Hepatitis C virus (HCV) has infected an estimated 130 million people worldwide, and most of them are chronically infected. HCV-infected people serve as a reservoir for transmission and have a high risk of developing cirrhosis and hepatocellular carcinoma. The development of methods for the prevention and treatment of HCV infection is strongly inhibited by viral factors. In fact, HCV has extensive nucleotide sequence diversity. Sequence comparisons of variants from different geographic areas have led to the identification and classification of various genotypes and subtypes. Thus far, sequencing of HCV isolates has identified 6 major genotypes and more than 83 subtypes. Genotypes 1, 2, and 3 are widely distributed throughout the world, whereas genotypes 4 and 5 have mainly been identified in Africa. In contrast, genotype 6 has been found in limited geographic regions, mainly in Southeast Asia (1).
Accurate HCV genotyping is important for predicting the response to antiviral therapy since genotypes 1 and 4 are less likely to respond to interferon than genotypes 2 and 3. Since HCV genotypes vary according to the epidemic history in different geographic regions, genotyping is also an essential tool for epidemiological studies and for estimating the infection route (2,3). In addition, epidemiological studies of HCV strains from blood donors, drug addicts, and hospital patients have demonstrated a correlation between subtypes and risk factors (4,5).
Asia is an area of high endemicity for HCV infection, which is a substantial risk factor for serious liver diseases. Although a small-scale survey has suggested a relatively restricted distribution of HCV genotype 6 variants in a limited region of Southeast Asia, larger studies are required to explore their distribution in other geographical regions. It is known that HCV genotype 6 viruses have the greatest genetic diversity. Recently, we reported isolating a novel subtype from Vietnamese patients (6), and thus far, this genotype has been found to contain at least 22 (6a–6v) subtypes. To explore the exact genotypic distribution and genetic variation of HCV in Vietnam, we conducted a large-scale survey based on direct sequence analysis, which is a gold standard method.
A total of 842 HCV-positive serum samples collected from Vietnamese blood donors and liver disease patients between 2005 and 2011 were analyzed. All serum samples were collected at Cho Ray Hospital, the Hospital for Tropical Diseases, and the Hospital at the University of Medicine and Pharmacy in Ho Chi Minh City, Vietnam. Serum samples were stored at −30° C until analysis. Informed consent for participation in this study was obtained from each individual. This study conforms to ethical guidelines and was approved by the ethics committees of the University of Medicine and Pharmacy, Cho Ray Hospital, the Hospital for Tropical Diseases, Vietnam, and National Institute of Infectious Diseases, Japan.
For the detection of HCV RNA, nested reverse transcriptase (RT)-PCR was performed using primers designed to amplify the NS5B gene of HCV. The primer sequences of the HCV used in this study were as follows: 5′-GAG YHT TCA CGG ADG CTA TGA CYA GGT A-3′ (HC3, sense, nt 8623–8650, with Y:C/T, H:A/C/T, D:A/G/T), and 5′-GAC ASG CTG TGA WAW ATG TCB CCC CCG-3′ (HC3R, antisense, nt 9307–9281, with S:G/C, W:A/T, B:G/C/T) for the outer primer pairs (685 bases), and 5′-GAC YTS GAG YTS ATA ACA TC-3′ (HC4, sense, nt 8688–8707, with Y:C/T, S:G/C), and 5′-ADT GGA GTG AGT TTK AGC TT-3′ (HC4R, antisense, nt 9229–9210, with D:A/G/T, K:G/T) for the inner primer pairs (542 bases). Nucleotide position was based on HCV-TV241 (GenBank accession no. EF632069). Briefly, total RNA was extracted from 150 μl of serum using an RNA extraction kit (NKRNAPREP kit; Nam Khoa Biotek Co., Ho Chi Minh City, Vietnam). Then, cDNA synthesis was performed using cDNA synthesis kit (NKcDNA synthesis kits; Nam Khoa Biotek Co.). Viral cDNA was synthesized by using random primers with the following cycle conditions: 25° C for 5 min, 42° C for 30 min, and 85° C for 5 min. HCV cDNA was amplified by nested PCR with the following cycle conditions: 40 cycles of 94° C for 20 s, 45° C for 20 s, and 72° C for 40 s for the first PCR, and 94° C for 20 s, 50° C for 20 s, and 72° C for 30 s for the seconf PCR. The PCR products were analyzed by electrophoresis on 2% agarose gels stained with ethidium bromide and recovered using the QIAquick gel extraction kit (Qiagen Inc., Chatsworth, Calif., USA). Purified PCR products were subjected to direct sequencing using the ABI PRISM™ Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, Calif., USA). To obtain genotyping results, the nucleotide data obtained by direct sequencing was submitted to the HCV sequence database at Los Alamos National Laboratory in USA (http://hcv.lanl.gov/content/sequence/BASIC_BLAST/basic_blast.html).
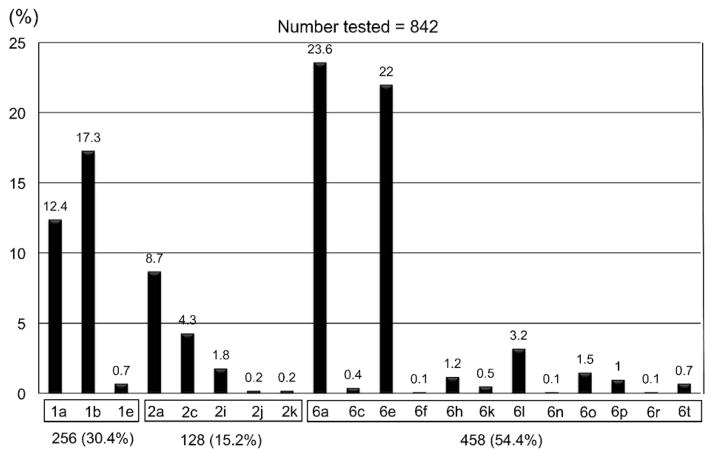
As a result, 3 different genotypes in the 842 Vietnamese patients tested were found. Genotype 6 variants were most predominant (458 patients; 54.4%) followed by genotype 1 (256; 30.4%) and genotype 2 (128; 15.2%) (Fig. 1). We did not detect any other genotypes in this study. Furthermore, the subtyping results obtained were as follows: 1a (104; 12.4%), 1b (146; 17.3%), 1e (6; 0.7%), 2a (73; 8.7%), 2c (36; 4.3%), 2i (15; 1.8%), 2j (2; 0.2%), 2k (2; 0.2%), 6a (199; 23.6%), 6c (3; 0.4%), 6e (185; 22%), 6f (1; 0.1%), 6h (10; 1.2%), 6k (4; 0.5%), 6l (27; 3.2%), 6n (1; 0.1%), 6o (13; 1.5%), 6p (8; 1%), 6r (1; 0.1%), and 6t (6; 0.7%). There was no significant difference in genotypic distribution between the blood donor group and the liver disease group.
Fig. 1.
Genotypic distribution of HCV in Ho Chi Minh City, Vietnam.
Genotyping of HCV is important to clarify the infection route and pathogenesis of the virus. In particular, examination of the sequence diversity among different isolates of the virus is important because variants may differ in their patterns of serologic reactivity, pathogenicity, virulence, and response to therapy. HCV has genetic variation, which corresponds to its geographic distribution, and it has been proposed that HCV can be classified into at least 6 major genotypes.
In this study, we confirmed the genotypic distribution of HCV in the southern part of Vietnam. To our knowledge, this is the largest survey on the genotypic distribution of HCV in Vietnam examined by the gold standard method of direct sequence analysis. Although HCV genotype 6 has been reported in patients from Southeast Asia, its prevalence and clinical characteristics have not been well described in a large patient sample by means of an accurate genotyping method, such as the 5′UTR-core and/or NS5B sequencing test. Surprisingly, our study showed that more than half of the Vietnamese patients who lived in Ho Chi Minh City were infected with genotype 6 variants. The reason why such peculiar HCV variants are circulating in this limited geographical region remains unknown.
Accurate HCV genotyping is clinically important for predicting the response to and determining the duration of antiviral therapy, and for the development of effective vaccines and testing kits. This is illustrated by the fact that genotypes 1 and 4 are more resistant to treatment with pegylated alpha interferon and ribavirin than genotypes 2 and 3 (2,7). Moreover, it has been suggested that patients with chronic HCV subtype 1b infection have more serious liver disease than patients infected with other genotypes (8,9). However, only a few studies on the response to antiviral treatment of genotype 6-infected patients have been reported so far (5,10–13). The reason for this is that genotype 6 virus is only seen in very limited regions of Southeast Asia where long-term observation of patients is difficult for economic reasons. However, fortunately, patients infected with HCV genotype 6 respond better to interferon-based therapy than those infected with genotype 1, although patient baseline clinical characteristics and side effect profiles are similar between HCV genotype 6 and other HCV genotypes (5,13). Further studies are required to address this issue. To resolve this issue, we are now investigating the different clinical and treatment outcomes between genotype 6 and other genotypes. In addition, we are conducting a prospective survey of the risk factors for HCV acquisition to explore other potential routes of viral transmission in this population.
Acknowledgments
We want to thank Hideki Hasegawa at Department of Pathology, National Institute of Infectious Diseases, for his continuous encouragement during this study, and An X.D. Vo, Ngoc H. Hoang, Trang T.T. Phan at Nam Khoa Biotek Laboratory, for their skillful techniques.
This study was supported by grants from Science Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 21406007 for Dr. Kenji Abe) and National Institute of Allergy and Infectious Diseases of the USA (No. 1R01AI080734–01A for Dr. Ling Lu). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of interest None to declare.
References
- 1.Simmonds P, Bukh J, Combet C, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 2.McHutchison JG, Gordon SC, Schiff ER, et al. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 3.Fried MW, Hadziyannis SJ, Shiffman ML, et al. Rapid virological response is the most important predictor of sustained virological response across genotypes in patients with chronic hepatitis C virus infection. J Hepatol. 2011;55:69–75. doi: 10.1016/j.jhep.2010.10.032. [DOI] [PubMed] [Google Scholar]
- 4.Roman F, Hawotte K, Struck D, et al. Hepatitis C virus genotypes distribution and transmission risk factors in Luxembourg from 1991 to 2006. World J Gastroenterol. 2008;28:1237–1243. doi: 10.3748/wjg.14.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen NH, Vutien P, Trinh HN, et al. Risk factors, genotype 6 prevalence, and clinical characteristics of chronic hepatitis C in Southeast Asian Americans. Hepatol Int. 2010;4:523–529. doi: 10.1007/s12072-010-9181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu L, Murphy D, Li C, et al. Complete genomes of three subtype 6t isolates and analysis of many novel HCV variants within genotype 6. J Gen Virol. 2008;89:444–452. doi: 10.1099/vir.0.83460-0. [DOI] [PubMed] [Google Scholar]
- 7.Zylberberg H, Chaix ML, Brechot C. Infection with hepatitis C virus genotype 4 is associated with a poor response to interferon-alpha. Ann Intern Med. 2000;132:845–846. doi: 10.7326/0003-4819-132-10-200005160-00029. [DOI] [PubMed] [Google Scholar]
- 8.Silini E, Bono F, Cividini A, et al. Differential distribution of hepatitis C virus genotypes in patients with and without liver function abnormalities. Hepatology. 1995;21:285–290. [PubMed] [Google Scholar]
- 9.Abe K, Edamoto Y, Park YN, et al. In situ detection of hepatitis B, C and G virus nucleic acids in human hepatocellular carcinoma tissues from different geographic regions. Hepatology. 1998;28:568–572. doi: 10.1002/hep.510280239. [DOI] [PubMed] [Google Scholar]
- 10.Hui CK, Yuen MF, Sablon E, et al. Interferon and ribavirin therapy for chronic hepatitis C virus genotype 6: a comparison with genotype 1. J Infect Dis. 2003;187:1071–1074. doi: 10.1086/368217. [DOI] [PubMed] [Google Scholar]
- 11.Fung J, Lai CL, Hung I, et al. Chronic hepatitis C virus genotype 6 infection: response to pegylated interferon and ribavirin. J Infect Dis. 2008;198:808–812. doi: 10.1086/591252. [DOI] [PubMed] [Google Scholar]
- 12.Seto WK, Lai CL, Fung J, et al. Natural history of chronic hepatitis C: genotype 1 versus genotype 6. J Hepatol. 2010;53:444–448. doi: 10.1016/j.jhep.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Chao DT, Abe K, Nguyen MH. Systematic review: epidemiology of hepatitis C genotype 6 and its management. Aliment Pharmacol Ther. 2011;34:286–296. doi: 10.1111/j.1365-2036.2011.04714.x. [DOI] [PubMed] [Google Scholar]