Background: Whether ubiquitination of endoplasmic reticulum-associated degradation substrate is required for dislocation remains unresolved.
Results: Mutant major histocompatibility complex I (MHCI) heavy chains lacking tails or lysine residues are dislocated in a deubiquitinating enzyme-dependent manner.
Conclusion: Ubiquitination of MHCI heavy chains is not required for dislocation as it is required for degradation.
Significance: Our data support a model that dislocation and degradation require two mechanistically distinct ubiquitination events.
Keywords: Antigen Presentation, Deubiquitination, ER Quality Control, ER-associated Degradation, Ubiquitination
Abstract
Aberrantly or excessively expressed proteins in the endoplasmic reticulum are identified by quality control mechanisms and dislocated to the cytosol for proteasome-mediated, ubiquitin-dependent degradation by a process termed endoplasmic reticulum-associated degradation (ERAD). In addition to its role in degradation, ubiquitination has also been implicated in substrate dislocation, although whether direct ubiquitin conjugation of ERAD substrates is required for dislocation has been difficult to ascertain. An obstacle in probing the mechanism of quality control-induced ERAD is the paucity of ERAD substrates being dislocated and detected at any given time. To obviate this problem, we report here the use of a sensitive biotinylation system to probe the dislocation of major histocompatibility complex I (MHCI) heavy chain substrates in the absence of immune evasion proteins. Using this assay system the dislocation of MHCI heavy chains was found not to require potential ubiquitin conjugation sites in the cytoplasmic tail or Lys residues in the ectodomain. By contrast, dislocation of MHCI heavy chains did require deubiquitinating enzyme activity and rapid proteasome-mediated degradation required Lys residues in MHCI heavy chain ectodomain. These combined findings support the model that the endoplasmic reticulum quality control-induced dislocation of MHCI heavy chains may not require direct ubiquitination/deubiquitination as is required for proteasome-mediated degradation post dislocation.
Introduction
Endoplasmic reticulum-associated degradation (ERAD)3 is the major mechanism by which cells get rid of unwanted proteins that fail to pass quality controls in the secretory pathway or are in an excess in metabolic regulation. Mechanistically the ERAD of substrates can be divided into four steps: recognition, retrotranslocation (dislocation), ubiquitination, and degradation. Significant progress has been made in identifying critical components for each of these four steps of ERAD. Notably, however, the molecular mechanism of substrate dislocation remains largely unknown (1, 2). Of particular recent interest has been defining the specific role substrate ubiquitination plays in dislocation (3).
Using a highly evolutionarily conserved enzymatic cascade of an E1 activating enzyme, an E2 conjugating enzyme, and an E3 ligase, substrates are typically coupled with ubiquitin (Ub) on a Lys residue (4). Extending this initial Ub conjugation, a polyubiquitin chain can be built by E2/E3 pairs adding sequential Ub moieties with selected linkage. Substrates with polyubiquitin chains of length greater than 4 moieties typically coupled via Lys48- or Lys11-linkage are targeted to the proteasome for degradation (5, 6). Once they reach the proteasome, the Ub-tagged proteins are first deubiquitinated by deubiquitinating enzymes (DUBs) in or associated with the 19S regulatory particle that releases Ub for recycling and allows the substrate to enter the narrow pore of the 20S core for degradation (7–10). In addition to proteasome targeting, Ub conjugation on substrate is thought to be a prerequisite for dislocation. Of note, although different ERAD substrates are associated with different multimeric dislocation protein complexes, an ER membrane-associated E3 ligase is always found as a central component of the dislocation complex (11, 12). The major role of the E3 ligase within the dislocation complex is to connect the ER luminal chaperone responsible for substrate recognition with its cognate E2 enzyme for substrate ubiquitination. The cytosolic p97 (Cdc48 in yeast) ATPase is then recruited with its cofactors Ufd1 and Npl4 to provide the pulling force for dislocation of ubiquitinated substrates from diverse dislocation complexes (13–16). Binding of a polyubiquitin chain via Ufd1 and Npl4 is required for dislocation of substrate (17). The polyubiquitin chain on the substrate therefore was speculated to serve as a binding handle for association with the p97 complex, whereas hydrolysis of ATP takes place.
Taken together these findings suggest that substrate ubiquitination may be required for complete dislocation. Curiously, however, direct ubiquitination of substrates during dislocation has been difficult to demonstrate (18, 19). Also, it is not clear whether ubiquitination plays the same role in dislocation of disparate substrates that may use different molecular mechanisms of dislocation. For example, it is conceptually easy to envisage how the cytoplasmic domain of a membrane protein can be ubiquitinated by the cytosolic ubiquitination machinery and how this modification could play a critical role in recruiting the p97 complex to extract the substrate from the ER membrane. However, in some cases potential Ub acceptor sites in the cytosolic domain or even a cytoplasmic tail itself are dispensable for dislocation of certain model ERAD substrates (20–22). In these scenarios as well as for soluble substrates in the ER lumen, an initial movement (a so-called partial dislocation) prior to their complete extraction has been proposed to allow ubiquitination to occur in the cytosol. In support of this partial dislocation model, Kopito and co-workers (23) recently found that Derlin proteins are inactive members of the rhomboid family of intramembrane proteases. Mutations in the rhomboid domain of Derlin1 stabilized mutant α1-antitrypsin, a soluble model ERAD substrate, on the cytosolic face of the ER membrane without interfering with p97 binding (23). These findings suggested that a soluble substrate is first transferred from the ER lumen to the membrane after which Derlin1 and p97 combine to complete dislocation. The role of substrate ubiquitination in this model remained undefined.
Intriguingly, several recent studies suggest that a p97-associated deubiquitinating step is required for dislocation of aberrant soluble or membrane proteins from the ER. For example, the newly discovered chemical inhibitor Eeyarestatin 1 (Eer1) was found to interfere with p97-associated DUB activity (24) and this drug could block dislocation of orphan TCRα and US11-induced dislocation of major histocompatibility complex class I (MHCI) heavy chain (25). Additionally, enzymatically inactive YOD1, a p97 associated DUB, stabilized truncated ribophorin (RI332, a soluble substrate of ERAD), and TCRα in the ER, whereas expression of a chimeric protein with the UBX domain of YOD1 for p97 association and the DUB domain of EBV facilitated the release of stalled RI332 and TCRα into the cytosol. However, this chimeric protein also blocked proteasome-mediated degradation suggesting p97-associated deubiquitination is necessary for protein dislocation but premature removal of a polyubiquitin chain from the substrate before proteasome impairs proteasome-mediated degradation (26–28). These combined observations led to the proposal that two sequential rounds of ubiquitination may be required for ERAD of at least a subset of substrates. The initial round of ubiquitination/deubiquitination would be required for substrate to pass through the narrow pore of p97 to complete dislocation and the second round of ubiquitination/deubiquitination is required for substrate to pass through the proteasome to be degraded (8). Although appealing, this model remains to be experimentally proven. A key question to test in this model or any ERAD model is whether the substrate is the direct target of ubiquitination and deubiquitination prior to or during dislocation.
One of the most extensively studied substrates used to probe the mechanisms underlining ERAD in mammals is MHCI heavy chain (HC), a type I integral membrane protein (29, 30). These investigations have been facilitated by viral immune evasion proteins that induce rapid ERAD of HC, such as US2 and US11 of human cytomegalovirus and mK3 of murine γ herpesvirus 68 (29, 30). Interestingly, although all three of these viral proteins target HC for degradation by the proteasome, the mechanisms by which HCs are recognized, ubiquitinated, and dislocated are distinct. For example, US2-induced ERAD of HC depends on TRC8 E3 ligase and signal peptide peptidase (31, 32), whereas US11-mediated dislocation of HCs is dependent upon SEL1L and Derlin1 (33, 34). Although associated with the HRD1 E3 ligase complex, the functional dependence of US11 on HRD1 or any other ER-associated E3 ligase has not been demonstrated (35–37). Furthermore, based on studies of Lys-less HC, direct Ub conjugation on HC appears to be required for dislocation of HC by US2 but not by US11 (18). Given that US11 can directly interact with Derlin1 that is essential for recruiting p97 and dislocation (23, 33, 34), US11-induced dislocation could conceivably bypass the need for substrate ubiquitination. Different from US2- and US11-induced ERAD of HC, mK3 facilitates the Ub conjugation on the cytoplasmic tail of HCs (38). This finding implies that mK3 induces the extraction of HC from the ER vectorially by its C-terminal tail. Thus studies of these 3 viral proteins targeting ERAD of HC accentuate current dogma that there are multiple mechanisms to induce dislocation. But after dislocation all ERAD substrates are thought to use a similar mechanism for degradation (1).
Interestingly, recent studies of physiologic ERAD of HC demonstrate that it uses a mechanism distinct from that induced by any of the above viral immune evasion proteins. Quality control-induced ERAD of incompletely assembled HC molecules has been studied using cells deficient in β2m, the MHCI light chain, or cells deficient in the transporter associated with antigen processing (35, 39, 40). For example, Burr et al. (35) recently showed that HRD1 and UBE2J1 play an essential role in the ERAD of non-β2m assembled HC. However, in this study whether direct ubiquitination of HC is required for dislocation was not addressed. A challenge in determining the role substrate ubiquitination plays in dislocation versus degradation results from the limited substrates undergoing ERAD at a given time relative to the total pool of HCs. However, the study by Burr et al. (35) clearly demonstrates that viral proteins do not faithfully mimic early mechanisms of substrate ubiquitination and dislocation induced by physiologic ERAD.
To selectively probe the mechanism of dislocation versus degradation of HC under physiological conditions, we adapted a cytosolic site-specific biotinylation system that allows the N terminus of HC to be biotinylated once it is exposed to the cytosol (41). Using this system, combined with pharmacological inhibition of p97 and/or proteasome, we were able to quantitatively monitor the dislocation of membrane-bound, glycosylated HCs to the cytosol as well as the degradation of post-dislocated HCs in the cytosol. We found that inhibition of DUB activity leads to an accumulation of biotinylated MHCI molecules in the ER membrane, however, neither the tail of HC nor the Lys residues in the luminal site are required for the dislocation of these HCs. Nevertheless, their ectodomain lysines are clearly required for degradation of the HCs by the proteasome. These findings support the model that during quality control induced ERAD, direct ubiquitination of HC substrates is not required for dislocation, but is clearly required for degradation.
EXPERIMENTAL PROCEDURES
Cell Lines
Murine B6/WT3 (WT3, H-2b) embryonic fibroblasts and triple knock-out fibroblasts (Kb−/− Db−/− β2m−/−, 3KO) were described previously (42). 293T cells (43) were used for production of retrovirus. All cells were maintained in complete RPMI 1640 or DMEM supplemented with 10% fetal calf serum (HyClone) as described (44). Retrovirus-containing supernatants were produced with transient transfection of 293T cells using the pVpack vector system (Stratagene).
DNA Constructs
Retroviral BirA expressing vector pMIP.cyt-BirA was constructed by subcloning the coding sequence of an Escherichia coli-derived BirA ligase from pDisplay.BirA ligase (kindly provided by Dr. Mike Diamond, Washington University) into a pMSCV.IRES.Puro (pMIP) retroviral expression vector (gift of Dr. L. Lybarger, University of Arizona). The ER-BirA expression vector pMIP.ER-BirA was generated by fusing a signal peptide from the murine Ighv14-3 gene to the N terminus and a KDEL ER-retention signal to the C terminus of the birA ligase gene in the construct pMIP.cyt-BirA. A-15 amino acid (GLNDIFEAQKIWHE) biotin acceptor peptide (BAP) (45) + an NheI site (encoding AS) were placed between the Ld signal peptide and the 1st residue of the mature Ld polypeptide in the pMIN.Ld vector described previously (44) to generate the retroviral BAP-Ld expression construct. The Ld mutants including Ld tailΔK (with all 3 Lys residues on the cytoplasmic tail replaced by Arg), Ld tailΔKCST (with KCST residues on the tail mutated), LdΔtail (with tail sequence after the basic cluster RRRRNT deleted), and LdΔK (with all 13 Lys residues on the entire Ld molecule replaced by Arg) have been described previously (38). The correct sequences for all of the constructs were confirmed by DNA sequencing.
Antibodies and Reagents
Rabbit antisera to the cytoplasmic tail of Ld (Ra20873) and mAbs to the ectodomain of Ld (30-5-7 and 64-3-7) were described previously (44, 46). Antibody to Ub (P4D1), β-actin (AC-74), and rabbit anti-calnexin (SPA-860) were purchased from Santa Cruz, Sigma, and Enzo, respectively. PR-619, DBeQ, and Eer1 were obtained from LifeSensors, Sigma, and EMD Millipore, respectively.
Cell Permeabilization and Fractionation
After treatment with proteasome or DUB inhibitors as indicated and detachment by trypsin-EDTA, the cells were washed with PBS twice and resuspended in PBS containing 20 mm iodoacetamide (Sigma), protease inhibitors (Complete Mini, Roche Applied Science), and 0.02% digitonin (Wako Chemicals, Richmond, VA) at 0.5–1.0 × 107/ml and incubated on ice for 20 min. Soluble cytosolic proteins in the supernatant and membrane proteins in the pellet were separated by centrifugation of digitonin-treated cells at 18,000 × g, 4 °C for 15–20 min.
Immunoprecipitation and Immunoblot
Cells or pellet fractions were lysed in PBS buffer containing 1% Nonidet P-40, 20 mm iodoacetamide and protease inhibitors (Complete Mini, Roche). Postnuclear lysates or supernatant fractions were then incubated at 4 °C overnight with Ld mAbs and protein A-Sepharose (Sigma). After washing 4 times with 0.15% Nonidet P-40/PBS/iodoacetamide, precipitates were eluted with 1× LDS sample loading buffer (Invitrogen), and β-mercaptoethanol was added to a final concentration of 5% where indicated. Immunoblot was performed following SDS-PAGE separation of precipitated proteins or cell lysates. Specific proteins were visualized by standard enhanced chemiluminescence HRP substrate (ECL, Thermo) or supersensitive ECL (Immun-Star WesternC, Bio-Rad) as indicated. For PNGase F treatment, after washing, the immunoprecipitates were eluted in the denaturing buffer (0.5% SDS, 1% β-mercaptoethanol) at 95 °C for 5 min and then incubated at 37 °C for 1 h in 50 mm sodium phosphate, pH 7.5, containing 1% Nonidet P-40 and 5 units of PNGase F (New England Biolabs). For proteinase K treatment, cells were incubated on ice with 5 μg/ml of proteinase K (Invitrogen) for 20 min. After the digestion was stopped by addition of 1 mm PMSF the cells were lysed and subjected to immunoprecipitation. Densitometry was done using ImageJ (NIH).
RESULTS
A Biotinylation System Enables Detection of a Small Fraction of HCs Subjected to ERAD
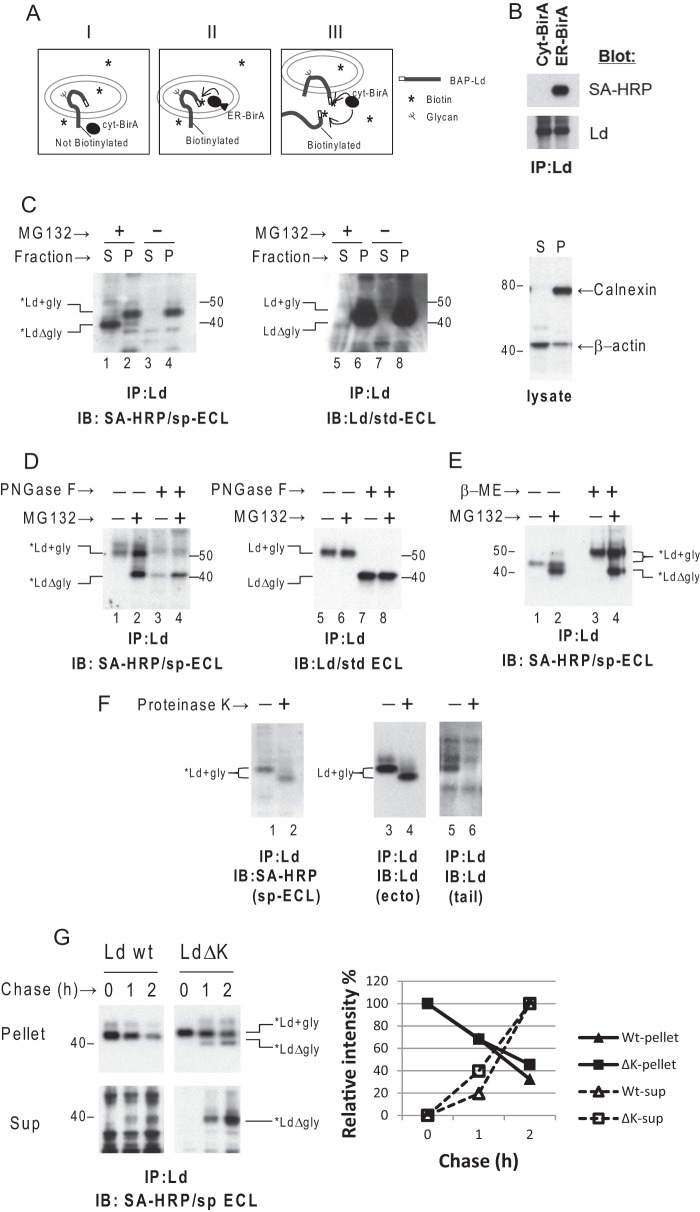
The dislocation of HC proteins, like other ERAD substrates, is typically monitored by blocking proteasome activity and detecting a small fraction of the total HCs as soluble, deglycosylated forms in the cytosol by immunoblot. Prior to deglycosylation by cytosolic N-glycanase this small fraction of the HCs in the ER membrane cannot be distinguished from the majority of HC molecules destined for surface expression along the secretory pathway. To directly and quantitatively monitor dislocation of HC, an in vivo cytosolic site-specific biotinylation system was developed. In this system E. coli-derived BirA biotin ligase (cyt-BirA) (47) was co-expressed in mouse embryonic fibroblast cells with the HC molecule (Ld) having a BAP (45) fused to its N terminus. Because BirA without a signal peptide is expressed exclusively in the cytosol, the N-terminal BAP-tagged Ld molecules (BAP-Ld) can be biotinylated only when their N termini are exposed to the cytosol after being dislocated (or partial dislocated) (Fig. 1A, panels I and III). Of note the BAP tag on the Ld molecules does not disrupt its native folding or surface expression.
FIGURE 1.
A cytosolic site-specific biotinylation system allows detection of limited MHCI HCs subject to ERAD. A, schematic depiction of site-specific biotinylation of N-terminal BAP-tagged Ld HC in cells: panel I, a type I transmembrane protein, BAP-Ld, cannot be biotinylated by cytosolic expressed BirA ligase (cyt-BirA); panel II, BAP-Ld can be biotinylated by ER expressed BirA (ER-BirA); panel III, only partially or completely dislocated BAP-Ld can be biotinylated by cyt-BirA. B, mouse 3KO cells were first transduced with the BAP-Ld then co-transduced with either cyt-BirA or ER-BirA. BAP-Ld molecules were immunoprecipitated (IP) using mAb 30-5-7 and 64-3-7. The precipitates were separated by SDS-PAGE and blotted to detect biotinylated BAP-Ld (*Ld) by HRP-conjugated streptavidin (SA-HRP) or total BAP-Lds by rabbit anti-Ld (Ra20873). C, mouse WT3 (H-2b) cells co-transduced with cyt-BirA and BAP-Ld were treated with dimethyl sulfoxide or 60 μm MG132 for 2 h before permeabilization and fractionation. Left panel, *Ld molecules were recovered by anti-Ld mAbs from the supernatant fraction (S) or the pellet fraction (P), and visualized in the gel by SA-HRP and supersensitive chemiluminescence HRP substrate (sp-ECL). Middle panel, total BAP-Ld including *Ld in the supernatant and pellet fractions was detected by blotting with anti-Ld mAb 64-3-7 and visualized by standard chemiluminescence HRP substrate (std-ECL). Right panel, lysates from the supernatant or pellet fractions were blotted for ER membrane protein calnexin or for cytosolic protein β-actin that served as a control for fractionation. D, WT3 cells, expressing cyt-BirA and BAP-Ld and treated with or without MG132, were lysed in Nonidet P-40 and immunoprecipitated (IP) for Ld. An aliquot of the precipitates was treated by PNGase F before blotting for *Ld and total BAP-Ld as described in C. The total Ld blot here serves as a control for PNGase F treatment. E, cells treated with or without MG132 as in C and D were immunoprecipitated for Ld. An aliquot of the precipitates of each treatment was reduced by 5% β-mercaptoethanol before SDS-PAGE along with the non-reduced aliquots. F, cells used in D were permeabilized and fractionated. The resuspended pellet fraction was incubated on ice with 5 μg/ml of proteinase K for 20 min before immunoprecipitation of Ld. *Ld and total Ld molecules in the precipitates were detected by blotting with SA-HRP or anti-Ld antibodies (64-3-7 to the ectodomain of Ld or Ra20873 to the tail). The total Ld blots served as a control for proteinase K cleavage. G, WT3 cells expressing cyt-BirA and BAP-Ld (wild type or ΔK mutant) were incubated with 60 μm MG132 and 100 μm CHX for the indicated time before permeabilization, fractionation, and immunoprecipitation of Ld. Levels of glycosylated *Ld (*Ld+gly) in the membrane (pellet) and dislocated deglycosylated *Ld (*LdΔgly) in the cytosol (sup) at different time points were detected. Quantification of the bands is shown in the chart as relative intensity of time 0 of *Ld+gly in the upper gel or as relative intensity of 2 h of *LdΔgly in the lower gel. Similar findings were observed in at least one additional independent experiment. IB, immunoblot; IP, immunoprecipitation.
To validate this system, an ER expressing BirA (ER-BirA) with a signal peptide fused to the N-terminal end and a KDEL ER-retrieval signal at the C-terminal end was engineered and co-expressed with BAP-Ld in murine fibroblasts lacking classical HC and β2m. Ld molecules were retrieved from the cell lysates by precipitation with anti-Ld antibodies. Total Ld HCs and biotinylated Ld HCs (*Ld) in the precipitates were then determined by blotting with anti-Ld and streptavidin-HRP conjugates (SA-HRP), respectively, and visualized by the standard enhanced chemiluminescence substrate (ECL) for HRP. Under such conditions the BAP-Ld molecules were clearly biotinylated by ER-BirA, but not by cyt-BirA, although they are equally expressed (Fig. 1, A, panel II, and B). To detect the steady-state level of HCs being removed from the ER to the cytosol under more physiological conditions, WT3 cells with β2m and endogenous MHCI proteins were co-transduced with cyt-BirA and BAP-Ld. These cells were treated with proteasome inhibitor MG132 followed by permeabilization to separate the cytosolic fraction in the supernatant (S) from the membrane fraction in the pellet (P). The lysate blots for the ER membrane protein calnexin and the cytosolic protein β-actin verified the fractionation (Fig. 1C, right panel). *Ld molecules captured by immunoprecipitation of Ld from the two fractions were blotted by SA-HRP and visualized using supersensitive ECL. Using this approach, a band corresponding to glycosylated *Ld molecules was readily detected in the membrane fraction with or without MG132 treatment (Fig. 1C, lanes 2 and 4). These molecules likely represent HCs whose N termini have been exposed to the cytosol, but remain membrane associated (i.e. having undergone partial dislocation). Additionally in the presence of proteasome inhibitor, more than half of the *Ld molecules in the cytosol displayed a faster migration rate (Fig. 1C, lane 1); and these faster migrating *Ld molecules were deglycosylated HCs based on their unchanged position in the gel after PNGase F treatment (Fig. 1D, lane 4 versus lane 2). Thus these molecules are HCs that have been fully dislocated to the cytosol. Of note, some of the deglycosylated *Ld molecules were detected in the P fraction (Fig. 1C, lane 2) as well. These P-associated *Ld molecules are likely residual cytosolic proteins resulting from incomplete depletion during fractionation because residues of typical cytosolic proteins such as β-actin could also be observed in the P fraction (Fig. 1C, right panel). The amount of these residual deglycosylated *Ld forms in the P fraction varied depending on whether a wash step was conducted following permeabilization and centrifugation.
To determine the fraction of HC having undergone dislocation, the same samples were also blotted for Ld rather than SA-HRP. Densitometry comparisons of Fig. 1C, lanes 5 and 6, indicated that less than 1% of total Ld was detected as soluble forms in the cytosol. Unfortunately, it was not possible to estimate the percentage of membrane-bound Ld that was biotinylated because glycosylated forms of *Ld and Ld could not be distinguished (Fig. 1C, lanes 2 and 4 versus lanes 6 and 8). Furthermore, by treating Ld molecules with β-mercaptoethanol, the presence of intracellular disulfide bonds in *Ld was determined. Although the glycosylated *Ld band shifted up under reducing conditions (Fig. 1E, lanes 3 and 4), the band corresponding to deglycosylated *Ld molecules remained unchanged after β-mercaptoethanol treatment (Fig. 1E, lane 4 versus lane 2). These findings demonstrated that glycosylated *Ld forms remain folded, but the deglycosylated *Ld species are unfolded. Thus reduction of disulfide bonds may be a prerequisite for complete dislocation of HC as implicated by studies by other authors (48–50).
To determine whether the glycosylated *Ld molecules have their N termini continuously extended in the cytosol, proteinase K sensitivity was tested. As shown in Fig. 1F, the cytoplasmic tail of membrane-bound *Ld molecules was sensitive to proteinase K treatment as evidenced by a smaller band that could be detected by antibodies specific for the ectodomain of Ld but not by antibodies specific for the tail of Ld (Fig. 1F, lane 4 versus lane 6). By contrast, N termini of membrane-bound *Ld appeared to be resistant to treatment because the strength of the SA-HRP signal for the N-terminal biotin was unchanged (Fig. 1F, lane 2 versus 1). This result suggests that at steady-state the majority of the membrane-bound *Ld have their N termini in the ER lumen and the C termini in the cytosol. How these HCs expose their N termini to the cytosol to get biotinylated by the cyt-BirA and subsequently retrieve their type I integral membrane protein topology is intriguing and may reflect repeated refolding attempts during ER quality control. Alternatively, the N terminus of membrane-bound *Ld may reside on the cytosolic side of the ER membrane, but be protected against proteinase K cleavage by associated proteins involved in dislocation. Regardless of the membrane orientation of glycosylated *Ld, most of these molecules are fated for ERAD. This conclusion is supported by steady-state data showing that more than half of the *Ld molecules are dislocated within a short time after blockade of proteasome function (Fig. 1C). In addition, kinetically, in a cycloheximide (CHX) chase experiment, the decrease over time in the level of membrane-bound, glycosylated *Ld was correlated with the increase in the level of deglycosylated *Ld in the cytosol (Fig. 1G). Thus most membrane-bound *Ld molecules appear to be HCs sentenced to ERAD that are continuously removed from the ER.
Dislocation of BAP-Ld Is Dependent of DUB Activity
As mentioned in the Introduction, DUB function has been implicated in the dislocation of several ERAD substrates including US11-induced dislocation of HC. However, whether DUB activity is required for dislocation of HC by quality control induced ERAD remains undetermined. To address this question here we used two different DUB inhibitors to probe the role of deubiquitination in dislocation of HC not induced by viral immune evasion proteins.
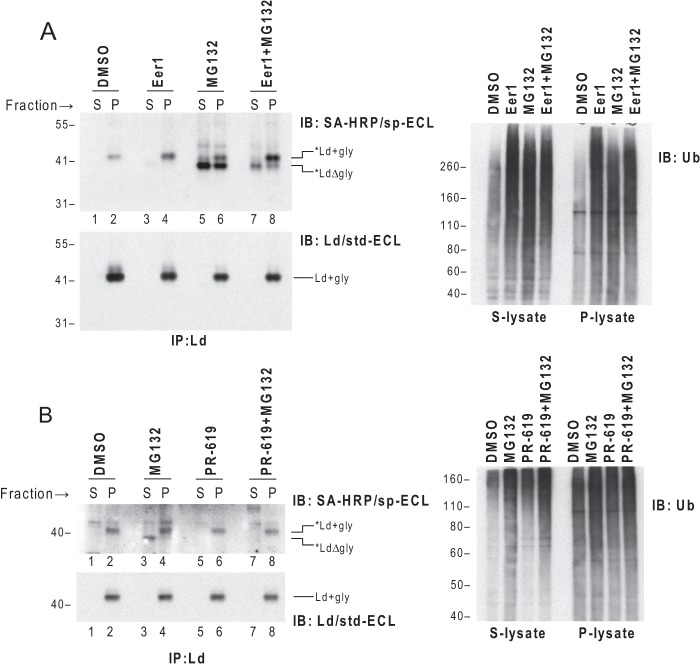
Eer1 is a newly discovered small molecule inhibitor that blocks dislocation of certain ERAD substrates, but does not prevent proteasome-mediated degradation. More specifically, Eer1 has been proposed to block p97-associated DUB activity (24). To test whether Eer1 can impair dislocation of *Ld, cells co-expressing cyt-BirA and BAP-Ld were treated with Eer1 or MG132 before being permeabilized and fractionated. *Ld molecules in the supernatant or pellet fractions were recovered with anti-Ld and blotted with SA-HRP. As expected, when the cells were treated with MG132, deglycosylated *Ld HCs accumulated in the cytosol (Fig. 2A, left upper panel, lane 5). In contrast, when the cells were treated with Eer1, *Ld molecules were only detected in the membrane fraction in glycosylated forms (Fig. 2A, left upper panel, lane 4). This latter finding demonstrates that Eer1 blocks HC dislocation. This conclusion was further supported by treatment with both Eer1 and MG132. When co-treated with both metabolic inhibitors, the majority of *Ld molecules remained glycosylated in the membrane with fewer deglycosylated forms appearing in the cytosol compared with MG132 only treatment (Fig. 2A, left upper panel, lanes 7 and 8 versus lanes 5 and 6).
FIGURE 2.
Inhibition of DUB activity by Eer1 or PR-619 impairs dislocation of *Ld. A, left panel: WT3 cells co-expressing cyt-BirA and BAP-Ld were incubated with Eer1 (10 μm), MG132 (15 μm), or a combination of the two for 5.5 h before collection, permeabilization, and fractionation. *Ld and total Ld from the supernatant (S) and pellet (P) fractions of each treatment were detected by IP of Ld followed by blotting with SA-HRP/sp-ECL or Ld mAb 64-3-7/std-ECL. Right panel, lysates from the supernatant or pellet fractions before immunoprecipitation (IP) were blotted (IB) for ubiquitinated proteins to show the effect of DUB or proteasome inhibition. B, cells used in A were incubated with 40 μm MG132 or PR-619, or a combination of the two for 1 h before being subjected to permeabilization and fractionation. The subsequent detections were the same as in A. Similar findings were observed in at least one additional independent experiment.
Knowing the proposed targets of Eer1 is p97-associated DUB, an Ub blot of lysates from the supernatant or pellet fractions was used as a positive control for drug effect. Indeed, upon Eer1 treatment ubiquitinated proteins accumulated in both the supernatant and pellet lysates (Fig. 2A, right panel), which is in agreement with p97-associated DUB inhibition given that only a fraction of p97 molecules are membrane-bound (51, 52).
To further implicate DUBs in the dislocation of HC under physiological conditions, we tested the effect of PR-619, a non-selective, reversible DUB inhibitor (53), on the dislocation of *Ld. As a control for DUB inhibition, the amount of ubiquitinated proteins in both the supernatant and pellet fractions of the cells treated with PR-619 was augmented (Fig. 2B, right panel). To avoid a secondary effect of depletion of free Ub by prolonged inhibition of DUB activity, treatment was kept short for only 1 h. As shown in Fig. 2B, incubation of cells with PR-619 led to a stall of glycosylated *Ld in the pellet fraction. Thus, although it is known to also block proteasome degradation, the effect of PR-619 on ERAD of *Ld was strikingly similar to that of Eer1 and not MG132 that resulted in accumulation of deglycosylated *Ld primarily in the cytosol. The mechanistic implications of these findings are that Eer1 and PR-619 both affect dislocation by inhibiting DUB activity, implying a deubiquitination step is required for dislocation of HC in the absence of viral evasins.
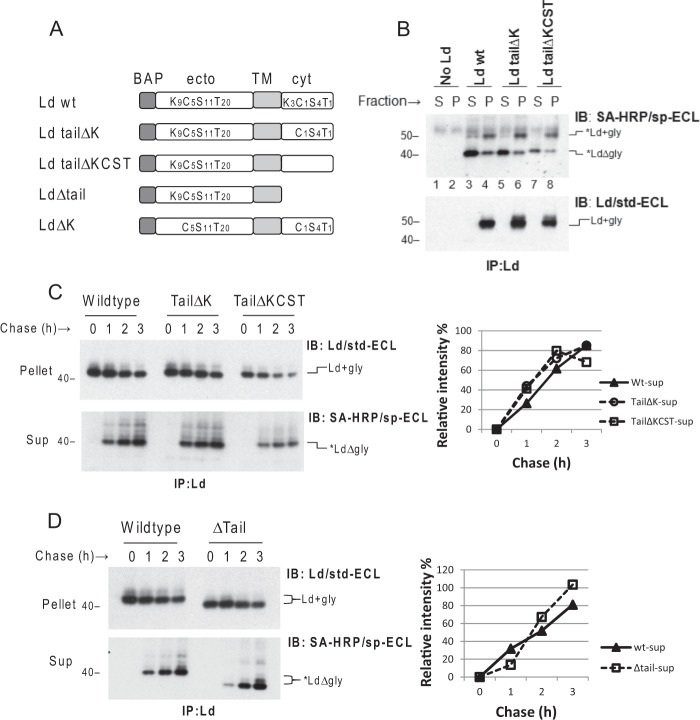
Ubiquitination on the Tail of Ld Is Not Required for Its Dislocation
We next used Ld mutants to address the question of whether direct ubiquitination of substrate is required for dislocation. To this end, a set of N-terminal BAP-tagged Ld mutants that lack Ub conjugation sites were generated (Fig. 3A) and transduced into WT3 cells expressing cyt-BirA. To determine whether dislocation requires conventional Ub coupling on Lys residues in the cytoplasmic tail, a mutant BAP-Ld with a Lys-less tail (Ld tailΔK) was tested. In addition, to test whether dislocation might also require unconventional Ub coupling on hydroxyl- or thiol-containing amino acids, a mutant BAP-Ld with a KCST-less tail (Ld tailΔKCST) was also tested. Following treatment with MG132, like what was observed with wt *Ld, the majority of *Ld forms of both mutants were deglycosylated and detected in the cytosol (Fig. 3B, lanes 5 and 7 versus lane 3). This data suggests that ubiquitination on the tail is not required for HC dislocation. However, it remained possible that tail ubiquitination could promote dislocation efficiency perhaps by strengthening p97 association as suggested by studies of US11-induced dislocation of HC (17). To test the rates of dislocation, cells co-expressing cyt-BirA with Ld tailΔK, Ld tailΔKCST, or wt Ld molecules were treated with CHX and MG132 to stop protein synthesis and proteasome degradation for 0 to 3 h. Increases in the amounts of soluble, deglycosylated forms of *Ld tailΔK, *Ld tailΔKCST, or wt *Ld detected in the cytosol were monitored over 3 h to compare the relative rates of dislocation. Similar to what was seen in steady-state comparisons, none of the mutants displayed obviously altered rates of substrate dislocation (Fig. 3C). This data demonstrates that direct tail ubiquitination does not contribute to the dislocation efficiency. Although tail Ub modification was not required, it remained possible that a tail was still required for dislocation. Indeed this is the case with US11-induced dislocation that requires a tail, but not where tail Lys resides (18). To test whether a tail was required for HC dislocation, a BAP-Ld mutant lacking a tail (LdΔtail) was also tested. As shown in Fig. 3D, the LdΔtail was still dislocated into the cytosol with similar kinetics as wt Ld. Thus, neither tail ubiquitination nor a tail itself is required for complete dislocation of HC substrates.
FIGURE 3.
Direct ubiquitination of the cytoplasmic tail of Ld as well as the tail itself is not required for dislocation of *Ld. A, nomenclature and schematic depiction of variant Ld molecules used in this study. Subscripted number shown with Lys, Cys, Ser, or Thr represents the number of each amino acid residue substituted in the ectodomain (ecto) and cytoplasmic (cyt) domains of Ld; TM, transmembrane. B, WT3 cells were co-transduced with cyt-BirA and one of the following BAP-tagged Ld: Ld wt, Ld tailΔK, or Ld tailΔKCST. Each of the above cells was incubated with 8.5 μm MG132 for 6 h before permeabilization, fractionation, and immunoprecipitation (IP) of Ld. *Ld molecules in the gel were detected as indicated. The total Ld blot shown in the lower panel served as a control for expression of different BAP-Ld molecules. C, left panel: cells in B were treated with 100 μg/ml of CHX and 60 μm MG132 for the indicated time before permeabilization, fractionation, and IP to recover BAP-Ld. Levels of dislocated deglycosylated *Ld in the cytosol fraction (sup) and total Ld in the membrane fraction (pellet) at different time point were detected as in B. Right panel, the rate of dislocation was quantified as the relative band intensities of dislocated, deglycosylated *Ld molecules at each time point in the lower gel to the intensity of time 0 of glycosylated Ld in the upper gel. D, cells co-expressing cyt-BirA and BAP-Ld (wt or Δtail) were used in a CHX chase experiment as in C. Similar findings were observed in at least one additional independent experiment.
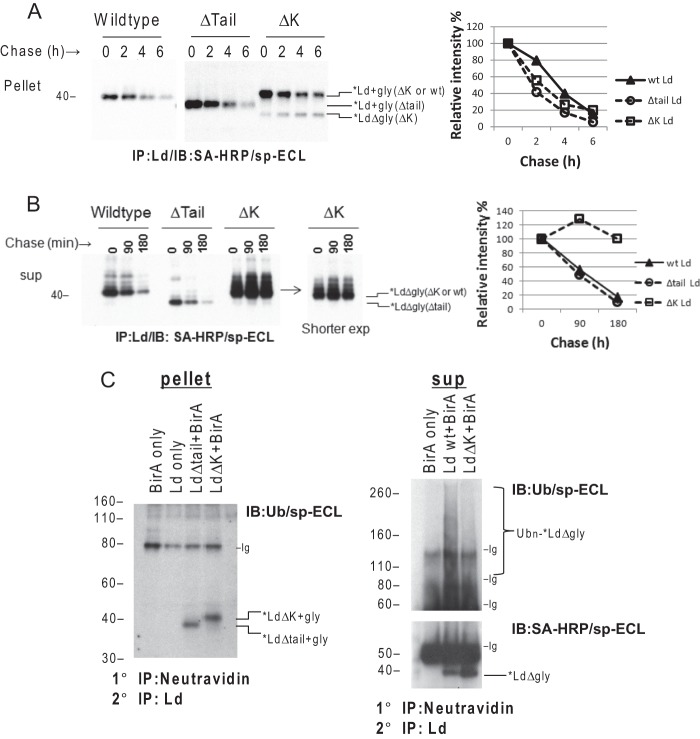
Ubiquitination of Lys Resides on the Ectodomain of Ld Is Not Required for Dislocation but Is Required for Rapid Degradation by the Proteasome
An alternative hypothesis to tail-mediated dislocation of type I integral membrane protein is a partial dislocation model. In a partial dislocation model, ERAD substrates could expose Lys residues on the lumen portion of the substrate to the ubiquitination machinery in the cytosol. If this mechanism is used by physiological ERAD of MHCI molecules, then the Lys residues in their ectodomains would be important for complete dislocation. To test this possibility, a CHX chase experiment (as above) was conducted with cells co-expressing cyt-BirA and a BAP-Ld mutant having all 13 Lys residues mutated to Arg (LdΔK). By monitoring the disappearance of glycosylated *Ld species during the chase without MG132 treatment, glycosylated *LdΔK mutants were found to decay at a similar rate as the glycosylated wt *Ld and *LdΔtail molecules (Fig. 4A). This result implies that Lys resides in the ectodomain of HC are not required for dislocation. However, accumulation of deglycosylated forms was only seen with *LdΔK molecules (Fig. 4A). Of note, deglycosylated ERAD substrates are typically only found in the cytosol of cells treated with proteasome inhibitors and this experiment was carried out in the absence of MG132. Thus the unique detection of deglycosylated forms of *LdΔK was likely attributed to poor degradation. To confirm this conclusion, cells were treated with the reversible proteasome inhibitor MG132 for 3 h to allow accumulation of post-dislocated deglycosylated *Ld. These cells were then chased with Eer1 to prevent continuous dislocation while monitoring the disappearance of the deglycosylated *Ld forms in the cytosol. The results in Fig. 4B show that deglycosylated wt *Ld and *LdΔtail have comparable turnover rates, with half-lives of 90 min. By contrast, deglycosylated *LdΔK molecules were remarkably stable during the chase. These findings thus demonstrate that direct ubiquitination on lysines of the ectodomain of HC is not required for dislocation but is critical for proteasome-mediated degradation. In support of this conclusion, the post-dislocated wt *Ld molecules in the cytosol, but not *LΔK, were heavily ubiquitinated (Fig. 4C, right panel), whereas the membrane-bound *Ld molecules with or without ectodomain lysines had no detectable Ub conjugation (Fig. 4C, left panel).
FIGURE 4.
Direct ubiquitination of Lys residues on the ectodomain of Ld is not required for dislocation but is critical for degradation by proteasome. A, left panel: cells co-expressing cyt-BirA and BAP-Ld (wt, Δtail, or ΔK) were incubated with CHX (100 μg/ml) for the indicated times before being subjected to immunoprecipitation (IP) for Ld and detection of *Ld with SA-HRP/sp-ECL. Right panel, the dislocation rate was quantified for each type of Ld molecules by plotting the relative band intensities of glycosylated *Ld as a percentage of the intensity of time 0. B, left panel: cells in A were preincubated with 60 μm MG132 for 2 h to allow accumulation of dislocated deglycosylated *Ld in the cytosol. After extensive washing, these cells were subjected to chase in the presence of 10 μm Eer1 for the indicated time. The *Ld molecules were recovered from the supernatant of permeabilized cells by IP of Ld and visualized by blotting with SA-HRP/sp-ECL. A shorter exposure of *LdΔK blot is shown for better resolution. Right panel: quantification of the decay rate of each type of *Ld molecules is conducted by plotting the relative band intensities of deglycosylated *Ld in the gel as a percentage of the intensity of time 0. C, left panel: cells co-expressing cyt-BirA and BAP-Ld (Δtail or ΔK), or expressing cyt-BirA only/BAP-Ld only as controls, were permeabilized and fractionated. The membrane-bound *Ld molecules in the pellet fractions were retrieved by two rounds of IPs: 1° IP with NeutrAvidin-agarose beads followed by denaturing with 0.5% SDS Tris-HCl buffer and then the 2° IP with anti-Ld (64-3-7). The precipitates were SDS-PAGE separated and blotted with anti-Ub (P4D1), biotinylated secondary antibody, and SA-HRP to visualize *Ld and ubiquitinated *Ld. Right panel: cells expressing cyt-BirA only or co-expressing BirA and BAP-Ld (wt or ΔK) were treated with 50 μm MG132 for 2 h before permeabilization and fractionation. Cytosolic *Ld was retrieved by 1° and 2° IPs as described above from the supernatant fractions. Ubiquitinated and non-ubiquitinated *Ld in the precipitates were detected by Ub or SA-HRP blotting of a reducing gel. Nonspecific antibody bands (Ig) in the gels are indicated.
The above findings were unexpected in light of our previous evidence that DUB activity is required for BAP-Ld dislocation. Indeed, we found that the DUB inhibitor PR-619 blocked the dislocation of *Ld mutants with no Lys residues in the entire molecule (Fig. 5A), although like a typical ERAD process, dislocation of *LdΔK depends on p97 function (Fig. 5B). Thus, these results demonstrate that direct ubiquitination/deubiquitination of Lys residues on HCs is not required for HC dislocation. In total our findings support the model that the processes of ubiquitination/deubiquitination required for dislocation and degradation of HC are distinct events and only the latter occurs directly on HC substrates during ERAD.
FIGURE 5.
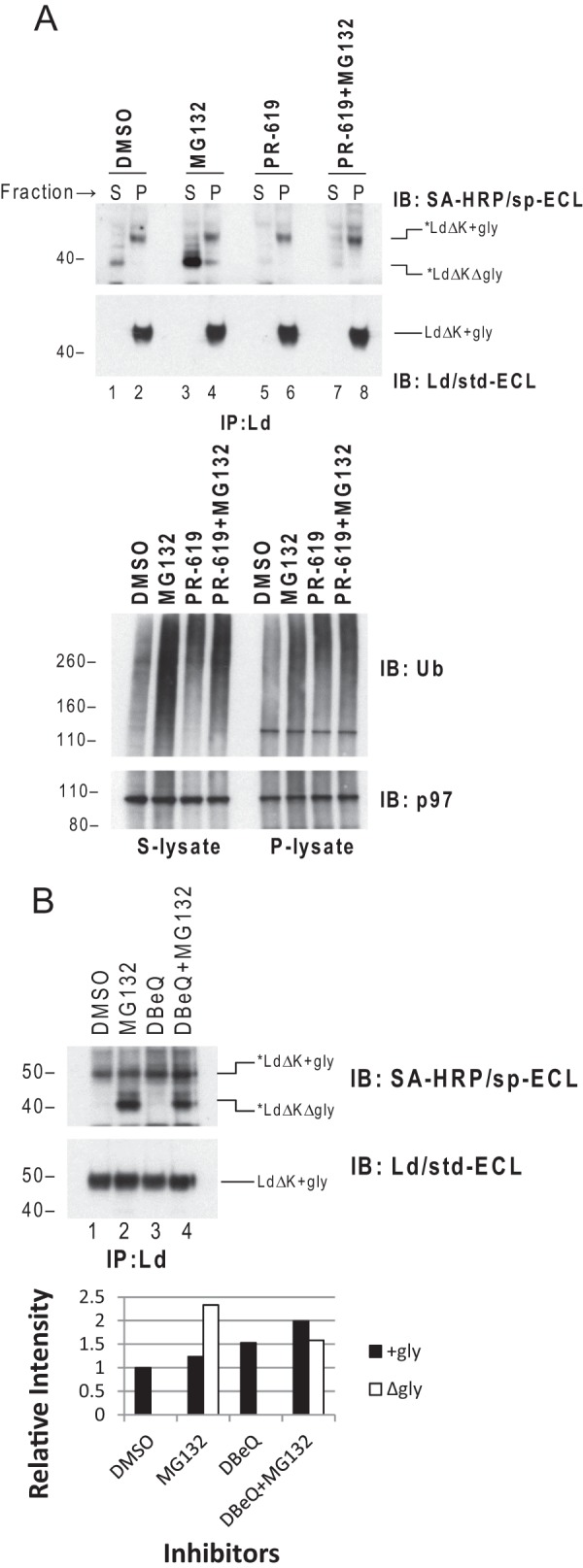
DUB activity is required by dislocation of K-less Ld. A, upper panel: WT3 cells co-transduced with cyt-BirA and BAP-LdΔK were incubated with 40 μm MG132, PR-619, or a combination of the two for 1 h before permeabilization and fractionation. *Ld and total Ld from the supernatant (S) and pellet (P) fractions of each treatment were detected by immunoprecipitation (IP) of Ld followed by blotting (IB) with SA-HRP/sp-ECL or Ld mAb 64-3-7/std-ECL. Lower panel, lysates from the supernatant or pellet fractions before IP were blotted for ubiquitinated proteins to show the effect of DUB or proteasome inhibition by PR-619 or MG132. None of the treatments significantly altered the level of p97 in the supernatant or pellet fraction as shown in p97 blot. B, cells used in A were incubated with 20 μm MG132, 20 μm DBeQ, or a combination of the two for 2 h before permeabilization and fractionation. *Ld and total Ld in the lysates of the cells were determined as in A. DBeQ is a selective p97 inhibitor known to impair ERAD of orphan TCRα by ATP competition (66). Relative intensity of the bands of glycosylated and deglycosylated *LdΔK is shown in below.
DISCUSSION
In this study we report using a sensitive site-specific biotinylation system to probe the dislocation of HC substrates subjected to ERAD in the absence of immune evasion proteins. Using this assay system evidence is presented that dislocation of HCs does not require potential Ub sites in its cytoplasmic tail, or Lys residues in its ectodomain. However, dislocation of HC does require DUB activity, and rapid degradation of HC post-dislocation requires Lys resides in the HC ectodomain. Our combined findings support the model that physiologic dislocation and degradation during ERAD of HC require two separate and mechanistically distinct ubiquitination/deubiquitination events.
Much progress has been made in understanding how different ERAD substrates are dislocated. However, two remaining questions are when an ERAD substrate gets ubiquitinated and what specific role ubiquitination plays in dislocation? Past dogma has been that ubiquitination of substrates is required for proteasome-mediated degradation and occurs prior to the complete substrate extraction. In this model, Ub conjugates on substrates were proposed to function as either a handle or ratchet for p97-mediated extraction. However, the fact that the enzymatic domains of the ERAD-associated E2/E3 complexes are located in the cytosol raises the question of how soluble ERAD substrates or membrane substrates with no cytosolic Lys residues are initially dislocated for exposure of their ectodomain to the ubiquitination machinery. A partial dislocation model is a potential solution and it is supported by the observation that ubiquitinated soluble substrates are sometimes caught on the ER membrane after blocking p97 activity (13, 15). However, some ERAD substrates stall inside of the ER in a non-ubiquitinated form under comparable conditions (14, 54). Most surprisingly, Ub conjugation prior to complete dislocation has been difficult to detect even for transmembrane proteins that have potential cytoplasmic Ub acceptor sites (18, 28).
Adding to the mystery of how ERAD substrates get ubiquitinated prior to dislocation, recent studies suggest that two rounds of ubiquitination/deubiquitination may be required, at least for a subset of ERAD substrates such as soluble protein RI331, as well as membrane proteins TCRα and MHCI HC (in the presence of US11) (26–28). The first round of ubiquitination/deubiquitination was hypothesized to be required for binding p97, after which Ub moieties have to be removed for substrates to pass through the narrow pore of p97 during dislocation. Analogously, the second round of ubiquitination/deubiquitination is required for proteasome association, after which Ub moieties are known to be removed prior to protein degradation (8). Our data reported here support the requirement for deubiquitination for dislocation of HC in the absence of immune evasins. However, the target of this deubiquitination appears not to be HC itself. Multiple viral proteins are known to target HC for ERAD via different strategies (29, 30). Although more efficient, evidence by others (35) and here suggest that ERAD induced by these viral proteins bypasses some of the critical steps of physiological pathway. More specifically, in the absence of viral proteins, we found less than 1% of the total HC pool is being dislocated and degraded. This observation likely reflects rate-limiting quality control mechanisms of substrate recognition that are bypassed by immune evasion proteins. Detection of the ubiquitination status on such a minor fraction of substrates is beyond the sensitivity range of the Ub blot. To circumvent this impediment, we adapted a cytosolic site-specific biotinylation system to quantitatively monitor the dislocation of wt HC and mutants lacking Ub acceptor sites. Indeed, by pharmacologically inhibiting dislocation or proteasome, this assay system allowed us to clearly identify and quantify the ER membrane-bound HCs destined for dislocation as well as the post-dislocated HCs destined for proteasome degradation. Surprisingly, we found that dislocation of the HC required neither a cytosolic tail nor ectodomain Lys residues. Nevertheless, the degradation of post-dislocated Lys-less HC was kinetically much slower than wt HCs suggesting that ubiquitination via its ectodomain Lys residues is important for the degradation of HCs by the proteasome. These findings also suggest that direct Ub conjugation of HC is not required for dislocation; and even if it is required, it occurs by a different mechanism than the Ub conjugation required for proteasome-mediated degradation of HC.
The likelihood that HCs are modified via non-Lys ubiquitination is low, but it is difficult to definitively rule out this possibility. For example, it has been reported that Ub conjugation of substrate could occur on an α-NH2 group of the N terminus (55). However, the degron for N-terminal conjugation is thought to be located on the N termini of the substrates because the conjugation is often disrupted by insertion of an N-terminal tag (56). In our study, the N terminus of the HC was fused with a 15-amino acid biotin acceptor peptide that could potentially interfere with N-terminal ubiquitination. In addition, to the best of our knowledge there have been no reports of N terminally Ub-modified ERAD substrates. Alternatively, non-Lys residues such as Ser, Thr, or Cys on the substrate may serve as Ub acceptor sites (57). Evidence reported by us and other groups suggest that this type of non-canonical ubiquitination can occur on selected ERAD substrates and is potentiated by viral E3 ligase mK3 or cellular E3 ligase Hrd1 (38, 58, 59). However, in these studies whether the ubiquitination occurs prior to the dislocation was not addressed.
Indeed an attractive model for ERAD of HC initiated by cellular quality control is an indirect (trans) ubiquitination model in which an accessory protein associated with substrate rather than substrate itself is ubiquitinated. This indirect ubiquitination model was first proposed by Kopito and co-workers (60) when they unexpectedly found that Lys-less TRCα molecules could be efficiently degraded by the proteasome. Lack of evidence of direct ubiquitination on the substrate during dislocation was also observed in US11-induced ERAD of HC. Wiertz and co-workers (18) demonstrated that Lys residues on the HC are dispensable in US11-induced HC dislocation although a functional ubiquitination system was required for the dislocation to occur. In the current study, substrates undergoing ERAD were specifically detected and tracked during dislocation versus degradation. In contrast to inhibition of proteasome, which resulted in accumulation of deglycosylated HCs in the cytosol, incubation of cells with a non-selective DUB inhibitor led to stabilization of K-less *Ld molecules primarily in the membrane. This is similar to what we saw with Eer1 treatment, which is known to block p97 by probably interfering with p97-associated DUB (24). These combined observations indicate that a deubiquitination step is required for substrate dislocation prior to proteasome degradation. The target of this deubiquitination, however, is not likely the substrate itself based on our mutagenesis data. Taken together, these data thus support a model in which an indirect ubiquitination is required by substrate dislocation and direct ubiquitination is needed by proteasome degradation.
Based on these accumulating findings by us and others, we speculate that dislocation of ER membrane-bound HC is facilitated by ubiquitination/deubiquitination of a component of the dislocation complex. Consistent with this model published reports have shown that p97-associated DUBs interact with the components of dislocation machinery and can regulate activity of ERAD-associated E3 ligase (26, 61–63). These findings suggest that the ubiquitination status of the components of the dislocation complex may be critical for its assembly or function (64). In addition, Derlins and UBAC2, a newly identified UBA-domain containing ERAD component interacting with gp78 and p97 recruiter UBXD8, were recently shown to be members of the rhomboid pseudoprotease family that may function similarly to rhomboid protease (23, 65). These authors proposed that Derlin might facilitate dislocation by unfolding a subset of ERAD substrates. If true, this conclusion also raises the possibility that direct substrate ubiquitination may not be required for their extraction. Thus these published finding of others and our substrate mutagenesis data reported here support a role of non-substrate ubiquitination/deubiquitination in dislocation. An attractiveness of this model is that it would explain why only a few E3 ligases out of more than 600 are located in the ER yet are potentially responsible for ubiquitination of 1/3 of synthesized proteins.
Acknowledgment
We thank Dr. Janet Connolly for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AI019687.
- ERAD
- endoplasmic reticulum-associated degradation
- BAP
- biotin acceptor peptide
- DUB
- deubiquitinating enzyme
- Eer1
- Eeyarestatin 1
- MHCI HC
- major histocompatibility complex class I heavy chain
- SA-HRP
- streptavidin-HRP
- CHX
- cycloheximide
- Ub
- ubiquitin
- β2m
- β2-microglobulin
- PNGase F
- peptide:N-glycosidase.
REFERENCES
- 1. Olzmann J. A., Kopito R. R., Christianson J. C. (2012) The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb. Perspect. Biol. 10.1101/cshperspect.a013185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hampton R. Y., Sommer T. (2012) Finding the will and the way of ERAD substrate retrotranslocation. Curr. Opin. Cell Biol. 24, 460–466 [DOI] [PubMed] [Google Scholar]
- 3. Tsai Y. C., Weissman A. M. (2011) Ubiquitylation in ERAD. Reversing to go forward? PLoS Biol. 9, e1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 5. Pickart C. M. (2000) Ubiquitin in chains. Trends Biochem. Sci. 25, 544–548 [DOI] [PubMed] [Google Scholar]
- 6. Ye Y., Rape M. (2009) Building ubiquitin chains. E2 enzymes at work. Nat. Rev. Mol. Cell. Biol. 10, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amerik A. Y., Hochstrasser M. (2004) Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695, 189–207 [DOI] [PubMed] [Google Scholar]
- 8. Pickart C. M., Cohen R. E. (2004) Proteasomes and their kin. Proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5, 177–187 [DOI] [PubMed] [Google Scholar]
- 9. Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., 3rd, Koonin E. V., Deshaies R. J. (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615 [DOI] [PubMed] [Google Scholar]
- 10. Yao T., Cohen R. E. (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 [DOI] [PubMed] [Google Scholar]
- 11. Kostova Z., Tsai Y. C., Weissman A. M. (2007) Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin. Cell Dev. Biol. 18, 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mehnert M., Sommer T., Jarosch E. (2010) ERAD ubiquitin ligases. Multifunctional tools for protein quality control and waste disposal in the endoplasmic reticulum. Bioessays 32, 905–913 [DOI] [PubMed] [Google Scholar]
- 13. Bays N. W., Wilhovsky S. K., Goradia A., Hodgkiss-Harlow K., Hampton R. Y. (2001) HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell 12, 4114–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ye Y., Meyer H. H., Rapoport T. A. (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652–656 [DOI] [PubMed] [Google Scholar]
- 15. Jarosch E., Taxis C., Volkwein C., Bordallo J., Finley D., Wolf D. H., Sommer T. (2002) Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4, 134–139 [DOI] [PubMed] [Google Scholar]
- 16. Rabinovich E., Kerem A., Fröhlich K. U., Diamant N., Bar-Nun S. (2002) AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell Biol. 22, 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ye Y., Meyer H. H., Rapoport T. A. (2003) Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol. Dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hassink G. C., Barel M. T., Van Voorden S. B., Kikkert M., Wiertz E. J. (2006) Ubiquitination of MHC class I heavy chains is essential for dislocation by human cytomegalovirus-encoded US2 but not US11. J. Biol. Chem. 281, 30063–30071 [DOI] [PubMed] [Google Scholar]
- 19. Burr M. L., Boname J. M., Lehner P. J. (2013) Studying ubiquitination of MHC class I molecules. Methods Mol. Biol. 960, 109–125 [DOI] [PubMed] [Google Scholar]
- 20. Barel M. T., Pizzato N., van Leeuwen D., Bouteiller P. L., Wiertz E. J., Lenfant F. (2003) Amino acid composition of α1/α2 domains and cytoplasmic tail of MHC class I molecules determine their susceptibility to human cytomegalovirus US11-mediated down-regulation. Eur. J. Immunol. 33, 1707–1716 [DOI] [PubMed] [Google Scholar]
- 21. Furman M. H., Loureiro J., Ploegh H. L., Tortorella D. (2003) Ubiquitinylation of the cytosolic domain of a type I membrane protein is not required to initiate its dislocation from the endoplasmic reticulum. J. Biol. Chem. 278, 34804–34811 [DOI] [PubMed] [Google Scholar]
- 22. Shamu C. E., Story C. M., Rapoport T. A., Ploegh H. L. (1999) The pathway of US11-dependent degradation of MHC class I heavy chains involves a ubiquitin-conjugated intermediate. J. Cell Biol. 147, 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Greenblatt E. J., Olzmann J. A., Kopito R. R. (2011) Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α-1 antitrypsin from the endoplasmic reticulum. Nat. Struct. Mol. Biol. 18, 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Q., Li L., Ye Y. (2008) Inhibition of p97-dependent protein degradation by Eeyarestatin I. J. Biol. Chem. 283, 7445–7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fiebiger E., Hirsch C., Vyas J. M., Gordon E., Ploegh H. L., Tortorella D. (2004) Dissection of the dislocation pathway for type I membrane proteins with a new small molecule inhibitor, eeyarestatin. Mol. Biol. Cell 15, 1635–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ernst R., Mueller B., Ploegh H. L., Schlieker C. (2009) The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol. Cell 36, 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ernst R., Claessen J. H., Mueller B., Sanyal S., Spooner E., van der Veen A. G., Kirak O., Schlieker C. D., Weihofen W. A., Ploegh H. L. (2011) Enzymatic blockade of the ubiquitin-proteasome pathway. PLoS Biol. 8, e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sanyal S., Claessen J. H., Ploegh H. L. (2012) A viral deubiquitylating enzyme restores dislocation of substrates from the endoplasmic reticulum (ER) in semi-intact cells. J. Biol. Chem. 287, 23594–23603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hansen T. H., Bouvier M. (2009) MHC class I antigen presentation. Learning from viral evasion strategies. Nat. Rev. Immunol. 9, 503–513 [DOI] [PubMed] [Google Scholar]
- 30. Lilley B. N., Ploegh H. L. (2005) Viral modulation of antigen presentation. Manipulation of cellular targets in the ER and beyond. Immunol. Rev. 207, 126–144 [DOI] [PubMed] [Google Scholar]
- 31. Loureiro J., Lilley B. N., Spooner E., Noriega V., Tortorella D., Ploegh H. L. (2006) Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature 441, 894–897 [DOI] [PubMed] [Google Scholar]
- 32. Stagg H. R., Thomas M., van den Boomen D., Wiertz E. J., Drabkin H. A., Gemmill R. M., Lehner P. J. (2009) The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J. Cell Biol. 186, 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lilley B. N., Ploegh H. L. (2004) A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429, 834–840 [DOI] [PubMed] [Google Scholar]
- 34. Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 35. Burr M. L., Cano F., Svobodova S., Boyle L. H., Boname J. M., Lehner P. J. (2011) HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc. Natl. Acad. Sci. U.S.A. 108, 2034–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lilley B. N., Ploegh H. L. (2005) Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. U.S.A. 102, 14296–14301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mueller B., Klemm E. J., Spooner E., Claessen J. H., Ploegh H. L. (2008) SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 105, 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang X., Herr R. A., Chua W. J., Lybarger L., Wiertz E. J., Hansen T. H. (2007) Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 177, 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Albring J., Koopmann J. O., Hämmerling G. J., Momburg F. (2004) Retrotranslocation of MHC class I heavy chain from the endoplasmic reticulum to the cytosol is dependent on ATP supply to the ER lumen. Mol. Immunol. 40, 733–741 [DOI] [PubMed] [Google Scholar]
- 40. Hughes E. A., Hammond C., Cresswell P. (1997) Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome. Proc. Natl. Acad. Sci. U.S.A. 94, 1896–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Petris G., Vecchi L., Bestagno M., Burrone O. R. (2011) Efficient detection of proteins retro-translocated from the ER to the cytosol by in vivo biotinylation. PLoS One. 6, e23712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang X., Lybarger L., Connors R., Harris M. R., Hansen T. H. (2004) Model for the interaction of gammaherpesvirus 68 RING-CH finger protein mK3 with major histocompatibility complex class I and the peptide-loading complex. J. Virol. 78, 8673–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. DuBridge R. B., Tang P., Hsia H. C., Leong P. M., Miller J. H., Calos M. P. (1987) Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell Biol. 7, 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lybarger L., Wang X., Harris M. R., Virgin H. W., 4th, Hansen T. H. (2003) Virus subversion of the MHC class I peptide-loading complex. Immunity 18, 121–130 [DOI] [PubMed] [Google Scholar]
- 45. Beckett D., Kovaleva E., Schatz P. J. (1999) A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith J. D., Solheim J. C., Carreno B. M., Hansen T. H. (1995) Characterization of class I MHC folding intermediates and their disparate interactions with peptide and β2-microglobulin. Mol. Immunol. 32, 531–540 [DOI] [PubMed] [Google Scholar]
- 47. Barker D. F., Campbell A. M. (1981) The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. J. Mol. Biol. 146, 451–467 [DOI] [PubMed] [Google Scholar]
- 48. Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D. Y., Nagata K. (2008) ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321, 569–572 [DOI] [PubMed] [Google Scholar]
- 49. Dong M., Bridges J. P., Apsley K., Xu Y., Weaver T. E. (2008) ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol. Biol. Cell 19, 2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Riemer J., Hansen H. G., Appenzeller-Herzog C., Johansson L., Ellgaard L. (2011) Identification of the PDI-family member ERp90 as an interaction partner of ERFAD. PLoS One. 6, e17037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T. A. (2005) Inaugural article. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. U.S.A. 102, 14132–14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Latterich M., Fröhlich K. U., Schekman R. (1995) Membrane fusion and the cell cycle. Cdc48p participates in the fusion of ER membranes. Cell 82, 885–893 [DOI] [PubMed] [Google Scholar]
- 53. Seiberlich V., Goldbaum O., Zhukareva V., Richter-Landsberg C. (2012) The small molecule inhibitor PR-619 of deubiquitinating enzymes affects the microtubule network and causes protein aggregate formation in neural cells. Implications for neurodegenerative diseases. Biochim. Biophys. Acta 1823, 2057–2068 [DOI] [PubMed] [Google Scholar]
- 54. Elkabetz Y., Shapira I., Rabinovich E., Bar-Nun S. (2004) Distinct steps in dislocation of luminal endoplasmic reticulum-associated degradation substrates. Roles of endoplamic reticulum-bound p97/Cdc48p and proteasome. J. Biol. Chem. 279, 3980–3989 [DOI] [PubMed] [Google Scholar]
- 55. Ciechanover A., Ben-Saadon R. (2004) N-terminal ubiquitination. More protein substrates join. Trends Cell Biol. 14, 103–106 [DOI] [PubMed] [Google Scholar]
- 56. Aviel S., Winberg G., Massucci M., Ciechanover A. (2000) Degradation of the Epstein-Barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J. Biol. Chem. 275, 23491–23499 [DOI] [PubMed] [Google Scholar]
- 57. Wang X., Herr R. A., Hansen T. H. (2012) Ubiquitination of substrates by esterification. Traffic 13, 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ishikura S., Weissman A. M., Bonifacino J. S. (2010) Serine residues in the cytosolic tail of the T-cell antigen receptor α-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J. Biol. Chem. 285, 23916–23924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shimizu Y., Okuda-Shimizu Y., Hendershot L. M. (2010) Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol. Cell 40, 917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yu H., Kopito R. R. (1999) The role of multiubiquitination in dislocation and degradation of the alpha subunit of the T cell antigen receptor. J. Biol. Chem. 274, 36852–36858 [DOI] [PubMed] [Google Scholar]
- 61. Hassink G. C., Zhao B., Sompallae R., Altun M., Gastaldello S., Zinin N. V., Masucci M. G., Lindsten K. (2009) The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Rep. 10, 755–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Scaglione K. M., Zavodszky E., Todi S. V., Patury S., Xu P., Rodríguez-Lebrón E., Fischer S., Konen J., Djarmati A., Peng J., Gestwicki J. E., Paulson H. L. (2011) Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Mol. Cell 43, 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu Y., Ye Y. (2012) Roles of p97-associated deubiquitinases in protein quality control at the endoplasmic reticulum. Curr. Protein Pept. Sci. 13, 436–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Christianson J. C., Olzmann J. A., Shaler T. A., Sowa M. E., Bennett E. J., Richter C. M., Tyler R. E., Greenblatt E. J., Harper J. W., Kopito R. R. (2012) Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 14, 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chou T. F., Brown S. J., Minond D., Nordin B. E., Li K., Jones A. C., Chase P., Porubsky P. R., Stoltz B. M., Schoenen F. J., Patricelli M. P., Hodder P., Rosen H., Deshaies R. J. (2011) Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc. Natl. Acad. Sci. U.S.A. 108, 4834–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]