Abstract
Repurposing has the objective of targeting existing drugs and failed, abandoned, or yet-to-be-pursued clinical candidates to new disease areas. The open-source model permits for the sharing of data, resources, compounds, clinical molecules, small libraries, and screening platforms to cost-effectively advance old drugs and/or candidates into clinical re-development. Clearly, at the core of drug-repurposing activities is collaboration, in many cases progressing beyond the open sharing of resources, technology, and intellectual property, to the sharing of facilities and joint program development to foster drug-repurposing human-capacity development. A variety of initiatives under way for drug repurposing, including those targeting rare and neglected diseases, are discussed in this review and provide insight into the stakeholders engaged in drug-repurposing discovery, the models of collaboration used, the intellectual property-management policies crafted, and human capacity developed. In the case of neglected tropical diseases, it is suggested that the development of human capital be a central aspect of drug-repurposing programs. Open-source models can support human-capital development through collaborative data generation, open compound access, open and collaborative screening, preclinical and possibly clinical studies. Given the urgency of drug development for neglected tropical diseases, the review suggests elements from current repurposing programs be extended to the neglected tropical diseases arena.
Keywords: repurposing, open source, rare diseases, neglected tropical diseases, models of collaboration, human-capacity development
Introduction
In May 2012, the National Institutes of Health (NIH) National Center for Advancing Translational Sciences (NCATS) initiative for drug repurposing was announced. The objective of the program – “Discovering new therapeutic uses for existing molecules” – is to foster collaboration between academic scientists and pharmaceutical companies to find new uses for previously shelved compounds. Pfizer, AstraZeneca, Eli Lilly, Abbott, Bristol-Myers Squibb, GlaxoSmithKline, Janssen Pharmaceuticals, and Sanofi are among the pharmaceutical organizations that joined the initiative.1,2 Collectively, these companies agreed to make 58 of these compounds available for the pilot program. The compounds have undergone significant research and development, including safety testing in humans, providing a starting point for scientists and permitting the clinical development process to move more rapidly. Not only does the program seek to match novel indications to existing compounds, but also provides template agreements to effectively manage the legal and administrative processes for partnering across multiple organizations.1,2 While the organizations will retain intellectual property (IP) on the original compound, a new idea will belong to the corresponding academic center. Companies will have an option to license the IP from their academic partners; in turn, academic partners can request a license for additional studies after the conclusion of the project. Furthermore, academic partners are not restricted from publishing their results or making their IP available to other nonprofit partners for research and teaching.1
The NCATS initiative parallels the initiative launched in 2011 in the UK. The UK’s Medical Research Council (MRC) and London-based AstraZeneca launched a mechanisms-of-disease grant program providing academics access to 22 AstraZeneca compounds. Alongside the compounds, AstraZeneca has deposited vast amounts of information regarding the compounds into the public domain. Through this partnership with the MRC, AstraZeneca has been able to efficiently manage more than 100 proposals submitted from 37 UK institutions. The goal once again is to progress some of these studies to more advanced trials.3 The rights to IP generated using the compounds will vary from project to project, but generally AstraZeneca will retain its existing rights relating to the compounds, and any new research findings will be owned by the academic institution.4
Several academic and nonprofit initiatives have similarly launched drug-repurposing programs to address rare and neglected diseases. In these programs, the open-source model permits for the sharing of data, resources, compounds, clinical molecules, and small libraries, as well as screening platforms in the search for new indications for old drugs and/or failed candidates. Clearly, at the core of drug-repurposing activities is collaboration. A variety of initiatives underway for drug repurposing, including those targeting rare and neglected diseases, are discussed in this review and provide insight into the stakeholders engaged in drug-repurposing discovery, the models of collaboration used, and the open sharing of knowledge, tools, and I P. In the case of neglected tropical diseases (NTDs), it is suggested that the development of human capital be a central aspect of drug-repurposing programs. Open-source models can support human-capital development through collaborative data generation, open-compound access, open and collaborative screening, preclinical and possibly clinical studies. Given the urgency of drug development for NTDs, this review suggests mechanisms to extend the collaborative, IP-management, and human-capital development elements from current repurposing programs to the NTD arena.
Open-source model deployment across the drug-repurposing value chain
Drug repurposing involves finding new indications for existing drugs or potential drug candidates. Drugs or candidates include those in clinical development whose mechanism of action is relevant to multiple diseases; drugs that have failed to demonstrate efficacy for a particular indication during phase II or III trials but have no major safety concerns; drugs that have been discontinued for commercial reasons; marketed drugs for which patents are close to expiry; and drug candidates from academic institutions and public sector laboratories not yet fully pursued.5 These opportunities for drug repositioning can arise from observations, discussions, and other collaborations, including the purposeful development of platforms for drug identification. With the selection of a repurposing approach, a viable target profile is generated for the drug or candidate, and the screening of compounds can occur. Data mining for the identification of potential targets, accessing compounds, and then screening these compounds all provide opportunities for the exploitation of the open-source model (Table 1).
Table 1.
Open-source model use across drug-repurposing activities
| Drug-repurposing activity | Employment of open-source model | Examples of initiatives |
|---|---|---|
| Data mining for potential targets | Open databases providing target and drug profiles | DrugBank, Potential Drug Target Database, Therapeutic Target Database, SuperTarget, Open PHACTS |
| Data mining for potential candidates | Open databases providing chemical structural information | PubChem, ChMBL, ChemSpider, Orange Book – Discontinued Drug Products, FDA Rare Disease Repurposing Database |
| Search for potential compounds | Open compound repositories/chemical libraries | NCATS initiative; MRC/AstraZeneca partnership; Johns Hopkins Malaria Research Institute collaboration, CWHM, TRND, Wisdom Re:Search, OSDD |
| Compound screening | Shared screening tools/platforms | JHCCSI, NCATS-Lilly partnership, Sanford-Burnham Medical Research Institute screening program |
Abbreviations: Open PHACTS, Open Pharmacological Space; FDA,US Food and Drug Administration; NCATS, National Center for Advancing Translational Sciences; MRC, Medical Research Council; CWHM, Center for World Health and Medicine; TRND,Therapeutics for Rare and Neglected Diseases; OSDD, Open Source Drug Discovery; JHCCSI, Johns Hopkins Clinical Compound Screening Initiative.
Data mining
Open-source databases such as DrugBank, the Potential Drug Target Database, Therapeutic Target Database, and SuperTarget provide target and drug profiles.6–9 These databases feature drug targets, including protein and active-site structures, association with related diseases, biological functions, and associated signaling pathways.7 SuperTarget provides the additional connectivity between target protein and relations to drugs: specifically, 7,300 relations to 1,500 drugs.8 Similarly, DrugBank, featuring both small- and large-molecule drugs in its database, provides comprehensive information on target diseases, proteins, genes, and organisms on which these drugs act.9
The search for potential compounds in parallel involves a review of various scientific databases to identify candidates within the public domain and/or in the development pipelines of private sector organizations.10 Compound specific databases comprise PubChem, ChEMBL, and ChemSpider, the US FDA’s electronic Orange Book’s Discontinued Drug Products list, and IDMap, with the capacity to reposition marketed drugs to novel targets, as well as provide supportive information on a compound’s chemistry and pharmacology (confirmation of the freedom to operate in a particular arena is provided through linked patent searches).11,12 IDMap allows researchers to map commercial chemicals to possible drug targets, bypassing to some extent the need to query multiple databases, and is freely available to noncommercial users.13
As an integrative platform, the Open Phacts initiative likewise permits multiple sources of publicly available pharmacological and physicochemical data to be intuitively queried. The software and data are available for download under an open-source and open-access model.14 The Open Phacts interface enables searching of compounds by name and chemical structure, in addition to the investigation of pharmacological data by compound, target, and enzyme family. Noteworthy is the fact that the Open Phacts consortium comprises 28 partners from the public and private sectors.14 Ultimately, a fully integrative platform will connect target and compound data repositories with literature and patent databases, screening and collaborative tools, encouraging an understanding of the business and technical landscape for drug repurposing and the dynamic capacity to engage in joint target validation and compound-screening activities.
Finally, the Promiscuous database provides an exhaustive set of drugs (25,000), including withdrawn or experimental drugs, annotated with drug–protein and protein–protein relationships compiled from public resources. Measures of structural similarity for drugs and known side effects can be easily connected to protein–protein interactions to establish and analyze biological networks (as well as the cross-specificity of molecules for a variety of targets), thereby providing an opportunity for drug repositioning based on a network-based approach.15 Emig et al discuss a network-based approach for the prediction of drug targets for a given disease. Their method allows both repositioning drug targets known for other diseases to the given disease and the prediction of unexploited drug targets that are not used for the treatment of any disease. These researchers further demonstrate the ability to identify promising candidates for such drug-target repositioning.16 The links between seemingly unrelated diseases can be identified, leading to the emergence of unanticipated drug-target candidates.17 Wu et al extend the discussion to the use of a network-based strategy from single-drug repositioning to the interaction effects discovered from the usage of drug combinations.15
Compound access
Beyond the databases is the need to access drugs or candidates. To address this challenge, several initiatives are under way with the intention of creating chemical libraries of existing drugs/candidate compounds and providing access to interested researchers.5 In the area of rare and/or neglected diseases, these initiatives include the Johns Hopkins Pharmacology and the Malaria Research Institute collaboration, the Center for World Health and Medicine (CWHM) at Saint Louis University, the Institute for Rare and Neglected Diseases Drug Discovery (iRND3), the FDA’s Rare Disease Repurposing Database initiative, the NIH collection available through the Therapeutics for Rare and Neglected Diseases (TRND) program, Wisdom Re:Search, the nonprofit Cures Within Reach (formerly known as Partnership for Cures), and the Learning Collaborative.
Compound screening
With access to compounds, several platforms can be used for screening out the best candidates. Padhy and Gupta5 discuss the fact that the platforms can be novel medium- or high-throughput systems using in vitro binding assays, cell-based assays including those genetically engineered to mimic diseases, proprietary multiplexing in vivo assays, or small organism-based screening systems.18 Here, the open-source model can provide value not only in terms of permitting access to technological platforms and the standardization of data generation but also collaborative screening as a means of developing the requisite screening skills.
The NCATS Pharmaceutical Collection (NPC) is a comprehensive publicly available database and is a physical sample collection. In an attempt to support new drug discovery and extend the value of the collection, the NCATS and Lilly Research Laboratories agreed in 2012 that the NCATS Pharmaceutical Collection of 3,800 approved and investigational medicines would be screened using Lilly’s Phenotypic Drug Discovery (PD2) panel.19 The PD2 assay panel, part of Lilly’s Open Innovation Drug Discovery platform, consists of human disease pathway-related assays for a variety of diseases. The assays are designed to reveal novel mechanisms or pathways for prospective medicines and – as part of this collaboration – approved medicines.19 The intention is to make the results available to the public. However, a medicine demonstrating activity against a disease pathway of interest to an investigator could be the impetus for a new partnership between that investigator and the owner of the medicine. Specifically, an agent with activity in a particular disease pathway might serve as a starting point for additional research to create a new treatment.2
Roche equivalently wants to find external researchers who can suggest screening experiments for its set of compounds. Collaborators will first receive the compounds and their molecular weights. If they uncover any significant findings, more information will be shared. Roche and the partner can then agree to the next steps, which might include publishing results, further experimentation, and/or a development plan. Projects may target rare or orphan diseases where potential partners would have expertise in disease biology and disease modulation in a target-patient population.20 In 2012, Roche announced that it would make available to the Broad Institute a collection of 300 compounds to identify new uses for compounds (using the institute’s screening technology) that originally failed to meet critical phase II milestones or whose development was halted for strategic reasons.21 While seemingly geared toward an open-innovation model, such public–private partnerships are relevant for NTD drug repurposing where the open-access model dominates.
For instance, the Johns Hopkins Clinical Compound Screening Initiative (JHCCSI) supports collaborative drug screening, including for NTDs. The JHCCSI was launched in 2002 as a joint collaboration between Johns Hopkins Pharmacology and the Malaria Research Institute at the Johns Hopkins Bloomberg School of Public Health. The collection of existing drugs is being screened by several collaborators at Johns Hopkins and globally on a variety of diseases, including malaria. To accelerate drug discovery, researchers at Johns Hopkins have created a library of 2,687 existing drugs and have screened for inhibitors of the human malaria parasite Plasmodium falciparum.22 The initiative has the long-term goal of acquiring and adding to its collection each of the 11,000 drugs ever used in medicine for possible repurposing.22
Learning from current drug-repurposing initiatives in the public and private sectors
With looming patent expiries for several blockbusters, drug developers are looking to drug repurposing, including for rare diseases, to augment their pipelines. To this effect, we see the formation of dedicated research-and-development (R&D) centers, programs for repurposing drugs, and the use of public resources and programs for drug repurposing. Given the number of programs in place in both the private and public sectors for drug repurposing, it is expected that valuable lessons with respect to incentives for participation, collaboration, open-source model usage, IP management, and human-capacity development can be transferred to repurposing for NTDs (Table 2–3). In many cases, initiatives targeting rarer diseases are discussed as a means of lessons transfer and ecosystem development for NTDs.
Table 2.
Drug-repurposing initiatives sponsored by public sector, private sector, and nonprofit organizations
| Public sector-driven initiative for drug repurposing | Private sector-driven initiative for drug repurposing | Nonprofit-driven initiative for drug repurposing |
|---|---|---|
| ➢ NCATS initiative* | ➢ New Indications Discovery Unit at the Novartis Institute for Biomedical Sciences | ➢ Center for World Health and Medicine at Saint Louis University* |
| ➢ MRC/AstraZeneca partnership* | ||
| ➢ Clinical and Translational Science Award (CTSA) Pharmaceutical Assets Portal* | ➢ Bayer Healthcare’s Common Mechanism Research | ➢ WIPO Re:Search consortium* |
| ➢ The Johns Hopkins Clinical Compound Screening Initiative* | ||
| ➢ NCATS-Lilly partnership | ➢ Pfizer’s Indications Discovery Unit (IDU) | |
| ➢ CSIR-OSDD Initiative* | ➢ Roche-Broad Institute partnership | ➢ Cures Within Research* |
| ➢ The Learning Collaborative* | ||
| ➢ Sanford-Burnham Medical Research Institute* |
Note:
Programs that also focus on drug repurposing for rare and/or neglected diseases.
Abbreviations: NCATS, National Center for Advancing Translational Sciences; MRC, Medical Research Council; CSIR, Council of Scientific and Industrial Research; OSDD, Open Source Drug Discovery; WIPO, World Intellectual Property Organization.
Table 3.
Models of collaboration, IP management, and human-capacity development employed by public and private sector-driven repurposing programs
| Models of collaboration | Initiatives | Models of IP management | Initiatives | Models of human-capacity development | Initiatives |
|---|---|---|---|---|---|
| Data generation | NCATS initiative; IDMap; Open PHACTS; International; FDA’s Rare Disease Repurposing Database (RDRD) initiative; OSDD | Open source | MRC/AstraZeneca partnership; Open PHACTS; Johns Hopkins Clinical Compound Screening Initiative | Collaborative compound synthesis | CWHM pilot program; D3 |
| Sharing and/or joint development of compounds | NCATS initiative; MRC/AstraZeneca partnership; Center for World Health and Medicine (CVVHM); Johns Hopkins Clinical Compound Screening; Sanford-Burnham Medical Research Institute; OSDD | IP separation based on usage | NCATS initiative | Collaborative opportunities at academic institutions | Via the Clinical and Translational Science Award (CTSA) Pharmaceutical Assets Portal |
| Joint model development | NCATS initiative; MRC/AstraZeneca partnership; Sanford-Burnham Medical Research Institute | Collaboration as an option to offer license | NCATS initiative; MRC-AstraZeneca Partnership; Lilly Phenotypic Drug Discovery (PD2); Roche-Broad Institute partnership; Learning Collaborative | Collaborative research at pharmaceutical organizations | WIPO Re:Search |
| Collaborative drug screening | Lilly Phenotypic Drug Discovery (PD2); Johns Hopkins Clinical Compound Screening Initiative; Sanford-Burnham Medical Research Institute; OSDD | Patent pool | WIPO Re:Search | ||
| Clinical development | MRC/AstraZeneca partnership; Cures Within Reach; OSDD | Royalty-free, open licensing | WIPO Re:Search |
Abbreviations: Open PHACTS, Open Pharmacological Space; FDA, US Food and Drug Administration; NCATS, National Center for Advancing Translational Sciences; MRC; Medical Research Council; OSDD; Open Source Drug Discovery; CWHM, Center for World Health and Medicine; WIPO, World Intellectual Property Organization.
Public sector initiatives
The NPC, also known as the NIH Chemical Genomics Center (NCGC) Pharmaceutical Collection, is a publicly accessible collection of approved and investigational molecular entities for high-throughput screening. For rare and neglected diseases, NCATS provides access to its set of approved drugs and bioactives through the TRND program. Of the nearly 2,750 small-molecule entities that have been approved for clinical use by the US, European Union, Japanese, and Canadian authorities, NCATS currently possesses 2,500, with approximately 1,000 additional investigational molecular entities as part of its screening collection. The collection was sourced from chemical suppliers, specialty collections, and through custom synthesis. All data generated through this effort will be deposited openly into PubChem.23 Alongside the compounds, the NPC has over 200 assays for drug targets.24 Virtual screening can be performed by any investigator worldwide; additionally, researchers are encouraged to inform the NCGC of their successes (and failures) using the NPC resource whilst depositing their results to PubChem.25 Laboratory-based screening is conducted at the NCGC in conjunction with any researcher who has a disease-relevant assay.23
The International Rare Disease Research Consortium, sponsored by the NIH, includes 170 research sites with the purpose of producing 200 treatments (new and repurposed therapies targeting rare diseases) by 2020.24,25 Members have established and provide access to harmonized data and samples, and collaboratively perform molecular and clinical characterization of rare diseases with the expressed outcome of joint diagnostic and therapy development.25 While the data included in the FDA’s Rare Disease Repurposing Database is a reconfiguration of already FDA-released information, it offers researchers a useful tool for finding opportunities to develop niche therapies that are already well advanced through development.26 Notably, the drugs have already been subjected to preclinical testing and are deemed to be pharmacologically active, effective, and safe in some clinical context, including as market-approved products for the treatment of other diseases.27
At the end of 2012, the MRC/AstraZeneca alliance, in which AstraZeneca made 22 of its chemical compounds available free of charge to scientists, announced funding for 15 research projects. Scientists will use the compounds to study a variety of conditions, also comprising rarer conditions such as motor neuron disease and muscular dystrophies. Eight of the projects will involve clinical (human) trials of potential new therapies, and seven will focus on earlier work in laboratory and animal models.4
The Clinical and Translational Science Award (CTSA) Pharmaceutical Assets Portal project provides a unique model for linking global stakeholders engaged in drug repurposing, providing a replicable model for NTD drug-repurposing activities. The Pharmaceutical Assets Portal facilitates industry–academic collaborations for discovery of new indications for compounds no longer being developed by pharmaceutical companies.28 The portal provides access to collaborative and resource-management tools, such as a tool for connecting partners and members (Foci of Expertise [FoX]), a funding arm (Partnership for Cures), an IP arm (the University–Industry Demonstration Partnership), and facilities to house, maintain, and distribute the discontinued compounds (with the opportunity to house and distribute the NCGC’s pharmaceutical collection through the CWHM).28 FoX links three search categories (target gene/protein, authors, disease) to enable users to readily identify potential collaborators, relying on information derived from Medline (publications), NIH Research Portfolio Online Reporting Tool – Expeditures and Results (funded grant abstracts), and Reactome (gene–gene interactions).28 Through its Indications Discovery Unit, Pfizer has contributed significantly to the Pharmaceutical Assets Portal as a means of leveraging the expertise of external researchers. The management of IP and material transfer agreements, the selection of projects for funding, and the housing and distribution of discontinued compound libraries have been effectively addressed through the portal and its partners, with the objective of eliminating the barriers associated with access to compounds.28 The portal and its network of members will continue to engage the pharmaceutical community to determine the compounds that may be available to the research community.28
Nonprofit initiatives
Several nonprofit organizations have joined the effort to repurpose drugs, such as the CWHM at Saint Louis University, the World Intellectual Property Organization (WIPO) Re:Search consortium, the JHCCSI, Cures Within Research, the Learning Collaborative, and the Sanford-Burnham Medical Research Institute.
The CWHM at Saint Louis University was founded in 2010 by former Pfizer scientists. The CWHM and others repurposing for rare and neglected diseases are looking for access to compound databases and libraries or are creating their own libraries in partnership with researchers in China, South Africa, and the US.14 Interestingly, through pilot programs at universities, students enrolled in advanced chemistry courses participate in the synthesis of compounds.20 The CWHM is among several nonprofit organizations, companies, and academic institutions participating in the WIPO Re:Search consortium. The WIPO has created a database of available IP assets – including compounds, technology, and regulatory data – to support research on neglected diseases. The CWHM is leveraging the agreements established by the WIPO to discuss and encourage pharmaceutical organizations to share data, advanced clinical molecules, or even small diverse libraries.20
WIPO Re:Search provides the opportunity to repurpose available IP assets for use in treating NTDs. AstraZeneca, for instance, has made its entire patent portfolio accessible through the platform. While much of the AstraZeneca IP relates to projects slated for commercialization, other material is not under development. AstraZeneca hopes to use the platform to review past projects and rework past assets. Under the WIPO Re:Search guiding principles, members agree to make IP assets and know-how available to qualified researchers of NTDs, with no licensing fee and on a royalty-free basis. Any products resulting from this research will also be available via royalty-free sales in least developed countries.29
The JHCCSI was launched in 2002 as a joint collaboration between Johns Hopkins Pharmacology and the Malaria Research Institute at the Johns Hopkins Bloomberg School of Public Health. The JHCCSI library is currently expanding to include every available drug used clinically in phase II trials or approved for sale on the market. The JHCCSI library will be made available to any researcher interested in screening existing drugs for targeting diseases of the developing world.22,30 Chong et al, for example, have demonstrated in two mouse models of malaria that the antihistamine astemizole and its principal human metabolite are promising new inhibitors of chloroquine-sensitive and multidrug-resistant parasites.22
Cures Within Reach works to repurpose drugs and devices for both common and rare disorders that currently do not have effective treatments. Through a network of partnerships with research institutions and funders, Cures Within Reach has been able to source, select, and fund pilot clinical research projects that test existing and approved drugs and devices for new purposes, or combine existing treatments in novel ways.31 Cures Within Reach partners total 30, comprising those from academia, patient-advocacy groups, the pharmaceutical and biotech industries. Specifically, Cures Within Reach finds drugs using this network, matches projects with its funders, conducts clinical testing in patients, and works alongside physicians to determine clinical usage.20 Cures Within Reach provides a noteworthy framework for network-model usage for rare and NTD drug-repurposing discovery and downstream commercialization activities.
Exploiting the network model is the Learning Collaborative, a collaboration between the NCGC and its TRND program, the Leukemia and Lymphoma Society, and the University of Kansas Cancer Center. From the outset, the founding partners established a memorandum of understanding to clearly define collective objectives, expectations, and deliverables. Partnerships with pharmaceutical and biotechnology companies provide a unique avenue to license drug therapies developed by the Learning Collaborative to for-profit partners.32 While repurposing by the Learning Collaborative does not revolve around the open-source model, Weir et al discuss that the lessons learned are of value, as the network model is scaled up and applied across a range of diseases, taking into account funding sources, technology transfer, IP management through repurposing off-patent and abandoned drugs, and developing licensing agreements for interested for-profit partners.32
Finally, scientists at Sanford-Burnham Medical Research Institute (San Diego, CA, USA and Orlando, FL, USA) have the objective of creating the world’s most comprehensive drug library. As in the case of the CWHM, Sanford-Burnham will create, house, maintain, and share the library with other institutions. In addition, researchers are using “disease in a dish” stem cell models to test the efficacy of these compounds against a number of human diseases.33 Through this methodology, researchers use skin samples from a patient or healthy volunteer, revert them back developmentally to stem cells, and induce the desired cell-type differentiation, with the desired genetic and epigenetic makeup.33 These differentiated cells are then used to screen and repurpose drugs. Scientists envision the greatest potential from the “disease in a dish” model for screening drugs targeting genetic diseases. A network of experts across a variety of disease arenas, clinical partnerships, and patient sample-access opportunities enable the institute to be among the most advanced nonprofit drug-screening programs.
Private sector initiatives
Through the NCATS and MRC drug-repurposing initiatives, we see the active participation of the private sector in open and collaborative discovery. Pfizer, AstraZeneca, Eli Lilly, Abbott, Bristol-Myers Squibb, GlaxoSmithKline, Janssen Pharmaceuticals, Sanofi, and Roche are among the larger pharmaceutical organizations sharing knowledge, compounds, and other technology with their public sector counterparts. Independently, Novartis, Bayer, and Pfizer have established dedicated units: the New Indications Discovery Unit at the Novartis Institute for Biomedical Sciences, Bayer Healthcare’s Common Mechanism Research, and Pfizer’s Indications Discovery Unit. All of the companies seek collaboration opportunities with academic and external partners, consisting of the joint sharing of resources and expertise between partners.
Of importance, however, is the need to consider the downstream value of such collaborations for private sector stakeholders. The drivers for repurposing a drug or a failed advanced-stage candidate drug are varied, from discovery, to validation, to commercial value creation. The commercial opportunities will depend on the origin of the drug, how much public and/or accessible data exist, and how well the repurposer can exploit the new value chain created by a successfully repurposed drug.20 Exploitation will commence with the filing of IP rights, which in itself is a process fraught with difficulties associated with prior art examination, type of patent, scope of patent, the existence of other blocking patents, and policy with respect to new-use patents. Regardless, in the case of downstream commercial exploitation, transparency of intentions with respect to appropriation of knowledge and IP assignment must be clarified, particularly when engaging in public–private partnerships.
Signaling its commitment to open-drug repurposing, Pfizer has provided scientists at Washington University School of Medicine in St Louis with access to information regarding more than 500 pharmaceuticals and pharmaceutical candidates in a partnership that focuses on discovering new uses for these compounds. To encourage the exchange of ideas, Pfizer has developed an online portal through which certain investigators will have access to information about Pfizer’s proprietary compounds. The compounds have been extensively studied, and their mechanisms of action are well-understood.34 In 2011, Pfizer and the Children’s Hospital in Boston announced a new collaboration to identify potential drug therapies for Duchenne muscular dystrophy. Through this partnership, Pfizer’s Orphan and Genetic Diseases Research Unit provided researchers at the Children’s Hospital access to select proprietary compounds, together with relevant data about these compounds. Pfizer’s medicinal chemistry expertise is used in a supportive capacity.35 Here, we see the potential for open collaborations via Pfizer’s Indications Discovery Unit and Orphan and Genetics Diseases Research Unit aimed at repurposing drugs for rare and orphan diseases.
Alongside the need for oversight of commercial interests is the need of oversight of repurposing to take advantage of the incentives currently offered to encourage rare and orphan disease drug development. Grants, research-design support, FDA fee waivers, tax incentives, and orphan drug-market exclusivity are the main incentives for orphan drug R&D.36 The US Orphan Drug Act also permits freedom for sponsors to recycle previously discontinued products. As a consequence, drugs previously discontinued due to safety concerns can be given a new life as orphan drugs with much more restricted applications. For example, Wellman-Labadie and Zhou found in their study that 26 discontinued products were found to have later obtained orphan designations, with 14 of these achieving orphan drug approval. In such cases, the authors contend that old pharmaceutical agents could potentially benefit from drug-development incentives as later orphan drugs and demand higher market prices, even though these pharmaceutically active agents were once available either as branded or generic drugs.37 To the suggested list of reforms comprising price regulation, subsidy paybacks for profitable drugs, the establishment of an international orphan drug office to harmonize regulations across markets, and reforms to the US Orphan Drug Act should be added the establishment of an oversight function for drug repurposing targeting orphan diseases.36–39 While the intention of this article is to review models of open-source drug repurposing, with the possibility of downstream commercial value creation, oversight beyond data management and access will become paramount.
Models of collaborative and open-source repurposing
Moving from early biological research to compound development and then clinical testing, the notion of openness changes: beginning with open access to information, materials, tools, and compounds, including public sector entities, private sector stakeholders, and uniquely patients providing sample donations; evolving to dissemination of newly created resources via open licensing; and emerging as open innovation between stakeholders acting on the periphery of what can be coined as the open health commons.40
With respect to drug-repurposing initiatives, the usage of the open-source model is increasingly being employed to ensure broad access to data, compounds, patient samples, and in some cases technological platforms. Predominantly, we see the sharing and the joint development of compounds, the joint task of drug screening through shared technological platforms, the creation of patent pools, and collaboration-enabling platforms. The public–private partnership structural and large-scale network models allow public and private sector stakeholders to provide and leverage the necessary resources in support of open-source models for drug repurposing. The transition then to the open-innovation model in many cases arises through public–private partnerships, with the expressed intent of licensing or offering an option to license from or to partners.
IP model development
A variety of models for IP management have been explored by the initiatives. The open-source model has typically been employed for data regardless of source – public or private sector. In the case of compound access and screening, the public sector initiatives tend to broadly apply the open-source model by encouraging the sharing of compounds, screening platforms, and/or models. For the public–private partnerships, a variety of options are employed, such as IP ownership on new uses of compounds for academic partners, patent pools for compounds, technology and data deposited into the pools, and the royalty-free, open licensing for drug development targeting NTDs. In the case of drug repurposing, pooling of off-patent compounds and the donation of abandoned molecules to a common pool have been suggested by a variety of stakeholders.41
According to NIH director Francis S Collins, NIH has undertaken the suggested task of finding appropriate abandoned compounds, making data resources and expertise available, creating master agreements, and fostering partnerships. Oprea et al advocate that for orphan and NTDs, a network strategy should continue to be encouraged to develop a “global” system with open-access databases and compound repositories, alongside agreed-upon strategies with respect to IP protection and downstream development activities.42 Muthyala43 offers the novel suggestion that patient organizations should be able to take a lead in assembling drug-discovery patents for rare diseases. Indeed, for NTDs, nongovernmental organizations have typically adopted this role.
Human-capacity development
Similar to the CWHM pilot program at universities to permit students enrolled in advanced chemistry courses to participate in the synthesis of compounds is the Distributed Drug Discovery initiative (D3). D3 is a low-cost strategy to accelerate the discovery of drugs to treat NTDs, such as tuberculosis, leprosy, leishmaniasis, dengue fever, and Chagas’s disease.44 D3 uses a distributed educational approach to synthesize the candidate molecules. Undergraduate and graduate chemistry students from around the world synthesize subsets of these candidate molecules as part of their normal training in synthetic chemistry (providing both education and employment opportunities to those in developed and developing markets). Currently, students at IUPUI, the University of Indianapolis, and universities in Poland, Russia, and Spain have demonstrated their ability to make the molecules (or portions of the molecules) identified as potential candidates for drug discovery. The next step involves the evaluation of the molecules made by the students. As of 2011, 91 molecules created by students at IUPUI had been deposited at the NIH Small Molecule Repository for potential screening to confirm if they merit further movement into the drug-development pipeline. Such opportunities can and should be extended to those researchers in disease-endemic countries for the purposes of NTD drug repurposing. The open deposit of such molecules can not only ensure broad accessibility to other researchers but also serve as a strategy of governance, ensuring that students are involved in a transparent educational process and not a commercialization process with the associated legal and economic implications.45
Using the Pharmaceutical Assets Portal’s FoX system, researchers are able to start the process of organizing a symposium for collaborative research on a given disease at a specific institution, and can find key opinion leaders, including those that would be interested in repurposing compounds.28 One can envision an extension of this platform to NTDs, with membership consisting of universities from disease-endemic countries, a tool for connecting researchers from the public and private sectors focused on a particular disease arena, or with specific candidate-development interests, nongovernmental organizations with the expressed function of funding drug-repurposing projects and with the expertise to negotiate appropriate IP rights, and a public or non-profit sector partner that can house, maintain, and distribute compounds for screening and development activities. One possibility is the Malaria Research and Reference Reagent Resource Center (MR4), established in 1998 to provide a central source of quality-controlled malaria-related reagents and information to the international malaria-research community. MR4 serves to: provide improved access to malaria-associated reagents; provide improved authentication of materials; participate in the standardization of materials and their subsequent usage; act as manager for the distribution and quality control for reagents deposited by investigators; serve as a knowledge resource for malaria-research information and protocols; and generate new reagents as well as materials as knowledge advances.46 Special arrangements are possible for in-house reagent development or for the coordination of research collaborations. Aside from providing reagents, MR4 promotes technology transfer and fosters scientific exchange between new and established malaria researchers.47 The suggestion can be put forward that the merged models offered by the Pharmaceutical Assets Portal and the MR4 repository should be scaled across neglected tropical diseases.
Finally, WIPO Re:Search offers the opportunity for NTD researchers to work directly with scientists at pharmaceutical companies to advance R&D on these diseases. By providing a searchable, public database of available IP assets, information, and resources, WIPO Re:Search has the goal to facilitate new partnerships with organizations that conduct research on treatments for NTDs.29 Worth noting is that WIPO Re:Search has from the outset attempted to involve institutions from emerging and developing countries. A closer examination of the IP rights and downstream drug-access policies from such collaborations is warranted to ensure a transparent collaboration process.
The current state of affairs for neglected tropical diseases
The landscape of open-source initiatives in NTDs is changing fast, with the current common pool of assets increasing in value. Across the NTD discovery value chain, we see the sharing of publications, biological data, chemical structures, tools, software, and I P. From the open-access journal PLOS Neglected Tropical Diseases, to ChEMBL – a repository for open access to primary screening and medicinal chemistry data directed at NTDs (chemical structure deposits have been made by GlaxoSmithKline, Novartis, and the Drugs for Neglected Diseases Initiative to ChEMBL) – collaboration tools used by the Indian Open Source Drug Discovery (OSDD) initiative, to WIPO Re:Search, the expectation is that the open-source strategy will encourage multiple stakeholders to enter this arena to validate knowledge and generate downstream partnerships.48,49
Drug repurposing for NTDs
With reference to drug repurposing, the WHO Special Programme for Research and Training in Tropical Diseases, the Medicines for Malaria Venture, the Global Alliance for TB Drug Development, Drugs for Neglected Diseases, and the OSDD initiative all view drug repositioning as an attractive option for providing cost-effective and timely access to drugs to patients of the developing world.5,50,51
Several cases exist of both ongoing repurposing programs and completely repurposed drugs that target NTDs (Table 4). Researchers at the University of British Columbia (UBC) recently demonstrated that the avermectin commonly used in the developing world to eliminate the parasitic worms that cause river blindness and elephantiasis actually killed the bacteria that cause tuberculosis, including drug-resistant forms.51 Three members of the drug family – ivermectin, selamectin, and moxidectin – are active against mycobacterial species, including multi- and extensively drug resistant Mycobacterium tuberculosis.52 The international collaboration was led by scientists in UBC’s Department of Microbiology and Immunology associated with UBC’s Centre for Tuberculosis Research and the Neglected Global Diseases Initiative.
Table 4.
Examples of repurposed drugs for NTDs
| Drug | Original indication | Repurposing/repurposed for NTD |
|---|---|---|
| Astemizole | Antihistamine | Malaria |
| Avermectin | River blindness and elephantiasis | Tuberculosis |
| Miltefosine | Antineoplastic | Leishmaniasis |
| Tamoxifen | Anticancer | Leishmaniasis |
| Amphotericin B | Antifungal | Leishmaniasis |
| Eflornithine | Anticancer | African sleeping sickness |
| Phosphodiesterase-inhibitor analogues | Erectile dysfunction | African sleeping sickness; Chagas’s disease |
Abbreviation: NTDs, neglected tropical diseases.
Miltefosine was developed as an antineoplastic drug in the early 1980s, but its antileishmanial properties led to its approval as the first oral drug for the treatment of visceral leishmaniasis in India.53 Tamoxifen, a drug used to treat breast cancer, has been shown to be effective in experimental models of cutaneous and visceral leishmaniasis.54 Amphotericin B, used to treat fungal infections, has similarly been repurposed for leishmaniasis.55
Eflornithine, which was developed as an anticancer drug, has been successfully introduced for treating African trypanosomiasis.56 The FDA approved eflornithine for the treatment of sleeping sickness in 1990. Additionally, eflornithine is used for the treatment of Pneumocystis carinii pneumonia in AIDS patients and for treatment of anaplastic glioma.57 Repurposing of phosphodiesterase inhibitors against trypanosomal phosphodiesterases represents an approach for the discovery of drugs for African sleeping sickness and Chagas’s disease.42,58
The Sandler Center, in collaboration with the University of California San Francisco Small Molecule Discovery Center, focuses on parasite targets that are homologous to human targets. The intention is to repurpose leads rather than drugs. The driving factors for the repurposing of leads rather than drugs are the increased optimization and clinical efficacy of leads that may not always be possible for drugs already approved for other indications.59 Medicinal chemistry is used to optimize the leads specifically for an antiparasitic indication. Recently, however, the center made the decision to expand its library to include both FDA-approved drugs as well as compounds that have passed phase I human clinical trials but have failed to reach market for various reasons other than safety issues.59
The open-source model and NTD drug repurposing
OSDD is a Council of Scientific and Industrial Research (CSIR) Team India consortium that provides a global platform based for collaborative discovery work into novel therapies for NTDs, including malaria, tuberculosis, and leishmaniasis. OSDD brings together open-source, crowd-source, open-science, open-innovation, and product-development partnership concepts on the same platform.60,61 OSDD allows for the sharing of information, including but not limited to ideas, articles, papers, and other literary work, data, software, applications, notes, results of experiments, patented inventions, and other materials submitted by users. Participants formally register and agree to the terms outlined by OSDD, including those referring to the management of knowledge assets and the associated I P. These participants post innovative projects online, actively engage in ongoing scientific projects, share positive and negative results, and participate in the review process of other projects posted on the OSDD portal. All project documents and tracking are done through the OSDD portal.60,61
OSDD has designed a collaborative platform consisting of a wiki-based genome-annotation service, a platform for open-source tools for drug discovery, integrative genomic maps, an open-access document repository, and a metadata archive and search engine for open access to theses and dissertations. Contributions can be made in the form of database, molecular library, or other biological resource donations, laboratory access or the sharing of technological capabilities, computing time, bandwidth, or other computation-resource access, and monetary or in-kind rewards for the contributions themselves. Collaboration spaces essentially serve to develop human capital as participants post ideas, discuss methodologies and possible solutions to the problems, bookmark web-based resources for drug discovery, and share experimental data.60,61
The CSIR laboratories are currently engaging in large-scale screening of molecules against tuberculosis. As a consequence, CSIR laboratories have opened up their small-molecule libraries, from synthetic as well as natural sources. In parallel, contributions of small molecules to the OSDD Open Access Repository are encouraged. Data have been shared with Jubilant Chemsys, TCG Lifesciences, Sugen Life Sciences, PREMAS Biotech, and Vimta Labs to further the drug-development process by especially focusing on repositioning of patented or off-patent drugs.62
Drug repurposing for NTDs: an ecosystem of partners, assets, and policies
Drug repurposing for NTDs can be best described as an ecosystem of partners, assets, and strategic policies and technologies.63–66 Coles and Cloyd discuss that academic institutions are increasingly taking advantage of the innovation gap on rare and orphan disease drug development by taking a more central role across the drug-discovery and development value chain.65 For example, many academic institutions are making substantial investments in research infrastructure, establishing new programs or restructuring established units, adding faculty and staff with expertise in drug discovery and development and regulatory support, and streamlining licensing procedures to create incentives for faculty to commercialize their discoveries. Coles and Cloyd provide a typology of academic models based on the focus of research comprising the disease-focused model, the discovery-focused model, the industry collaboration-focused model, and the commercialization-focused model. Associated with these models are supporting mechanisms provided by an ecosystem of partners, from institutional to government to industry-funded resources and infrastructure, and the increased use of research consortia to connect these partners.67 While we contend that these models have their relevance for drug research and repurposing for NTDs, as Coles and Cloyd discuss, a parallel need will arise for increased funding to enhance both infrastructure and human-capacity development to support the increased role of academia in the repurposing arena (not only in terms of research capacity but also policy-development capacity to manage the commercialization aspects of NTD drug-repurposing programs).67
Allarakhia and Walsh68 discuss the fact that several models currently exist in the literature to describe such an ecosystem namely: the National Systems of Innovation model and the Institutional Analysis Development model. Models such as National Systems of Innovation do inform us of the interactions between the public and private sectors as determining the innovative performance of such organizations, including the import, modification, and diffusion of new technologies, and provide the framework within which governments form and implement policies to influence the innovation process.63,64,69 The Institutional Analysis Development framework describes such interactions, the diffusion of knowledge and technology, and the policies at the operational level to manage the interactions and at a collective level to enable the formation of collaborations alongside permitting the effective management of knowledge and technology.70 Noteworthy also is the emphasis in National Systems of Innovation models on mapping the knowledge flows of both tacit knowledge via the movement of human capital and of codified knowledge via publications and patents.71 In the Institutional Analysis Development model, the level of analysis is rather at the resource-attribute level, with the correlation between participant type and resource type as impacting such resource-governance strategy as IP-management strategy.
Regardless of the framework utilized to describe the interactions between participants and the level of analysis of flow of knowledge-based assets, it is imperative to consider the impact of these interactions on both technology development and – as emphasized in this review – skill development for NTD drug repurposing. Figure 1 provides a map of the stakeholders engaging in drug repurposing for NTDs, the overarching strategic policies to manage drug-repurposing programs, the outcomes from drug repurposing, and the reinvestment of outcomes into new drug-repurposing program development. Here, we see the interactions once again between resource sharing and knowledge sharing (notably through the open-source model), candidate advancement, and human-capacity development. While the model can be generalized across repurposing initiatives, the investment into and then reinvestment of human capacity, specifically NTD repurposing skill sets, is particularly salient to those researchers in disease-endemic countries. Knowledge ultimately moves cyclically to and from various programs in the proposed ecosystem.72
Figure 1.
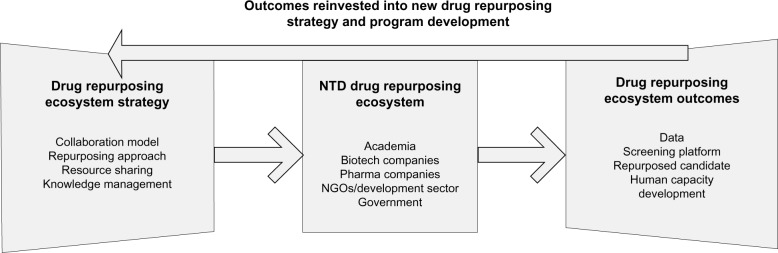
Neglected and Tropical Diseases Drug Repurposing Ecosystem.
Conclusion
An attempt has been made in this article to provide an overview of current drug-repurposing initiatives across the public, nonprofit, and private sectors, with special attention given to those focused on rare disease and NTD drug repurposing. The purpose has been to reveal how the open-source model has been or can be implemented for drug rediscovery. The variety of initiatives using the open-source model, the deposits made, and collaboration models used are all encouraging signs for stakeholders in this arena. Particularly, the connection made between information and communication-technology platforms used to permit open, collaborative drug repurposing and human-capacity development are paramount for the cost- and time-effective advancement of new chemical entities targeting rare and neglected diseases. We see the use of university-based graduate-level collaborative synthesis programs, opportunities for researchers in disease-endemic countries to work collaboratively at pharmaceutical organization sites, the suggested extension of the CTSA Pharmaceutical Assets Portal to neglected diseases with membership including those from universities in disease-endemic countries, and the OSDD initiative merging the open-source, crowd-source, open-science, open-innovation and product-development partnership concepts on the same platform. Recent studies focused on the network-based drug-repositioning strategy not only provide an indication of the value of analyzing the role of targets across unsuspected diseases and the cross-specificity of drugs for multiple targets but also re-analyzing drug-combination usage (ie, drug-cocktail usage to address drug resistance) for drug-repositioning opportunities. Here, the opportunity exists for the discovery of new and off-label uses of drugs when treating patients for multiple diseases, including comorbid diseases, as researchers attempt to understand the underlying biological interactions of comorbidities and target-based interactions between drugs.
Of increased significance is the inclusion of patients and physicians earlier in the drug-repurposing process for clinical usage determination and sample access, respectively. With the transition from the open-source model, the possibility still exists to engage in large-scale open innovation. Transparency Life Sciences (TLS), often cited as the first biopharmaceutical organization based on global open innovation, recently launched its Indication Finder, a survey-based crowd-sourcing tool designed to identify new indications for existing drug candidates. The TLS Indication Finder is initially focused on the 58 compounds currently included in the NCATS endeavor. TLS provides links to background information on the compounds and to relevant external resources to facilitate participation.72 The Indication Finder essentially taps into the scientific crowd to identify new uses for compounds. Survey responses are aggregated, presented to the crowd for analysis and discussion, and then the resulting data are curated by experts.72 Selected candidates have the potential of then being developed using the TLS Protocol Builder, a crowd-sourcing platform utilized for designing more effective clinical trials in partnership with researchers, physicians, and patients. Ultimately, trials will be conducted using remote patient-monitoring and data-capture technologies that minimize the need for inconvenient and costly patient site visits. As in the case of the CTSA Pharmaceutical Assets Portal, we can envision the extension of the TLS crowd-sourcing platforms to neglected disease drug repurposing: using the wisdom of researchers in NTD-endemic countries to suggest new indications for existing compounds or failed drugs, tapping into the crowd to collectively screen compounds for NTD indications, uniquely utilizing the crowd to permit skill development by matching students and researchers to partner research sites, and then crowd-sourcing clinical trial design in disease-endemic countries. The possibilities are endless, keeping in mind the need for openness both in terms of asset (including IP) management and stakeholder engagement.
Footnotes
Disclosure
Dr Minna Allarakhia is a director of the company BioEndeavor, a nonprofit organization with the objective of fostering health innovation-capacity development in emerging and BoP markets.
References
- 1.Alison M. NCATS launches drug repurposing program. Nat Biotechnol. 2012;30(7):571–572. doi: 10.1038/nbt0712-571a. [DOI] [PubMed] [Google Scholar]
- 2.National Center for Advancing Translational Sciences Rescuing and repurposing drugs Available from: http://www.ncats.nih.gov/research/reengineering/rescue-repurpose/rescue-repurpose.htmlAccessed March 21, 2013
- 3.Philippidis A.NCATS MRC take aim at teaching old drug new tricks 2012Available from: http://www.genengnews.com/insight-and-intelligenceand153/ncats-mrc-take-aim-at-teaching-old-drugs-new-tricks/77899618Accessed June 16, 2013
- 4.Medical Research Council Alzheimer’s, cancer and rare disease research to benefit from landmark MRC-AstraZeneca compound collaboration [press release] London: MRC; 2012[Oct 31]. Available from: http://www.mrc.ac.uk/Newspublications/News/MRC008918Accessed March 21, 2013 [Google Scholar]
- 5.Padhy BM, Gupta YK. Drug repositioning: re-investigating existing drugs for new therapeutic indications. J Postgrad Med. 2011;57(2):153–160. doi: 10.4103/0022-3859.81870. [DOI] [PubMed] [Google Scholar]
- 6.Zhu S, Stavrovskaya IG, Drozda M, et al. Minocycline inhibits cytochrome c release and delays progression of amyotrophic lateral sclerosis in mice. Nature. 2002;417(6684):74–78. doi: 10.1038/417074a. [DOI] [PubMed] [Google Scholar]
- 7.Gao Z, Li H, Zhang H, et al. PDTD: a web-accessible protein database for drug target identification. BMC Bioinformatics. 2008;9:104. doi: 10.1186/1471-2105-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Günther S, Kuhn M, Dunkel M, et al. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008;36(Database issue):D919–D922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knox C, Law V, Jewison T, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39(Database issue):D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 11.Ma’ayan A, Jenkins SL, Goldfarb J, Iyengar R. Network analysis of FDA approved drugs and their targets. Mt Sinai J Med. 2007;74(1):27–32. doi: 10.1002/msj.20002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poh A.Breathing new life into old drugs 2008Available from: http://www.bio-itworld.com/issues/2008/nov-dec/chantest.htmlAccessed July 19, 2013
- 13.Ha S, Seo YJ, Kwon MS, Chang BH, Han CK, Yoon JH. IDMap: facilitating the detection of potential leads with therapeutic targets. Bioinformatics. 2008;24(11):1413–1415. doi: 10.1093/bioinformatics/btn138. [DOI] [PubMed] [Google Scholar]
- 14.Open PHACTS [website on the Internet] Available from: http://www.openphacts.orgAccessed March 21, 2013
- 15.Wu Z, Wang Y, Chen L. Network-based drug repositioning. Mol Biosyst. 2013;9(6):1268–1281. doi: 10.1039/c3mb25382a. [DOI] [PubMed] [Google Scholar]
- 16.Emig D, Ivliev A, Pustovalova O, et al. Drug target prediction and repositioning using an integrated network-based approach. PLoS One. 2013;8(4):e60618. doi: 10.1371/journal.pone.0060618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Eichborn J, Murgueitio MS, Dunkel M, Koerner S, Bourne PE, Preissner R. PROMISCUOUS: a database for network-based drug-repositioning. Nucleic Acids Res. 2011;39(Database issue):D1060–D1066. doi: 10.1093/nar/gkq1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giacomotto J, Ségalat L. High-throughput screening and small animal models, where are we? Br J Pharmacol. 2010;160(2):204–216. doi: 10.1111/j.1476-5381.2010.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Institutes of Health NIH and Lilly to generate public resource of approved and investigational medicines [press release] Bethesda(MD)NIH; 2012[Mar 13]. Available from: http://www.nih.gov/news/health/mar2012/ncats-13.htmAccessed March 21, 2013 [Google Scholar]
- 20.Thayer AM. Drug repurposing. Chem Eng News. 2012;90(40):15–25. [Google Scholar]
- 21.Genetic Engineering and Biotechnology News Roche, Broad Institute seek new uses for old drugs 2012Available from: http://www.genengnews.com/gen-news-highlights/roche-broad-institute-seek-new-uses-for-old-drugs/81247689Accessed June 16, 2013
- 22.Chong CR, Chen X, Shi L, Liu OJ, Sullivan DJ., Jr A clinical drug library screen identifies astemizole as an antimalarial agent. Nat Chem Biol. 2006;2(8):415–416. doi: 10.1038/nchembio806. [DOI] [PubMed] [Google Scholar]
- 23.Huang R, Southall N, Wang Y, et al. The NCGC Pharmaceutical Collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci Transl Med. 2011;3(80):80ps16. doi: 10.1126/scitranslmed.3001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muthyala R. Orphan/rare drug discovery through drug repositioning. Drug Discov Today Ther Strateg. 2011;8(3–4):71–76. [Google Scholar]
- 25.International Rare Diseases Research Consortium [website on the Internet] Available from: http://www.irdirc.orgAccessed March 21, 2013
- 26.Xu K, Coté TR. Database identifies FDA-approved drugs with potential to be repurposed for treatment of orphan diseases. Brief Bioinform. 2011;12(4):341–345. doi: 10.1093/bib/bbr006. [DOI] [PubMed] [Google Scholar]
- 27.US Food and Drug Administration A valuable resource for drug developers: the Rare Disease Repurposing Database (RDRD) 2011Available from: http://www.fda.gov/ForIndustry/DevelopingProductsforRareDiseasesConditions/HowtoapplyforOrphanProductDesignation/ucm216147.htmAccessed March 21, 2013
- 28.Marusina K, Welsch DJ, Lynn R, Brock D, Bahr N. The CTSA Pharmaceutical Assets Portal – a public-private partnership model for drug repositioning. Drug Discov Today Ther Strateg. 2011;8(3–4):77–83. doi: 10.1016/j.ddstr.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Intellectual Property Organization Re:Search [website on the Internet] Available from: http://www.wipo.int/research/enAccessed March 21, 2013
- 30.Johns Hopkins Clinical Compound Screening Initiative [website on the Internet] Available from: http://www.jhccsi.org/background.htmlAccessed March 21, 2013
- 31.Cures Within Reach [website on the Internet] Available from: http://www.cureswithinreach.org/about-usAccessed March 21, 2013
- 32.Weir SJ, DeGennaro LJ, Austin C P. Repurposing approved and abandoned drugs for the treatment and prevention of cancer through public-private partnerships. Cancer Res. 2012;72(5):1055–1058. doi: 10.1158/0008-5472.CAN-11-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassig CA, Smith LH, Jackson M.Creating the world’s most comprehensive drug repurposing library 2012Available from: http://www.americanlaboratory.com/913-Technical-Articles/114059-Creating-the-World-s-Most-Comprehensive-Drug-Repurposing-LibraryAccessed June 16, 2013
- 34.Arbanas C.Washington University, Pfizer announce groundbreaking research collaboration 2010Available from: http://news.wustl.edu/news/Pages/20770.aspxAccessed June 16, 2013
- 35.Boston’s Children Hospital Children’s Hospital Boston and Pfizer enter into novel collaborative research agreement for muscular dystrophy therapeutics 2011Available from: http://childrenshospital.org/newsroom/Site1339/mainpageS1339P795.htmlAccessed June 16, 2013
- 36.Seoane-Vazquez E, Rodriguez-Monguio R, Szeinbach SL, Visaria J. Incentives for orphan drug research and development in the United States. Orphanet J Rare Dis. 2008;3:33. doi: 10.1186/1750-1172-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wellman-Labadie O, Zhou Y. The US Orphan Drug Act: rare disease research stimulator or commercial opportunity? Health Policy. 2010;95(2–3):216–228. doi: 10.1016/j.healthpol.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Tambuyzer E. Rare diseases, orphan drugs and their regulation: questions and misconceptions. Nat Rev Drug Discov. 2010;9(12):921–929. doi: 10.1038/nrd3275. [DOI] [PubMed] [Google Scholar]
- 39.Peabody J W, Ruby A, Cannon P. The economics of orphan drug policy in the US. Can the legislation be improved? Pharmacoeconomics. 1995;8(5):374–384. doi: 10.2165/00019053-199508050-00002. [DOI] [PubMed] [Google Scholar]
- 40.Allarakhia M.Evolving concept of openness in the health commons: engaging global stakeholders in technology development for malaria 2011Available from: http://www.bioendeavor.net/CommonData/Resources/ECHC.pdfAccessed March 21, 2013
- 41.Agres T.New life for old drugs 2011Available from: http://www.dddmag.com/articles/2011/07/new-life-old-drugsAccessed June 16, 2013
- 42.Oprea TI, Bauman JE, Bologa CG, et al. Drug repurposing from an academic perspective. Drug Discov Today Ther Strateg. 2011;8(3–4):61–69. doi: 10.1016/j.ddstr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muthyala R. Orphan/rare drug discovery through drug repositioning. Drug Discovery Today. 2011;8(3–4):71–76. [Google Scholar]
- 44.Scott C, Núñez M.Grid computing guiding malaria drug discovery 2009Available from: http://www.scidev.net/en/news/grid-computing-guiding-malaria-drug-discovery.htmlAccessed June 16, 2013
- 45.Indiana University-Purdue University Indianapolis Molecules made by IUPUI students may have potential to cure diseases 2011Available from: http://newscenter.iupui.edu/4997/Molecules-Made-by-IUPUI-Students-May-Have-Potential-To-Cure-Diseases110124Accessed June 16, 2013
- 46.American Type Culture Collection What we offer Available from: http://www.atcc.org/About/About_ATCC/What_We_Offer.aspxAccessed June 16, 2013
- 47.Wu Y, Fairfield AS, Oduola A, Cypess RH. The Malaria Research and Reference Reagent Resource (MR4) Center – creating African opportunities. Afr J Med Sci. 2001;30( Suppl):52–54. [PubMed] [Google Scholar]
- 48.Allarakhia M, Ajuwon L. Understanding and creating value from open source drug discovery for neglected tropical diseases. Expert Opin Drug Discov. 2012;7(8):643–657. doi: 10.1517/17460441.2012.690390. [DOI] [PubMed] [Google Scholar]
- 49.Allarakhia M. Developing a framework for understanding and enabling open source drug discovery. Expert Opin Drug Discov. 2010;5(8):709–714. doi: 10.1517/17460441.2010.498815. [DOI] [PubMed] [Google Scholar]
- 50.Kinnings SL, Liu N, Buchmeier N, Tonge PJ, Xie L, Bourne PE. Drug discovery using chemical systems biology: repositioning the safe medicine Comtan to treat multi-drug and extensively drug resistant tuberculosis. PLoS Comput Biol. 2009;5:e1000423. doi: 10.1371/journal.pcbi.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gloeckner C, Garner AL, Mersha F, et al. Repositioning of an existing drug for the neglected tropical disease onchocerciasis. Proc Natl Acad Sci U S A. 2010;107(8):3424–3429. doi: 10.1073/pnas.0915125107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim LE, Vilchèze C, Ng C, Jacobs WR, Jr, Ramón-García S, Thompson CJ. Anthelmintic avermectins kill Mycobacterium tuberculosis, including multidrug-resistant clinical strains. Antimicrob Agents Chemother. 2013;57(2):1040–1046. doi: 10.1128/AAC.01696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sundar S, Jha TK, Thakur C P, Bhattacharya SK, Rai M. Oral miltefosine for the treatment of Indian visceral leishmaniasis. Trans R Soc Trop Med Hyg. 2006;100( Suppl 1):S26–S33. doi: 10.1016/j.trstmh.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 54.Miguel DC, Yokoyama-Yasunaka JK, Andreoli WK, Mortara RA, Uliana SR. Tamoxifen is effective against Leishmania and induces a rapid alkalinization of parasitophorous vacuoles harbouring Leishmania (Leishmania) amazonensis amastigotes. J Antimicrob Chemother. 2007;60(3):526–534. doi: 10.1093/jac/dkm219. [DOI] [PubMed] [Google Scholar]
- 55.Freitas-Junior LH, Chatelain E, Kim HA, Siqueira-Neto JL. Visceral leishmaniasis treatment: what do we have, what do we need and how to deliver it? Int J Parasitol Drugs Drug Resist. 2012;2:11–19. doi: 10.1016/j.ijpddr.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.One world problem, one world solution? Nature Rev Drug Discov. 2005;4:701. [No authors listed] [Google Scholar]
- 57.Sardana D, Zhu C, Zhang M, Gudivada RC, Yang L, Jegga AG. Drug repositioning for orphan diseases. Brief Bioinform. 2011;12(4):346–356. doi: 10.1093/bib/bbr021. [DOI] [PubMed] [Google Scholar]
- 58.Pollastri MP, Campbell RK. Target repurposing for neglected diseases. Future Med Chem. 2011;3(10):1307–1315. doi: 10.4155/fmc.11.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robertson SA, Renslo AR. Drug discovery for neglected tropical diseases at the Sandler Center. Future Med Chem. 2011;3(10):1279–1288. doi: 10.4155/fmc.11.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhardwaj A, Scaria V, Raghava GP, et al. Open source drug discovery – a new paradigm of collaborative research in tuberculosis drug development. Tuberculosis (Edinb) 2011;91(5):479–486. doi: 10.1016/j.tube.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 61.Open Source Drug Discovery [website on the Internet] Available from: http://www.osdd.netAccessed February 15, 2012
- 62.Sreelata M.Open source TB megaproject yields first fruits 2010Available from: http://www.scidev.net/en/news/open-source-tb-mega-project-yields-first-fruits.htmlAccessed June 16, 2013
- 63.Nelson R, editor. National Innovation Systems: A Comparative Analysis. Oxford: Oxford University Press; 1993. [Google Scholar]
- 64.Freeman C. Technology and Economic Performance: Lessons from Japan. London: Pinter; 1987. [Google Scholar]
- 65.Coles LD, Cloyd JC. The role of academic institutions in the development of drugs for rare and neglected diseases. Clin Pharmacol Ther. 2012;92(2):193–202. doi: 10.1038/clpt.2012.83. [DOI] [PubMed] [Google Scholar]
- 66.Ekins S, Williams AJ, Krasowski MD, Freundlich JS. In silico repositioning of approved drugs for rare and neglected diseases. Drug Discov Today. 2011;16(7–8):298–310. doi: 10.1016/j.drudis.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 67.Sardana D, Zhu C, Zhang M, Gudivada RC, Yang L, Jegga AG. Drug repositioning for orphan diseases. Brief Bioinform. 2011;12(4):346–356. doi: 10.1093/bib/bbr021. [DOI] [PubMed] [Google Scholar]
- 68.Allarakhia M, Walsh S. Managing knowledge assets under conditions of radical change: The case of the pharmaceutical industry. Technovation. 2011;31(2):105–117. [Google Scholar]
- 69.Metcalfe S. The economic foundations of technology policy: equilibrium and evolutionary perspectives. In: Stoneman P, editor. Handbook of the Economics of Innovation and Technological Change. Oxford: Blackwell; 1995. [Google Scholar]
- 70.Hess C, Ostrom E. Understanding Knowledge as a Commons: From Theory to Practice. Cambridge (MA): Blackwell; 2006. [Google Scholar]
- 71.Organisation for Economic Co-operation and Development National systems of innovation 1997Available from: http://www.oecd.org/science/inno/2101733.pdfAccessed March 21, 2013
- 72.PR Newswire.Transparency Life Sciences launches Indication Finder crowdsourcing tool for drug repurposing 2012Available from: http://www.prnewswire.com/news-releases/transparency-life-sciences-launches-indication-finder-crowdsourcing-tool-for-drug-repurposing-175572351.htmlAccessed April 2, 2013


