Abstract
Objectives
In vivo experimentation is costly and time-consuming, and presents a major bottleneck in anti-tuberculosis drug development. Conventional methods rely on the enumeration of bacterial colonies, and it can take up to 4 weeks for Mycobacterium tuberculosis to grow on agar plates. Light produced by recombinant bacteria expressing luciferase enzymes can be used as a marker of bacterial load, and disease progression can be easily followed non-invasively in live animals by using the appropriate imaging equipment. The objective of this work was to develop a bioluminescence-based mouse model of tuberculosis to assess antibiotic efficacy against M. tuberculosis in vivo.
Methods
We used an M. tuberculosis strain carrying a red-shifted derivative of the firefly luciferase gene (FFlucRT) to infect mice, and monitored disease progression in living animals by bioluminescence imaging before and after treatment with the frontline anti-tuberculosis drug isoniazid. The resulting images were analysed and the bioluminescence was correlated with bacterial counts.
Results
Using bioluminescence imaging we detected as few as 1.7 × 103 and 7.5 × 104 reporter bacteria ex vivo and in vivo, respectively, in the lungs of mice. A good correlation was found between bioluminescence and bacterial load in both cases. Furthermore, a marked reduction in luminescence was observed in living mice given isoniazid treatment.
Conclusions
We have shown that an improved bioluminescent strain of M. tuberculosis can be visualized by non-invasive imaging in live mice during an acute, progressive infection and that this technique can be used to rapidly visualize and quantify the effect of antibiotic treatment. We believe that the model presented here will be of great benefit in early drug discovery as an easy and rapid way to identify active compounds in vivo.
Keywords: drug testing, bioluminescence, optical imaging, mouse model
Introduction
Mycobacterium tuberculosis remains one of the most successful bacterial pathogens, despite ongoing efforts to curb the tuberculosis (TB) epidemic, and poses a severe threat to public health. Confounding factors include the concurrent HIV epidemic, and the increasing incidence of infections with multidrug-resistant (MDR) and extremely drug-resistant (XDR) strains.1 It is evident that more effective control measures are urgently needed, and in vivo experimentation is an essential tool in addressing this goal. At a basic research level, the appropriate use of animal models can help to improve our understanding of host–pathogen interactions. In translational research, in vivo testing is an absolute requirement for pre-clinical evaluation of new drug and vaccine candidates before these can advance along the development pipeline. However, in vivo experimentation is costly and time-consuming, and presents a major bottleneck in drug and vaccine development. This is partly due to the long replication time of M. tuberculosis; conventional methods rely on the enumeration of bacterial colonies, which can take 3–4 weeks to form on solid medium. Therefore, novel approaches to accelerate efficacy assessment of new control measures are urgently required.
Bioluminescence, the production of light by the action of luciferase enzymes on their cognate substrates, offers the potential to accelerate the development and testing of new TB control measures. Light produced by recombinant bacteria expressing such luciferase enzymes can be used as a marker of bacterial load, and disease progression can be easily followed non-invasively in live animals by using the appropriate imaging equipment. Indeed, this approach has been successfully used for in vivo studies of a wide variety of bacteria.2 The most extensively used luciferase is LuxAB, naturally found in luminous bacteria such as Photorhabdus luminescens, Vibrio sp. and Photobacterium sp. The luciferase is encoded by a five-gene operon, which also includes genes for the synthesis of the luciferase substrate, a long-chain aldehyde. Therefore, recombinant expression of the whole operon renders the bacterial host autoluminescent. The firefly luciferase (FFluc) has occasionally been used in bacterial research and is very popular in viral and eukaryotic studies.2 The genes for the synthesis of its substrate, d-luciferin, are unknown and therefore it has to be administered exogenously before imaging. Fortunately the substrate is innocuous and the light produced by FFluc is of a wavelength more appropriate for in vivo imaging than that from LuxAB.3,4
In TB research, bioluminescent reporter technology has been successfully exploited for in vitro applications for more than two decades. In particular, bioluminescence has been harnessed as a surrogate biomarker for mycobacterial numbers and viability,5,6 and bioluminescent mycobacterial reporter strains have been utilized for high-throughput, real-time screening of antimycobacterial agents.7–10 More recently this powerful technology has also been applied to real-time, non-invasive in vivo imaging of mycobacterial infection. The first demonstration of in vivo imaging of bioluminescent Mycobacterium bovis BCG in immunodeficient mice was provided in 2009.11 However, as this strain only carried the luxAB genes encoding the luciferase, it required the administration of the toxic aldehyde substrate, decanal, for visualization of the mycobacteria. Furthermore, this work was carried out with the attenuated vaccine strain M. bovis BCG, and not with virulent M. tuberculosis. Other researchers have utilized bioluminescent Mycobacterium ulcerans in a mouse footpad model.12 In 2010 we published the first report of the use of bioluminescent M. tuberculosis,13 demonstrating the successful detection of mycobacteria expressing either FFluc or Lux in the lungs of infected mice. It was also the first time that the complete lux operon had been successfully expressed in mycobacteria. More recently, autoluminescent M. tuberculosis (expressing the complete bacterial luciferase operon) has been exploited in proof-of-concept studies to monitor drug and vaccine efficacy using a tube luminometer to measure bioluminescence from organ homogenates and from mice.14
Here, we report on the development of an improved FFluc reporter (FFlucRT) for in vivo imaging of M. tuberculosis. We demonstrate the in vivo visualization of the improved FFlucRT reporter strain in severe combined immunodeficiency (SCID) mice, and provide evidence that this system could be applied to high-throughput in vivo testing of drug efficacy. In addition, we explore ways to further enhance the reporter system.
Materials and methods
Bacterial strains and growth conditions
M. tuberculosis H37Rv and Mycobacterium smegmatis mc2155 were grown on 7H11 agar (BD Diagnostics) supplemented with 0.5% glycerol, 10% oleic acid/albumin/dextrose/catalase (OADC) (BD Diagnostics) and appropriate antibiotics. Liquid cultures of M. tuberculosis were grown with shaking in 7H9 broth (BD Diagnostics) supplemented with 0.05% Tween-80, 0.2% glycerol, 10% OADC and appropriate antibiotics. Luria–Bertani (LB) medium was used for culturing Escherichia coli. All the strains were grown at 37°C. The following antibiotics were added when appropriate: ampicillin [100 mg/L (Sigma)] and kanamycin [25 mg/L for mycobacteria and 50 mg/L for E. coli (Sigma)].
Construction of FFlucRT reporter plasmids and strains
The plasmids used in this study are described in Table S1 (available as Supplementary data at JAC Online). We designed a modified version of FFluc, designated FFlucRT (GenBank accession number KC688279), containing nine amino acid substitutions. The corresponding M. tuberculosis codon-optimized DNA fragment was synthesized by DNA 2.0. We then used the synthetic DNA construct as a PCR template to incorporate a STOP codon and an XbaI site at the 3′ end of fflucRT (sense primer: 5′- CTTTCGCCCGGGCTAATTAG-3′; antisense primer: 5′-AGGCTTCTAGATCACAATTTCGACTTGCCACC-3′. The restriction site is underlined and the STOP codon is in bold). An EcoRI-XbaI PCR product was cloned into pUC18 and the sequence was confirmed by DNA sequencing. To obtain pMV306G13 + FFlucRT, fflucRT was cloned into pMV306hsp as an EcoRI-SalI insert and the hsp60 promoter replaced with the Mycobacterium marinum G13 promoter by digestion with NotI-EcoRI as previously described.13 The integrase-free reporter plasmid pMV306DIG13 + FFlucRT was produced by deleting the int gene from pMV306G13 + FFlucRT by inverted PCR using the Phusion® Site-Directed Mutagenesis Kit (Thermo Scientific) and primers IntUp (5′-TCTTGTCAGTACGCGAAGAACCAC-3′) and IntLw (5′-GTCCATCTTGTTGTCGTAGGTCTG-3′). The deletion was confirmed by digesting with SphI and the sequence of PG13 and fflucRT partially checked by DNA sequencing.
The reporter strain M. tuberculosis pMV306G13 + FFlucRT was produced by electroporation of M. tuberculosis with the corresponding vector. To obtain the integrase-free reporter strain, M. tuberculosis was co-transformed with pMV306DIG13 + FFlucRT and pBS-Int,15 a suicide plasmid that carries the integrase gene but no attachment site, and is therefore lost from the bacterium. The presence of the reporter plasmid in both strains and the absence of the integrase gene from pMV306DIG13 + FFlucRT were confirmed by PCR.
In vitro bioluminescence assays
d-Luciferin (Gold BioTechnology®) was prepared in distilled water at 3 × 104 mg/L. The stock was stored at −20°C and diluted in broth medium or Dulbecco's PBS (D-PBS) (without calcium or magnesium) immediately before use. Working solutions were kept on ice in the dark during preparation. d-Luciferin was added at a final concentration of 150 mg/L and bioluminescence was measured for 10 s using a microplate reader (Luminoskan Ascent, Thermo Scientific). Bioluminescence was expressed as relative light units (RLUs).
Bioluminescence emission spectra
Bioluminescence emission spectra were obtained by imaging M. smegmatis cultures expressing FFluc and FFlucRT with the IVIS® Spectrum system (Caliper Life Sciences). Briefly, 50 μL of each culture was inoculated in triplicate in a 96-well opaque black plate, 50 μL of 300 mg/L luciferin was added to each well, and the plate was imaged using a set of filters (20 nm bandpass) from 500 to 800 nm on the auto-exposure setting. Bioluminescence in each well was quantified using the region of interest (ROI) tool in the Living Image software program (reported as photons/s).
In vivo studies
Experiments were performed in accordance with the Animals (Scientific Procedures) Act (1986) and were approved by the local ethics review committee. Barrier-bred female 6–12 week old CB-17 SCID mice (Charles River UK Ltd) were anaesthetized by intraperitoneal injection of a mixture of ketamine (100 mg/kg body weight; Ketaset; Fort Dodge Animal Health) and xylazine (10 mg/kg body weight; Rompun; Bayer) and infected with wild-type (WT) or bioluminescent M. tuberculosis in 20 μL of PBS via the intranasal route. In each experiment one group of mice was treated with isoniazid starting on day 19 post-infection. Isoniazid was dissolved in 40% sucrose at a final concentration of 5000 mg/L and filter sterilized. Mice were given daily doses of 25 mg/kg body weight of isoniazid by oral gavage. Control mice were given daily doses of 40% sucrose by oral gavage.
Assessment of bioluminescence [photons/s/cm2/steradian (sr)] from living animals was performed using an IVIS® Spectrum system. Prior to bioluminescent imaging, mice were injected via the intraperitoneal route with 500 mg/kg body weight of d-luciferin dissolved in sterile D-PBS together with ketamine and xylazine as above. Mice were contained in a large airtight box for safety considerations and placed into the imaging chamber of the IVIS® Spectrum imaging system with the stage heated to 37°C. A greyscale reference image was taken under low illumination prior to quantification of emitted photons over 5 min using the software Living Image version 3.2. Bioluminescence within specific regions of individual mice was also quantified using the ROI tool in the Living Image software program (given as photons/s).
At specific timepoints and immediately after performing the in vivo imaging, animals were culled by cervical dislocation while still under anaesthesia and the lungs and spleens were removed aseptically. Ex vivo imaging of the lungs and spleens as intact organs was performed as above, placing the organs in a sterile, black, 24-well plate with clear bottom contained in the airtight box. Finally, to determine the bacterial burden, the organs were homogenized by mechanical disruption in PBS with 0.05% Tween and viable counts were determined by serial dilution plating on Middlebrook 7H11 agar.
Plasmid stability
To assess the stability of the reporter vectors in vitro, the bioluminescent M. tuberculosis strains were grown in liquid medium with no antibiotic selection and passaged into fresh medium fortnightly. Every 4 weeks appropriate dilutions of the culture were plated on 7H11 to obtain at least 100 isolated colonies. These colonies were then inoculated in 200 μL of 7H9 medium in opaque, white, 96-well plates and the luminescence was read after 5–7 days of incubation, as described above. To prevent sample evaporation during incubation, 200 μL of sterile water was added to all outer perimeter wells. A sterile control without bacteria was prepared for each assay and the luminescence reading from this control was used as the background reading. Bacterial viability was determined by adding 30 μL of sterile 0.01% resazurin to all wells and incubating for a further 24 h. A change in colour from blue (oxidized state) to pink (reduced state) indicated growth of the bacteria. Plasmid stability in vivo was evaluated in a similar way, but using colonies isolated from organ homogenates.
Statistical analysis
Statistical analysis was performed with GraphPad Prism 5 software. For both cfu and luminescence, data from different groups at specific timepoints were analysed by a Mann–Whitney test. To assess the relationship between cfu and luminescence, a Pearson correlation calculation was performed and the resulting Pearson r values and two-tailed P values were reported. Linear regression analysis was used to determine the relationship between cfu and bioluminescence. P values <0.05 were considered to be statistically significant.
Results
Development of an improved FFluc reporter
We had previously developed FFluc-based reporters for use in mycobacteria.13 To enhance the limit of detection of FFluc activity in vivo, in the present work we modified the protein in two ways. First, we introduced previously described mutations shown to increase bioluminescence intensity, namely Ile423Leu, Asp436Gly and Leu530Arg.16 Second, to produce a thermostable red-emitting luciferase, we incorporated the following mutations: Ser284Thr, Thr214Ala, Ala215Leu, Ile232Ala, Phe295Leu and Glu354Lys.17 The resulting reporter, named FFlucRT (GenBank accession number KC688279), was not significantly brighter than the original FFluc, but it demonstrated an emission spectrum shift towards the red in comparison with the FFluc reporter; the maximum emission shifted from 560 nm for FFluc to 620 nm for FFlucRT (Figure S1, available as Supplementary data at JAC Online).
In order to maximize the expression of the new reporter we used an M. tuberculosis codon-optimized synthetic gene together with an improved Shine Dalgarno sequence, both of which have proved useful in increasing FFluc signal in mycobacteria.13 The reporter was expressed under control of the PG13 promoter,18,19 which provided the optimal combination of signal strength and reporter plasmid stability (data not shown).
We next assessed the growth kinetics and luminescence of M. tuberculosis expressing the FFlucRT reporter. M. tuberculosis pMV306G13 + FFlucRT demonstrated identical in vitro growth kinetics to M. tuberculosis WT (Figure 1). The luminescence levels of the FFlucRT-expressing M. tuberculosis strain were similar to that produced by the FFluc-carrying strain (Figure 1). However, whereas the FFluc reporter signal decreased upon entry into stationary phase (Figure 1), this was not the case for the FFlucRT reporter for which luminescence remained stable in the stationary phase (Figure 1). FFlucRT therefore demonstrated a better overall correlation between luminescence and bacterial numbers [as measured by optical density at 600 nm (OD600)].
Figure 1.
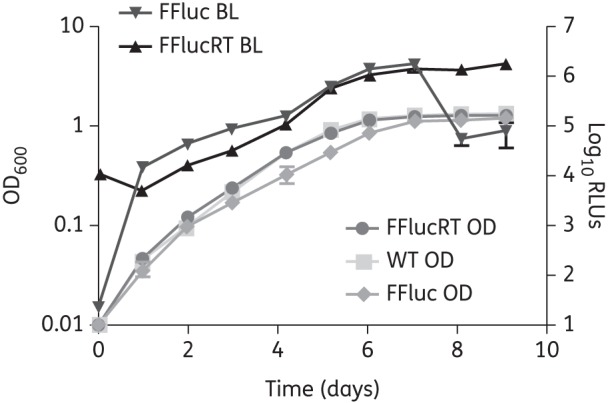
FFlucRT bioluminescence correlates with cell density. Cultures of M. tuberculosis pMV306G13 + FFlucRT, M. tuberculosis pMV306G13 + FFluc and M. tuberculosis H37Rv (WT) were inoculated at an OD of 0.01, and the OD and luminescence were measured over 9 days. The plots represent the means and standard deviations of values for three independent cultures, and values are plotted on a logarithmic scale. BL, bioluminescence.
Evaluation of the reporter in vivo
Having established that the M. tuberculosis FFlucRT reporter strain has similar in vitro growth kinetics to M. tuberculosis WT, we went on to investigate in vivo growth kinetics. To this end, immunodeficient mice were intranasally infected with parental M. tuberculosis WT and FFlucRT reporter strains and the bacterial load in the lungs was determined by serial dilution plating of organ homogenates. Both strains showed similar growth kinetics with an ∼1000-fold increase in lung bacterial burden within 3 weeks (Figure 2), at which point the experiment was terminated because the humane endpoint (20% weight loss) was reached. Furthermore, both WT and FFlucRT strains disseminated to the spleens to a similar extent, reaching levels of 6.1 × 104 (±3.7 × 104) and 2.5 × 104 (±1.7 × 104) cfu/spleen, respectively, by the end of the experiment. Altogether these results indicate that expression of the FFlucRT reporter does not incur a fitness cost.
Figure 2.
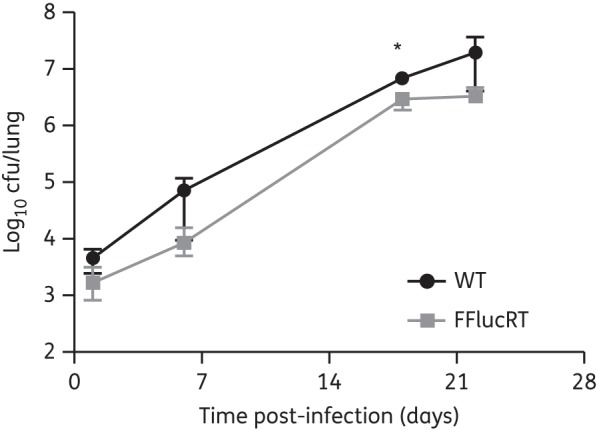
Growth of M. tuberculosis in SCID mice is not affected by the expression of the FFlucRT reporter. SCID mice were infected with 1.1 × 104 cfu of M. tuberculosis pMV306G13 + FFlucRT or with 1.0 × 104 cfu of the parental M. tuberculosis WT via the intranasal route. Bacterial burden was determined by serial dilution plating of organ homogenates. Each point on the graph represents the median and range (n = 5 mice). This result is representative of two independent experiments. Statistical significance was evaluated by the Mann–Whitney test and those found to be significant (P < 0.05) are indicated with an asterisk.
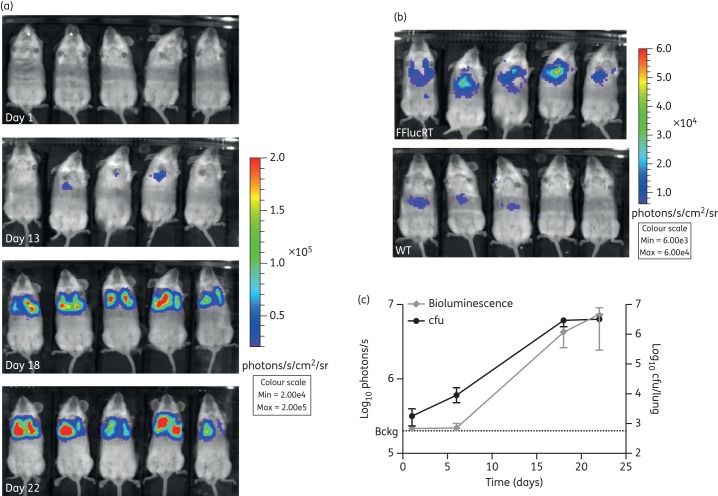
We next assessed the ability to detect bacteria expressing the FFlucRT reporter in vivo. Infected mice were imaged at multiple timepoints after administering intraperitoneal luciferin. The background level of luminescence was estimated by imaging mice inoculated with M. tuberculosis WT. Bioluminescence above background levels was detected in the thorax region of live animals infected with M. tuberculosis pMV306G13 + FFlucRT at day 13 post-infection, when the lung bacterial load was estimated to be ∼105 cfu (Figure 3a–c). The increase in thorax-localized luminescence paralleled the increase in lung cfu (Figure 3c). A weak signal was also observed in the abdomen of WT- and FFlucRT-infected mice when the scale of the images was adjusted to a low setting (Figure 3b). Upon dissection of the mice we confirmed that the signal came from the livers. This signal is probably the result of the high concentration of luciferin used together with the high uptake and probable metabolism of luciferin in the liver.20,21
Figure 3.
In vivo imaging of M. tuberculosis infection. SCID mice were infected with 1.1 × 104 cfu of M. tuberculosis pMV306G13 + FFlucRT or with 1.0 × 104 cfu of M. tuberculosis WT via the intranasal route. (a) Mice were injected intraperitoneally with 500 mg/kg d-luciferin, and images were acquired using an IVIS® Spectrum system. *M. tuberculosis WT-infected mice; all others infected with M. tuberculosis pMV306G13 + FFlucRT. Day 1 and 6 images were very similar. Only day 1 is shown here. (b) The scale on the images shown here has been adjusted to demonstrate that a detectable signal was observed in the lungs of all five mice as early as day 13. M. tuberculosis pMV306G13 + FFlucRT-infected mice are the same mice as shown in (a). The signal in the abdomen of the mice comes from the liver due to background luminescence from the luciferin substrate. (c) Bioluminescence in the thorax was quantified for each mouse at each timepoint and compared with cfu data at corresponding timepoints. Each point on the graph represents the median and error (n = 5 mice). This result is representative of two independent experiments. Bckg, background luminescence (2 × 105 photons/s). This figure appears in colour in the online version of JAC and in black and white in the printed version of JAC.
At selected timepoints we also determined the bioluminescent signal in lungs and spleens ex vivo. We detected bioluminescence in all lungs harvested from mice infected with M. tuberculosis pMV306G13 + FFlucRT as early as day 1 post-infection (Figure 4a), when the total lung bacterial load was 1.7 × 103 cfu. As expected, the bioluminescent signal increased over the course of the infection, corresponding to the increase in cfu (Figure 4b).
Figure 4.
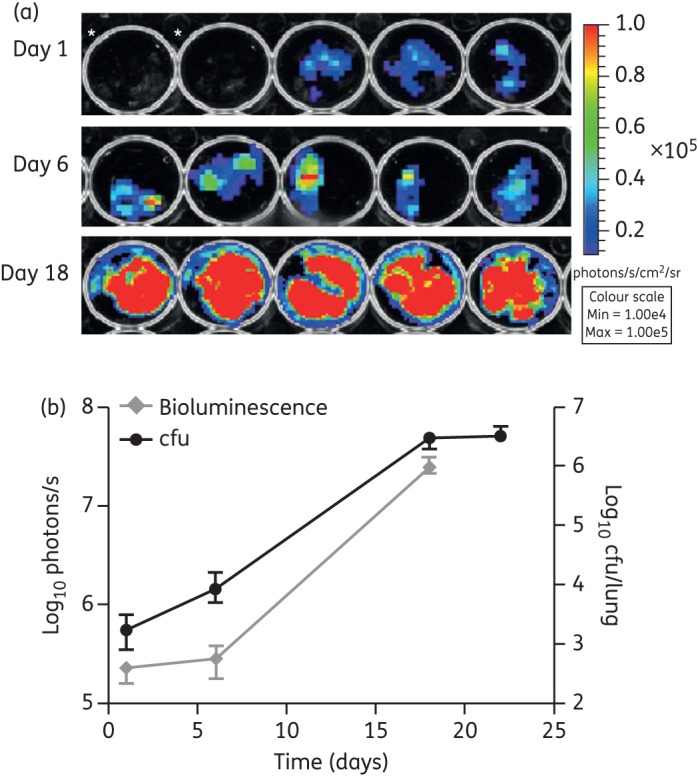
Ex vivo bioluminescence in infected lungs. (a) Images of lungs harvested from the mice in Figure 3 were acquired using an IVIS® Spectrum system at multiple timepoints following infection. *M. tuberculosis WT-infected mice; all others infected with M. tuberculosis pMV306G13 + FFlucRT. (b) Bioluminescence (measured as photons/s) compared with cfu data at corresponding timepoints. cfu data are the same data as in Figure 3, but are included here for comparison. The background luminescence (1.5 × 104 photons/s) is outside the axis limits. Each point on the graph represents the median and range (n = 5 mice). This result is representative of two independent experiments. This figure appears in colour in the online version of JAC and in black and white in the printed version of JAC.
Ex vivo bioluminescence in spleens was detected at 18 days post-infection (Figure 5a), which was also the earliest timepoint when bacteria could be cultured from this organ. The bacterial load in the spleens at this timepoint was 2.8 × 103 cfu (Figure 5b).
Figure 5.
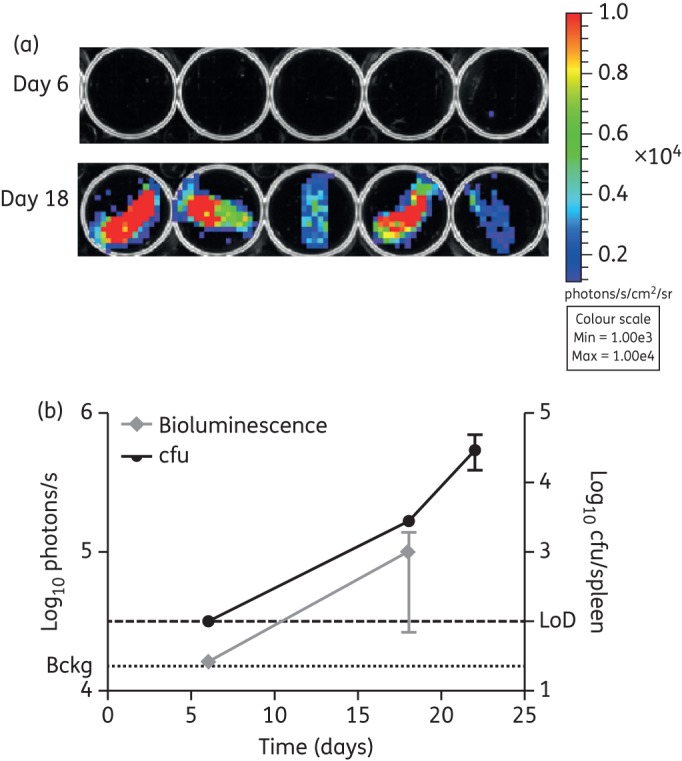
Ex vivo bioluminescence in infected spleens. (a) Images of spleens harvested from the mice in Figure 3 infected with M. tuberculosis pMV306G13 + FFlucRT were acquired using an IVIS® Spectrum system at multiple timepoints following infection. (b) Bioluminescence (measured as photons/s) was quantified for each spleen and compared with cfu data at corresponding timepoints. Each point on the graph represents the median and range (n = 4–5 mice). Bckg, background luminescence (1.3 × 104 photons/s); LoD, limit of detection (100 cfu). This figure appears in colour in the online version of JAC and in black and white in the printed version of JAC.
Taken together, these results demonstrated that as few as 103 and 105 cfu of bioluminescent M. tuberculosis could be detected ex vivo and in vivo, respectively, in the lungs of mice, and that both ex vivo and in vivo bioluminescence corresponded to bacterial load.
Application to drug testing
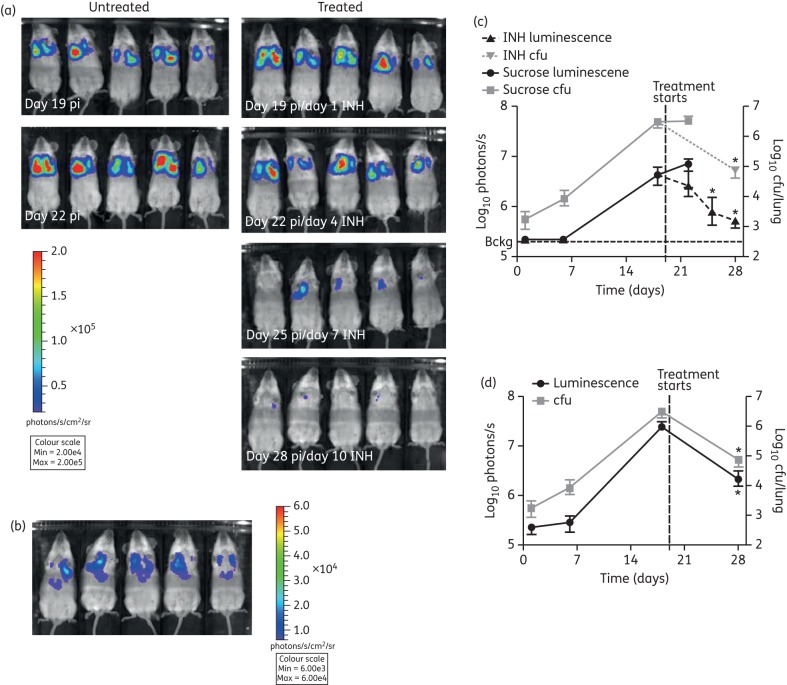
The results described above suggested that this technology might be a useful tool to accelerate in vivo drug testing. As proof-of-concept, we treated infected mice with isoniazid, part of frontline anti-TB drug treatment, or sucrose as the carrier control, and followed the response to treatment in real time using bioluminescence imaging. In vivo imaging of live mice revealed a marked reduction in luminescence as early as day 4 of isoniazid treatment in three out of five mice (day 22 post-infection, Figure 6a). By day 10 of treatment (day 28 post-infection), we observed an 8.8-fold reduction in luminescence in the thorax of all infected mice. The differences in luminescence levels were statistically significant between the beginning and end of treatment (Figure 6a and c). The observed reduction in luminescence was corroborated by plating lung homogenates after 10 days of isoniazid treatment, which confirmed a reduction in bacterial burden in line with the luminescent measurements for this organ (Figure 6c). At this point the bacterial load was just 7.5 × 104 cfu/lung and yet bioluminescence was clearly detectable non-invasively in the thorax of live mice (Figure 6b and c), representing the lowest number of bacteria that we were able to detect in vivo.
Figure 6.
Effects of drug treatment were detected non-invasively in live mice by bioluminescent imaging. M. tuberculosis pMV306G13 + FFlucRT-infected SCID mice were treated daily with 25 mg/kg isoniazid (INH) by oral gavage from day 19 post-infection (pi). A control group was treated with sucrose. (a) To visualize the effect of INH treatment, mice were injected intraperitoneally with 500 mg/kg d-luciferin, and images were acquired using an IVIS® Spectrum system. (b) The scale on the images shown here has been adjusted to demonstrate that a detectable signal was observed in the lungs of all five mice on day 28 post-infection, when the bacterial load was just 7.5 × 104 cfu/lung. The mice are the same mice as shown in (a). The reduction in bacterial burden quantified in live mice (c) or lungs ex vivo (d) by bioluminescent imaging was confirmed by enumerating lung cfu. Each point on the graphs represents the median and range (n = 3–5 mice). Bckg, background luminescence in live mice (2 × 105 photons/s). The background luminescence in the lungs ex vivo (1.5 × 104 photons/s) is outside the axis limits. Statistical significance was evaluated by the Mann–Whitney test and those found to be significant (P < 0.05) are indicated with an asterisk. This figure appears in colour in the online version of JAC and in black and white in the printed version of JAC.
Similar results were obtained ex vivo; we observed a statistically significant reduction in lung luminescence levels over 10 days of isoniazid treatment. This was closely paralleled by the reduced bacterial organ burden as revealed by cfu counts (Figure 6d). These results provide proof-of-principle that the M. tuberculosis pMV306G13 + FFlucRT reporter strain can be applied to rapid in vivo testing of new anti-TB compounds.
Sensitivity of M. tuberculosis bioluminescence imaging and correlation with bacterial load
The correlation between bioluminescence and bacterial burden was statistically analysed by Pearson correlation. A good correlation was found between bioluminescence in the thorax of live mice and cfu in the lungs [r(9) = 0.958, two-tailed P value <0.0001; Figure 7a]. In addition, bioluminescence measured in the lungs ex vivo strongly correlated with bacterial burden in these organs [r(14) = 0.946, two-tailed P value <0.0001; Figure 7b]. A linear regression analysis was performed to calculate the detection limit of this assay (Figure 7a and b). Because any detectable amount of reporter bacteria should produce higher amounts of bioluminescence than WT bacteria, the signal (Y) was set based on the average amount of bioluminescence detected in the thorax and the lungs of mice infected with WT M. tuberculosis. The average background bioluminescence measured was 2 × 105 photons/s in live mice and 1.5 × 104 photons/s in the lungs ex vivo; therefore, the threshold for the signal (Y) was arbitrarily set to 4 × 105 and 3 × 104 photons/s, respectively. From this, the detection limit in live mice was calculated as 4.4 × 104 cfu. Similarly the detection limit for excised lungs was calculated as 1.2 × 102 cfu.
Figure 7.
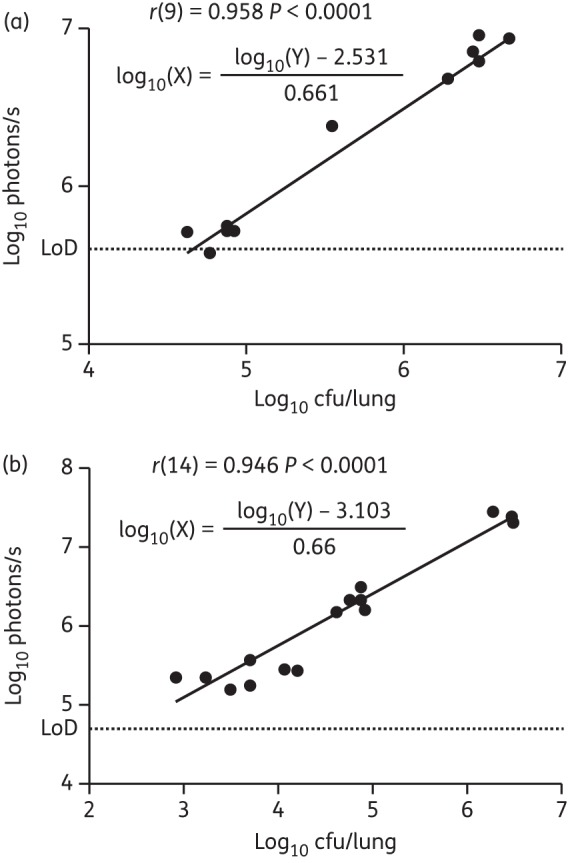
In vivo and ex vivo bioluminescence correlates with bacterial burden. Correlation of cfu in the lungs and bioluminescence in the thorax (a) or in the harvested lungs (b) of infected mice shown in Figures 3 and 6. The number of cfu X for a given bioluminescence measurement Y is described by the equation shown, which was obtained from linear regression analysis.
Optimization of plasmid stability
One parameter that could impact on the sensitivity of bioluminescent reporter strains is stability of the reporter plasmid. To determine whether this was a contributing factor in the experiments described here, we assessed the stability of integrated pMV306G13 + FFlucRT following in vivo growth. Bioluminescence of colonies recovered from organ homogenates was measured to calculate the percentage of colonies retaining the ability to express the FFlucRT reporter. First, we examined plasmid stability in colonies recovered from the lungs of SCID mice, and found that by the end of the experiment (day 22 post-infection), 87.2% of colonies isolated from lungs of sucrose-treated mice retained a functional FFlucRT reporter.
To determine whether drug treatment or different organ environments could influence plasmid stability, we compared colonies recovered from isoniazid-treated versus sucrose-treated mice as well as different organs. Bacteria recovered from isoniazid-treated mice demonstrated similar plasmid stability (94.9%) to those recovered from sucrose-treated control animals (87.2%). We also observed similar levels of plasmid stability in bacteria recovered from spleens (data not shown).
In summary, we found that within the first 3 weeks of infection ∼10% of bacteria had lost the ability to express the FFlucRT reporter. PCR amplification of selected colonies indicated that non-bioluminescent colonies had lost the integrated plasmid (data not shown). To address this shortcoming we developed more stable reporters by constructing integrase-free reporter plasmids. We subsequently introduced an integrase-free reporter into M. tuberculosis, creating the M. tuberculosis pMV306DIG13 + FFlucRT strain (DI FFlucRT). We confirmed that the DI FFlucRT strain grew identically to both WT M. tuberculosis and M. tuberculosis pMV306G13 + FFlucRT in vitro, and that the DI FFlucRT strain produced equivalent levels of bioluminescence to the FFlucRT counterpart. Crucially, in vitro the DI FFlucRT strain exhibited substantially superior reporter stability in comparison with the FFlucRT parent strain. Following 3 months of in vitro growth (with fortnightly subculturing), ∼40% of colonies recovered from the M. tuberculosis pMV306G13 + FFlucRT strain had lost the ability to luminesce. This was in sharp contrast to the pMV306DIG13 + FFlucRT strain, where only 1 of 118 colonies did not luminesce, demonstrating that >99% of colonies retained a functional bioluminescent reporter after 3 months of in vitro growth (P < 0.0001).
Discussion
The urgent need for new anti-TB drugs requires innovative approaches for accelerating the drug development pipeline. Efficacy testing in animal models constitutes an early and significant bottleneck partly because bacterial burden is measured as cfu recovered from organ homogenates and it can take up to 4 weeks for M. tuberculosis to grow on agar plates. Molecular imaging can provide an alternative readout for disease progression and treatment efficacy. Bioluminescence imaging is particularly suitable for the rapid screening of drug candidates in mice thanks to its short acquisition times, the fact that up to 5 mice can be imaged simultaneously, and that it is highly sensitive and quantitative. We have previously optimized three bioluminescent reporters for use in mycobacteria and proved that Lux and FFluc are suitable for in vivo imaging.13 While the former yields autoluminescent mycobacteria, the latter requires the administration of the substrate luciferin. However, this is non-toxic, easily administered and the signal obtained is much brighter than with Lux. Here, we demonstrated that bioluminescence imaging can be used as a non-invasive approach to visualize and quantify M. tuberculosis infection and drug treatment in live mice using a further improved FFluc reporter.
A key challenge in bioluminescence imaging is light absorption by tissues. This is mainly caused by haemoglobin, which absorbs in the blue and green part of the visible spectrum, whereas red light can travel through several centimetres of tissue.4 The WT FFluc produces yellow-green light. In order to improve FFluc for in vivo imaging, we introduced six mutations known to produce a thermostable red-emitting FFluc17 plus three mutations that had been previously shown to increase bioluminescence intensity more than 10-fold.16 The resulting reporter strain, M. tuberculosis FFlucRT, emitted red light as expected, but it was not significantly brighter in vitro than the strain expressing the original FFluc. Although the precise mechanism underlying this is unknown, it is possible that the ‘red mutations’ adversely affected the ‘bright phenotype’. Nonetheless, the 60 nm shift on the emission spectra of the luciferase constitutes a vast improvement for in vivo imaging.
Interestingly, the bioluminescence produced by FFlucRT-expressing M. tuberculosis correlated with cell density in vitro during both the exponential and stationary phases, whereas the signal produced by FFluc decreased during the stationary phase. It is thought that this decrease in FFluc bioluminescence reflects the metabolic state of the cells in the stationary phase since the luciferase uses ATP as a cofactor. In this sense, the unaltered bioluminescence of FFlucRT during the stationary phase could be explained by an increased affinity of the enzyme for ATP in comparison with FFluc. Alternatively, it could also be due to a longer half-life of the protein as a result of increased thermostability, a property that has been shown to correlate with imaging sensitivity.22 A more detailed study would be necessary to establish how the nine mutations may have affected the ATP affinity and the thermostability of the enzyme, but this is beyond the scope of this work.
The FFlucRT-expressing M. tuberculosis retained full virulence in vivo. Importantly, we were able to non-invasively detect the microorganisms in the lungs of live mice as early as 2 weeks after infection, when the bacterial load was estimated to be around 105 cfu per lung and the signal was 5.8 × 105 photons/s. A similar level of bioluminescence (5.2 × 105 photons/s) was measured in the thorax of live mice after 10 days of treatment with isoniazid 4 weeks post-infection, when the bacterial burden was 7.5 × 104 cfu/lung. This is the lowest number of bacteria that we were able to detect in live mice and is very close to the calculated limit of detection of 4.4 × 104 cfu/lung. To our knowledge, this is by far the lowest limit of detection of M. tuberculosis infection achieved so far using bioluminescence imaging. Furthermore, as few as 103 cfu were detected ex vivo in lungs and spleens with a calculated limit of detection in the lungs of just 1.2 × 102 cfu.
Significantly, both in vivo and ex vivo bioluminescence imaging allowed the rapid assessment of antibiotic treatment efficacy, with a decrease in bioluminescence observed in vivo after just 7 days of treatment, giving a total turnaround time for the assay of 3–4 weeks. In this study, we used immunocompromised mice since they allow a faster growth of M. tuberculosis and therefore a high bacterial load can be achieved in a shorter time. In addition, using the immunocompromised mouse model allows measurement of the anti-tubercular efficacy exclusively due to the drug action and independent of the immune system contribution. Results could be obtained in an even shorter time if bioluminescence imaging is used together with previously described rapid drug-testing assays.23 In these assays, treatment is started 1 day after infection with a high dose (105 cfu), and results are assessed after 7 days of treatment. Using such a high infection dose of the reporter strain described here would potentially allow assessment of treatment efficacy non-invasively in real time from day 1. If immunocompetent mice were used, an infection dose of around 103 cfu would be needed to detect the bacteria non-invasively, considering that M. tuberculosis typically grows 3–4 log10 before the onset of adaptive immunity. In low-dose infection models in which mice are challenged with 50–100 cfu, the highest bacterial load achieved in the lungs would be very close to the limit of detection, but we would expect the infection to be visualized and quantified in the dissected lungs. Another parameter that would need to be assessed is the effect of the immune response on bioluminescence production in terms of metabolic stress caused by the immune system or substrate availability in granulomatous lesions.
We have used isoniazid, which is often utilized as a control in drug screening experiments,24 to provide a proof-of-concept of the feasibility of our bioluminescence–mouse model to assess drug efficacy. With this drug we have observed a similar effect on bioluminescence and cfu; however, other drugs with different mechanisms of action could affect these two readouts to different degrees, as has previously been reported for ethambutol, linezolid and moxifloxacin.14 Even so, any compound with an effect on cfu will very likely also affect bioluminescence. Therefore, bioluminescence imaging could be used for the initial rapid screening of compounds, with the best candidates selected for further studies using cfu analysis.
We have previously utilized M. tuberculosis expressing fluorescent reporters in vitro and in vivo.25,26 The advantage of using fluorescent reporters is that, in contrast to bioluminescent reporter systems, samples can be analysed by fluorescence microscopy and flow cytometry. Furthermore, as fluorescent reporters do not use ATP or FMNH2 as cofactors, they are not as tightly linked to bacterial metabolism. This should prove advantageous for studying dormant M. tuberculosis in vivo. However, fluorescent reporter proteins require excitation light to generate a photonic signal. Unfortunately, the autofluorescence of endogenously produced fluorophores such as keratin, porphyrins, NAD(P)H and collagen, and absorption by haemoglobin and melanin in some cases generally results in worse signal-to-noise ratios than observed for bioluminescence, making fluorescence imaging much less sensitive. Indeed, when we applied these fluorescent reporter strains to test anti-TB drugs in vivo, the limits of detection were 100–1000-fold higher (∼8 × 107 cfu/lung in vivo and 2 × 105 cfu/lung ex vivo).26 While bioluminescent and fluorescent reporters can be considered complementary, depending on the particular experimental question being addressed, the bioluminescence method presented here represents a major improvement for drug screening in vivo since it can be used for non-invasive imaging of live mice and has a much better sensitivity both in vivo and ex vivo.
A different approach for in vivo imaging of mycobacteria was described in 2010 by Kong et al.27 In this case a fluorogenic substrate for the mycobacterial β-lactamase was used. This reaction results in the production of fluorescence that can be detected in vivo. The authors claim that as few as 104 cfu/lung can be detected using this method. However, this particular experiment was done with the non-pathogenic BCG rather than with M. tuberculosis, and only a very dim signal could be seen close to the animal axilla. In addition, to get a good correlation between signal and cfu the imaging has to be performed 24 h post-substrate administration; at later timepoints substrate accumulation leads to a similar level of fluorescence independent of bacterial numbers. In fact, it takes 72–96 h for the signal to completely fade, which means that repeated imaging cannot be performed at shorter intervals. In contrast, using our reporter strain the imaging can be performed every few hours if required, and the signal is stronger and can be clearly localized in the lungs.
One parameter that could negatively affect the limit of detection for our reporter strain is the loss of the reporter vector in vivo in the absence of antibiotic selection. In fact, we found that >10% of the colonies recovered from the lungs of infected SCID mice 3 weeks post-infection had lost the plasmid. It is known that the loss of L5-based integration vectors is due to site-specific excision catalysed by the integrase present in these vectors.15 Plasmid loss can be prevented by providing the integrase gene separately on a non-replicating suicide vector. Using this strategy we managed to increase the stability of the vector from 60% to >99% in vitro following 3 months of culture with no antibiotic selection. Future work will evaluate the use of this strain in vivo regarding virulence and the limit of detection using bioluminescence imaging.
In summary, we have shown that an improved M. tuberculosis bioluminescent strain can be visualized by non-invasive imaging in live mice during an acute, progressive infection. Furthermore, we prove that this method can be used to visualize and quantify the effect of antibiotic treatment. If necessary, the sensitivity of the technique can be increased by imaging dissected organs ex vivo, offering a more rapid readout than traditional growth-based assays. We believe that the model presented here can be of great use in early drug discovery as an easy and rapid way to identify active compounds in vivo.
Funding
This work was supported by a Bill and Melinda Gates Foundation TB Drug Accelerator Program grant. S. W. is supported by a Sir Charles Hercus Fellowship from the Health Research Council of New Zealand. The funders had no role in study design.
Transparency declarations
None to declare.
Supplementary data
References
- 1.WHO. Tuberculosis Fact Sheet. http://www.who.int/mediacentre/factsheets/fs104/en/index.html. (10 April 2013, date last accessed)
- 2.Andreu N, Zelmer A, Wiles S. Noninvasive biophotonic imaging for studies of infectious disease. FEMS Microbiol Rev. 2011;35:360–94. doi: 10.1111/j.1574-6976.2010.00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rice BW, Cable MD, Nelson MB. In vivo imaging of light-emitting probes. J Biomed Opt. 2001;6:432–40. doi: 10.1117/1.1413210. [DOI] [PubMed] [Google Scholar]
- 4.Zhao H, Doyle TC, Coquoz O, et al. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J Biomed Opt. 2005;10:41210. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- 5.Prioli RP, Tanna A, Brown IN. Rapid methods for counting mycobacteria–comparison of methods for extraction of mycobacterial adenosine triphosphate (ATP) determined by firefly luciferase assay. Tubercle. 1985;66:99–108. doi: 10.1016/0041-3879(85)90074-1. [DOI] [PubMed] [Google Scholar]
- 6.Janaszek W, Aleksandrowicz J, Sitkiewicz D. The use of the firefly bioluminescent reaction for the rapid detection and counting of Mycobacterium BCG. J Biol Stand. 1987;15:11–6. doi: 10.1016/0092-1157(87)90012-6. [DOI] [PubMed] [Google Scholar]
- 7.Cooksey RC, Crawford JT, Jacobs WR, Jr, et al. A rapid method for screening antimicrobial agents for activities against a strain of Mycobacterium tuberculosis expressing firefly luciferase. Antimicrob Agents Chemother. 1993;37:1348–52. doi: 10.1128/aac.37.6.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arain TM, Resconi AE, Hickey MJ, et al. Bioluminescence screening in vitro (Bio-Siv) assays for high-volume antimycobacterial drug discovery. Antimicrob Agents Chemother. 1996;40:1536–41. doi: 10.1128/aac.40.6.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deb DK, Srivastava KK, Srivastava R, et al. Bioluminescent Mycobacterium aurum expressing firefly luciferase for rapid and high throughput screening of antimycobacterial drugs in vitro and in infected macrophages. Biochem Biophys Res Commun. 2000;279:457–61. doi: 10.1006/bbrc.2000.3957. [DOI] [PubMed] [Google Scholar]
- 10.Andreu N, Fletcher T, Krishnan N, et al. Rapid measurement of antituberculosis drug activity in vitro and in macrophages using bioluminescence. J Antimicrob Chemother. 2012;67:404–14. doi: 10.1093/jac/dkr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heuts F, Carow B, Wigzell H, et al. Use of non-invasive bioluminescent imaging to assess mycobacterial dissemination in mice, treatment with bactericidal drugs and protective immunity. Microbes Infect. 2009;11:1114–21. doi: 10.1016/j.micinf.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Zhang T, Li S-Y, Converse PJ, et al. Using bioluminescence to monitor treatment response in real time in mice with Mycobacterium ulcerans infection. Antimicrob Agents Chemother. 2011;55:56–61. doi: 10.1128/AAC.01260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreu N, Zelmer A, Fletcher T, et al. Optimisation of bioluminescent reporters for use with mycobacteria. PLoS One. 2010;5:e10777. doi: 10.1371/journal.pone.0010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang T, Li S-Y, Nuermberger EL. Autoluminescent Mycobacterium tuberculosis for rapid, real-time, non-invasive assessment of drug and vaccine efficacy. PLoS One. 2012;7:e29774. doi: 10.1371/journal.pone.0029774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Springer B, Sander P, Sedlacek L, et al. Instability and site-specific excision of integration-proficient mycobacteriophage L5 plasmids: development of stably maintained integrative vectors. Int J Med Microbiol. 2001;290:669–75. doi: 10.1016/S1438-4221(01)80004-7. [DOI] [PubMed] [Google Scholar]
- 16.Fujii H, Noda K, Asami Y, et al. Increase in bioluminescence intensity of firefly luciferase using genetic modification. Anal Biochem. 2007;366:131–6. doi: 10.1016/j.ab.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Branchini BR, Ablamsky DM, Murtiashaw MH, et al. Thermostable red and green light-producing firefly luciferase mutants for bioluminescent reporter applications. Anal Biochem. 2007;361:253–62. doi: 10.1016/j.ab.2006.10.043. [DOI] [PubMed] [Google Scholar]
- 18.Barker LP, Brooks DM, Small PL. The identification of Mycobacterium marinum genes differentially expressed in macrophage phagosomes using promoter fusions to green fluorescent protein. Mol Microbiol. 1998;29:1167–77. doi: 10.1046/j.1365-2958.1998.00996.x. [DOI] [PubMed] [Google Scholar]
- 19.Barker LP, Porcella SF, Wyatt RG, et al. The Mycobacterium marinum G13 promoter is a strong sigma 70-like promoter that is expressed in Escherichia coli and mycobacteria species. FEMS Microbiol Lett. 1999;175:79–85. doi: 10.1111/j.1574-6968.1999.tb13604.x. [DOI] [PubMed] [Google Scholar]
- 20.Rettig GR, McAnuff M, Liu D, et al. Quantitative bioluminescence imaging of transgene expression in vivo. Anal Biochem. 2006;355:90–4. doi: 10.1016/j.ab.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Lee KH, Byun SS, Paik JY, et al. Cell uptake and tissue distribution of radioiodine labelled D-luciferin: implications for luciferase based gene imaging. Nucl Med Commun. 2003;24:1003–9. doi: 10.1097/00006231-200309000-00009. [DOI] [PubMed] [Google Scholar]
- 22.Baggett B, Roy R, Momen S, et al. Thermostability of firefly luciferases affects efficiency of detection by in vivo bioluminescence. Mol Imaging. 2004;3:324–32. doi: 10.1162/15353500200403178. [DOI] [PubMed] [Google Scholar]
- 23.Rullas J, García JI, Beltrán M, et al. Fast standardized therapeutic-efficacy assay for drug discovery against tuberculosis. Antimicrob Agents Chemother. 2010;54:2262–4. doi: 10.1128/AAC.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franzblau SG, DeGroote MA, Cho SH, et al. Comprehensive analysis of methods used for the evaluation of compounds against Mycobacterium tuberculosis. Tuberculosis. 2012;92:453–88. doi: 10.1016/j.tube.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Carroll P, Schreuder LJ, Muwanguzi-Karugaba J, et al. Sensitive detection of gene expression in mycobacteria under replicating and non-replicating conditions using optimized far-red reporters. PLoS One. 2010;5:e9823. doi: 10.1371/journal.pone.0009823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zelmer A, Carroll P, Andreu N, et al. A new in vivo model to test anti-tuberculosis drugs using fluorescence imaging. J Antimicrob Chemother. 2012;67:1948–60. doi: 10.1093/jac/dks161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong Y, Yao H, Ren H, et al. Imaging tuberculosis with endogenous β-lactamase reporter enzyme fluorescence in live mice. Proc Natl Acad Sci USA. 2010;107:12239–44. doi: 10.1073/pnas.1000643107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.