Abstract
Purpose
Gene expression profiling classifies breast cancer into intrinsic subtypes based on the biology of the underlying disease pathways. We have used material from a prospective randomized trial of tamoxifen versus placebo in premenopausal women with primary breast cancer (NCIC CTG MA.12) to evaluate the prognostic and predictive significance of intrinsic subtypes identified by both the PAM50 gene set and by immunohistochemistry.
Experimental Design
Total RNA from 398 of 672 (59%) patients was available for intrinsic subtyping with a quantitative reverse transcriptase PCR (qRT-PCR) 50-gene predictor (PAM50) for luminal A, luminal B, HER-2–enriched, and basal-like subtypes. A tissue microarray was also constructed from 492 of 672 (73%) of the study population to assess a panel of six immunohistochemical IHC antibodies to define the same intrinsic subtypes.
Results
Classification into intrinsic subtypes by the PAM50 assay was prognostic for both disease-free survival (DFS; P = 0.0003) and overall survival (OS; P = 0.0002), whereas classification by the IHC panel was not. Luminal subtype by PAM50 was predictive of tamoxifen benefit [DFS: HR, 0.52; 95% confidence interval (CI), 0.32–0.86 vs. HR, 0.80; 95% CI, 0.50–1.29 for nonluminal subtypes], although the interaction test was not significant (P = 0.24), whereas neither subtyping by central immunohistochemistry nor by local estrogen receptor (ER) or progesterone receptor (PR) status were predictive. Risk of relapse (ROR) modeling with the PAM50 assay produced a continuous risk score in both node-negative and node-positive disease.
Conclusions
In the MA.12 study, intrinsic subtype classification by qRT-PCR with the PAM50 assay was superior to IHC profiling for both prognosis and prediction of benefit from adjuvant tamoxifen.
Introduction
Breast cancer is increasingly recognized as a heterogeneous disease, both on a molecular basis and in terms of clinical behavior. The initial molecular profiling studies of primary tumors showed that breast cancers could be segregated into biologic (or "intrinsic") subtypes based on hierarchical clustering analyses of gene expression (1). One of the most important distinctions recognized was that hormone receptor–positive breast cancers could be separated into at least 2 categories, luminal A and B, with significantly different clinical outcomes (2). Subsequent studies have further shown that gene expression profiles also contribute additional prognostic value to current standard clinicopathologic factors (3–9).
Adjuvant hormonal therapy is the backbone of systemic management in the majority of hormone receptor–positive early-stage breast cancers. Meta-analyses have confirmed the potential of tamoxifen and aromatase inhibitors to reduce the risk of relapse (ROR) by approximately 50% and the risk of breast cancer death by 30% in the study populations (10). Adjuvant hormonal therapies, however, are not devoid of toxicities, costs, and issues surrounding compliance with 5 years of therapy. At present although, other than the expression of the hormone receptor proteins, there are no fully validated predictive biomarker(s) to enhance the selection of patients for endocrine treatment. Furthermore, premenopausal women are frequently underrepresented in biomarker investigations, leaving uncertainty to the performance of genomic assays in younger patient populations.
The intent of this study was to use both an established immunohistochemical (IHC) classifier and a gene expression classifier (PAM50) of the breast cancer intrinsic subtypes to assess for both prognostic and predictive value to adjuvant hormonal therapy in a study population of pre-menopausal women, randomized to tamoxifen or placebo for 5 years. A ROR model, generated from the gene expression classifier data in addition to clinicopathologic data, was also applied to generate a continuous risk score for further investigation as a clinical test.
Materials and Methods
The NCIC Clinical Trials Group (CTG) MA.12 study was a randomized trial of 672 premenopausal women with stage I–III breast cancer after adjuvant chemotherapy, of any hormone receptor status, comparing tamoxifen 20 mg per os daily with placebo for a planned 5 years. The details of the conduct of this study and its final results have recently been published (11). Median follow-up on the study was 9.7 years.
Formalin-fixed, paraffin-embedded (FFPE) blocks were requested from local pathology departments for all patients on the study. Four hundred and ninety-two FFPE blocks with invasive breast cancer were available to extract duplicate 0.6-mm tissue cores, creating 4 MA.12 tissue microarray (TMA) blocks for IHC studies using methods previously described (12). An additional three 1-mm2 pathologistguided cores were successfully taken from the invasive component (from areas adjacent to the cores extracted for the TMA) within 484 FFPE blocks and used to isolate RNA for quantitative reverse transcriptase PCR (qRT-PCR).
Immunohistochemistry for estrogen receptor (ER), progesterone receptor (PR), HER-2, Ki67, CK 5/6, and EGF receptor (EGFR) were conducted concurrently on serial sections; the specific antibodies used, and the staining conditions have been described previously (13, 14) but are also outlined in Supplementary Table S1. Scoring for the 6 IHC antibodies using published criteria developed on other breast cancer cohorts (13–15) was conducted by 2 pathologists (T.O. Nielsen and K. Ung) blinded to the clinicopathologic characteristics and outcome. In brief, tumors were considered positive for ER or PR if immunostaining was observed in more than 1% of the tumor nuclei (15, 16). Tumors were considered HER-2 positive if immunostaining was scored as 3+ according to recent ASCO/CAP guidelines (17). For equivocal cases (2+), FISH was conducted with a ratio of 2.0 or more considered as positive. Ki67 scoring was conducted visually for percentage of tumor cell nuclei with positive immunostaining above the background level where a cutoff of more than 13% defined luminal B among ER+/HER-2 – cases. We have previously shown that more than 13% was the optimal cut-off to segregate luminal B vs. luminal A when correlating IHC-stained TMAs to gene expression profiling for subtype distribution in primary breast cancers (14). Only TMA samples, which contained 50 or greater tumor cells, were considered interpretable. The stained TMAs were digitally scanned with primary image data available for public access (http://www.gpecimage.ubc.ca; username:ma12; password:abc123).
Total RNA was extracted by use of the High Pure RNA Paraffin Kit (Roche Applied Science) as previously described (18). After treatment with TURBO DNase (Ambion), total RNA yield was assigned with the ND-1000 Spectrophotometer (NanoDrop Technologies). Of the 484 cases scored, 410 cases yielded sufficient RNA for gene expression analysis by qRT-PCR, determined as previously described (18–20). PCR primers optimized for use on FFPE materials were used to make quantitative measurements for a panel of 50 discriminatory genes (PAM50 bioclassifier panel) to segregate tumors into luminal A, luminal B, HER-2-enriched, basal-like, and normal breast–like expression profiles (19). 398 cases had sufficient complete measurements for subtype assignment (REMARK diagram;Supplementary Fig. S1).
Statistical analyses
The detailed statistical plan and analyses for the MA.12 study are published elsewhere (11). The following are the definitions for the clinical endpoints of the study; disease-free survival (DFS): time from randomization to earliest date of recurrence or death or censored on the last date the patient was known to be alive; overall survival (OS): time from randomization to date of death or censored on the last date the patient was known to be alive. Biomarker subtype definitions and hypotheses to be tested were prespecified in writing, and statistical analyses were then conducted independently by the NCIC CTG statistical centre. The primary objectives of this translational study were 2-fold: (i) to determine whether the intrinsic subtypes, as categorized by PAM50, had prognostic significance in the entire MA.12 cohort and (ii) to determine whether the luminal subtypes (A and B), as determined by PAM50, were predictors of greater efficacy to adjuvant tamoxifen than the hormone receptor–positive status in MA.12, as determined by biochemical or IHC methods. Both OS and DFS by ER status (determined locally), by subtypes based on the immunohistochemistry and by PAM50 classifiers, were described by Kaplan– Meier curves and compared with log-rank tests. A Cox model with an interaction term between treatment and subtype was used to assess for predictive significance based on differences between treatment arms and subtype classification. All statistical tests were 2 sided.
PAM50-derived ROR scores were calculated using formulas previously trained and described (19, 20). Harrell C-index was used to compare the prognostic and predictive capacity of clinical, IHC, and expression profile–derived models (21). A C-index higher than 0.5 implies a good prediction ability of the model, whereas a C-index equal to 0.5 implies no predictive ability (no better than random guessing). Bootstrap percentile method with 200 replications was used to calculate the 95% confidence interval (CI) for the predicted probability of disease at 10 years based on the ROR scores with the highest C-index. This study received research ethical board approval from the host institution (University of British Columbia, Vancouver, British Columbia, Canada).
Results
The NCIC MA.12 study enrolled 672 premenopausal women from 1993 to 2000: 338 randomized to tamoxifen and 334 to placebo (11). Baseline characteristics of all patients randomized (N = 672), patients within the TMA series for IHC classification (IHC cohort: n = 492), and the PAM50 cohort for intrinsic subtype classification (PAM50 cohort: n = 398) are outlined in Table 1. The standard clinicopathologic characteristics were similar among all 3 cohorts. Median age was approximately 45 years (range: 26–58 years), approximately 75% of the study cohorts were node-positive, and all patients received adjuvant chemotherapy (45% non-anthracycline and 55% anthracycline-based, as detailed in Table 1).
Table 1.
Cohort characteristics
| Characteristic Age (median and range) |
All randomized (n = 672) 45.7 y (26–58) |
IHC Cohort (n = 492) 45.6 y (27–58) |
PAM50 cohort (n = 398) 45.5 y (27–58) |
|---|---|---|---|
| Stage | |||
| I | 10% | 10% | 10% |
| II | 84% | 85% | 84% |
| III | 6% | 5% | 6% |
| Nodal status | |||
| 0 | 25% | 24% | 25% |
| 1–3 | 56% | 56% | 55% |
| ≥ 4 | 19% | 20% | 20% |
| Clinical ER/PR status | |||
| ER+ and/or PR+ | 75% | 74% | 73% |
| ER−/PR unknown | 14% | 14% | 15% |
| ER¯/PR¯ | 11% | 12% | 12% |
| Chemotherapy | |||
| Cyclophosphamide, methotrexate and fluorouracil | 45% | 44% | 43% |
| Doxorubicin and cyclophosphamide | 33% | 32% | 32% |
| Cyclophosphamide, epirubicin and fluorouracil | 22% | 24% | 25% |
| Grade | |||
| I | 19% | 26% | 24% |
| II | 30% | 42% | 42% |
| III | 22% | 31% | 34% |
| Missing | 29% | 2% | 0% |
In the original MA.12 study population, the ER assay was done by a biochemical method (quantitative ligand-binding assay) in 60% of patients randomized to tamoxifen and in 51% of the placebo group and the remainder by immu-nohistochemistry. All clinical progesterone receptor (PR) assays were done by the biochemical method. The hormone receptor status was deemed positive (ER- and/or PR-positive) in 75% (n = 505) of the original study population, as determined by local laboratories. By the centrally assessed IHC classifier conducted for this study, 71% were classified as luminal breast cancers (40% luminal A and 31% luminal B) with the specific breakdown across all the intrinsic subtypes detailed in Table 2. Among the 398 patient samples that had classification by PAM50, 53% were classified as luminal breast cancers (34% luminal A and 19% luminal B). The concordance for intrinsic subtypes among 348 patients who could be classified by both IHC and PAM50 classifiers (Table2) was 82.2%, 81.0%, 87.6%, and 97.4% for luminal A, luminal B, HER-2-enriched (defined by PAM50)/luminal B HER-2+ and ER−/PR−/HER-2+ (defined by immunohistochemistry), and basal-like breast cancers, respectively, with an overall κ 0.64 (95% CI, 0.58–0.71). Within these 348 patients: 209 (60%) were ER- and/ or PR-positive by central immunohistochemistry, 66 (19%) were HER-2–positive by central immunohistochemistry, and 75 (21%) were core basal by IHC definition. Other than assessing for concordance in the 348 patients with available data across classifiers, no further assessment of prognosis or prediction was conducted because of the limited sample size.
Table 2.
Intrinsic subtype distribution by immunohistochemistry and PAM50 (n = 348)
| Intrinsic subtypes by IHC/FISH |
||||||
|---|---|---|---|---|---|---|
| Intrinsic subtypes by PAM50 | Luminal A | Luminal B | Luminal/HER-2+ | HER-2+/ER PR | Core basal | Total |
| Luminal A | 101 (78%) | 20 (15%) | 8 (6%) | 0 | 1 (1%) | 130 |
| Luminal B | 23 (33%) | 39 (57%) | 7 (10%) | 0 | 0 | 69 |
| HER-2-E | 9 (12%) | 14 (19%) | 26 (35%) | 23 (31%) | 3 (4%) | 75 |
| Basal-like | 1 (1%) | 2 (3%) | 0 | 2 (3%) | 69 (93%) | 74 |
| Total | 134 | 75 | 41 | 25 | 73 | 348 |
Association with outcomes and prognostic values
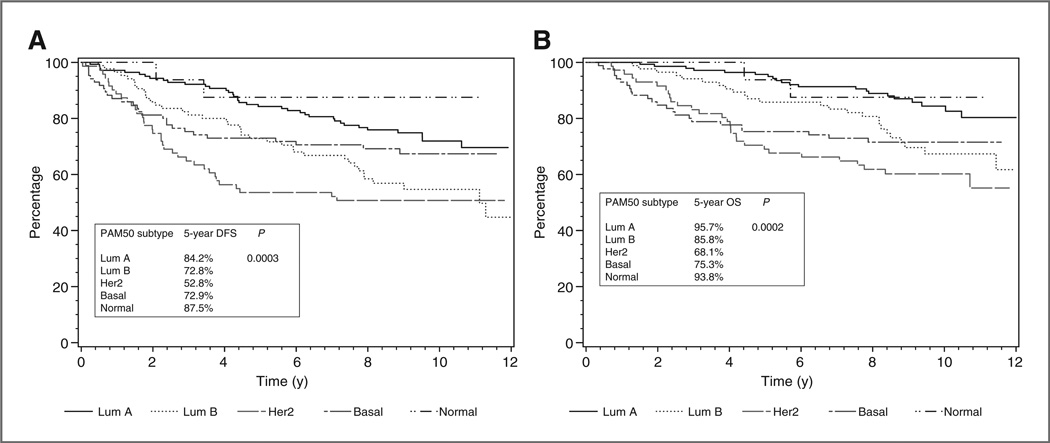
Among all randomized patients in both treatment groups, there was no significant difference between patients with positive and negative ER by clinical assay in either DFS or OS. Intrinsic subtype classification by the 6 antibody IHC panel created a larger discriminatory effect for DFS and OS with the worst prognostic group being the HER-2–positive nonluminal breast cancers for DFS and basal-like breast cancers for OS compared with the best prognostic group being luminal A for both DFS and OS, but none of these differences were statistically significant. However, with the PAM50 classification, both DFS and OS were statistically significantly different across intrinsic subtypes (Fig. 1A and B) with the HER-2– enriched subtype having the lowest and the luminal A breast cancers the highest 5-year survival values. In multivariate analysis (Table 3) adjusting for tumor grade, tamoxifen treatment, and clinical variables including ER status, nodal status, and size of tumor, intrinsic subtyping, as classified by PAM50, was still independently prognostic from the standard clinicopathologic variables and treatment for both DFS (P = 0.02) and OS (P = 0.02). The multivariable Cox models that included the subtype and clinicopathologic variables listed earlier were significantly better than either clinicopath-ologic alone or subtype alone.
Figure 1.
Prognosis based on PAM50 classifier for DFS (A) and OS (B).
Table 3.
Multivariate analysis of prognostic variables
| DFS |
OS |
||||
|---|---|---|---|---|---|
| Variable | Comparison | HR (95% CI) | P | HR (95% CI) | P |
| Subtype | Basal vs. Luminal A | 1.57 (0.75–3.29) | 0.02 | 2.19 (0.91–5.26) | 0.02 |
| Her2+ vs. Luminal A | 2.25 (1.29–3.90) | 2.74 (1.39–5.40) | |||
| Luminal B vs. Luminal A | 1.98 (1.16–3.39) | 2.40 (1.25–4.63) | |||
| Treatment | Tamoxifen vs. placebo | 0.58 (0.41–0.82) | 0.002 | 0.68 (0.45–1.02) | 0.06 |
| ER | Positive vs. negative | 0.82 (0.53–1.28) | 0.38 | 0.58 (0.35–0.97) | 0.04 |
| Nodal status | 1–3 vs. negative | 1.97 (1.20–3.22) | 0.02 | 1.61 (0.92–2.81) | 0.17 |
| >3 vs. negative | 1.97 (1.12–3.48) | 1.78 (0.94–3.37) | |||
| Tumor size | T stage 1 vs. >1 | 0.64 (0.44–0.92) | 0.02 | 0.62 (0.40–0.96) | 0.03 |
| Tumor grade | 1 vs. 2 vs. 3 | 0.96 (0.71–1.31) | 0.81 | 0.83 (0.58–1.19) | 0.30 |
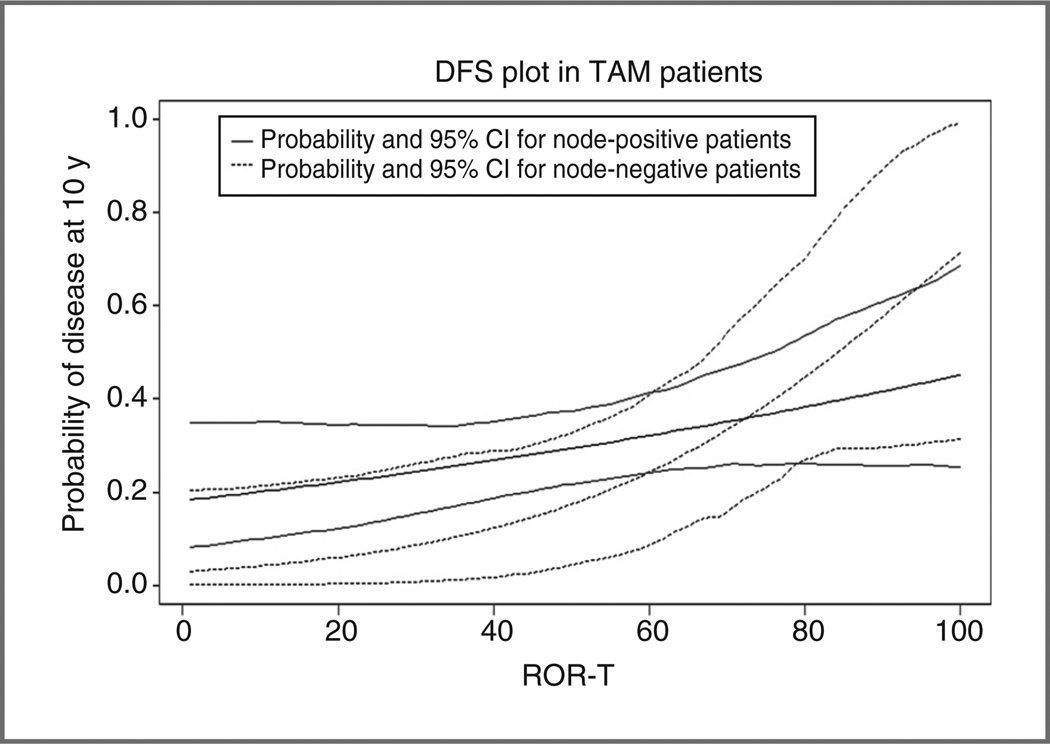
We also assessed the performance of ROR scores generated from PAM50 subtypes with and without clinical information based on algorithms developed previously (18, 19). The C-index for ROR-S (subtype only), ROR-P (subtype with proliferation signature index), ROR-T (subtype with tumor size), and for ROR-PT (subtype with both proliferation signature index and tumor size) are summarized in Supplementary Table S2. ROR-T had highest numerical prognostic capacity in both DFS and OS with a C-index of 59.8% (54.8%–64.2%) and 63.1% (57.9%–68.2%), respectively. Figure 2 shows that the ROR-T scores of tamoxifen-treated patients have a linear relationship with probability of 10-year relapse in both the node-negative and the node-positive cohorts.
Figure 2.
Probability of disease event at 10 years based on the ROR-T model.
Prediction of tamoxifen benefits
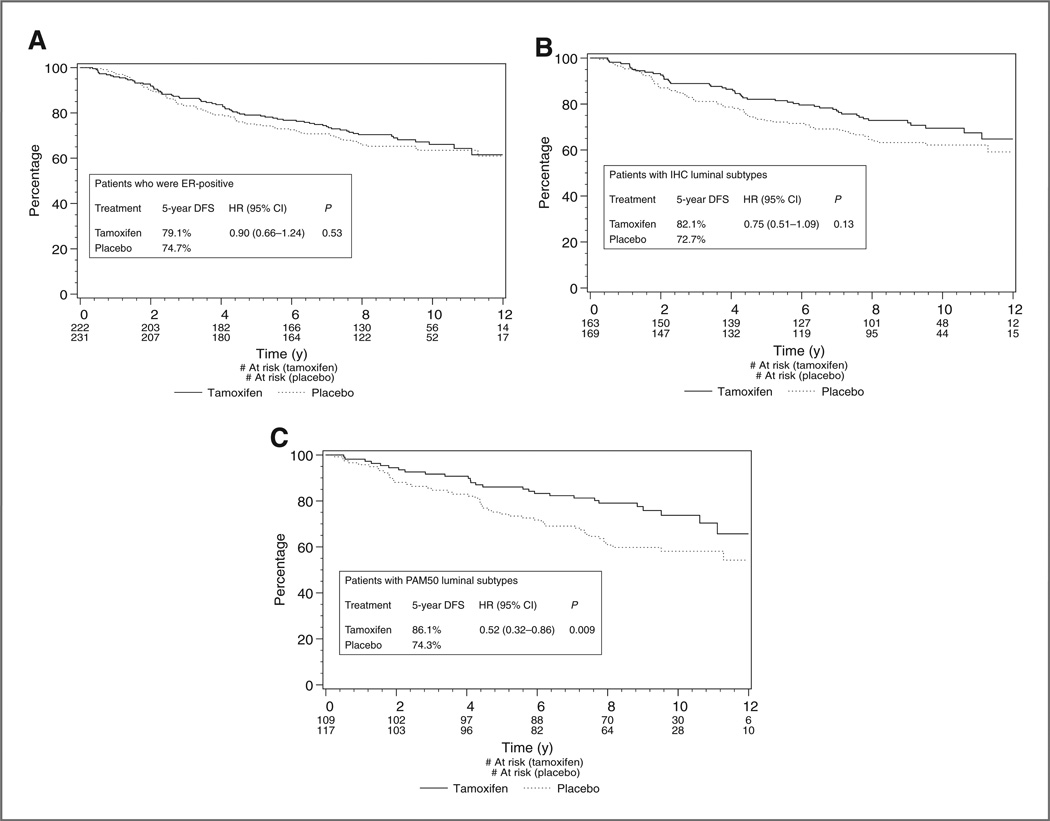
With all randomized patients included in the analysis, the original MA.12 study showed a trend for improved DFS for tamoxifen (HR, 0.77; 95% CI, 0.59–1.01; P = 0.06; ref. 11). Differences between tamoxifen and placebo were not statistically significant in OS for the trial as a whole (HR 0.78 with 95% CI 0.57–1.06 and P = 0.12) and, therefore, the following analyses were restricted to DFS only. Being ER-positive by a clinical test could not identify a group with statistically significant better outcomes on tamoxifen (Fig. 3A). Within the luminal cohorts (luminal A + B) based on the IHC classifier, there appeared a greater trend for benefit of adjuvant tamoxifen, though this was again not statistically significant (HR, 0.75; 95% CI,0.51–1.09;P = 0.13; Fig. 3B). However, using the PAM50 classifier to identify luminal subtypes, there was a statistically significant benefit in DFS for tamoxifen (HR 0.52; 95% CI, 0.32–0.86; P = 0.009; Fig. 3C), whereas no statistically significant benefit was found for patients with nonluminal subtypes (HR, 0.80; 95% CI, 0.50–1.29). However, the interaction test between luminal type by PAM50 and tamoxifen treatment was not statistically significant (P = 0.24). The corresponding absolute improvement in 5-year DFS rate was 11.8% for tamoxifen versus placebo in the hormone-sensitive cohort (luminal A and B), as defined by PAM 50 (5-year DFS rate = 86.1% vs. 74.3%, respectively; Fig. 3C). A forest plot comparing the efficacy of tamoxifen versus placebo across the various predictive classifiers is presented in Supplementary Fig. S2.
Figure 3.
A, Kaplan–Meier curves for DFS among ER+ (local assessment) patients for tamoxifen versus placebo. B, Kaplan–Meier curves for DFS among luminal subtypes by immunohistochemistry for tamoxifen versus placebo. C, Kaplan–Meier curves for DFS among luminal subtypes by PAM50 for tamoxifen versus placebo.
Discussion
The NCIC Clinical Trials Group MA.12 study was one of several randomized trials to assess the benefit of adjuvant tamoxifen for premenopausal early-stage breast cancer (11). The results of the main study showed a marginal, but nonsignificant, improvement in DFS for tamoxifen over placebo (HR, 0.77; 95% CI, 0.59–1.01). Somewhat surprising, the trend in benefit was irrespective of hormone receptor status, with the clinically ER-positive patients having a HR of 0.82. One possible explanation for this observation may be the limited accuracy of hormone receptor testing in community hospitals. When we conducted central immunohistochemistry-based testing to classify the luminal subtypes, the HR improved (HR 0.75), but this was still not statistically significant—in part perhaps due to the smaller sample size and subsequent limited power. Only when we used the RT-PCR approach with the PAM50 classifier was there a statistically significant benefit of adjuvant tamoxifen among patents with luminal subtypes (HR, 0.52; 95% CI, 0.32–0.86; P = 0.009). Furthermore, the benefit of adjuvant hormonal therapy in this premenopausal population is additive to the gains from adjuvant chemotherapy (all patients received adjuvant chemotherapy in MA.12), which is in keeping with the recent update from the EBCTCG overview of adjuvant tamoxifen (22). Our study illustrates the importance of accurate testing of predictive biomarkers, and the potential improvements that can be made with more advanced assay methodologies that also more comprehensively reflect pathway biology. The concordance between IHC assessment of limited biomarkers (ER and PR) versus the measurement globally of multiple activated genes in the luminal pathways, as with PAM50, was modest in our study at approximately 80%, suggesting the later approach is more reflective of the responsiveness of biologic pathways.
The luminal subtypes comprise the majority of breast cancer cases. Adjuvant hormonal therapies potentially have the greatest clinical impact (with relative risk reductions approximating 50%) in improving breast cancer outcomes, but the benefit is only in hormone receptor–positive breast cancers (10). Patients with hormone receptor–negative breast cancer would be at risk of treatment toxicities without potential benefit. Both the previous methodology of quantitative ligand-binding biochemical assay and current methodology using immunohistochemistry are subject to preanalytical and analytical variability, leading to estimates that, up to 20% of current hormone receptor testing worldwide, may be inaccurate (23). A recent guideline developed by a joint American Society of Clinical Oncology and College of American Pathologists taskforce recommended testing algorithms for more accurate and reproducible assays in hormone receptor testing (24).
qRT-PCR measures gene expression using RNA extracted from FFPE tumor tissue. Although this newer methodology has advantages in its ability to precisely quantitate multiple gene transcripts simultaneously, it could also potentially suffer from variabilities in RNA extraction, primer selection, PCR protocols, and measurement platforms. Recent studies, however, comparing PCR-based assays and IHC techniques have showed high correlation between these 2 methodologies (25–28).
The intrinsic subtypes luminal A, luminal B, HER-2– enriched, basal-like, and normal-like have been extensively studied by microarray-based gene expression profiling (1, 2, 6, 29, 30). These subtypes have been shown to not only segregate based on biology, but also are independently prognostic (2). The PAM50 assay was developed with the aim to identify these intrinsic subtypes, and has also been validated across multiple cohorts and platforms (19). An advantage of intrinsic subtype determination using the PAM50 assay over other gene expression signatures is its ability to provide outcome prediction not only in ER-positive but also in ER-negative tumors. By virtue of the design of MA.12, which permitted enrolment of patients with ER-negative cancers, we were able to show that PAM50 was superior to IHC classification for prognosis across the full spectrum of breast cancer subtypes. Furthermore, in a recently published study of 786 primary tumors linked to long-term outcome, the PAM50 assay was able to identify a very low-risk prognostic group (>95% DFS at 10 years) in a cohort of women treated with tamoxifen but no chemotherapy (18). These results were similar in discriminatory magnitude to those obtained from the 21-gene assay when it was applied to clinical trial material (4, 5).
In conclusion, intrinsic subtype classification by RT-PCR with the PAM50 assay is feasible on routinely collected FFPE tumor material. Within the context of the randomized MA.12 study, this methodology and assay was superior to conventional local and central IHC ER assessment for both prognosis and more importantly prediction of benefit to adjuvant tamoxifen compared with placebo in a population of younger women showing the PAM50 model is effective regardless of the age of the patient at presentation.
Supplementary Material
Translational Relevance.
Adjuvant hormonal therapy is the most commonly prescribed class of agents for early-stage breast cancer worldwide. The presence of the estrogen receptor (ER), even at a 1% threshold by immunohistochemistry, is the only validated predictive biomarker today. We investigated the prognostic and predictive value of classifying breast cancers into intrinsic subtypes by qRT-PCR with the PAM50 bioclassifier panel using material from a randomized controlled trial comparing tamoxifen with placebo in premenopausal women also treated with adjuvant chemotherapy (MA.12). The intrinsic subtypes were found to have independent prognostic value in a multivariate analysis that included standard clinical and pathologic variable. More importantly, the luminal subtypes, as discriminated by PAM50, had predictive ability to refine the population of patients who obtained a greater benefit from adjuvant tamoxifen.
Acknowledgments
The authors thank the Canadian women who participated in the NCIC MA.12 study and Central Tumour/Tissue Repository of the NCIC CTG (Kingston, ON, Canada).
Grant Support
This study was supported by a grant from the Canadian Breast Cancer Foundation: BC/Yukon (to S.K. Chia) and funding provided by a National Cancer Institute Strategic Partnering to Evaluate Cancer Signatures Grant No. U01 CA114722 (M.J. Ellis).
Footnotes
Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
J. Parker is an author of the patent (in part) for the PAM50 assay. P.S. Bernard, C.M. Perou, M.J. Ellis, and T.O. Nielsen have ownership interest (including patents) in Bioclassifier LLC. No potential conflict of interests were disclosed by other authors.
Authors' Contributions
Conception and design: S.K. Chia, V. Bramwell-Wesley, D. Tu, E. Mardis, K. I. Pritchard, C.M. Perou, M.J. Ellis,T.O. Nielsen
Development of methodology: S.K. Chia, E.Mardis, C.M. Perou, M.J. Ellis, T.O. Nielsen
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): S.K. Chia, V. Bramwell-Wesley, L. Shepherd, E. Mardis, S. Leung, K. Ung, M.J. Ellis, T.O. Nielsen
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S.K. Chia, V. Bramwell-Wesley, D. Tu, S. Jiang, S. Leung, K.I. Pritchard, J. Parker, P.S. Bernard, C.M. Perou, M.J. Ellis, T. O. Nielsen
Writing, review, and/or revision of the manuscript: S.K. Chia, V. Bram-well-Wesley, D. Tu, L. Shepherd, E. Mardis, K.I. Pritchard, P.S. Bernard, C.M. Perou, M.J. Ellis, T.O. Nielsen
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): S.K. Chia, L. Shepherd, T. Vickery, S. Leung, M.J. Ellis, T.O. Nielsen
Study supervision: E. Mardis, M.J. Ellis, T.O. Nielsen, S.K. Chia
References
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van't Veer LJ, Dai J, van de Vijver MJ, He YD, Hart AAM, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 4.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen treated node negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 5.Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, et al. Gene expression and benefit of chemotherapy in women with node negative estrogen receptor positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 6.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 8.Perreard L, Fan C, Quackenbush JF, Mullins M, Gauthier NP, Nelson E, et al. Classification and risk stratification of invasive breast carcinomas using a real time quantitative RT-PCR assay. Breast Cancer Res. 2006;8:R23. doi: 10.1186/bcr1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 10.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15 year survival: an overview of the randomized trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 11.Bramwell VHC, Pritchard KI, Tu D, Tonkin K, Vachhrajani H, Vanden-berg TA, et al. A randomized placebo controlled study of tamoxifen after adjuvant chemotherapy in premenopausal women with early breast cancer (National Cancer Institute of Canada – Clinical Trials Group Trial, MA.12) Ann Oncol. 2010;21:283–290. doi: 10.1093/annonc/mdp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voduc D, Cheang M, Nielsen T. GATA-3 expression in breast cancer has a strong association with estrogen receptor but lacks independent prognostic value. Cancer Epidemiol Biomarkers Prev. 2008;17:365–373. doi: 10.1158/1055-9965.EPI-06-1090. [DOI] [PubMed] [Google Scholar]
- 13.Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, et al. Basal like breast cancer defined by five biomarkers has superior prognostic value than triple negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 14.Cheang MCU, Chia SK, Voduc D, Gao D, Leung S, Snider J, et al. Ki67, HER2 status and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheang MD, Treaba DO, Speers CH, Olivotto IA, Bajdik CD, Chia SK, et al. Immunohistochemical detection using the new rabbit monoclonal antibody SP1 of estrogen receptor in breast cancer is superior to mouse monoclonal antibody 1D5 in predicting survival. J Clin Oncol. 2006;24:5637–5644. doi: 10.1200/JCO.2005.05.4155. [DOI] [PubMed] [Google Scholar]
- 16.Liu S, Chia SK, Mehl E, Leung S, Rajput A, Cheang MCU, et al. Progesterone receptor is a significant factor associated with clinical outcomes and effect of adjuvant tamoxifen therapy in breast cancer patients. Breast Cancer Res Treat. 2009;119:53–61. doi: 10.1007/s10549-009-0318-0. [DOI] [PubMed] [Google Scholar]
- 17.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen treated estrogen receptor positive breast cancer. Clin Cancer Res. 2010;16:5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullins M, Perreard L, Quackenbush JF, Gauthier N, Bayer S, Ellis M, et al. Agreement in breast cancer classification between microarray and quantitative reverse transcription PCR from fresh frozen and formalin fixed, paraffin embedded tissues. Clin Chem. 2007;53:1273–1279. doi: 10.1373/clinchem.2006.083725. [DOI] [PubMed] [Google Scholar]
- 21.Harrell FE, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 22.Early Breast Cancer Trialists' Collaborative Group. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient level meta-analysis of randomised trials. Lancet. 2011;378:771–784. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhodes A, Jasani B, Barnes DM, Bobrow LG, Miller KD. Reliability of immunohistochemical demonstration of estrogen receptors in routine practice: inter-laboratory variance in the sensitivity of detection and evaluation of scoring systems. J Clin Pathol. 2000;53:125–130. doi: 10.1136/jcp.53.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond MEH, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Badve SS, Baehner FL, Gray RP, Childs BH, Maddala T, Liu ML, et al. Estrogen and progesterone receptor status in ECOG 2197: comparison of immunohistochemistry by local and central laboratories and quantitative reverse transcription polymerase chain reaction by central laboratory. J Clin Oncol. 2008;26:2473–2481. doi: 10.1200/JCO.2007.13.6424. [DOI] [PubMed] [Google Scholar]
- 26.Cronin M, Sangli C, Liu ML, Pho M, Dutta D, Nguyen A, et al. Analytical validation of the Oncotype DX genomic diagnostic test for recurrence prognosis and therapeutic response prediction in node negative estrogen receptor positive breast cancer. Clin Chem. 2007;53:1084–1091. doi: 10.1373/clinchem.2006.076497. [DOI] [PubMed] [Google Scholar]
- 27.Shak S, Baehner FL, Palmer G, Ballard JT, Baker J, Watson D. Subtypes of breast cancer defined by standardized quantitative RT-PCR analysis of 10,618 tumors. Updated analysis with 20,050 tumors [abstract] San Antonio, TX: Proceedings of the 29th Annual San Antonio Breast Cancer Symposium; Dec 14–17, 2006. Abstract nr 6118. [Google Scholar]
- 28.Baehner FL, Habel LA, Quesenberry CP, Capra A, Tang G, Paik S, et al. Quantitative RT-PCR analysis of ER and PR by Oncotype DX indicates distinct and different associations with prognosis and prediction of tamoxifen benefit [abstract] San Antonio, TX: Proceedings of the 29 th Annual San Antonio BreastCancer Symposium; Dec 14–17, 2006. Abstract nr 45. [Google Scholar]
- 29.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel AB, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DSA, Nobel AB, et al. Concordance among gene expression based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.