Abstract
Objective
Single nucleotide polymorphisms (SNPs) accumulated frequently in the mitochondrial displacement loop (D-loop) in many cancers. We had identified cancer risk-associated SNPs in the D-loop of non-Hodgkin lymphoma (NHL) patients previously, in this study, we investigated the association of age at onset and D-loop SNPs in NHL patients.
Materials and methods
The D-loop region of mtDNA was sequenced for 133 NHL patients recorded at the Fourth Hospital of Hebei Medical University. The Kaplan–Meier method was used to identify age at onset-associated SNPs in the D-loop of NHL patients. The Cox proportional hazards model was used to identify independent risk factors for age at onset.
Results
The SNP sites of nucleotides 146C/T, 151T/C, 194T/C, 315C/C insert, 523Del/A, and 525Del/C were identified for their association with age at onset, by the logrank test. In an overall multivariate analysis, allele 146 (relative risk, 0.403; 95% confidence interval [CI]: 0.182–0.895) (P = 0.026), allele 151 (relative risk, 0.378; 95% CI: 0.165–0.868) (P = 0.022), and allele 315 (relative risk, 3.554; 95% CI: 1.344–9.400) (P = 0.011) were identified as independent predictors for age at onset in NHL patients.
Conclusion
SNPs in the D-loop can predict age at onset in NHL patients. Analysis of the D-loop SNPs can help identify NHL patient subgroups at high risk of early onset.
Keywords: Kaplan–Meier, Cox proportional hazards model, D-loop, single nucleotide polymorphisms
Introduction
Non-Hodgkin lymphoma (NHL) is the fifth most frequent form of cancer among men and women, accounting for 2.15% of all cancer-related deaths in the People’s Republic of China over the period 2003–2007.1,2 Although the etiology of NHL is poorly understood, the immune-related conditions, family histology of hematopoietic malignancy, and body mass index have been shown to have an association with the cancer risk.3,4 Some studies have also shown that genetic factors are important to development of this cancer.5–7 The polymorphism of genes in the oxidative stress pathway also was identified for their association with a risk of NHL. However, the true mechanism of this cancer remains unknown.8–10 Although some studies have focused on genetics associated with the age at onset of NHL, they have yet to demonstrate a genetic basis of this disease.11
The human mitochondrial genome is 16 kilobase (kb) in length and is formed as a closed-circular duplex molecule.12 Mitochondrial deoxyribonucleic acid (mtDNA) is believed to be more susceptible to DNA damage and so acquires mutations at a higher rate than nuclear DNA because of the high levels of reactive oxygen species (ROS) generation, lack of protective histones, and limited DNA repair capacity in the mitochondria.13,14
The mitochondrial displacement loop (D-loop) region is important for regulating both replication and expression of the mitochondrial genome because it contains the leading-strand origin of replication and the main promoter for transcription. In cancer patients, sequence changes accumulated extensively in this region. 15–18 Previously, we sequenced the D-loop region in a case-control study of NHL patients and identified 140 single nucleotide polymorphisms (SNPs), including 26 SNPs with frequency distribution of minor allele greater than 5%, and we also identified NHL risk-associated SNPs.19 In this study, we investigated the association of age at onset and SNPs of the D-loop, in NHL patients.
Materials and methods
Tissue specimens and DNA extraction
Diagnosis of lymphoma was made according to the WHO Classification of Tumors.20 Blood samples were collected from 133 NHL patients who received treatment in the Respiratory Department at the Fourth Hospital of Hebei University, Shijiazhuang, People’s Republic of China, between 1990 and 2011. The genomic deoxyribonucleic acid (DNA) was immediately extracted using the Wizard® Genomic DNA Purification Kit (Promega Corporation, Fitchburg, WI, USA) and stored at −20°C. All procedures were supervised and approved by the Human Tissue Research Committee at the hospital. Informed consent was obtained from all participants before enrollment.
Polymerase chain reaction (PCR) amplification and sequence analysis
A 982-base pair (bp) product of D-Loop was amplified with the forward primer 5′-CCCCATGCTTACAAGCAAGT-3′ (nucleotide 16190–16209) and reverse primer 5′-GCTTTGAGGAGGTAAGCTAC-3′ (nucleotide 602–583). PCR was run with a PCR Master Mix Kit (Promega Corporation) and purified prior to sequencing. Cycle sequencing was performed with the BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad CA, USA), and the products were then read with the ABI PRISM® 3100 Genetic Analyzer (Life Technologies). Polymorphisms were confirmed by repeated analyses from both strands.
Statistical analysis
The age at onset curve of NHL patients was calculated using the Kaplan–Meier method at each of the SNP sites, and compared with the logrank test. Multivariate analysis was performed with a Cox proportional hazards model. All of the statistical analysis was done with the SPSS Statistics for Windows, Version 17.0 software package (SPSS Inc, Chicago, IL, USA). A P-value of <0.05 was considered statistically significant.
Results
The age at onset distribution of the NHL patients was listed in Table 1. Those analyzed included: 25 patients at age less than 30 years old; 16 patients at age 30–40; 28 patients at age 41–50; 30 patients at age 51–60; 20 patients at age 61–70, and 14 patients at age older than 70. Age at onset was analyzed together with clinical characteristics, including gender and TNM (tumor node metastasis) classification, using the Kaplan–Meier method, and these were compared by the logrank test. TNM stage was associated with the age at onset, at statistically significant levels (Table 2 and Figure 1).
Table 1.
Onset age distribution in NHL patients
| Age | Cases (N)
|
|
|---|---|---|
| Male | Female | |
| ≤30 | 20 | 5 |
| 30~40 | 5 | 11 |
| 41~50 | 16 | 12 |
| 51~60 | 20 | 10 |
| 61~70 | 10 | 10 |
| >70 | 10 | 4 |
Abbreviation: NHL, non-Hodgkin lymphoma.
Table 2.
Clinical characteristics and their association with onset age in NHL patients
| Characteristics | Cases (N) | P-value |
|---|---|---|
| Sex | 0.443 | |
| Male | 81 | |
| Female | 52 | |
| TNM classification | 0.031 | |
| I–II | 43 | |
| III–IV | 90 |
Abbreviations: NHL, non-Hodgkin lymphoma; TNM, tumor node metastasis.
Figure 1.
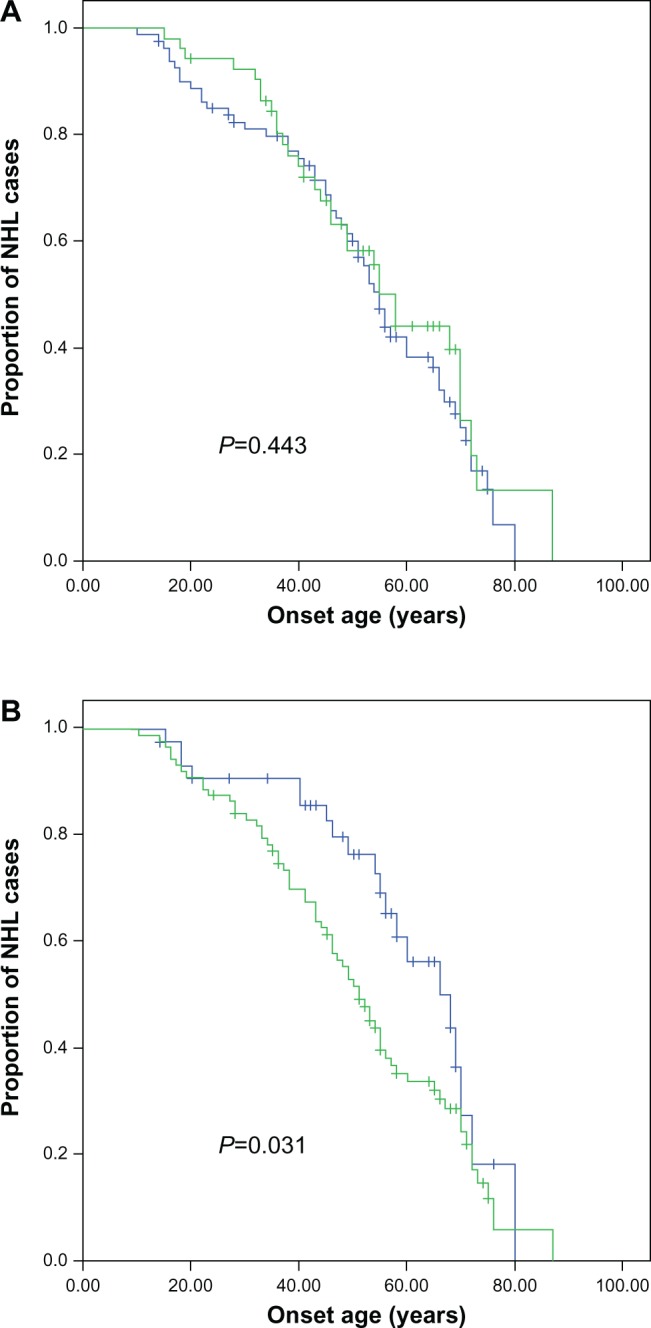
Comparison of age at onset for NHL patients according to (A) sex and (B) TNM classification with Kaplan–Meier methods.
Abbreviations: NHL, non-Hodgkin lymphoma; TNM, tumor node metastasis.
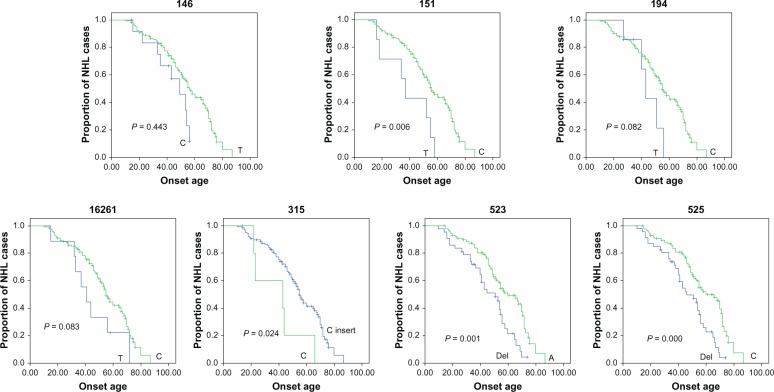
Twenty-six SNPs with minor allele frequency higher than 5% were subsequently evaluated for their association with age at onset. The NHL patients were divided into two groups on the basis of their genotype at each SNP site, and the association of SNPs in the D-loop and age at onset was calculated with Kaplan–Meier method. The SNP sites of nucleotides 146C/T, 151T/C, 194T/C, 315C/C insert, 523Del/A, and 525Del/C were identified for their association with age at onset by the logrank test (Table 3 and Figure 2); the SNP site of nucleotide 16261 was also identified for its association with age at onset but with borderline statistical significance (P = 0.083). We performed multivariate analysis for these predictors, including these SNPs and the clinical characteristics, with the Cox proportional hazards model. Allele 146 (relative risk, 0.403; 95% confidence interval [CI]: 0.182–0.895) (P = 0.026), allele 151 (relative risk, 0.378; 95% CI: 0.165–0.868) (P = 0.022), and allele 315 (relative risk, 3.554; 95% CI: 1.344–9.400) (P = 0.011) were identified as independent predictors for age at onset in the NHL patients (Table 4).
Table 3.
Polymorphic sites of the D-loop and their association with onset age in NHL patients
| Polymorphic sites | Cases (N) | P-value |
|---|---|---|
| 146 | 0.044 | |
| C | 13 | |
| T | 120 | |
| 151 | 0.006 | |
| T | 7 | |
| C | 126 | |
| 194 | 0.082 | |
| T | 7 | |
| C | 126 | |
| 16261 | 0.083 | |
| T | 9 | |
| C | 124 | |
| 315 | 0.024 | |
| C | 6 | |
| C inserta | 127 | |
| 523 | 0.001 | |
| Del | 43 | |
| A | 90 | |
| 525 | 0.000 | |
| Del | 46 | |
| C | 87 |
Note:
Including C and CC insertion.
Abbreviations: D-loop, displacement loop; NHL, non-Hodgkin lymphoma.
Figure 2.
Comparison of age at onset for NHL patients, according to the genotype of nucleotides 146, 151, 194, 16261, 315, 523, and 525 in the mitochondrial D-loop, using the Kaplan–Meier method.
Abbreviations: D-loop, displacement loop; NHL, non-Hodgkin lymphoma.
Table 4.
Multivariate analysis of predictors associated with age at onset of NHL patients
| Factors | Relative risk | 95% CI | P-value |
|---|---|---|---|
| 16261 | 0.714 | 0.329–1.548 | 0.393 |
| 146 | 0.403 | 0.182–0.895 | 0.026 |
| 151 | 0.378 | 0.165–0.868 | 0.022 |
| 194 | 0.515 | 0.192–1.381 | 0.187 |
| 315–316 | 3.554 | 1.344–9.400 | 0.011 |
| 523 | 1.330 | 0.392–4.515 | 0.648 |
| 525 | 0.400 | 0.121–1.322 | 0.133 |
| TNM classification | 1.601 | 0.927–2.764 | 0.091 |
Abbreviations: CI, confidence interval; NHL, non-Hodgkin lymphoma; TNM, tumor node metastasis.
Discussion
Many SNPs in the D-loop region have been identified for their association with cancer risk and disease outcome in cancers.21–27 The present study has extended those analyses to determine the relationship between age at onset and germline SNPs, in NHL patients. The alleles 146, 151, and 315 were identified for their association with age at onset, at statistically significant levels, by multivariate analysis.
We have previously identified age at onset-associated SNPs of the D-loop, in patients with hepatocellular carcinoma and esophageal squamous carcinoma.28,29 In this study, we also identified age at onset-associated SNPs in NHL patients.
All of the cancer risk-associated SNP sites are located in the hypervariable segment (HVII) region, mutational hotspots at which germline and tumor mtDNA mutations preferentially occur.30 We have identified outcome-associated SNPs of these regions in other cancers.21,27 The allele 315 was identified as a cancer risk-associated SNP for NHL, and allele 151 was identified as a cancer risk-associated SNP for non-small cell lung cancer in previous study.19,31 Our data imply the important role of HVII region in modifying the onset age of cancer.
Since the mitochondrial D-loop is important for replication and expression of the mitochondrial genome, SNPs in this region might alter the function of electron transport chains by modifying mtDNA replication and transcription, which is responsible for the generation of ROS, and could contribute to nuclear genome damage as well as cancer initiation and promotion.32–35 These ROS-mediated mechanisms may accelerate earlier onset of this disease.
In conclusion, SNPs in the D-loop can predict age at onset in NHL patients, and analysis of D-loop SNPs can help to identify NHL patient subgroups at high risk of early onset.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Geyer SM, Morton LM, Habermann TM, et al. Smoking, alcohol use, obesity, and overall survival from non-Hodgkin lymphoma: a population-based study. Cancer. 2010;116(12):2993–3000. doi: 10.1002/cncr.25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang M, Li GC, Zhang YL, Zhang SW, Yang NN. [An analysis of the incidence and mortality with malignant lymphoma in China during 2003–2007] China Cancer. 2012;21:190–196. Chinese. [Google Scholar]
- 3.Müller AM, Ihorst G, Mertelsmann R, Engelhardt M. Epidemiology of non-Hodgkin’s lymphoma (NHL): trends, geographic distribution, and etiology. Ann Hematol. 2005;84(1):1–12. doi: 10.1007/s00277-004-0939-7. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee N, Hartge P, Cerhan JR, et al. Risk of non-Hodgkin’s lymphoma and family history of lymphatic, hematologic, and other cancers. Cancer Epidemiol Biomarkers Prev. 2004;13(9):1415–1421. [PubMed] [Google Scholar]
- 5.Lan Q, Wang SS, Menashe I, et al. Genetic variation in Th1/Th2 pathway genes and risk of non-Hodgkin lymphoma: a pooled analysis of three population-based case-control studies. Br J Haematol. 2011;153(3):341–350. doi: 10.1111/j.1365-2141.2010.08424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diao LP, Ma H, Wei GC, et al. Matrix metalloproteinase-2 promoter and tissue inhibitor of metalloproteinase-2 gene polymorphisms in non-Hodgkin’s lymphoma. Int J Cancer. 2012;131(5):1095–1103. doi: 10.1002/ijc.26483. [DOI] [PubMed] [Google Scholar]
- 7.Hosgood HD, Purdue MP, Wang SS, et al. A pooled analysis of three studies evaluating genetic variation in innate immunity genes and non-Hodgkin lymphoma risk. Br J Haematol. 2011;152(6):721–726. doi: 10.1111/j.1365-2141.2010.08518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan Q, Zheng T, Shen M, et al. Genetic polymorphisms in the oxidative stress pathway and susceptibility to non-Hodgkin lymphoma. Hum Genet. 2007;121(2):161–168. doi: 10.1007/s00439-006-0288-9. [DOI] [PubMed] [Google Scholar]
- 9.Lightfoot TJ, Skibola CF, Smith AG, et al. Polymorphisms in the oxidative stress genes, superoxide dismutase, glutathione peroxidase and catalase and risk of non-Hodgkin’s lymphoma. Haematologica. 2006;91(9):1222–1227. [PubMed] [Google Scholar]
- 10.Wang SS, Davis S, Cerhan JR, et al. Polymorphisms in oxidative stress genes and risk for non-Hodgkin lymphoma. Carcinogenesis. 2006;27(9):1828–1834. doi: 10.1093/carcin/bgl013. [DOI] [PubMed] [Google Scholar]
- 11.Wiernik PH, Wang SQ, Hu XP, Marino P, Paietta E. Age of onset evidence for anticipation in familial non-Hodgkin’s lymphoma. Br J Haematol. 2000;108(1):72–79. doi: 10.1046/j.1365-2141.2000.01815.x. [DOI] [PubMed] [Google Scholar]
- 12.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 13.DiMauro S, Schon EA. Mitochondrial DNA mutations in human disease. Am J Med Genet. 2001;106(1):18–26. doi: 10.1002/ajmg.1392. [DOI] [PubMed] [Google Scholar]
- 14.Beal MF. Mitochondria, free radicals, and neurodegeneration. Curr Opin Neurobiol. 1996;6(5):661–666. doi: 10.1016/s0959-4388(96)80100-0. [DOI] [PubMed] [Google Scholar]
- 15.Yoneyama H, Hara T, Kato Y, Yamori T, Matsuura ET, Koike K. Nucleotide sequence variation is frequent in the mitochondrial DNA displacement loop region of individual human tumor cells. Mol Cancer Res. 2005;3(1):14–20. [PubMed] [Google Scholar]
- 16.Nishikawa M, Nishiguchi S, Shiomi S, et al. Somatic mutation of mitochondrial DNA in cancerous and noncancerous liver tissue in individuals with hepatocellular carcinoma. Cancer Res. 2001;61(5):1843–1845. [PubMed] [Google Scholar]
- 17.Sanchez-Cespedes M, Parrella P, Nomoto S, et al. Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors. Cancer Res. 2001;61(19):7015–7019. [PubMed] [Google Scholar]
- 18.Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410(2):103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 19.Gao Y, Zhao G, Diao L, Guo Z. Identification of sequence polymorphisms in the D-loop region of mitochondrial DNA as a risk factor for non-Hodgkin lymphoma. Mitochondrial DNA. 2013 doi: 10.3109/19401736.2013.796457. [DOI] [PubMed] [Google Scholar]
- 20.Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon: International Agency for Research on Cancer (IARC); 2008. [Google Scholar]
- 21.Zhang R, Wang R, Zhang F, et al. Single nucleotide polymorphisms in the mitochondrial displacement loop and outcome of esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2010;29:155. doi: 10.1186/1756-9966-29-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navaglia F, Basso D, Fogar P, et al. Mitochondrial DNA D-loop in pancreatic cancer: somatic mutations are epiphenomena while the germline 16519 T variant worsens metabolism and outcome. Am J Clin Pathol. 2006;126(4):593–601. doi: 10.1309/GQFCCJMH5KHNVX73. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Bamlet WR, de Andrade M, et al. Mitochondrial genetic polymorphisms and pancreatic cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16(7):1455–1459. doi: 10.1158/1055-9965.EPI-07-0119. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, McDonnell SK, Hebbring SJ, et al. Polymorphisms in mitochondrial genes and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17(12):3558–3566. doi: 10.1158/1055-9965.EPI-08-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007;67(10):4687–4694. doi: 10.1158/0008-5472.CAN-06-3554. [DOI] [PubMed] [Google Scholar]
- 26.Zhang R, Zhang F, Wang C, Wang S, Shiao YH, Guo Z. Identification of sequence polymorphism in the D-Loop region of mitochondrial DNA as a risk factor for hepatocellular carcinoma with distinct etiology. J Exp Clin Cancer Res. 2010;29:130. doi: 10.1186/1756-9966-29-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Zhang F, Fan H, et al. Sequence polymorphisms of mitochondrial D-loop and hepatocellular carcinoma outcome. Biochem Biophys Res Commun. 2011;406(3):493–496. doi: 10.1016/j.bbrc.2011.02.088. [DOI] [PubMed] [Google Scholar]
- 28.Guo Z, Yang H, Wang C, Liu S. Mitochondrial DNA haplogroup M is associated with late onset of hepatocellular carcinoma. Exp Ther Med. 2012;3(3):499–502. doi: 10.3892/etm.2011.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo Z, Yang H, Zhang F, Zhang R, Wang C. Single nucleotide polymorphisms in the mitochondrial displacement loop and age-at-onset of esophageal squamous cell carcinoma. Oncol Lett. 2012;3(2):482–484. doi: 10.3892/ol.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoneking M. Hypervariable sites in the mtDNA control region are mutational hotspots. Am J Hum Genet. 2000;67(4):1029–1032. doi: 10.1086/303092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding C, Li R, Wang P, Jin P, Li S, Guo Z. Identification of sequence polymorphisms in the D-loop region of mitochondrial DNA as a risk factor for lung cancer. Mitochondrial DNA. 2012;23(4):251–254. doi: 10.3109/19401736.2012.674120. [DOI] [PubMed] [Google Scholar]
- 32.Bandy B, Davison AJ. Mitochondrial mutations may increase oxidative stress: implications for carcinogenesis and aging? Free Radic Biol Med. 1990;8(6):523–539. doi: 10.1016/0891-5849(90)90152-9. [DOI] [PubMed] [Google Scholar]
- 33.Gille JJ, Joenje H. Cell culture models for oxidative stress: superoxide and hydrogen peroxide versus normobaric hyperoxia. Mutat Res. 1992;275(3–6):405–414. doi: 10.1016/0921-8734(92)90043-o. [DOI] [PubMed] [Google Scholar]
- 34.Shigenaga MK, Hagen TM, Ames BN. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci U S A. 1994;91(23):10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dement GA, Maloney SC, Reeves R. Nuclear HMGA1 nonhistone chromatin proteins directly influence mitochondrial transcription, maintenance, and function. Exp Cell Res. 2007;313(1):77–87. doi: 10.1016/j.yexcr.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]