Background: AtCRY2 forms photobodies in plants.
Results: AtCRY2 forms photobodies in human cells. By fusing AtCRY2 to a human checkpoint protein, the DNA damage response can be activated by light.
Conclusion: AtCRY2 and its fusion proteins can achieve light-induced protein multimerization independent of other plant proteins.
Significance: AtCRY2 can function as an optogenetic tool to modulate signaling pathways.
Keywords: Checkpoint Control, DNA Damage, FAD, Nucleus, Photoreceptors, ATR, Chk1, Cryptochrome, Nuclear Bodies, TopBP1
Abstract
Nuclear bodies are discrete suborganelle structures that perform specialized functions in eukaryotic cells. In plant cells, light can induce de novo formation of nuclear bodies called photobodies (PBs) composed of the photosensory pigments, phytochrome (PHY) or cryptochrome (CRY). The mechanisms of formation, the exact compositions, and the functions of plant PBs are not known. Here, we have expressed Arabidopsis CRY2 (AtCRY2) in mammalian cells and analyzed its fate after blue light exposure to understand the requirements for PB formation, the functions of PBs, and their potential use in cell biology. We found that light efficiently induces AtCRY2-PB formation in mammalian cells, indicating that, other than AtCRY2, no plant-specific proteins or nucleic acids are required for AtCRY2-PB formation. Irradiation of AtCRY2 led to its degradation; however, degradation was not dependent upon photobody formation. Furthermore, we found that AtCRY2 photobody formation is associated with light-stimulated interaction with mammalian COP1 E3 ligase. Finally, we demonstrate that by fusing AtCRY2 to the TopBP1 DNA damage checkpoint protein, light-induced AtCRY2 PBs can be used to activate DNA damage signaling pathway in the absence of DNA damage.
Introduction
Nuclear bodies (NBs)2 are nuclear structures similar to cytoplasmic organelles such as mitochondrion, Golgi organelles, and lysosomes. NBs differ from cytoplasmic organelles in one important aspect, in that although cytoplasmic organelles are enclosed by a membrane and have, to a significant extent, a relatively well defined composition, NBs are not encapsulated by a membrane, and as a result, they have a more dynamic composition that is influenced by intracellular as well as extracellular milieu (1–3). In mammalian cells, the best known NBs are nucleoli, Cajal bodies, and promyelocytic leukemia nuclear bodies (PML-NBs). Nucleoli and Cajal bodies are involved in ribosomal RNA and small nuclear RNA biosynthesis and processing, respectively. PML-NBs are involved in maintenance of genome stability under physiological conditions and in DNA repair and apoptosis under conditions of genotoxic stress.
Plant cells have nuclear bodies as well, and upon light exposure, plant proteins involved in photosensory functions form a unique class of nuclear bodies with properties quite similar to mammalian NBs (4–7). The plant NBs that are formed following light exposure are referred to as photobodies (PBs) (4). Among plant photosensory proteins, the red light receptor phytochromes (PHYs) are the most extensively studied with respect to photobody formation. More recently, it has been found that the blue light photosensory receptor cryptochrome (CRY) forms photobodies as well (8, 9).
The compositions of PHY-PB and CRY-PB and the role of PB formation in signaling by these photoreceptors are not known. A genetic screen has identified several genes that are required for PHY-PB formation, and one of these proteins, hemera, has been found to associate with PHY-PBs (10). CRY-PBs have been analyzed mostly by using AtCRY2 (8, 9). Recent work suggests that AtCRY2 controls signaling pathways by two mechanisms (11). In one mechanism, AtCRY2 binds to a basic helix-loop-helix family member, the CIB (cryptochrome-interacting basic helix-loop-helix 1) transcription factor, upon excitation by blue light and promotes transcription of cognate promoters (11). In the second mechanism, AtCRY2 binds to the COP1 (constitutively photomorphogenic 1) E3 ligase through a light-activated and SPA1 (suppressor of phytochrome A) protein-mediated mechanism, forming a ternary CRY2-SPA1-COP1 complex and photobodies. Within photobodies, the E3 ligase activity of COP1 on cognate transcription factors that mediate the blue light response, such as flowering, is inhibited, which leads to accumulation of these factors and up-regulation of blue light-responsive genes (12–14). In addition, within CRY2-PBs, COP1 ubiquitylates AtCRY2, leading to its eventual proteolysis and down-regulation of the blue light signaling pathway.
Humans have two CRY homologues (15, 16). Although both exhibit a high degree of sequence similarity with Arabidopsis CRYs, there is no convincing experimental evidence suggesting that human CRYs, which function as core circadian clock proteins, mediate any blue light sensory activity (15, 16). Hence, we reasoned that human cells could be used to determine the requirements for photobody formation by AtCRY2. We find that blue light stimulates hCOP1-AtCRY2 binding in the absence of SPA1 and leads to formation of CRY-PBs in which AtCRY2 is subject to proteolysis. We conclude that AtCRY2-PB formation does not depend on other plant proteins or nucleic acids. Using AtCRY2-human checkpoint fusion proteins, we demonstrate that we can regulate human DNA damage checkpoint responses by sequestering and concentrating the relevant proteins with blue light pulses.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Plasmids used in this work can be obtained from Addgene with detailed sequence information. To generate pEGFP-N1-AtCRY2, AtCRY2 was amplified by PCR and inserted into the pEGFP-N1 vector (Clontech). To generate pEGFP-N1-mCRY2, mCRY2 was amplified by PCR and inserted into the pEGFP-N1 vector. pEGFP-N1-DmCRY was generated by subcloning the SpeI-XhoI fragment from pFastBac-DmCRY (17) into the pEGFP-N1 vector. To generate pmCherry-N1-mCOP1, mCOP1 was amplified by PCR and inserted into the pmCherry-N1 (Clontech) vector. To generate pcDNA3.1-AtCRY2-GFP-TopBP1, TopBP1 from LacR-TopBP1 (18) was amplified by PCR and cloned into the NotI/PmeI sites of pcDNA3.1-AtCRY2-GFP plasmid. To generate pcDNA5/FRT/TO-AtCRY2-EGFP, AtCRY2-EGFP fragment was amplified from pEGFP-N1-AtCRY2 by PCR. Then the PCR product was inserted into the pcDNA5/FRT/TO vector (Invitrogen).
All plasmid sequences were verified by sequencing at the Genome Analysis Facility at the University of North Carolina at Chapel Hill. Primer sequences used for PCR will be provided upon request.
Cell Lines
HEK293T and FlpTM-In T-RExTM-293 cells (Invitrogen) were maintained in Dulbecco's minimal essential medium supplemented with 10% fetal bovine serum and penicillin-streptomycin. The Flp-In/FLAG-AtCRY2-EGFP cell line was generated according to the manufacturer's protocols (Invitrogen).
Immunoprecipitation
Flp-in/FLAG-AtCRY2-EGFP cells were treated with 0.5 μg/ml tetracycline and continuously cultured in the dark for 48 h. Then the cells were exposed to 366 nm of light (1 milliwatt/cm2) and harvested at the indicated time points. Cells were lysed with PBS (phosphate-buffered saline) buffer containing 0.5% Triton X-100 and protease inhibitors (Roche Applied Science) followed by two rounds of freeze-thaw cycles. Equal amounts of protein were incubated with anti-FLAG M2-agarose beads (Sigma) for 2 h at 4 °C. Beads were washed three times with lysis buffer; bound proteins were eluted in 2× SDS-sample buffer and analyzed by immunoblotting.
Fluorescence Microscopy
For GFP fluorescence microscopy, HEK293T cells were cultured on poly-d-lysine Cellware 12-mm round coverslips (BD Biosciences) placed in 35-mm dishes. Cells were transfected with the indicated plasmids using Lipofectamine 2000 reagent (Invitrogen). After a 24-h incubation at 37 °C, cells were exposed to 366 nm of light (2 milliwatts/cm2) from a Blak-Ray long wave UV lamp (UVP, LLC) for the indicated time points. We obtained the same results with a lamp emitting mainly at 470 nm (blue light), but we performed most of our experiments with the Blak-Ray long wave UV lamp emitting mainly at 366 nm, which we will refer to as blue light for simplicity. The cells were washed twice with PBS, fixed immediately with 3.7% methanol-free formaldehyde in PBS for 15 min, and washed twice with PBS. For GFP (FITC) and mCherry fluorescence imaging, the cells were washed three times and mounted in Prolong Gold antifade with DAPI reagent.
For PML body immunofluorescence staining, cells were permeabilized by PBT (0.1% Triton X-100 in PBS) after fixation, blocked with antibody blocking buffer (1% BSA in PBT) for 1 h, and incubated with anti-PML (2 μg/ml) antibody (Santa Cruz Biotechnology) for 45 min. After washing three times with PBT, the cells were incubated with Alexa Fluor 594 (1:500 dilution) (goat anti-mouse IgG, Invitrogen) secondary antibody for 30 min. Then the coverslip was washed three times and mounted in Prolong Gold antifade with DAPI reagent. Images were captured using a LeicaSP2 confocal microscope. The protein proximity index was calculated by using the protein proximity analyzer (PPA) software (24).
Light-induced in Vivo Proteolysis of CRYs
HEK293T cells (∼90% confluency) were transfected with 2 μg of appropriate plasmid DNA using Lipofectamine 2000 reagent (Invitrogen) in 35-mm dishes. The cells were kept in the dark for 24 h and then exposed to blue light at a fluence rate of 2 milliwatts/cm2 for the indicated periods (19). The extent of photoinduced proteolysis was determined by Western blotting using the appropriate antibodies, and the level of CRY proteolytic degradation was quantified using the ImageQuant 5.0 (Molecular Dynamics) software after scanning the immunoblots.
Checkpoint Response Assay
HEK293T cells (∼90% confluency) were transfected in 35-mm dishes using FuGENE HD (Promega) and increasing amounts of plasmid DNA expressing AtCRY2-GFP-TopBP1 fusions. Total transfected DNA was maintained constant in each dish by the addition of pcDNA3.1 empty vector. After incubation for 18 h, cells were exposed to blue light for 1 h and incubated for a further 1 h at 37 °C, and then the cells were harvested and lysed, and cell lysates were separated by SDS-PAGE. Chk1 phosphorylation was detected by immunoblotting using Ser-345 phospho-ChK1 antibodies (Cell Signaling), and the levels of total Chk1 protein were subsequently detected by immunoblotting with anti-Chk1 antibodies (Santa Cruz Biotechnology) using the same membrane. Levels of phosphorylation were quantified using the ImageQuant 5.0 (Molecular Dynamics) software after scanning the immunoblots. The highest level of Chk1 phosphorylation in each experiment was set equal to 100, and the levels of phosphorylated Chk1 in the other lanes were determined relative to this value. The averages from three independent experiments were graphed, and the error bars in Figs. 2, 3, and 5 indicate the S.D.
FIGURE 2.

Properties of AtCRY2-GFP photobodies in human cells. A, degradation of photosensory cryptochromes expressed in human cells by blue light. HEK293T cells, which were transfected with vectors expressing β-galactosidase as a control together with AtCRY2-GFP or DmCRY-GFP, were subjected to blue light irradiation for the indicated time periods and then analyzed by immunoblotting. Left panel, immunoblots. Right panel, quantitative analysis of three experiments. Values were normalized to β-galactosidase levels (internal control). Error bars indicate S.E. B, stability of photobodies in mammalian cells. HEK293T cells were transfected with AtCRY2-GFP and exposed to blue light for the indicated periods of time followed by incubation in dark for 60 (60'L:60'D), 120 (60'L:120'D), or 360 min (60'L:360'D) and visualized by fluorescence microscopy (GFP) and DAPI staining. C, effect of AtCRY2 levels on photobody formation. Flp-In/FLAG-AtCRY2-EGFP cells were treated with tetracycline (Tet) for the indicated periods of time. Top panel, a fraction of the cells was harvested and analyzed by immunoblotting. Bottom panel, the rest of the cells were exposed to blue light for 1 h, fixed, and visualized by DAPI staining and fluorescence microscopy. The numbers in parentheses indicate the percentage of cells that contained nuclear photobodies. These numbers were obtained from the lower magnification field (three left columns), and the last column shows selected frames to illustrate photobody formation under all three conditions. Bar = 5 μm.
FIGURE 3.

Mouse COP1 co-localizes with AtCRY2 in photobodies formed in human cells. A, schematic of the proteins used in this study. B and C, HEK293T cells were co-transfected with AtCRY2-GFP and mCOP1-mCherry expression vectors either individually (B) or in combination (C) and then exposed to blue light where indicated and analyzed by DAPI staining and GFP fluorescence. B, light induces formation of AtCRY2 photobodies but has no effect on subnuclear distribution of COP1 when cells are transfected with the individual expression vectors. C, when cells are co-transfected with AtCRY2-GFP and COP1-mCherry, both AtCRY2 and COP1 make photobodies, and the two photobodies overlap, indicating that AtCRY2 recruits COP1 to AtCRY2 photobodies. D, light stimulates binding of AtCRY2 to COP1. Flp-In/FLAG-AtCRY2-EGFP were induced with tetracycline (Tet) for 48 h where indicated. Cells were then irradiated with blue light (BL) for the indicated periods of time and lysed, and AtCRY2 was immunoprecipitated with FLAG antibodies. The immunoprecipitates (IP) were analyzed for AtCRY2 and COP1 by immunoblotting. The relative levels of COP1 in AtCRY2 immunoprecipitates were quantified and plotted as shown in the right panel, and the bar graph shows the average of three experiments ± S.E. COP1 levels were normalized to the AtCRY2 protein levels; normalized COP1 level at the 0-min time point was given a value of 1. Bar = 5 μm.
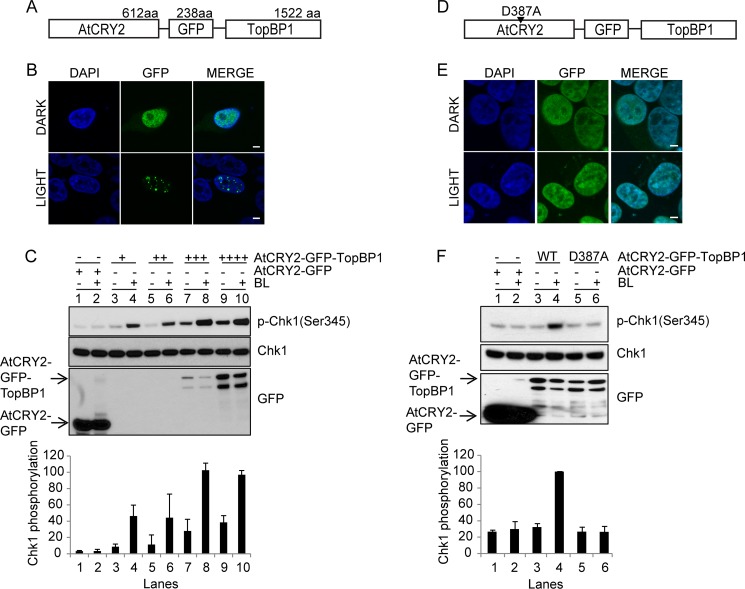
FIGURE 5.
Activation of the ATR-Chk1 signaling pathway by photobody formation. A, schematic of the AtCRY2-GFP-TopBP1 construct. aa, amino acids. B, formation of AtCRY2-GFP-TopBP1 photobodies probed by fluorescence microscopy imaging. C, in vivo checkpoint activation by AtCRY2-GFP-TopBP1. HEK293T cells were transfected with either AtCRY2-GFP or increasing amounts of AtCRY2-GFP-TopBP1 plasmids (0.1, 0.25, and 0.5, 1 μg) and kept in the dark or exposed to blue light (BL) as indicated. Cell lysates were prepared, and the expression of AtCRY2-GFP, AtCRY2-GFP-TopBP1, and Chk1 as well as the level of Ser-345 phospho-Chk1 (p-Chk1(Ser345)) were probed by Western blotting. The top panel shows a representative Western blot experiment; the bottom panel shows quantitative analysis of Chk1 phosphorylation. Error bars indicate S.D. from three independent experiments. D, schematic of the fusion protein construct AtCRY2(D387A)-GFP-TopBP1 with the AtCRY2 FAD binding site mutation. E, fluorescence microscopy imaging with AtCRY2(D387A)-GFP-TopBP1 mutant. F, in vivo checkpoint assay with AtCRY2(D387A)-GFP-TopBP1 mutant. Top panel, Western blotting; bottom panel, quantitative analysis of Chk1 phosphorylation, expressed relative to the highest level. HEK293T cells were transfected with AtCRY2(D387A)-GFP-TopBP1 plasmids (0.1 μg) and kept in the dark or exposed to blue light as indicated. Error bars indicate S.D. from three independent experiments. Bar = 5 μm.
RESULTS
Formation of Cryptochrome Photobodies in Mammalian Cells
Although cryptochromes from plants, insects, and mammals exhibit highly conserved structures, they play diverse roles in light signal transduction pathways. Human cells possess cryptochromes 1 and 2, which are core circadian clock proteins, but are not known to have a photosensory function; they are not degraded following blue light exposure and are not known to form photobodies (16, 19). In contrast, insect Type 1 CRYs including Drosophila CRY (DmCRY) and plant cryptochromes including AtCRY2 are degraded following blue light exposure, and AtCRY2 forms photobodies prior to degradation in plant cells (11, 19). We have previously shown that light-induced DmCRY proteolysis does not require any Drosophila-specific protein as it occurs when mammalian cells ectopically expressing DmCRY are exposed to blue light (19). We wished to determine whether AtCRY2 would be similarly degraded by blue light in mammalian cells and whether the degradation of both DmCRY and AtCRY2 proceeds through a prerequisite photobody formation as AtCRY2 does in plant cells.
To this end, we constructed plasmids expressing AtCRY2-GFP, DmCRY-GFP, and mouse mCRY2-GFP fusion proteins (Fig. 1A) and transfected human HEK293T cells with these plasmids, which were then exposed to blue light and analyzed by fluorescence microscopy. As shown in Fig. 1, B–D, both AtCRY2-GFP and DmCRY-GFP are predominantly located in the nucleus, and prior to blue light exposure, exhibit uniform nuclear distribution. Similarly, mCRY2-GFP also appears to be exclusively in the nuclei (Fig. 1C). Exposure to blue light does not affect the nuclear localization patterns of DmCRY-GFP and mCRY2-GFP. In contrast, AtCRY2-GFP forms distinct photobodies following blue light exposure (Fig. 1B), quite similar to photobodies that form in plant cells. Two conclusions emerge from these findings. First, as evident from the behavior of DmCRY, not all photosensory CRYs make photobodies following light exposure. Second, and of special relevance, AtCRY2 forms photobodies in mammalian cells much in the same way as it does in plant cells, indicating that photobody formation does not require any plant-specific protein other than cryptochrome itself.
FIGURE 1.

Effect of light on subnuclear distribution of Arabidopsis, Drosophila, and mouse cryptochromes expressed in human cells. A, schematic of the proteins used in this study. B–D, HEK293T cells were transfected with the indicated expression vectors kept in the dark or exposed to blue light, fixed, and visualized by DAPI staining and/or fluorescence microscopy. Bar = 5 μm.
Light-induced Proteolysis of AtCRY2 in Mammalian Cells
As in all signal transduction systems, the phototransduction signal must be eventually down-regulated to maintain the organismic homeostasis. Photosignaling initiated by DmCRY, as well as AtCRY2, is turned off by light-induced proteolysis of these cryptochromes (15, 19). Previously, we showed that light promotes DmCRY degradation in mammalian cells, indicating that no specific Drosophila protein is required for DmCRY proteolysis (19). We wished to find out whether AtCRY2 behaved in a similar manner. HEK293T cells expressing either AtCRY2-GFP or DmCRY-GFP were exposed to blue light for 1–4 h, and the levels of these proteins were analyzed by immunoblotting. As seen in Fig. 2A, both cryptochromes are degraded with similar kinetics, whereas mouse CRY2, which is not known to be a photosensory protein, is not affected under similar conditions (19), indicating that the light-induced proteolysis of certain cryptochromes represents a physiological step in their signaling pathways and is not caused by nonspecific protein degradation by toxic light doses. We also note that because DmCRY, which does not form PBs, and the photobody-forming AtCRY2 are both subject to light-induced proteolysis, these data indicate that PB formation is not an essential step on the pathway to proteolysis.
Kinetics of Formation and Resolution of AtCRY2 Photobodies
In plant cells, AtCRY2-GFP photobodies form in a light-dose-dependent manner and gradually disappear upon transfer from a light to a dark environment (1, 20–22). We wished to know whether AtCRY2-GFP followed the same pattern in human cells. As seen in Fig. 2B, after 30 min of light exposure, a substantial amount of AtCRY2-GFP is in photobodies under our illumination conditions. After 60 min of blue light exposure, the majority of AtCRY2-GFP is in photobodies. When cells were then incubated in the dark, the number of photobodies gradually decreased over the course of 6 h, as has been observed in plant cells.
Because photobody formation entails clustering of the photosensory protein, and this process is expected to be dependent on the amount of the photoreceptor, we next investigated the effect of AtCRY2-GFP protein level on photobody formation. Flp-In/FLAG-AtCRY2-EGFP cells stably expressing AtCRY2-GFP under a tetracycline-on promoter were treated with tetracycline for 12, 24, or 48 h and exposed to blue light for 1 h, and then analyzed by immunoblotting for AtCRY2-GFP expression and for photobody formation by fluorescence microscopy. As seen in Fig. 2C, AtCRY2-GFP steadily increases over the course of the experiment, and the number of photobody-positive cells increases in parallel after blue light exposure. All these findings suggest that AtCRY2-GFP follows the same path for photobody formation in human cells as it does in plant cells.
Composition of AtCRY2 Photobodies in Mammalian Cells
All NBs characterized to date contain many types of proteins, up to 40 different proteins in PML-NBs (2, 3). However, in general, the roles of the different types of proteins other than the defining member (PML in case of PML-NBs) in NB formation and function are not known. A genetic screen in Arabidopsis identified several proteins that were essential for phytochrome B photobody formation (10). No such screen has been reported for AtCRY2. However, it has been found that light promotes binding of AtCRY2 to SPA1, which leads to formation of AtCRY2-SPA1-COP1 ternary complex and inhibition of E3 ligase activity of COP1 on transcription factors leading to the accumulation of these transcription factors and hence to blue light-specific gene expression pattern and phenotype (11, 20–22). Humans do not possess an SPA1 homolog, and hence the pathway operative in plant cells for AtCRY2-COP1 interaction is not relevant in human cells. However, humans do have a COP1 homolog (23), which could potentially directly interact with AtCRY2. Thus, we wished to determine whether AtCRY2 interacted with human COP1 and whether this interaction was affected by light. To this end, we analyzed the effect of AtCRY2 on the subcellular distribution of COP1 in HEK293T cells. The cells were transfected with either mouse COP1-Cherry or AtCRY2-GFP plasmids (Fig. 3A), alone or in combination, and the cellular localizations of the two proteins before and after blue light exposure were analyzed by fluorescence microscopy. As seen in Fig. 3B, COP1 is uniformly distributed in the nucleus and cytoplasm, and light exposure does not affect its distribution in the absence of AtCRY2. In contrast, AtCRY2-GFP, which is uniformly distributed in the nucleus in the dark, forms photobodies upon light exposure, as expected. Importantly, in cells transfected with both plasmids, although in the dark both COP1 and AtCRY2 exhibit distribution similar to that when each protein is expressed alone, after light exposure most of AtCRY2 and a significant fraction of COP1 are in nuclear bodies and, within the resolution of our assay, all of the COP1 nuclear bodies overlap with AtCRY2 photobodies (Fig. 3C). This is supported by the protein proximity index analysis (24), which yields a value of 0.8, consistent with co-localization. Thus, it appears that even in the absence of SPA1, photoexcitation of AtCRY2 promotes its interaction with COP1. Because it has been reported that AtCRY2 interacts with COP1 only through intermediacy of SPA1 (14), we decided to analyze the AtCRY2-COP1 interaction identified by photobody analysis by an additional method, using immunoprecipitation.
Flp-In/FLAG-AtCRY2-EGFP cells were induced with tetracycline for 48 h, irradiated with blue light for 5–60 min, and then lysed, AtCRY2 was immunoprecipitated, and the immunoprecipitate was analyzed for COP1 by immunoblotting. As seen in Fig. 3D, AtCRY2 binds to COP1 even in the dark, and importantly, this binding is stimulated by blue light by up to 2.5-fold. This finding suggests that light can stimulate direct AtCRY2-COP1 interaction in the absence of SPA1. However, the possibility that this interaction is aided by a human protein that can substitute for SPA1 cannot be eliminated.
Specificity of Protein-Protein Interactions within AtCRY2 Photobodies
To ascertain that the AtCRY2-COP1 interaction we observe in AtCRY2 photobodies represents light-promoted specific protein-protein interactions and not blue light stress-induced protein aggregation, we analyzed AtCRY2 photobodies for PML protein because this protein has been implicated in NBs that contribute to cellular responses to various stress conditions (2, 3). PML-NBs are constitutively present in most human cell lines including HEK293T, which was used in our study. To address a potential role for PML in formation of AtCRY2 PBs, HEK293T cells were transfected with AtCRY2-GFP plasmids and either kept in the dark or exposed to blue light and analyzed for AtCRY2 by immunofluorescence staining and for PML by immunohistochemical staining. The results are shown in Fig. 4. In the dark, AtCRY2-GFP exhibits uniform nuclear distribution as expected. In contrast, PML localizes to nuclear bodies, consistent with its known properties. Upon light exposure, AtCRY2-GFP forms nuclear bodies, and hence the nuclei of these cells contain two types of NBs, PML-NBs and AtCRY2-PB. Merging of the two images by using protein proximity index analysis (24) yields a value of 0.4, which indicates that this overlap is within the limit of random overlap. Thus, we conclude that the AtCRY2-PBs are specific structures that form by light-induced association of AtCRY2 with itself and COP1 E3 ligase and represent a step on the AtCRY2 signaling pathway.
FIGURE 4.

PML bodies do not co-localize with AtCRY2 photobodies in human cells. HEK293T cells were transfected with AtCRY2-GFP, and 48 h later, they were either kept in the dark or exposed to blue light for 1 h and analyzed by immunofluorescence staining (GFP) or immunohistochemical staining (PML). PML bodies are apparent in both dark and light samples, whereas AtCRY2 is uniformly distributed in the nuclei of the dark samples and forms nuclear bodies (photobodies) upon light exposure. The AtCRY2 photobodies and PML-NBs do not overlap, indicating that PML is not required for formation nor is it recruited to AtCRY2 photobodies. Bar = 5 μm.
Use of Photobodies to Regulate Intranuclear Signaling Pathways
The sequestration of AtCRY2 by light in discreet nuclear bodies raised the potential that this property of cryptochrome could be used to control certain signaling pathways by light. Here, we attempted to initiate the ATR-mediated DNA damage checkpoint signaling pathway by blue light in the absence of DNA damage. ATR (ATM and Rad3-related) initiates the DNA damage checkpoint response by phosphorylating downstream targets in response to DNA damage by UV and UV-mimetic agents (25). The TopBP1 protein, which contains multiple BRCT domains that are involved in protein-protein interactions, functions as an activator/mediator of ATR kinase function (26, 27). Previously, it was shown that by fusing TopBP1 to the Lac repressor and targeting it to an array of tandem Lac operators stably inserted into the genome of NIH3T3 cells, ATR kinase was activated in the absence of DNA damage by sequestering it into a subnuclear region defined by the tandem array of Lac operators (18). We reasoned that it might be possible to activate the ATR kinase by sequestering TopBP1 in the nucleus of mammalian cells in the form of photobodies.
To establish a system in which the ATR-initiated checkpoint is regulated by light, TopBP1 was fused to AtCRY2-GFP (Fig. 5A). HEK293T cells were transfected with this plasmid and exposed to blue light to induce photobody formation. As in the case of AtCRY2-GFP, the AtCRY2-GFP-TopBP1 fusion protein efficiently forms photobodies upon blue light exposure (Fig. 5B). We hypothesized that the clustering of TopBP1 within these photobodies artificially increases the local concentration of the checkpoint protein complex and may activate the DNA damage checkpoint in the absence of DNA damage. As a readout, we measured the phosphorylation of Chk1 checkpoint signaling kinase, a major target of ATR in DNA damage checkpoint response (25). As seen in Fig. 5C, blue light alone, in the absence of DNA damage, causes up to 5-fold increase in Chk1 phosphorylation in cells expressing AtCRY2-GFP-TopBP1 but not in AtCRY2-GFP-expressing cells, indicating that AtCRY2-mediated clustering of TopBP1 is sufficient to activate the checkpoint response.
In conducting these experiments, we were concerned about two potential sources of artifacts. First, it has been shown that transfecting cells with high levels of TopBP1-expressing plasmid can activate ATR in the absence of DNA damage (18, 27). Secondly, high doses of blue light could generate reactive oxygen species from overexpressed flavoproteins, such as AtCRY2, which could cause DNA damage and activate DNA damage checkpoints (28). To address these potential problems, we carried out two types of controls. First, we used high levels of AtCRY2-GFP in transfection assays in parallel with the AtCRY2-GFP-TopBP1 transfections. As seen in Fig. 5C, lanes 1 and 2, transfection with AtCRY2-GFP with and without light exposure does not cause Chk1 phosphorylation. In contrast, transfection with AtCRY2-GFP-TopBP1 induces Chk1 phosphorylation even with low level of DNA in the transfection assay and in a manner that is strongly dependent upon blue light exposure. At relatively high plasmid levels, some Chk1 phosphorylation is observed in the absence of light exposure (Fig. 5C, lanes 7 and 9), in agreement with previous studies indicating that high level expression of TopBP1 induces Chk1 phosphorylation (18, 27). However, even under these conditions, light exposure increases Chk1 phosphorylation markedly over the dark controls (compare lanes 7 and 8 with lanes 9 and 10). Secondly, to demonstrate that the blue light-induced checkpoint activation was due to clustering of TopBP1 in photobodies, we used the mutated AtCRY2 (D387A) as a control. The D387A mutation in the FAD binding pocket completely eliminates FAD binding to AtCRY2 and photobody formation (8). We constructed AtCRY2(D387A)-GFP-TopBP1 (Fig. 5D), transfected HEK293T cells with this plasmid, exposed cells to blue light, and examined photobody formation by immunofluorescence and checkpoint activation by Western blotting against Chk1 phosphorylation. The mutant protein, like wild type, exhibits uniform nuclear localization in the dark control, but in contrast to wild type, it does not form photobodies upon blue light exposure (Fig. 5E) and does not activate Chk1 phosphorylation (Fig. 5F) in the absence or presence of blue light (Fig. 5F, lanes 5 and 6) under conditions conducive to activation by the wild type (Fig. 5F, lanes 3 and 4). Taken together, our data lead us to conclude that by fusing the ATR co-activator TopBP1 to AtCRY2, we have developed a unique system of activating the ATR-Chk1 checkpoint signaling pathway with blue light in the absence of DNA damage.
DISCUSSION
In recent years, a number of methods have been developed that use photosensory proteins for regulating biological functions at the cellular and even organismal levels (29, 30). These include channelopsin, which employs retinal phytochromes, which employ a linear bilin, and LOV (light-oxygen-voltage-sensing) domain proteins, which employ FMN as chromophores. With all these systems, light-induced conformational changes in the photosensory protein itself, or in the target protein such as Raf fused to the chromophore binding module of the photosensory protein, initiate a chain of events that leads to activation of a particular signal transduction pathway or regulation of a specific biochemical network.
In addition to conformational change-induced regulation of activity, light is also known to induce complex formation of two plant photosensory proteins, the red light sensor phytochrome and the blue light sensor cryptochrome with their effector target proteins (4, 11). In both cases, light also induced the formation of large protein aggregates (0.1–0.5 μm in diameter), which have many features in common with other nuclear bodies, and because their formation is strictly light-dependent, they have been named photobodies (4). Although it is unclear at present whether photobody formation is an essential step on the activation pathway of these photoreceptors, in both cases, COP1 E3 ligase is associated with photobodies, and photobody formation is followed by degradation of the photosensory protein by the proteasome. Hence, currently available evidence is consistent with photobodies consisting of proteins in aggregates destined for dismantling and thus the deactivation of the light-activated signaling pathway. Formation of phytochrome photobodies appears to depend on participation of several other proteins, one of which is a polyubiquitin-binding protein-like protein, hemera (10). Whether or not cryptochrome photobody formation requires other proteins is not known. Previously, AtCRY2 was expressed in human cells, and upon blue light excitation, it was shown to interact with the co-expressed plant transcription factor, CIB1, in a manner similar to what is observed during blue light signaling in Arabidopsis (31). However, in that study, neither the association of AtCRY2 with endogenous human COP1 nor its proteolysis was investigated.
Here, we show 1) that AtCRY2 forms photobodies in the nuclei of human cells, 2) that photobody formation is followed by proteolysis, 3) that photobodies of AtCRY2 contain COP1 in mammalian cells as observed in plant cells, and 4) importantly, that by fusing AtCRY2 to a checkpoint protein, we can activate the DNA damage checkpoint signaling pathway in the absence of DNA damage. It is likely that recruitment of other checkpoint proteins to TopBP1 leads to high local concentration of ATR checkpoint complex in the photobodies, which is sufficient to activate the downstream signaling events. Hence, it will be informative to use our optogenetic system to quantitatively study the ATR-mediated cell cycle checkpoints at high spatial resolution in further studies.
We note that while this manuscript was in preparation, another group reported AtCRY2 photobody formation in human cells (32). In that study, AtCRY2 with a deletion of the nuclear localization signal in the C-terminal extension of the cryptochrome was used. As a result, cytoplasmic photobodies were formed, and by fusing this form of AtCRY2 with the appropriate proteins, specific membrane/cytoplasmic signaling pathways could be regulated. That study did not address the issue of photobody composition or the issue of light-induced proteolysis of AtCRY2 in mammalian cells. Nevertheless, technologically, the two studies are complementary in terms of one providing a method for activation of intranuclear pathways (this work) and the other providing a method for activation of cytoplasmic/membrane-associated (32) signaling pathways by light pulses. Because protein clustering regulates numerous signaling pathways, light-induced AtCYR2 photobody will be a powerful tool to study various cellular events.
Acknowledgment
We thank Dr. Chentao Lin (UCLA) for useful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM31082 and GM032833 (to A. S.).
- NB
- nuclear body
- PB
- photobody
- PHY
- phytochrome
- CRY
- cryptochrome
- AtCRY2
- Arabidopsis CRY2
- DmCRY
- Drosophila CRY
- mCRY2
- mouse CRY2
- PML-NB
- promyelocytic leukemia nuclear body
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1. Dundr M., Misteli T. (2010) Biogenesis of nuclear bodies. Cold Spring Harb. Perspect. Biol. 2, a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dellaire G., Bazett-Jones D. P. (2004) PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. BioEssays 26, 963–977 [DOI] [PubMed] [Google Scholar]
- 3. Bernardi R., Pandolfi P. P. (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 8, 1006–1016 [DOI] [PubMed] [Google Scholar]
- 4. Chen M., Chory J. (2011) Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21, 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Van Buskirk E. K., Decker P. V., Chen M. (2012) Photobodies in light signaling. Plant Physiol. 158, 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamaguchi R., Nakamura M., Mochizuki N., Kay S. A., Nagatani A. (1999) Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145, 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kircher S., Kozma-Bognar L., Kim L., Adam E., Harter K., Schafer E., Nagy F. (1999) Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zuo Z. C., Meng Y. Y., Yu X. H., Zhang Z. L., Feng D. S., Sun S. F., Liu B., Lin C. T. (2012) A study of the blue-light-dependent phosphorylation, degradation, and photobody formation of Arabidopsis CRY2. Mol. Plant 5, 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yu X., Sayegh R., Maymon M., Warpeha K., Klejnot J., Yang H., Huang J., Lee J., Kaufman L., Lin C. (2009) Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21, 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen M., Galvão R. M., Li M., Burger B., Bugea J., Bolado J., Chory J. (2010) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141, 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu H., Liu B., Zhao C., Pepper M., Lin C. (2011) The action mechanisms of plant cryptochromes. Trends Plant Sci. 16, 684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu B., Zuo Z., Liu H., Liu X., Lin C. (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25, 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zuo Z., Liu H., Liu B., Liu X., Lin C. (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21, 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lian H. L., He S. B., Zhang Y. C., Zhu D. M., Zhang J. Y., Jia K. P., Sun S. X., Li L., Yang H. Q. (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25, 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin C., Shalitin D. (2003) Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54, 469–496 [DOI] [PubMed] [Google Scholar]
- 16. Sancar A. (2003) Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103, 2203–2237 [DOI] [PubMed] [Google Scholar]
- 17. Ozturk N., Selby C. P., Annayev Y., Zhong D., Sancar A. (2011) Reaction mechanism of Drosophila cryptochrome. Proc. Natl. Acad. Sci. U.S.A. 108, 516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lindsey-Boltz L. A., Sancar A. (2011) Tethering DNA damage checkpoint mediator proteins topoisomerase IIβ-binding protein 1 (TopBP1) and Claspin to DNA activates ataxia-telangiectasia mutated and RAD3-related (ATR) phosphorylation of checkpoint kinase 1 (Chk1). J. Biol. Chem. 286, 19229–19236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ozturk N., Selby C. P., Song S. H., Ye R., Tan C., Kao Y. T., Zhong D., Sancar A. (2009) Comparative photochemistry of animal type 1 and type 4 cryptochromes. Biochemistry 48, 8585–8593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu N. N., Zhang Y. C., Yang H. Q. (2012) Substitution of a conserved glycine in the PHR domain of Arabidopsis cryptochrome 1 confers a constitutive light response. Mol. Plant 5, 85–97 [DOI] [PubMed] [Google Scholar]
- 21. Weidler G., Zur Oven-Krockhaus S., Heunemann M., Orth C., Schleifenbaum F., Harter K., Hoecker U., Batschauer A. (2012) Degradation of Arabidopsis CRY2 is regulated by SPA proteins and phytochrome A. Plant Cell 24, 2610–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sang Y., Li Q. H., Rubio V., Zhang Y. C., Mao J., Deng X. W., Yang H. Q. (2005) N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17, 1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang H., Kang D., Deng X. W., Wei N. (1999) Evidence for functional conservation of a mammalian homologue of the light-responsive plant protein COP1. Curr. Biol. 9, 711–714 [DOI] [PubMed] [Google Scholar]
- 24. Zinchuk V., Wu Y., Grossenbacher-Zinchuk O., Stefani E. (2011) Quantifying spatial correlations of fluorescent markers using enhanced background reduction with protein proximity index and correlation coefficient estimations. Nat. Protoc. 6, 1554–1567 [DOI] [PubMed] [Google Scholar]
- 25. Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 26. Choi J. H., Lindsey-Boltz L. A., Sancar A. (2007) Reconstitution of a human ATR-mediated checkpoint response to damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 104, 13301–13306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kumagai A., Lee J., Yoo H. Y., Dunphy W. G. (2006) TopBP1 activates the ATR-ATRIP complex. Cell 124, 943–955 [DOI] [PubMed] [Google Scholar]
- 28. Hassan B. H., Lindsey-Boltz L. A., Kemp M. G., Sancar A. (2013) Direct role for the replication protein Treslin (Ticrr) in the ATR kinase-mediated checkpoint response. J. Biol. Chem. 288, 18903–18910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Möglich A., Moffat K. (2010) Engineered photoreceptors as novel optogenetic tools. Photochem. Photobiol. Sci. 9, 1286–1300 [DOI] [PubMed] [Google Scholar]
- 30. Szobota S., Isacoff E. Y. (2010) Optical control of neuronal activity. Annu. Rev. Biophys. 39, 329–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kennedy M. J., Hughes R. M., Peteya L. A., Schwartz J. W., Ehlers M. D., Tucker C. L. (2010) Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bugaj L. J., Choksi A. T., Mesuda C. K., Kane R. S., Schaffer D. V. (2013) Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249–252 [DOI] [PubMed] [Google Scholar]