Abstract
Two major pathways of recombination-dependent DNA replication, “join-copy” and “join-cut-copy,” can be distinguished in phage T4: join-copy requires only early and middle genes, but two late proteins, endonuclease VII and terminase, are uniquely important in the join-cut-copy pathway. In wild-type T4, timing of these pathways is integrated with the developmental program and related to transcription and packaging of DNA. In primase mutants, which are defective in origin-dependent lagging-strand DNA synthesis, the late pathway can bypass the lack of primers for lagging-strand DNA synthesis. The exquisitely regulated synthesis of endo VII, and of two proteins from its gene, explains the delay of recombination-dependent DNA replication in primase (as well as topoisomerase) mutants, and the temperature-dependence of the delay. Other proteins (e.g., the single-stranded DNA binding protein and the products of genes 46 and 47) are important in all recombination pathways, but they interact differently with other proteins in different pathways. These homologous recombination pathways contribute to evolution because they facilitate acquisition of any foreign DNA with limited sequence homology during horizontal gene transfer, without requiring transposition or site-specific recombination functions. Partial heteroduplex repair can generate what appears to be multiple mutations from a single recombinational intermediate. The resulting sequence divergence generates barriers to formation of viable recombinants. The multiple sequence changes can also lead to erroneous estimates in phylogenetic analyses.
Keywords: homologous recombination, evolution, transcription, translation
The purpose of looking back is not, of course, merely to obtain satisfaction from reflecting on past triumphs; rather, it is to discover as many clues as possible to the likely developments of the future.
Glenn T. Seaborg
The tight interrelationship between homologous recombination and DNA replication was first evident in T4 and the related T-even phages. Because DNA of T4 and its host E. coli differ in base composition and modifications and because the host DNA is rapidly degraded after phage infection, molecular aspects of T4 replication and recombination could be readily investigated by biochemical, biophysical, and genetic methods. Early characterization of mutations in most essential genes (1) and the almost complete dependence of replication and recombination on phage-encoded proteins (2) allowed analyses of recombination and replication proteins, as well as “reality checks” of results obtained with genetic and biochemical methods (3). The following idiosyncrasies of T4 chromosomes revealed the importance of DNA ends and recombination-dependent DNA replication. Ends of T4 chromosomes are cut during packaging from branched concatemers, which are generated by recombination-dependent replication. A “headful mechanism” packages a complete genome and ≈3% DNA repeated at each end as “terminal redundancy,” thereby generating the random circular permutation of chromosomal ends (4). Some smaller T4 particles, formed because of assembly errors, package incomplete genomes whose ends are also randomly circularly permuted (5). Multifactor crosses revealed stimulation of recombination by their DNA ends, regardless of map positions (5, 6). Moreover, different segregation patterns of alleles in patch vs. splice recombinants strongly suggested that 3′-ended single-stranded (ss) termini, invading a homologous double-stranded (ds) region, prime recombination-dependent DNA replication (6). These suggestions were later confirmed by mutational analyses combined with electron microscopy and density-labeling experiments (7–9). Origin-dependent replication facilitates early recombination by generating ss termini and ds interruptions, which make ds DNA more prone to invasion by ss termini (9). Priming of DNA replication from these recombinational intermediates is essential for T4 growth, because origin initiation ceases during T4's development (7, 10–12). Thus, recombination-deficient T4 mutants that can initiate origin-dependent DNA replication have a so-called DNA-arrest phenotype.
Many subsequent studies revealed multiple redundant pathways and enzymes with overlapping functions for homologous recombination and recombination-dependent DNA replication (11–15). Although redundancy may appear complicated or confusing to us as investigators, it makes any organism thrive. Most seemingly paradoxical results on recombination (in T4 as well as other organisms) can be reconciled by the fact that different pathways are preferred under certain conditions, or with certain model substrates, and that elimination of one pathway channels intermediates into another pathway.
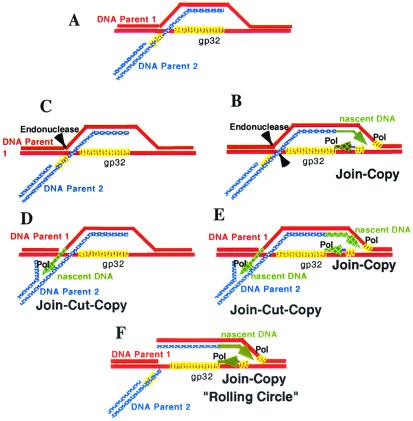
Keeping in mind the caveats inherent in definitions of recombination pathways, five T4 pathways can be distinguished (11). All of these pathways include pairing of complementary ss DNA segments in so-called heteroduplexes, but they “mix and match” different ways to generate ss DNA and resolve the recombinational intermediates (11). A ss annealing pathway can initiate recombination in the absence of DNA replication late after infection (16). When DNA polymerase is active, recombination starts much earlier, and at least two different pathways can initiate DNA replication from recombinational “D-loops” (Fig. 1; refs. 7, 9, and 14). “Ds break repair” (17) and “synthesis-dependent DNA annealing” pathways, in addition to those depicted in Figs. 1 and 3, can contribute to DNA repair and explain “homing” of intron DNA (15, 18, 19). Most of the proteins involved in these pathways have been biochemically characterized in exquisite detail (2, 12, 20–35).
Figure 1.
Different ways to initiate DNA replication from intermediates of homologous recombination. The ss end of parent 2 DNA invades homologous ds DNA of parent 1. Details are explained in the text.
Figure 3.
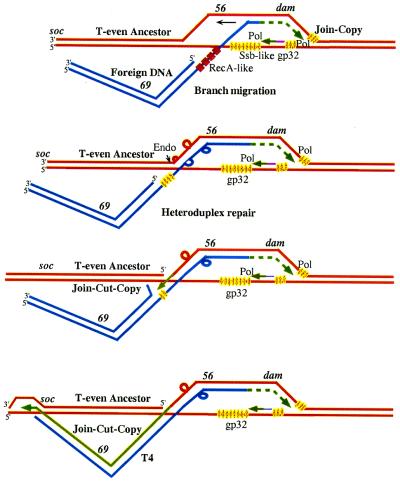
A recombination model for acquisition of a DNA (gene 69 in T4 or soc.1 and soc.2 in T2) by a T-even phage (e.g., RB15) lacking a gene between genes 56 and soc. Details are explained in the text.
Here we focus on two D-loop-dependent pathways that we have called “join-copy” and “join-cut-copy,” the roles of a complex containing gp46 (gene product 46)—the 46 complex—and two late T4 proteins, endonuclease VII (endoVII, gp49) and terminase (gp17), and on implications for horizontal gene transfer, mutagenesis, and evolution.
Results and Discussion
In the now classical example of recombination-induced DNA replication, a 3′ end of a ss DNA segment that invades a homologous ds DNA (Fig. 1A) primes DNA synthesis in the rightward direction as drawn in Figs. 1 A and B. We have called this pathway “join-copy,” because in undamaged wild-type (wt) T4 it occurs mainly at the unreplicated termini of parental chromosomes (which have been cut during the previous growth cycle), and no additional breaks are required (7). This mechanism depends only on genes expressed from early or middle promoters, and it can start as soon as an origin-initiated replication fork has reached an end (9, 36). This mechanism is equivalent to “break-copy recombination” or “break-induced replication,” and can also repair ds breaks in T4 DNA (19).
Another pathway, which we have dubbed “join-cut-copy,” uses a ss nick in a D-loop intermediate to prime DNA replication in the leftward direction, as drawn in Fig. 1D (11, 14, 37). This pathway depends on at least two late genes and therefore operates mainly late after infection. Priming of DNA synthesis requires that the nick occur in one of the invaded DNA strands. If the invading strand is nicked, parent 2 cannot be copied by this mechanism, but its ss terminus generates a patch recombinant with parent 1. Combined join-copy and join-cut-copy recombination can initiate two diverging replication forks (Fig. 1E). If a ds break cuts the (backward) recombination junction (as indicated in Fig. 1B), join-copy recombinants appear like rolling circles initiated from origins (Fig. 1F).
Late Synthesis of Endo VII and Terminase Make the Join-Cut-Copy Pathway a Late Pathway.
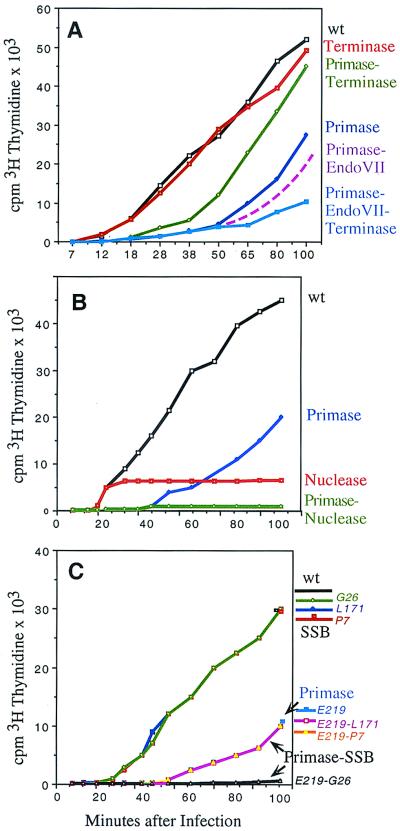
In T4 primase (gene 61) mutants, origin-initiated leading-strand DNA synthesis (primed by origin-specific transcripts) proceeds normally, but Okazaki fragments, primed by primase, are not synthesized. Primase mutants are viable, because the unreplicated ss regions can later be copied by the join-cut-copy pathway (11, 14). Either endo VII or terminase are required for this pathway, and additional defects in both corresponding genes (49 and 17) drastically reduce and delay (recombination-dependent) DNA replication of primase mutants (Fig. 2A). The residual late replication in the triple mutants may be due to nicks caused by other late nucleases, or by other agents.
Figure 2.
(A) Cumulative T4 DNA synthesis, measured by incorporation of 3H-labeled thymidine (45) at 30°C in E. coli B of a primase-defective mutant (E219), alone or in combination with endo VII (C9)- or terminase (NG178)- defective mutations. The E219-C9 incorporation relative to wt and E219 represents the average of three experiments done on different days. At first (14) we had mistakenly assumed that only endo VII produces the nicks required for the join-cut-copy pathway. Subsequently we found that the primase-endo VII double mutant used in those studies had acquired a third mutation. (B and C) Cumulative phage DNA synthesis at 25°C in E. coli B. (B) The primase mutant E219, the gene 46 mutant N130, and the double mutant are compared with wild-type T4. (C) The primase mutant E219, three gene 32 ts mutants (L171, P7, and G26), and the corresponding double mutants were compared with wild-type T4.
Both genes 49 and 17 are predominantly expressed late, and this late expression is responsible for the so-called DNA-delay phenotype of primase or topoisomerase mutants (11, 14, 37). Replicating parental DNA of topoisomerase mutants also contains large ss regions at termini and elsewhere (38), probably because leading- and lagging-strand DNA synthesis are uncoupled. Recombination-dependent DNA synthesis of primase or topoisomerase mutants is more severely delayed at decreasing growth temperatures because of the intricate regulation of genes 49 (14, 39) and 17 (40, 41). Remarkably, genes 49 and 17 produce two or more proteins by initiation of translation from internal ribosome binding sites, and the synthesis of the different products is temporally regulated by transcriptional and translational mechanisms. For example, the endo VII gene (49) is transcribed early and late (from different promoters), and the first ribosome binding site, from which a full-length 18-kDa protein is initiated, is sequestered in a hairpin in the early, but not in the late, transcripts. A shorter 12-kDa protein, initiated from an internal GUG, is the major early gene product at 25°C. At higher temperatures there is more early 18-kDa protein, presumably because the sequestering hairpin is less stable. In the crystal structure of endo VII (33), the full-length 18-kDa protein forms antiparallel dimers with two active sites each at the interphase of the C-terminal and N-terminal ends of different subunits. These two active sites are ideally placed to cut two strands of DNA across a Holliday junction. Our results with several N-terminal deletions of a cloned gene 49 showed that all deletions that eliminate the first AUG codon and leave the internal GUG initiation site intact complement internal or C-terminal gene 49 mutants, but none of these deletions could be incorporated into viable phage (39). Together these results indicate that the 12-kDa homodimers have no endo VII activity, 18-kDa and 12-kDa heterodimers can singly nick (sufficient for viable phage production) and 18-kDa homodimers make concerted ds breaks. The temporal regulation of these proteins is ideally suited for T4 growth. If too much 18-kDa endo VII peptide is produced too early, growth of primase mutants is impaired, because the chance to copy ss templates by join-cut-copy is reduced.
The significance of the multiple gene 17 (terminase) proteins is less well understood. A ss DNA-binding activity of purified wt or nuclease-defective mutant proteins resides in the N-terminal region (41), which is present in am (amber) mutants, but missing in the internally initiated peptides. Our results (Fig. 2A and data not shown) suggest that the ss DNA binding activity of the nuclease-defective terminase am mutants NG178 and N56 can protect the ss DNA of primase mutants from degradation by other nucleases, allowing more replication of the primase-terminase double mutants. However, triple mutants, defective in primase and the nuclease activities of terminase and endo VII, are more delayed and defective in DNA replication than any of the single or double mutant combinations (Fig. 2A and similar results with the terminase amN56 mutation in combination with primase and endo VII mutations not shown).
Protein Complexes Containing the Products of T4 Genes 46 and 47 Are Important in Several T4 Recombination Pathways.
Mutations in genes 46 and 47 cause the most severe T4 recombination and repair deficiencies (42, 43). These genes are important not only for T4 recombination, but also for degradation of dC-containing host DNA to nucleotides that are reprocessed for phage DNA synthesis (44). Consistent with the recombination deficiencies, mutants of these genes have a severe “DNA arrest” phenotype in E. coli B (1, 45), and they are defective in forming branched recombinational intermediates (16). The arrest of DNA replication is less complete in E. coli K strains (46). Possibly, a P2-like prophage in E. coli B (47) is responsible for this difference.
Surprisingly, the T4 46 and 47 proteins are not required in the current in vitro pairing (28) or recombination-dependent replication systems (12, 13). One possible reason for this apparent paradox is that all in vitro systems use simple ss DNA fragments for invasions, whereas in vivo most recombination intermediates are formed from molecules containing ss and ds segments, which impose more constraints.
Nuclease activities that were absent in 46-47 mutants have been found in membrane fractions (48), but proteins with the expected nuclease activities have only recently been purified from overproducing clones (49).
It has been suggested that the 46 complex of T4 has a similar role as the RecBCD nuclease of E. coli. (e.g., processing ds DNA ends to ss tails that can invade homologous DNA; ref. 23). However, replication generates ss tails, making it unlikely that this is the most essential common function of the T4 46 complex. Moreover, T4 primase-gene 46 double mutants, in which the lagging strand templates are displaced during origin-dependent replication as large ss tails of forked molecules (data not shown), are completely defective in all recombination-dependent DNA replication (Fig. 2B). If processing of ds DNA to ss tails were the most important function of the 46 complex, it should be dispensable in the primase mutants. However, in contrast to forked wt chromosomes, in which all ss termini were found invading other DNA molecules (9), the ss termini of primase-46 double mutant chromosomes were free, suggesting that their most important defect is impaired pairing of ss tails.
Electron micrographs of single primase mutants revealed D-loops of varying lengths, generated by displacement synthesis (14, 50). However, we found no forked molecules with equal-sized ss and ds tails, suggesting that the displaced ss DNA is rapidly assimilated into triple-stranded DNA or degraded once the primase-deficient replisomes have reached an end. Earlier results (51) had indicated that unmodulated nuclease activities of the 46 complex cut or degrade ss tails, when interacting proteins, most importantly the ssDNA binding protein gp32, are defective. Therefore, we combined primase mutations with mutations in other genes, whose products we suspected to modulate excessive degradation of ss regions during D-loop formation. The interaction with gp32 is allele-specific. It is defective in the am (amber) mutants lacking the C-terminal segment of gp32, and in the ts (temperature-sensitive) mutant G26, but it is not defective in two other gene 32 ts mutants, P7 and L171 (amino acid changes are depicted in ref. 11). DNA synthesis of the single gene 32 mutants G26, L171, and P7 at the permissive temperature (25°C) is indistinguishable from wt, and synthesis of the double mutants E219-L171 and E219-P7 is indistinguishable from E219 alone. In contrast, the E219-G26 double mutant is severely impaired (Fig. 2C). Double mutants containing the same gene 32 mutations in combination with another primase mutation, HL 627, showed identical patterns. Similarly, among double mutants containing the (DNA-delay) topoisomerase mutation H17 in combination with the same three gene 32 mutations, only H17-G26 was more defective than H17 alone (data not shown). We surmise that ts G26 is already partially defective at otherwise permissive temperatures in taming the nuclease activity of the 46 complex. This defect is masked when all other members of the complex are present, but it becomes apparent when one additional member is missing or defective. Taken together, our results suggest that primase, topoisomerase, and probably RNaseH, together with gp32, modulate the nuclease activity of the 46 complex. All these proteins are important for lagging-strand DNA synthesis. If they remain bound to DNA when the replication fork has reached an end, they may help to protect any ss tails from degradation, allowing join-cut-copy recombination as indicated in Fig. 1 C and D. If one of them is missing (e.g., in primase and topoisomerase mutants), the invading ss termini (e.g., of parent 2 in Fig. 1C) are expected to be cut frequently, resulting in excessive patch-type recombinants, which are preferentially formed by these mutants (50, 52).
The gene 46-47 proteins share similarities with the E. coli SbcCD nuclease and with the eukaryotic Mre11 complex (53), which is essential for ds break repair (54, 55) in yeast and in mammals. Ogawa and coworkers (56) have shown that the Mre11 complex has multiple modes of binding to DNA and interaction sites with other proteins, and that it can unwind DNA ends or help the annealing of complementary DNA. Thus, the homology of the T4 46 and the Mre11 complex probably reflects similarities of proteins that have different activities depending on the larger protein complexes in which they participate.
Mutagenic and Evolutionary Potential of Homologous Recombination Pathways.
Recombination-dependent replication pathways can be mutagenic or antimutagenic in several ways: (i) Replisomes assembled at origins, or by join-copy or join-cut-copy pathways, may differ in mutagenic potential because of different protein composition. T4 encodes only one DNA polymerase, whose mutagenicity is strongly affected by certain mutations in its gene (57, 58), and may be modulated by mutations affecting accessory proteins (59). For example, use of the dda helicase in the join-cut-copy pathway (14) might selectively alter the mutagenicity of this pathway. (ii) If ectopic base pairing initiates strand invasion, which is extended by branch migration (in the leftward direction in Fig. 1) into an adjacent heterologous region, mismatches and bulges are formed in vitro and in vivo (refs. 60–62; G.M., unpublished results). Subsequent partial mismatch repair is bound to generate novel sequence combinations (e.g., mutations; Fig. 3). Our previous results (63) suggested that such sequence variations can be generated during horizontal gene transfer. In T4, mismatch repair depends on endo VII in vivo and in vitro (refs. 30 and 62; G.M., unpublished results). The host mismatch repair genes have little or no effect, perhaps because T4's dCs are modified (described below) and/or because endo VII masks any possible contribution of the host system.
Many viruses share a common gene pool, from which individual genes can be exchanged (64) or acquired by horizontal transfer (65). Illegitimate site-specific recombination or transposition have been proposed to exchange entire gene modules of related functions (66). However, gene segments can also be exchanged or added (67–71). We have suggested that a combination of join-copy and join-cut-copy pathways, together with heteroduplex repair (Fig. 3), can explain otherwise paradoxical aspects of horizontal gene transfer. We postulate that it generates multiple mutations in a single heteroduplex overlap (11), which was initiated by homologous pairing between incidentally similar, short sequences and extended into heterologous regions.
To test this working model, we have focused our analyses on gene 56 of T4 and related (T-even) phages. This gene encodes a dCTPase/dCDPase/dUTPase/dUDPase (72), hereafter called dCTPase, which is essential for growth of T-even phages because it prevents incorporation of dCTP or dUTP into T-even DNA. T-even DNA contains hydroxymethyl cytosine, further modified by glycosyl residues, which prevent degradation by phage and host restriction enzymes (73). In crosses between T2 and T4, T2 gene 56 alleles are severely excluded from the progeny (74, 75), but the mechanism has been elusive. Our working model (Fig. 3) has been confirmed and extended by sequence and recombinational analyses of different T-even phages (summarized below). Details of these experiments will be reported elsewhere.
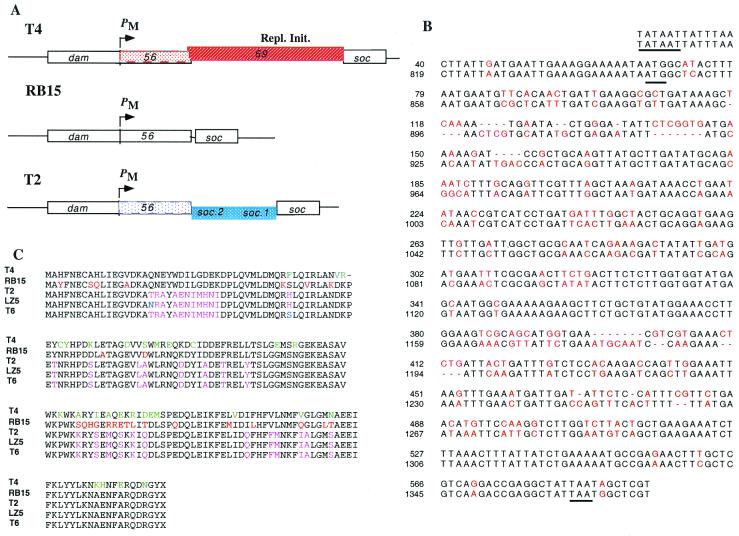
PCR analyses using primers corresponding to sequences shared by T2 and T4 in genes 56 and soc (small outer capsid protein) showed that different T-even phages contain different amounts of DNA between genes 56 and soc, corresponding to three size classes. DNA sequencing showed that class A, which includes T4, contains a gene-69-like sequence, and that class B, which includes T2, contains two small ORFs (soc.1 and soc.2); class C, which includes RB15, has no gene between 56 and soc (Fig. 4A). Most significantly in terms of our hypothesis, both base and amino acid sequences of different T-even classes differ considerably, including apparent multiple compensating frameshifts (Fig. 4B and ref. 61), which result in differences of blocks of amino acids (Fig. 4C). In contrast, sequences of phages belonging to the same class differ by less than 3%. We surmise that the large differences between classes were established concomitantly with acquisition by a T-even ancestor of different genes (69 vs. soc.1 and soc.2) adjacent to gene 56. The model depicted in Fig. 3 provides the simplest interpretation.
Figure 4.
Comparison of dCTPases and adjacent genes of different T-even phages. (A) Positions of genes 56 and soc in classes A, B, and C. (B) Alignment of the gene 56 sequences of T2 and RB15. The −10 region of the promoter PM and the initiation and termination codons are underlined. (C) dCTPase sequences of different phages. Green, different in T4 from all others; red, different in RB15 from all others; purple, identical among class B (T2, LZ5, and T6), but different in T4 and RB15; blue, different in only one phage.
Specifically, we propose that limited (“ectopic”) homology between a foreign DNA fragment and the dCTPase gene 56 allowed ss DNA invasion by the fragment's end, and that this recombinational intermediate was stabilized by join-copy replication from the 3′-invading end. Branch migration in the opposite direction generated heteroduplexes with multiple mismatches and loops. When an endonuclease made a single cut in the invaded strand of the recombinational intermediate (Fig. 1 C and D), the remainder of the foreign DNA fragment was copied by the join-cut-copy pathway. Limited homology of the DNA fragment with the soc promoter region allowed recombination at the other end of the foreign fragment. Partial mismatch repair within the heteroduplex region (e.g., by endo VII; refs. 30 and 62) generated multiple mutations of gene 56, but only those combinations that regenerated a functional dCTPase survived. Because invasion by different foreign DNA fragments resulted in different functional dCTPases (e.g., in T4 and T2), these genes now appear to have diverged considerably.
The model predicts, and we found, that the present-day extensive sequence divergence between certain genes of different T-even phages is the major reason for the lack of viable recombinants in such genes. Sequence divergence does not inhibit heteroduplex formation in T4 DNA (ref. 62; G.M., unpublished observation), but the corresponding heteroduplexes must contain multiple mismatches and loops, and partial heteroduplex repair is unlikely to reconstitute a functional essential gene such as the dCTPase gene 56, except when the DNA sequences are made similar (e.g., in chimeric phages; ref. 63).
Conclusions
Two major recombination pathways of T4, join-copy and join-cut-copy (11), are maintained during evolution, because they integrate recombination with developmental changes in other DNA transactions, mainly transcription and packaging, and because they are important for repairing DNA damages during different developmental stages. The join-copy pathway, which requires only genes expressed in the early and middle mode, is active within a minute after the onset of origin-dependent DNA replication. In contrast, the late join-cut-copy pathway depends on at least two late proteins: endo VII and terminase. This late pathway can bypass deficiencies of T4 primase or topoisomerase mutants, which have a so-called DNA-delay phenotype.
Even within a given pathway, some enzyme activities (e.g., nuclease activities of T4 endo VII and terminase) can substitute for each other. These enzymes, which are required for packaging DNA (76), interact with each other (77). Other proteins, such as the ss DNA binding protein gp 32, or T4 gene 46-47 proteins, appear to be important in all recombination pathways, but different interactions with other proteins are important for their roles in different pathways. In at least one case, that of T4 endo VII, different functions of a multifunctional gene can be related to the exquisitely regulated synthesis of two proteins from two ribosome binding sites of the same gene.
These homologous recombination pathways also contribute to evolution, because they facilitate acquisition of any foreign DNA with limited sequence homology (≈20 bp) during horizontal gene transfer, without requiring transposition or site-specific recombination functions. The example of the dCTPase genes shows that in this situation, horizontal gene transfer can generate mosaic sequence divergence within essential genes, in turn, establishing species barriers. Similar mosaic arrangements in other genes of the T-even (69, 71) and other phages (67) are also most readily explained by the mechanism illustrated in Fig. 3. In such situations, cladistic analyses to deduce timing of divergence of viruses or their genes become ambiguous.
Acknowledgments
We thank Betty Kutter for a collection of T-even phages, Mamie Wood-Blankenship for electron microscopy, Jeff and Dana Franklin for critical reading of the manuscript, the Vanderbilt Ingram Cancer Center, supported by Public Health Service Award CA68485, for facilities, and the National Science Foundation (MCB 9983568) and the Natural Science Fund of Vanderbilt University for financial support.
Abbreviations
- ss
single-stranded
- ds
double-stranded
- wt
wild type
- ts
temperature sensitive
- gp
gene product
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Epstein R H, Bolle A, Steinberg C, Kellenberger E, Boy de la Tour E, Chevalley R, Edgar R, Susman M, Denhardt C, Lielausis I. Cold Spring Harbor Symp Quant Biol. 1964;28:375–392. [Google Scholar]
- 2.Nossal N G. In: Molecular Biology of Bacteriophage T4. Karam J, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, et al., editors. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 43–53. [Google Scholar]
- 3.Alberts B M, Barry J, Bedinger B P, Burke R L, Hibner U, Liu C C, Sheridan R. UCLA Symp Mol Cell Biol. 1980;19:449–473. [Google Scholar]
- 4.Streisinger G. In: Phage and the Origins of Molecular Biology. Cairns J, Stent G S, Watson J D, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1966. pp. 335–340. [Google Scholar]
- 5.Mosig G. Cold Spring Harbor Symp Quant Biol. 1964;28:35–42. [Google Scholar]
- 6.Mosig G, Ehring R, Schliewen W, Bock S. Mol Gen Genet. 1971;113:51–91. doi: 10.1007/BF00335007. [DOI] [PubMed] [Google Scholar]
- 7.Luder A, Mosig G. Proc Natl Acad Sci USA. 1982;79:1101–1105. doi: 10.1073/pnas.79.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dannenberg R, Mosig G. J Virol. 1981;40:890–900. doi: 10.1128/jvi.40.3.890-900.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dannenberg R, Mosig G. J Virol. 1983;45:813–831. doi: 10.1128/jvi.45.2.813-831.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosig G, Colowick N, Gruidl M E, Chang A, Harvey A J. FEMS Microbiol Rev. 1995;17:83–98. doi: 10.1111/j.1574-6976.1995.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 11.Mosig G. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 12.Kreuzer K N. Trends Biochem Sci. 2000;25:165–173. doi: 10.1016/s0968-0004(00)01559-0. [DOI] [PubMed] [Google Scholar]
- 13.Formosa T, Alberts B M. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- 14.Mosig G, Luder A, Ernst A, Canan N. New Biol. 1991;3:1195–1205. [PubMed] [Google Scholar]
- 15.Mueller J E, Clyman J, Huang Y-J, Parker M M, Belfort M. Genes Dev. 1996;10:351–364. doi: 10.1101/gad.10.3.351. [DOI] [PubMed] [Google Scholar]
- 16.Broker T R. J Mol Biol. 1973;81:1–16. doi: 10.1016/0022-2836(73)90243-x. [DOI] [PubMed] [Google Scholar]
- 17.Kreuzer K N, Saunders M, Weislo L J, Kreuzer H W E. J Bacteriol. 1995;177:6844–6853. doi: 10.1128/jb.177.23.6844-6853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belfort M, Perlman P S. J Biol Chem. 1995;270:30237–30240. doi: 10.1074/jbc.270.51.30237. [DOI] [PubMed] [Google Scholar]
- 19.George J W, Kreuzer K N. Genetics. 1996;143:1507–1520. doi: 10.1093/genetics/143.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alberts B M, Frey L. Nature (London) 1970;227:1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- 21.Alberts B M, Miake-Lye R. Cell. 1992;68:415–420. doi: 10.1016/0092-8674(92)90179-g. [DOI] [PubMed] [Google Scholar]
- 22.Nossal N G, Hinton D M, Hobbs L J, Spacciapoli P. Methods Enzymol. 1995;262:560–584. doi: 10.1016/0076-6879(95)62045-1. [DOI] [PubMed] [Google Scholar]
- 23.Kreuzer K N, Morrical S W. In: Molecular Biology of Bacteriophage T4. Karam J, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, et al., editors. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 28–42. [Google Scholar]
- 24.Morrical S, Hempstead K, Morrical M, Chou K M, Ando R, Grigorieva O. Ann NY Acad Sci. 1994;726:349–350. doi: 10.1111/j.1749-6632.1994.tb52848.x. [DOI] [PubMed] [Google Scholar]
- 25.von Hippel P H, Kowalczykowski S C, Lonberg N, Newport J W, Paul L S, Stormo G D, Gold L. In: Bacteriophage T4. Mathews C K, Kutter E M, Mosig G, Berget P B, editors. Washington, DC: Am. Soc. Microbiol.; 1983. pp. 202–207. [Google Scholar]
- 26.Salinas F, Benkovic S J. Proc Natl Acad Sci USA. 2000;97:7196–7201. doi: 10.1073/pnas.97.13.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beernink H T, Morrical S W. Biochemistry. 1998;37:5673–5681. doi: 10.1021/bi9800956. [DOI] [PubMed] [Google Scholar]
- 28.Beernink H T, Morrical S W. Trends Biochem Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- 29.Kemper B, Jensch F, Depka-Prondzynski M U, Fritz H J, Borgmeyer R U, Mizuuchi M. Cold Spring Harbor Symp Quant Biol. 1984;49:815–825. doi: 10.1101/sqb.1984.049.01.092. [DOI] [PubMed] [Google Scholar]
- 30.Kemper B. In: DNA Damage and Repair. Nickoloff J A, Hoekstra M, editors. Vol. 1. Totowa, NJ: Humana Press; 1998. pp. 179–204. [Google Scholar]
- 31.Shamoo Y, Friedman A M, Parsons M R, Konigsberg W H, Steitz T A. Nature (London) 1995;376:362–366. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 32.Shamoo Y, Steitz T A. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 33.Raaijmakers H, Vix O, Törõ I, Golz S, Kemper B, Suck D. EMBO J. 1999;18:1447–1458. doi: 10.1093/emboj/18.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueser T C, Nossal N G, Hyde C C. Cell. 1996;85:1101–1112. doi: 10.1016/s0092-8674(00)81310-0. [DOI] [PubMed] [Google Scholar]
- 35.Mueser T C, Jones C E, Nossal N G, Hyde C C. J Mol Biol. 2000;296:597–612. doi: 10.1006/jmbi.1999.3438. [DOI] [PubMed] [Google Scholar]
- 36.Mosig G. In: Bacteriophage T4. Mathews C K, Kutter E M, Mosig G, Berget P B, editors. Washington, DC: Am. Soc. Microbiol.; 1983. pp. 120–130. [Google Scholar]
- 37.Mosig G. In: Molecular Biology of Bacteriophage T4. Karam J, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, et al., editors. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 54–82. [Google Scholar]
- 38.Thompson R J, Davies J P, Lin G, Mosig G. In: The Bacterial Chromosome. Drlica K, Riley M, editors. Washington, DC: Am. Soc. Microbiol.; 1990. pp. 227–240. [Google Scholar]
- 39.Barth K A, Powell D, Trupin M, Mosig G. Genetics. 1988;120:329–343. doi: 10.1093/genetics/120.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franklin J L, Mosig G. Gene. 1996;177:179–189. doi: 10.1016/0378-1119(96)00299-5. [DOI] [PubMed] [Google Scholar]
- 41.Franklin J L, Haseltine D, Davenport L, Mosig G. J Mol Biol. 1998;277:541–557. doi: 10.1006/jmbi.1998.1619. [DOI] [PubMed] [Google Scholar]
- 42.Berger H, Warren A J, Fry K E. J Virol. 1969;3:171–175. doi: 10.1128/jvi.3.2.171-175.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernstein H. Cold Spring Harbor Symp Quant Biol. 1968;33:325–331. doi: 10.1101/sqb.1968.033.01.037. [DOI] [PubMed] [Google Scholar]
- 44.Kutter E M, Wiberg J S. J Mol Biol. 1968;38:395–411. doi: 10.1016/0022-2836(68)90394-x. [DOI] [PubMed] [Google Scholar]
- 45.Mosig G, Colowick N. Methods Enzymol. 1995;262:587–604. doi: 10.1016/0076-6879(95)62046-x. [DOI] [PubMed] [Google Scholar]
- 46.Stitt B L, Mosig G. J Bacteriol. 1989;171:3872–3880. doi: 10.1128/jb.171.7.3872-3880.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rutberg B, Rutberg L. J Bacteriol. 1965;90:891–894. doi: 10.1128/jb.90.4.891-894.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mickelson C, Wiberg J S. J Virol. 1981;40:65–77. doi: 10.1128/jvi.40.1.65-77.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bleuit J S, Xu H, Ma Y, Wang T, Liv J, Morrical S W. Proc Natl Acad Sci USA. 2001;98:8298–8305. doi: 10.1073/pnas.131007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luder A. Ph.D. thesis. Nashville, TN: Vanderbilt University; 1981. [Google Scholar]
- 51.Mosig G, Bock S. J Virol. 1976;17:756–761. doi: 10.1128/jvi.17.3.756-761.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamlett N V, Berger H. Virology. 1975;63:539–567. doi: 10.1016/0042-6822(75)90326-8. [DOI] [PubMed] [Google Scholar]
- 53.Sharples G J, Leach D R F. Mol Microbiol. 1995;17:1215–1220. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- 54.Paques F, Haber J E. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nelms B E, Maser R S, MacKay J F, Lagally M G, Petrini J H. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 56.Yu X, Jacobs S A, West S C, Ogawa T, Egelman E H. Proc Natl Acad Sci USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reha-Krantz L J. Genetics. 1998;148:1551–1557. doi: 10.1093/genetics/148.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nossal N G. Genetics. 1998;148:1535–1538. doi: 10.1093/genetics/148.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drake J W, Ripley L S. In: Molecular Biology of Bacteriophage T4. Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, et al., editors. Washington, DC: Am. Ssoc. Microbiol.; 1994. pp. 98–124. [Google Scholar]
- 60.Bianchi M E, Radding C M. Cell. 1983;35:511–520. doi: 10.1016/0092-8674(83)90185-x. [DOI] [PubMed] [Google Scholar]
- 61.Lichten M, Fox M S. Proc Natl Acad Sci USA. 1984;81:7180–7184. doi: 10.1073/pnas.81.22.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mosig G, Shaw M, Garcia G M. Cold Spring Harbor Symp Quant Biol. 1984;49:371–382. doi: 10.1101/sqb.1984.049.01.044. [DOI] [PubMed] [Google Scholar]
- 63.Gary T P, Colowick N E, Mosig G. Genetics. 1998;148:1461–1473. doi: 10.1093/genetics/148.4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cohen D. Virology. 1959;7:112–126. doi: 10.1016/0042-6822(59)90180-1. [DOI] [PubMed] [Google Scholar]
- 65.Hendrix R W, Smith M C, Burns R N, Ford M E, Hatfull G F. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Botstein D, Herskowitz I. Nature (London) 1974;251:584–589. doi: 10.1038/251584a0. [DOI] [PubMed] [Google Scholar]
- 67.Haggard-Ljungquist E C, Halling C, Calendar R. J Bacteriol. 1992;174:1462–1477. doi: 10.1128/jb.174.5.1462-1477.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sandmeier H. Mol Microbiol. 1994;12:343–350. doi: 10.1111/j.1365-2958.1994.tb01023.x. [DOI] [PubMed] [Google Scholar]
- 69.Henning U, Hashemol-Hosseini S. In: Molecular Biology of Bacteriophage T4. Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, et al., editors. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 291–298. [Google Scholar]
- 70.Tetart F, Desplats C, Krisch H M. J Mol Biol. 1998;282:543–556. doi: 10.1006/jmbi.1998.2047. [DOI] [PubMed] [Google Scholar]
- 71.Repoila F, Tétart F, Bouet J-Y, Krisch H M. EMBO J. 1994;13:4181–4192. doi: 10.1002/j.1460-2075.1994.tb06736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wiberg J S, Dirksen M-L, Epstein R H, Luria S E, Buchanan J M. Proc Natl Acad Sci USA. 1962;48:293–302. doi: 10.1073/pnas.48.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carlson K, Raleigh E A, Hattman S. In: Molecular Biology of Bacteriophage T4. Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, et al., editors. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 369–381. [Google Scholar]
- 74.Streisinger G, Weigle J. Proc Natl Acad Sci USA. 1956;42:504–510. doi: 10.1073/pnas.42.8.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Russell R L, Huskey R J. Genetics. 1974;78:989–1014. doi: 10.1093/genetics/78.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Black L W. BioEssays. 1995;17:1025–1030. doi: 10.1002/bies.950171206. [DOI] [PubMed] [Google Scholar]
- 77.Golz S, Kemper B. J Mol Biol. 1999;285:1131–1144. doi: 10.1006/jmbi.1998.2399. [DOI] [PubMed] [Google Scholar]