which FOXO1 decreases TXNIP expression, we analyzed the TXNIP promoter sequence of human, rat, and mouse to identify areas of conservation. Conserved sequences included the two E-box motifs, which comprise the ChoRE and have been shown to be involved in glucose-induced TXNIP transcription (12), and a putative FOXO binding site (Fig. 2A). To determine the promoter region involved in the regulation of TXNIP by FOXO1 we performed luciferase reporter assays using constructs containing the full-length TXNIP promoter or a number of TXNIP promoter deletions (Fig. 2B). FOXO1 overexpression led to a significant decrease in full-length TXNIP promoter (FL) activity (Fig. 2B), confirming that the effect occurs at the transcriptional level. Although we expected that the effect would be lost with the D3 promoter deletion construct, in which the putative FOXO binding site was deleted, the FOXO1 effect was only lost when the first E-box motif of the ChoRE was mutated (mutD4) (Fig. 2B). Moreover, the ChoRE was also able to confer FOXO1-mediated transcriptional inhibition to a heterologous SV40 promoter (SV40-ChoRE) (Fig. 2C). These results suggest that the ChoRE is not only necessary, but also sufficient for FOXO1-mediated inhibition of TXNIP transcription.
FIGURE 2.
Analysis of TXNIP promoter region conferring FOXO1 effects. A, alignment of the proximal human, rat, and mouse TXNIP promoter sequence reveals a highly conserved ChoRE with an E-box repeat (black) and putative FOXO binding site (white). B, INS-1 cells were co-transfected with FOXO1 expression plasmid or LacZ control plasmid and luciferase reporter constructs driven by the full-length TXNIP promoter or a number of TXNIP promoter deletions (D1-mutD4). C, INS-1 cells were co-transfected with FOXO1 expression plasmid or LacZ control plasmid and reporter constructs containing luciferase driven by the heterologous SV40 promoter with or without upstream ChoRE repeat (SV40-ChoRE). Luciferase activities are expressed as percentage of the LacZ control; bars represent means ± S.E. (error bars); n = 3–4.
FOXO1 and ChREBP Bind to the Same Region in the TXNIP Promoter
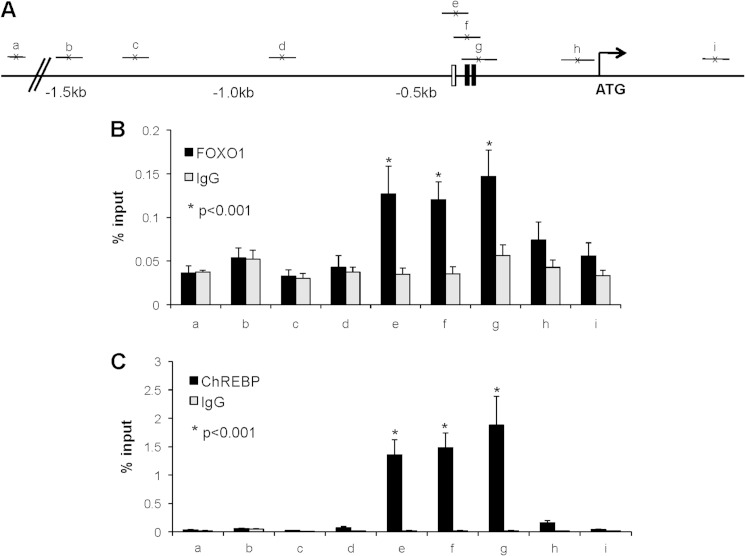
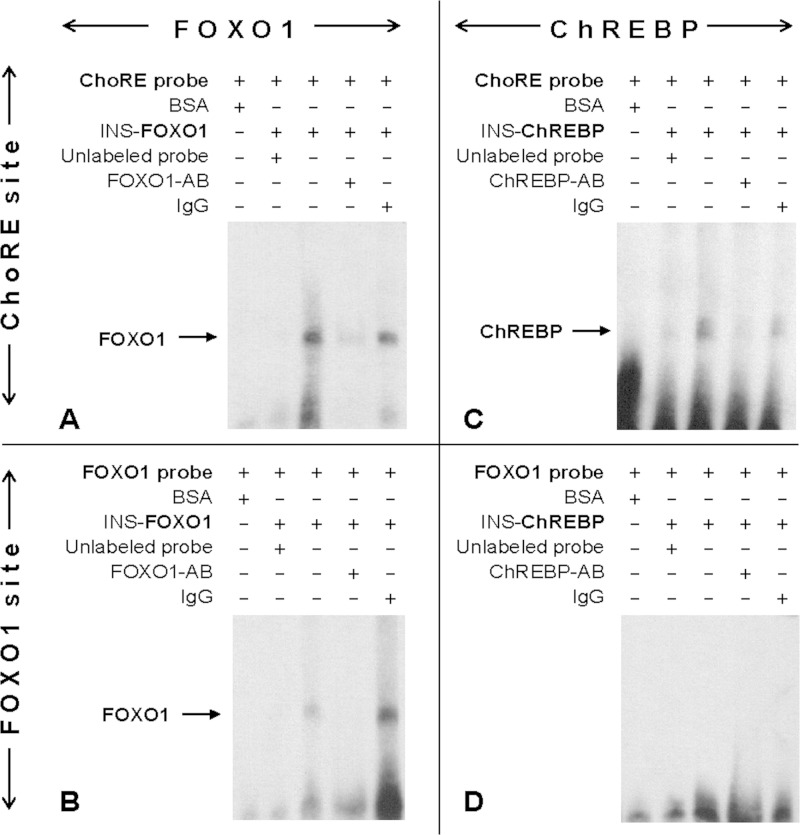
Promoter deletion studies provided functional evidence that FOXO1 and ChREBP use the same element in the TXNIP promoter to confer their effects. To analyze and compare the in vivo binding patterns of FOXO1 and ChREBP at the TXNIP promoter in greater detail, we fine-mapped the binding of these transcription factors by a series of ChIP assays using a variety of primer sets near the ChoRE as well as ranging from 300 bp downstream of the ATG start codon to −1.5 kb upstream (Fig. 3A). INS-1 beta cells were cross-linked and ChIP assays performed using FOXO1 and ChREBP antibodies or control IgG. Interestingly, FOXO1 was significantly enriched at the three promoter regions near the ChoRE (e–g), but not enriched at any of the other sites in the TXNIP promoter or at a noncoding site 40 kb upstream of the TXNIP gene, run as an additional negative control (Fig. 3B). ChREBP enrichment followed the same pattern as FOXO1, demonstrating binding exclusively to the regions encompassing the ChoRE (Fig. 3C). Because the putative FOXO1 binding site is within <40 bp of the ChoRE, these ChIP data cannot exclude involvement of this site in FOXO1 binding. However, together with the promoter deletion and mutation studies, these results strongly suggested that FOXO1 and ChREBP bind to a common region within the proximal TXNIP promoter. To obtain direct evidence for this notion, we also performed electromobility shift assays (EMSA) using a probe containing the TXNIP promoter ChoRE sequence, but lacking the FOXO1 site. Indeed, these studies revealed that FOXO1 was able to effectively bind to the TXNIP-ChoRE (Fig. 4A). Moreover, this effect was not restricted to TXNIP as EMSA also revealed FOXO1 binding to the ChoRE sequence found in the L-PK promoter (supplemental Fig. S1). As expected, FOXO1 also bound to its consensus binding site within the TXNIP promoter (Fig. 4B), and ChREBP bound to the TXNIP ChoRE (Fig. 4C). However, ChREBP was not able to bind to the FOXO1 site (Fig. 4D), demonstrating that the cross-interaction was not reciprocal.
FIGURE 3.
TXNIP promoter binding patterns of FOXO1 and ChREBP. INS-1 cells were maintained in regular growth medium, cells were cross-linked, and ChIP assays were performed as described under “Experimental Procedures.” A, schematic of the location of the primer sets to amplify various regions along the 2 kb of the TXNIP promoter, as well as an upstream noncoding region on chromosome 2. B and C, FOXO1 (B) or ChREBP (C) binding to the TXNIP promoter was assessed by ChIP using FOXO1, ChREBP, or rabbit IgG antibodies; bars represent mean percent input ± S.E. (error bars); n = 3–4.
FIGURE 4.
FOXO1 binding to TXNIP-ChoRE sequence. EMSAs were performed as described under “Experimental Procedures” using whole cell extract of INS-1 cells transfected with FOXO1 expression plasmid (INS-FOXO1) (A and B) or with ChREBP expression plasmid (INS-ChREBP) (C and D) and DIG-labeled oligonucleotide probes for the TXNIP-ChoRE (A–C) or TXNIP-FOXO1 site (B–D) (third lane). First lane, BSA negative control; second lane, competition with unlabeled probe; fourth lane, with anti-FOXO1 or anti-ChREBP antibody; fifth lane, control IgG.
FOXO1 Inhibits ChREBP Binding to the TXNIP Promoter
The fact that the ChoRE E-box motif, which serves as the ChREBP binding site in the TXNIP promoter, is essential for FOXO1-mediated inhibition of TXNIP transcription (Fig. 2, B and C) raised the question of whether FOXO1 might regulate TXNIP expression through ChREBP. However, we found that FOXO1 had no effect on ChREBP mRNA expression (supplemental Fig. S2A), nor did it reduce nuclear ChREBP even though efficient FOXO1 overexpression was achieved (supplemental Fig. S2, B and C). In contrast, FOXO1 did lead to a small but highly significant reduction in ChREBP binding to the TXNIP promoter as assessed by ChIP (supplemental Fig. S2D). These results suggest that FOXO1 inhibits TXNIP transcription by interfering with ChREBP DNA binding rather than by altering its expression or nuclear translocation. To address this question further and to determine whether DNA binding is necessary for FOXO1-mediated inhibition of TXNIP, we employed a FOXO1 DNA-binding mutant (FOXO1-H215R). This mutant contains a point mutation of histidine 215 to arginine in the DNA binding domain, rendering the mutant unable to bind DNA (31). Furthermore, because the ChoRE E-box motif and ChREBP play such a critical role in glucose-induced gene transcription in general, and beta cell TXNIP transcription in particular (12), we hypothesized that FOXO1 might regulate not only basal, but also glucose-induced TXNIP expression. To test these possibilities, we first confirmed that we were able to achieve efficient and comparable overexpression and nuclear localization of both wild-type FOXO1 and mutant FOXO1-H215R proteins (supplemental Fig. S3A).
FOXO1 Inhibits Glucose-induced TXNIP Expression, and This Effect Is Dependent on FOXO1 DNA Binding
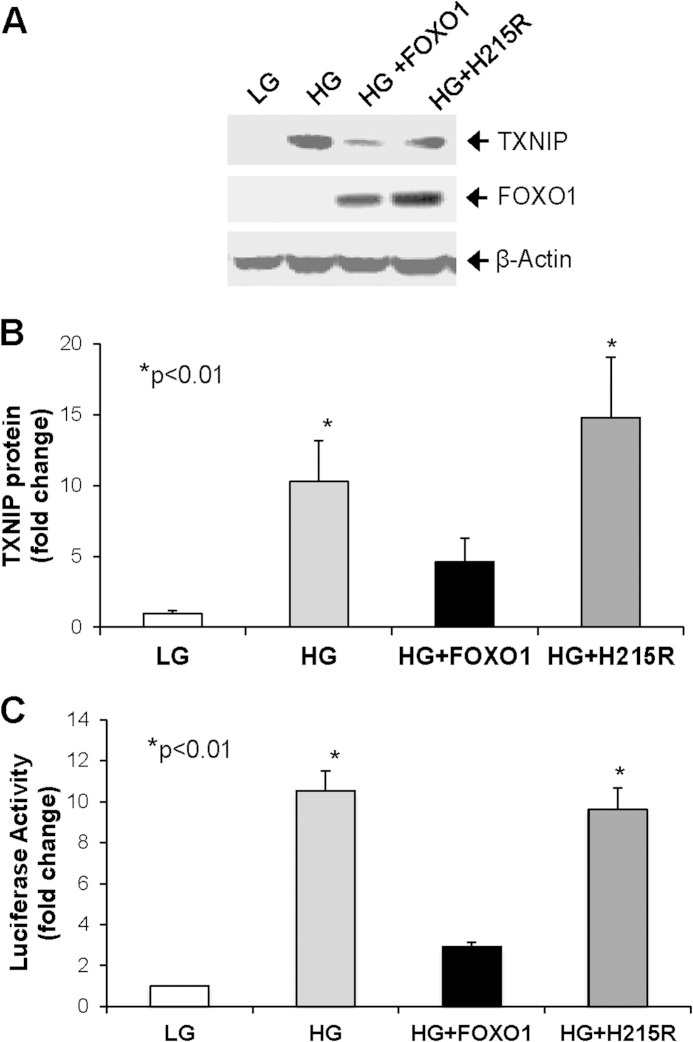
Next, we investigated the FOXO1 effects on glucose-induced TXNIP mRNA expression. To this end, INS-1 cells were transfected with control LacZ, wild-type FOXO1, or mutant FOXO1-H215R at basal glucose levels and changed to low (5 mm) or high (25 mm) glucose medium 24 h after transfection. Consistent with previous findings, we observed a dramatic 10-fold induction in endogenous TXNIP mRNA expression in response to incubation at high glucose as opposed to low glucose (supplemental Fig. S3B). Interestingly, overexpression of wild-type FOXO1 was able to reduce this effect significantly, whereas the FOXO1-H215R DNA-binding mutant was not (supplemental Fig. S3B). In addition, Western blotting revealed that FOXO1 overexpression also markedly reduced the dramatic >10-fold increase in TXNIP protein levels observed in response to high glucose (Fig. 5, A and B). In contrast, TXNIP protein levels remained increased >10-fold in response to overexpression of the FOXO1-H215R DNA-binding mutant (Fig. 5, A and B). Very similar results were also observed in terms of TXNIP promoter-driven luciferase activity. Whereas high glucose resulted in a ∼10-fold increase in TXNIP promoter activity in the context of control LacZ and mutant FOXO-H215R overexpression, wild-type FOXO1 blocked glucose-induced transcriptional activity of the TXNIP promoter (Fig. 5C). Together, these results show that FOXO1 is able to inhibit glucose-induced TXNIP expression and suggest that DNA binding is required for FOXO1-mediated inhibition of TXNIP. Using co-immunoprecipitation studies we also discovered that ChREBP and FOXO1 physically interact (supplemental Fig. S4). However, this interaction also occurred with the FOXO1-H215R DNA-binding mutant that failed to inhibit TXNIP expression. These findings therefore further suggest that it is the FOXO1-DNA binding that confers the observed effects rather than the protein-protein interaction.
FIGURE 5.
FOXO1 effects on glucose-induced TXNIP expression. A, INS-1 cells were transiently transfected with FOXO1 expression plasmid, FOXO1 DNA-binding mutant (H215R), or LacZ control plasmid and incubated in 5 mm low glucose (LG) or 25 mm high glucose (HG) medium 24 h after transfection. Cells were harvested 72 h after transfection, and TXNIP protein levels were measured by Western blotting. B, TXNIP protein levels were corrected for β-actin in six independent experiments and quantified. C, INS-1 cells co-transfected with FOXO1 expression plasmid, FOXO1 DNA-binding mutant (H215R) or LacZ control plasmid and the full-length TXNIP promoter luciferase reporter construct were incubated in 5 mm low glucose or 25 mm high glucose medium 24 h after transfection. Cells were harvested 48 h after transfection, and luciferase activity was measured. Bars represent mean -fold change ± S.E. (error bars); n = 3–6.
FOXO1 Blocks ChREBP Binding to Glucose-induced Target Gene Promoters
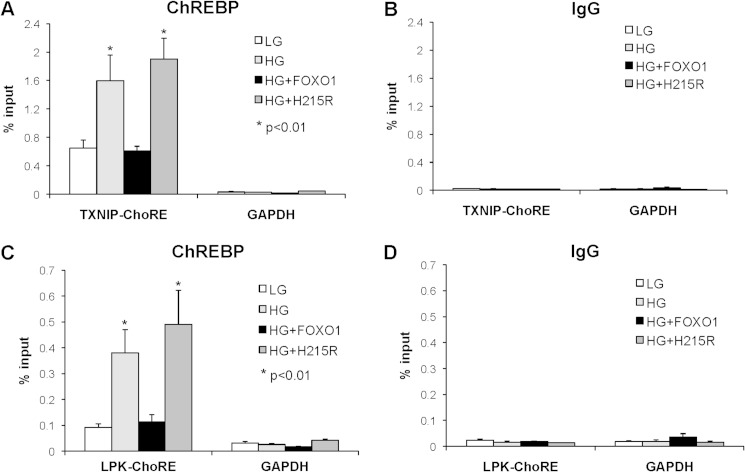
To understand better the mechanisms by which FOXO1 blunts glucose-induced TXNIP transcription, we used ChIP assays to assess ChREBP occupancy at the TXNIP promoter in cells transfected with LacZ, wild-type FOXO1, or mutant FOXO1-H215R and grown at low and high glucose. We found that ChREBP enrichment at the TXNIP promoter increased, as expected, from low to high glucose in control cells (Fig. 6A), whereas no enrichment was seen at the GAPDH internal control (Fig. 6A) or with the IgG control (Fig. 6B). FOXO1 overexpression completely blunted glucose-induced ChREBP enrichment (Fig. 6A), consistent with the observed decrease in TXNIP transcription and expression. In contrast, the FOXO1-H215R DNA-binding mutant failed to reduce ChREBP binding to the TXNIP promoter. Taking into consideration that FOXO1 binds to the TXNIP ChoRE (Fig. 4), FOXO1 DNA binding is required for FOXO1-mediated inhibition of glucose-induced TXNIP expression (Fig. 5), FOXO1 and ChREBP bind to the same region in the TXNIP promoter (Fig. 3), and that the inhibitory FOXO1 effect is dependent on the ChoRE E-box motif, which also serves as the ChREBP binding site (Fig. 2), these results suggest that FOXO1 regulates TXNIP expression by competing with and reducing binding of ChREBP to the ChoRE region of the TXNIP promoter. The ChIP data now provide further support for this notion demonstrating that glucose-induced in vivo ChREBP binding is dramatically reduced by FOXO1 (Fig. 6A). In addition, these findings also raise the question of whether FOXO1 may regulate the expression of other ChREBP target genes using the same mechanism.
FIGURE 6.
FOXO1 effects on ChREBP occupancy of target gene promoters. INS-1 cells, transfected with FOXO1 expression plasmid, FOXO1 DNA-binding mutant (H215R), or LacZ control plasmid were incubated at 5 mm low glucose (LG) or 25 mm high glucose (HG). ChIP assays were performed 48 h after transfection using ChREBP antibodies or rabbit IgG. The GAPDH coding region and TXNIP ChoRE region (A and B) or L-PK ChoRE region (C and D) were amplified by quantitative real-time PCR, and the percentage of input was calculated. Bars represent means ± S.E. (error bars); n = 3–5.
The L-PK gene is the best studied and established ChREBP target gene (15), and we previously used the L-PK promoter successfully as a positive control to assess glucose-induced DNA binding of ChREBP in beta cell ChIP assays (12). Indeed, FOXO1 was also able to bind to the L-PK ChoRE (supplemental Fig. S1), and parallel ChIP experiments revealed that FOXO1 overexpression decreased glucose-induced ChREBP binding to the L-PK promoter (Fig. 6C) whereas mutant FOXO1-H215R had no effect. There was no enrichment at the GAPDH internal control (Fig. 6C) or with the IgG control (Fig. 6D). These results suggest that the effects of FOXO1 are not restricted to TXNIP and that FOXO1 may interfere with ChREBP binding to promoters of many more glucose-induced target genes, underlining the potential implications of this newly discovered gene regulatory mechanism.
Having found that FOXO1 overexpression inhibits ChREBP function and TXNIP expression, we hypothesized that on the other hand FOXO1 knockdown might promote ChREBP-induced TXNIP expression. In fact, this is exactly what we observed. Transfection of INS-1 beta cells with siFOXO1 led to effective knockdown of FOXO1 (supplemental Fig. S5A) and to a small but significant increase in TXNIP expression (supplemental Fig. S5B). Moreover, FOXO1 knockdown clearly promoted ChREBP occupancy at the TXNIP-ChoRE as assessed by ChIP (supplemental Fig. S5, C and D). This provides additional strong support for the notion that FOXO1 is a novel regulator of TXNIP expression and ChREBP function.
DISCUSSION
Our results reveal for the first time cross-talk between the FOXO1 and ChREBP signaling pathways in the beta cell and suggest that FOXO1 competes with ChREBP for DNA binding, thereby inhibiting transcription of ChREBP target genes such as TXNIP and L-PK. FOXO1 has previously been shown to differentially regulate TXNIP expression in liver (28), neurons (29), and glucose-treated endothelial cells (30). However, until now nothing has been known about the regulation of TXNIP by FOXO1 in beta cells. FOXO1 has been shown to have a critical role in beta cell physiology. Inhibition and nuclear exclusion of FOXO1, in response to insulin and PI3K signaling, promote beta cell proliferation by allowing nuclear entry of Pdx1 and by increasing the activity of this key transcription factor (24–26). Because, on the other hand, increased FOXO1 inhibits Pdx1 function, FOXO1 has largely been associated with detrimental effects in regard to beta cell growth. However, activation and shuttling of FOXO1 into the nucleus, in response to oxidative stress and JNK pathway activation, also activate genes involved in insulin transcription such as MafA and NeuroD and thereby help preserve beta cell function (22, 23). In addition, recent work also demonstrated that FOXO1 ablation leads to dedifferentiation, dysfunction, and loss of beta cells resulting in hyperglycemia (32). This indicates that FOXO1 also has positive effects on the beta cell, especially in terms of beta cell function, beta cell survival, and in the context of oxidative stress. Our current findings demonstrate that FOXO1 inhibits TXNIP expression in primary human islets as well as INS-1 beta cells (Fig. 1), which is consistent with its effects in liver (28). They further suggest that by inhibiting TXNIP, which promotes oxidative stress (3–6) and beta cell apoptosis (7–9), FOXO1 may have additional beneficial effects on beta cell biology. Moreover, the results shed new light on the regulation of beta cell TXNIP expression and reveal FOXO1 as yet another transcription factor controlling expression of this important protein.
ChREBP is the major regulator of glucose-induced transcription and controls expression of a large number of glucose and lipid metabolism genes in liver, adipose tissue, and pancreatic beta cells, including L-PK as well as other tissue-specific targets such as acetyl-CoA carboxylase, fatty acid synthase, or peroxisome proliferator-activated receptor α (15, 17, 18). Using gain- as well as loss-of-function experiments, we have recently shown that ChREBP confers glucose-induced transcription of TXNIP in pancreatic beta cells (12), and similar findings were observed in the liver (33). In the present study, we have discovered that FOXO1 controls this ChREBP-mediated TXNIP expression in the beta cell. The results show that FOXO1 binds to the TXNIP promoter in vivo (Figs. 1 and 3) and prevents the binding of ChREBP, leading to an inhibition of TXNIP transcription. These inhibitory FOXO1 effects are especially apparent in the context of glucose-induced TXNIP transcription (Figs. 5 and 6), consistent with the critical role ChREBP plays in glucose-induced gene expression.
Interestingly, a FOXO1 DNA-binding mutant (FOXO1-H215R) was unable to inhibit TXNIP expression, suggesting that FOXO1 must bind the TXNIP promoter to exert its effect. Using fine-mapping ChIP experiments, we have shown that in beta cells FOXO1 binds to the same TXNIP promoter region, containing the ChoRE E-box motif that serves as the ChREBP binding site (Fig. 3). However, given the proximity of the putative FOXO1 consensus binding site, just 40 bp upstream of the ChoRE, these ChIP experiments cannot exclude the possibility that FOXO1 also binds to the FOXO1 site. On the other hand, luciferase reporter assays using TXNIP promoter deletions showed that FOXO1 is able to exert its inhibitory effects on TXNIP without this sequence (D2 and D3 deletion constructs), suggesting that this site is not critical for FOXO1-mediated TXNIP inhibition (Fig. 2B). Moreover, EMSA studies using DNA probes containing TXNIP or L-PK promoter ChoRE sequences but no consensus FOXO1 binding sites confirmed that FOXO1 was able to bind to these ChoRE sequences (Fig. 4A and supplemental Fig. S1). In addition, the ChoRE E-box motif was both necessary and sufficient for FOXO1-mediated inhibition of beta cell TXNIP transcription (Fig. 2). Although we discovered that ChREBP and FOXO1 also physically interact (supplemental Fig. S4), this interaction occurred with both wild-type and the FOXO1-H215R DNA-binding mutant. Because the H215R DNA-binding mutant failed to inhibit TXNIP expression, it is very unlikely that this protein-protein interaction confers the observed inhibition of TXNIP expression, and the results further support the notion that FOXO1-DNA binding is required for this effect. Because we found that FOXO1 was able to bind to the ChoRE site, we also wondered whether the reverse might be true too, i.e. that ChREBP could bind to the consensus FOXO1 site. However, EMSA studies revealed that ChREBP was only able to bind to its own consensus ChoRE and not to the FOXO1 site (Fig. 4, C and D). These results suggest that FOXO1 might be able to modulate DNA binding and transcriptional activity of ChREBP, but not vice versa.
Taken together with our observation that FOXO1 overexpression leads to decreased ChREBP binding at the TXNIP promoter (without affecting ChREBP expression or nuclear localization), these findings further suggest that the two transcription factors may be competing for binding to the ChoRE of the TXNIP promoter. In fact, such a mechanism could explain how FOXO1 inhibited glucose-induced TXNIP expression and why FOXO1 DNA binding was critical for this effect. If this scenario were true, one would expect that FOXO1 could also inhibit expression of other ChREBP target genes by preventing ChREBP occupancy of the target promoters. Indeed, this is exactly what we found as FOXO1 overexpression also decreased ChREBP binding at the L-PK promoter, another well known target of ChREBP (Fig. 6C). These results not only strongly support the notion of this newly discovered FOXO1-ChREBP cross-talk and gene regulatory mechanism, but also suggest that the implications go far beyond the regulation of beta cell TXNIP expression. FOXO1 may interfere with ChREBP binding at a variety of other target gene promoters, therefore affecting the expression of a large number of important glucose-induced proteins involved in metabolism in beta cells as well as other organs such as liver. The fact that FOXO1 has also been shown to down-regulate TXNIP in liver cells (28) lends further support to this idea and suggests that this novel regulatory mechanism is not restricted to one gene or one tissue.
Of note, liver and, to a lesser degree, pancreatic beta cells are considered major sites of ChREBP expression and function and both tissues demonstrated analogous effects in terms of FOXO1-mediated inhibition of TXNIP expression (Ref. 28 and Fig. 1, respectively). In contrast, in other tissues with less prominent or negligible ChREBP expression, FOXO1 seems to increase TXNIP expression (29, 30) and act as a classical activating transcription factor. Combined, these observations suggest that the presence of ChREBP may be an important determinant for the effects of FOXO1 as a transcriptional inhibitor and further underline the central role ChREBP seems to play in this newly identified gene regulatory mechanism.
In summary, we have discovered that FOXO1 inhibits beta cell TXNIP transcription by interfering with ChREBP binding to the TXNIP promoter. TXNIP is a key factor involved in glucotoxic beta cell death and diabetes development, and this control of TXNIP expression by two major transcription factors involved in insulin and glucose signaling further illustrates the importance a tight regulation of TXNIP levels seems to play for beta cell biology and metabolism. Interestingly though, the effects are not restricted to TXNIP, as FOXO1 also interferes with ChREBP binding at the L-PK promoter, suggesting that FOXO1 may be involved in the regulation of other glucose-induced ChREBP target genes. Together, these studies have identified a novel FOXO1-mediated gene regulatory mechanism in the beta cell and have revealed previously unappreciated cross-talk between FOXO1 and ChREBP, two major metabolic signaling pathways.
Supplementary Material
Acknowledgment
We thank Dr. Emery H. Bresnick for support and expert advice throughout these studies.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK-078752. This work was also supported by American Diabetes Association Grants 7-07-CD-22 and 7-12-BS-167 and Juvenile Diabetes Research Foundation/JNJSI Grant 40-2011-1.

This article contains supplemental Table S1 and Figs S1–S5.
- TXNIP
- thioredoxin-interacting protein
- ChoRE
- carbohydrate response element
- ChREBP
- carbohydrate response element-binding protein
- FOXO1
- forkhead boxO1
- L-PK
- liver pyruvate kinase.
REFERENCES
- 1. Shalev A., Pise-Masison C. A., Radonovich M., Hoffmann S. C., Hirshberg B., Brady J. N., Harlan D. M. (2002) Oligonucleotide microarray analysis of intact human pancreatic islets: identification of glucose-responsive genes and a highly regulated TGFβ signaling pathway. Endocrinology 143, 3695–3698 [DOI] [PubMed] [Google Scholar]
- 2. Chen K. S., DeLuca H. F. (1994) Isolation and characterization of a novel cDNA from HL-60 cells treated with 1,25-dihydroxyvitamin D3. Biochim. Biophys. Acta 1219, 26–32 [DOI] [PubMed] [Google Scholar]
- 3. Junn E., Han S. H., Im J. Y., Yang Y., Cho E. W., Um H. D., Kim D. K., Lee K. W., Han P. L., Rhee S. G., Choi I. (2000) Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J. Immunol. 164, 6287–6295 [DOI] [PubMed] [Google Scholar]
- 4. Nishiyama A., Masutani H., Nakamura H., Nishinaka Y., Yodoi J. (2001) Redox regulation by thioredoxin and thioredoxin-binding proteins. IUBMB Life 52, 29–33 [DOI] [PubMed] [Google Scholar]
- 5. Nishiyama A., Matsui M., Iwata S., Hirota K., Masutani H., Nakamura H., Takagi Y., Sono H., Gon Y., Yodoi J. (1999) Identification of thioredoxin-binding protein-2/vitamin D3 up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J. Biol. Chem. 274, 21645–21650 [DOI] [PubMed] [Google Scholar]
- 6. Yamanaka H., Maehira F., Oshiro M., Asato T., Yanagawa Y., Takei H., Nakashima Y. (2000) A possible interaction of thioredoxin with VDUP1 in HeLa cells detected in a yeast two-hybrid system. Biochem. Biophys. Res. Commun. 271, 796–800 [DOI] [PubMed] [Google Scholar]
- 7. Chen J., Saxena G., Mungrue I. N., Lusis A. J., Shalev A. (2008) Thioredoxin-interacting protein: a critical link between glucose toxicity and beta cell apoptosis. Diabetes 57, 938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Minn A. H., Hafele C., Shalev A. (2005) Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta cell apoptosis. Endocrinology 146, 2397–2405 [DOI] [PubMed] [Google Scholar]
- 9. Minn A. H., Pise-Masison C. A., Radonovich M., Brady J. N., Wang P., Kendziorski C., Shalev A. (2005) Gene expression profiling in INS-1 cells overexpressing thioredoxin-interacting protein. Biochem. Biophys. Res. Commun. 336, 770–778 [DOI] [PubMed] [Google Scholar]
- 10. Chen J., Hui S. T., Couto F. M., Mungrue I. N., Davis D. B., Attie A. D., Lusis A. J., Davis R. A., Shalev A. (2008) Thioredoxin-interacting protein deficiency induces Akt/Bcl-xL signaling and pancreatic beta cell mass and protects against diabetes. FASEB J. 22, 3581–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Masson E., Koren S., Razik F., Goldberg H., Kwan E. P., Sheu L., Gaisano H. Y., Fantus I. G. (2009) High beta cell mass prevents streptozotocin-induced diabetes in thioredoxin-interacting protein-deficient mice. Am. J. Physiol. Endocrinol. Metab. 296, E1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cha-Molstad H., Saxena G., Chen J., Shalev A. (2009) Glucose-stimulated expression of TXNIP is mediated by carbohydrate response element-binding protein, p300, and histone H4 acetylation in pancreatic beta cells. J. Biol. Chem. 284, 16898–16905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boergesen M., Poulsen L. L., Schmidt S. F., Frigerio F., Maechler P., Mandrup S. (2011) ChREBP mediates glucose repression of peroxisome proliferator-activated receptor α expression in pancreatic beta cells. J. Biol. Chem. 286, 13214–13225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poungvarin N., Lee J. K., Yechoor V. K., Li M. V., Assavapokee T., Suksaranjit P., Thepsongwajja J. J., Saha P. K., Oka K., Chan L. (2012) Carbohydrate response element-binding protein (ChREBP) plays a pivotal role in beta cell glucotoxicity. Diabetologia 55, 1783–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawaguchi T., Takenoshita M., Kabashima T., Uyeda K. (2001) Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc. Natl. Acad. Sci. U.S.A. 98, 13710–13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang H., Wollheim C. B. (2002) ChREBP rather than USF2 regulates glucose stimulation of endogenous L-pyruvate kinase expression in insulin-secreting cells. J. Biol. Chem. 277, 32746–32752 [DOI] [PubMed] [Google Scholar]
- 17. O'Callaghan B. L., Koo S. H., Wu Y., Freake H. C., Towle H. C. (2001) Glucose regulation of the acetyl-CoA carboxylase promoter PI in rat hepatocytes. J. Biol. Chem. 276, 16033–16039 [DOI] [PubMed] [Google Scholar]
- 18. Rufo C., Teran-Garcia M., Nakamura M. T., Koo S. H., Towle H. C., Clarke S. D. (2001) Involvement of a unique carbohydrate-responsive factor in the glucose regulation of rat liver fatty-acid synthase gene transcription. J. Biol. Chem. 276, 21969–21975 [DOI] [PubMed] [Google Scholar]
- 19. Glauser D. A., Schlegel W. (2007) The emerging role of FOXO transcription factors in pancreatic beta cells. J. Endocrinol. 193, 195–207 [DOI] [PubMed] [Google Scholar]
- 20. Kitamura T., Ido Kitamura Y. (2007) Role of FOXO proteins in pancreatic beta cells. Endocr. J. 54, 507–515 [DOI] [PubMed] [Google Scholar]
- 21. Maiese K., Chong Z. Z., Hou J., Shang Y. C. (2009) The “O” class: crafting clinical care with FOXO transcription factors. Adv. Exp. Med. Biol. 665, 242–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buteau J., Accili D. (2007) Regulation of pancreatic beta cell function by the forkhead protein FOXO1. Diabetes Obes. Metab. 9, 140–146 [DOI] [PubMed] [Google Scholar]
- 23. Kitamura Y. I., Kitamura T., Kruse J. P., Raum J. C., Stein R., Gu W., Accili D. (2005) FOXO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2, 153–163 [DOI] [PubMed] [Google Scholar]
- 24. Buteau J., Spatz M. L., Accili D. (2006) Transcription factor FOXO1 mediates glucagon-like peptide-1 effects on pancreatic beta cell mass. Diabetes 55, 1190–1196 [DOI] [PubMed] [Google Scholar]
- 25. Kawamori D., Kaneto H., Nakatani Y., Matsuoka T. A., Matsuhisa M., Hori M., Yamasaki Y. (2006) The forkhead transcription factor FOXO1 bridges the JNK pathway and the transcription factor PDX-1 through its intracellular translocation. J. Biol. Chem. 281, 1091–1098 [DOI] [PubMed] [Google Scholar]
- 26. Okamoto H., Hribal M. L., Lin H. V., Bennett W. R., Ward A., Accili D. (2006) Role of the forkhead protein FOXO1 in beta cell compensation to insulin resistance. J. Clin. Invest. 116, 775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kitamura T., Nakae J., Kitamura Y., Kido Y., Biggs W. H., 3rd, Wright C. V., White M. F., Arden K. C., Accili D. (2002) The forkhead transcription factor FOXO1 links insulin signaling to Pdx1 regulation of pancreatic beta cell growth. J. Clin. Invest. 110, 1839–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Candia P., Blekhman R., Chabot A. E., Oshlack A., Gilad Y. (2008) A combination of genomic approaches reveals the role of FOXO1a in regulating an oxidative stress response pathway. PLoS One 3, e1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Al-Mubarak B., Soriano F. X., Hardingham G. E. (2009) Synaptic NMDAR activity suppresses FOXO1 expression via a cis-acting FOXO binding site: FOXO1 is a FOXO target gene. Channels 3, 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li X., Rong Y., Zhang M., Wang X. L., LeMaire S. A., Coselli J. S., Zhang Y., Shen Y. H. (2009) Up-regulation of thioredoxin-interacting protein (TXNIP) by p38 MAPK and FOXO1 contributes to the impaired thioredoxin activity and increased ROS in glucose-treated endothelial cells. Biochem. Biophys. Res. Commun. 381, 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang E. D., Nuñez G., Barr F. G., Guan K. L. (1999) Negative regulation of the forkhead transcription factor FKHR by Akt. J. Biol. Chem. 274, 16741–16746 [DOI] [PubMed] [Google Scholar]
- 32. Talchai C., Xuan S., Lin H. V., Sussel L., Accili D. (2012) Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell 150, 1223–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ma L., Robinson L. N., Towle H. C. (2006) ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J. Biol. Chem. 281, 28721–28730 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.