Abstract
Although two major breast cancer susceptibility genes, BRCA1 and BRCA2, have been identified accounting for 20% of breast cancer genetic risk, identification of other susceptibility genes accounting for 80% risk remains a challenge due to the complex, multi-factorial nature of breast cancer. Complexity derives from multiple genetic determinants, permutations of gene-environment interactions, along with presumptive low-penetrance of breast cancer predisposing genes, and genetic heterogeneity of human populations. As with other complex diseases, dissection of genetic determinants in animal models provides key insight since genetic heterogeneity and environmental factors can be experimentally controlled, thus facilitating the detection of quantitative trait loci (QTL). We therefore, performed the first genome-wide scan for loci contributing to radiation-induced mammary tumorigenesis in female F2-(Dahl S x R)-intercross rats. Tumorigenesis was measured as tumor burden index (TBI) after induction of rat mammary tumors at forty days of age via 127Cs-radiation. We observed a spectrum of tumor latency, size-progression, and pathology from poorly differentiated ductal adenocarcinoma to fibroadenoma, indicating major effects of gene-environment interactions. We identified two mammary tumorigenesis susceptibility quantitative trait loci (Mts-QTLs) with significant linkage: Mts-1 on chromosome-9 (LOD-2.98) and Mts-2 on chromosome-1 (LOD-2.61), as well as two Mts-QTLs with suggestive linkage: Mts-3 on chromosome-5 (LOD-1.93) and Mts-4 on chromosome-18 (LOD-1.54). Interestingly, Chr9-Mts-1, Chr5-Mts-3 and Chr18-Mts-4 QTLs are unique to irradiation-induced mammary tumorigenesis, while Chr1-Mts-2 QTL overlaps with a mammary cancer susceptibility QTL (Mcs 3) reported for 7,12-dimethylbenz-[α]antracene (DMBA)-induced mammary tumorigenesis in F2[COP x Wistar-Furth]-intercross rats. Altogether, our results suggest at least three distinct susceptibility QTLs for irradiation-induced mammary tumorigenesis not detected in genetic studies of chemically-induced and hormone-induced mammary tumorigenesis. While more study is needed to identify the specific Mts-gene variants, elucidation of specific variant(s) could establish causal gene pathways involved in mammary tumorigenesis in humans, and hence novel pathways for therapy.
Introduction
Breast cancer is one of the most prevalent female cancers in the world, affecting at least 10% of women in industrialized nations [1], [2]. Breast cancer is a complex multifactorial trait encompassing genetic and environmental factors [3], [4]. To date few breast cancer susceptibility genes have been identified in human populations with BRCA1 and BRCA2 variants accounting for less than 20% of the genetic risk of breast cancer [5]. Due to the complex inheritance of this disorder and genetic heterogeneous nature of human populations it has been difficult to unravel novel breast cancer susceptibility/resistance genes that could elucidate novel pathways for diagnosis, treatment and prevention of breast cancer.
Two classes of rat models of mammary carcinogenesis have been frequently used; chemically-induced mammary carcinogenesis using compounds like the polycyclic aromatic hydrocarbon 7,12-dimethylbenz[a]anthracene (DMBA) [6], N-nitroso-N-methylurea (NMU) [7], 2-amino-1-methyl-6phenylimidazo[4,5-b]pyridine (PhIP) [8], estrogens [9] and radiation-induced mammary carcinogenesis [10]–[15]. Of the few reported genetic studies that have been performed in animal models of mammary carcinogenesis, all have utilized the chemically-induced model as the chosen animal model system [3]. Thus, QTLs affecting susceptibility to mammary tumors have been reported in rat intercrosses subjected to DMBA-induced [16] and 17β-Estradiol-induced [17] mammary carcinogenesis. However, there are no reports on genetic studies performed on radiation-induced mammary carcinogenesis, despite the fact that ionizing radiation is one of the few well-characterized etiologic factors of human breast cancer [18] and the well-established fact that the female breast is one of the most susceptible organs to radiation-induced cancer [19]–[22].
The objective of this study was to perform a genome scan for QTLs affecting radiation-induced tumorigenesis. The sample used was an F2 (Dahl S x R)-intercross female rat population phenotyped for radiation-induced mammary tumorigenesis after induction of rat mammary gland tumors at 40 days of age via 127Cs-radiation. We chose the Dahl S/Dahl R rat model because these two inbred rat lines were derived from the Sprague-Dawley rat [23], one of the rat strains most susceptible to radiation-induced mammary carcinogenesis [24]–[26].
Results
In order to assess potential differences in susceptibility to radiation-induced tumorigenesis we first analyzed inbred parental strains, Dahl S and Dahl R female rats, selected for the observed tendency to develop spontaneous mammary tumors in F2 [Dahl S x R]-intercross rats. We measured latency to tumor development and number of mammary gland tumors after a single dose of irradiation at 40 days of age in parental Dahl S and Dahl R female rats. As shown in Figure 1, Dahl R rats exhibited decreased tumor latency and increased tumor burden index compared with Dahl S rats (P<0.05), indicating increased radiation-induced breast cancer susceptibility in Dahl R female rats.
Figure 1. Tumor development in Dahl S and Dahl R female rats.
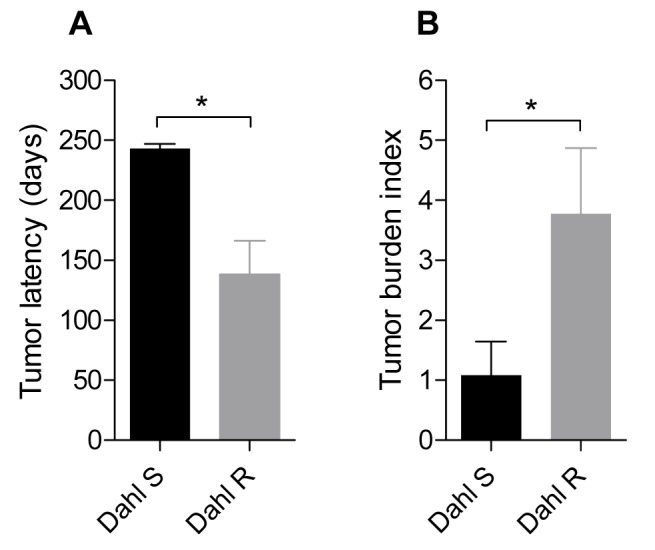
The tumor latency (A) and tumor burden index (B) for Dahl S (black bars) and Dahl R (gray bars) female rats are presented. *,P<0.05 unpaired t-test.
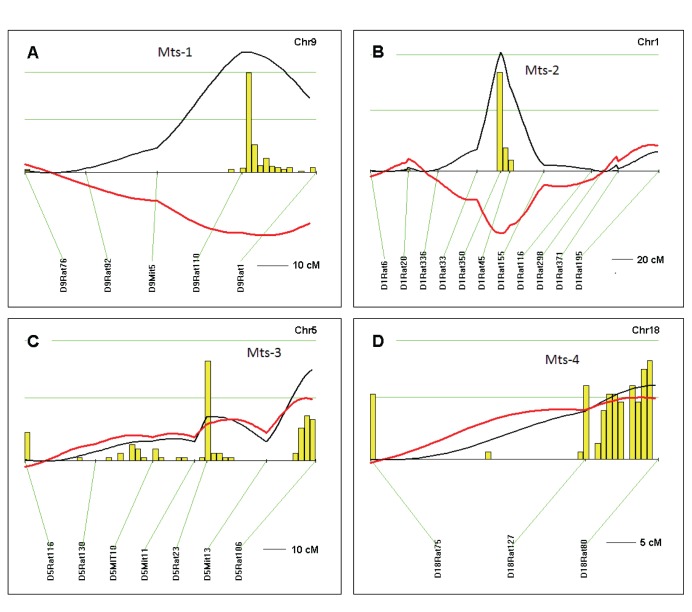
We next performed a total genome scan for QTLs affecting radiation-induced mammary tumorigenesis susceptibility (Mts) using 150 F2 (Dahl S x R)-intercross female rats phenotyped for tumor burden index (TBI) as the quantitative measure for tumorigenicity measuring both latency to tumor formation and number of tumors. We detected two Mts-QTLs with significant linkage: (Mts-1 on chromosome 9, LOD 2.98 and Mts-2 on chromosome 1, LOD 2.61; Table 1 and Figure 2). We also detected two Mts-QTLs with suggestive linkage (Mts-3 on chromosome 5, LOD 1.93 and Mts-4 on chromosome 18, LOD 1.54; Table 1 and Figure 2). Additional analysis for interactive effects on breast cancer susceptibility reveals no gene-gene interaction in this F2 (Dahl S x R)-intercross female rat cohort. Histopathological analysis of representative Hematoxylin and Eosin stained sections from seven rats detected mammary adenocarcinomas in five, including poorly differentiated adenocarcinoma (Figure 3), and fibroadenomas in two (Figure 3). This is similar to observations by Cronkite et al 1960 [27], and suggest the paradigm that tumorigenesis induced by a given environmental factor, irradiation, produces a spectrum of mammary tumor pathology.
Table 1. QTLs for mammary tumorigenesis susceptibility in F2 (Dahl S x R)-intercross female rats.
| QTL | Location | LOD | % | QTLa | Candidateb |
| Mts-1 | Chr9∶84–104 Mbp | 2.98 (S) | 9 (↓) | Prlh | |
| Mts-2 | Chr1∶125–145 Mbp | 2.61 (S) | 8 (↓) | Mcs3[16] | Anpep orBlm |
| Mts-3 | Chr5∶147–167 Mbp | 1.93 (Sug) | 6 (↑) | Ece1 | |
| Mts-4 | Chr18∶66–86 Mbp | 1.54 (Sug) | 5 (↑) | Smad2 |
QTL, quantitative trait locus; Mts, mammary tumorigenesis susceptibility; Chr, chromosome; %, the amount in % of total trait variance that would be explained by a QTL at these loci; Mbp, mega-base pair; LOD, logarithm of the odds score derived from the likelihood ratio statistic using a factor of 4.6; ↑, S-allele increases trait; ↓, S-allele decreases trait. Significance determined from 2000 permutations on data set: LOD 2.48 significant (S); LOD 1.30 suggestive (Sug).
Overlapping QTL detected in previous study: Mcs = mammary cancer susceptibility.
candidate genes in rat-human syntenic regions; Prlh, prolactin hormone; Anpep, alanyl (membrane) aminopeptidase; Blm, Bloom syndrome homolog; Ece1, endothelin-converting enzyme-1; Smad2, SMAD family member 2.
Figure 2. QTLs for mammary tumorigenesis susceptibility (Mts) in F2 [Dahl S x R]-intercross female rats.
Chromosomes with significant and suggestive QTLs were analyzed by interval mapping with bootstrap resampling method to estimate a confidence interval (QTXb19 Map Manager): Panel A, chromosome 9 (Mts-1); B, chromosome 1 (Mts-2); C, chromosome 5 (Mts-3); D, chromosome 18 (Mts-4). Yellow histograms represent the bootstrap-based confidence intervals for the detected QTLs. Orientation of chromosomes: left → right starting from lowest Mbp. Horizontal green lines mark LOD values for significance of linkage, from top to bottom: significant LOD ≥2.48; suggestive LOD ≥1.30; LOD (black line); regression coefficient for additive effect (red line).
Figure 3. Hematoxylin and Eosin stained digital photomicrographs of representative breast tumor phenotypes in the F2-cohort of radiation-induced breast tumors.
A, Adenoductal carcinoma phenotype, B, fibroadenoma. →, representative duct morphology in each section; *, hyaline (pink) fibrous tissue markedly increased in fibroadenoma.
Although QTLs still need to be narrowed to pinpoint causal genes for tumorigenicity, and specific variants identified, analysis of syntenic regions to Mts-1 to Mts-4 in humans reveals candidate genes previously implicated in breast cancer. On chromosome 9, Prlh (prolactin releasing hormone) is a candidate gene for Mts-1 QTL since Prlh maps to rat chromosome-9 at coordinate 90.15 Mbp within the Mts-1 QTL region (Table 1). Prlh modulates secretion of prolactin [28], a hormone that has been shown to be a risk factor for human breast cancer [28], [29]. Notably, for Mts-2 QTL on chromosome-1, the marker at the QTL-peak, D1Rat350, is located within the Anpep [alanyl (membrane) aminopeptidase] transcription unit, an enzyme that might be a candidate gene because it has been associated with invasive colorectal cancer [30] and prostate cancer [31], and Barrett’s adenocarcinoma [32]. Another candidate gene on chromosome 1, Blm localizes at 136.2 Mb also within the Mts-2 QTL interval (Table 1). Notably, Blm (Bloom syndrome homolog) has been implicated in breast cancer susceptibility in humans [33], [34]. The Mts-3 chromosome-5 QTL region (Table 1) harbors Ece1 (endothelin-converting enzyme-1 at 156.6 Mb), an enzyme that has also been implicated in human breast cancer [35], [36]. Finally, within the Mts-4 interval on chromosome 18 (Table 1) resides Smad2 (SMAD family member 2/mothers against decapentaplegic homolog 2, 73.18 Mb), a signaling protein whose phosphorylation mediates TGF-beta induced breast cancer invasiveness [37], [38].
Discussion
This is the first genome-wide scan for QTLs affecting radiation-induced mammary tumorigenesis in rodents. We detected two significant Mts-QTLs on chromosomes 9 and 1 and two suggestive Mts-QTLs on chromosomes 5 and 18. The chromosome 9 Mts-1, chromosome 5 -Mts-3 and chromosome 18 Mts-4 QTLs represent novel QTL regions associated with mammary tumorigenesis not previously observed in other rat intercrosses of DMBA-induced and estrogen-induced mammary tumorigenesis [3]. Mts-2 QTL spanning chromosome1 125–145 Mbp region overlaps with Mcs3 QTL that maps to chromosome1 109.1–138.8 Mbp region in a COP x WF intercross rat population phenotyped for DMBA-induced mammary carcinogenesis [16]. Interesting, both QTLs decrease susceptibility to tumorigenesis suggesting that the same gene might underlie both QTLs effects on mammary tumorigenesis. Mts-3 mapping to chromosome 5 147–167 Mbp region partially overlaps with Emca1 spanning chromosome 5 107–159 Mbp region detected in an F2 (COP x ACI)-intercross rat population characterized for estrogen-induced mammary carcinogenesis [39]. However, having different modes of inheritance, recessive for Emca1 and co-dominant or additive for Mts-3, data suggest that different genes might account for these two QTL effects on rat mammary tumorigenesis. Finally, Mts-4 at chromosome 18 66–86 Mbp region partially overlaps with Mcsta2 that peaks at 68 Mbp in a SPRD-Cu3 x WKY backcross rat population characterized for DMBA-induced mammary carcinogenesis [40]. However, Mts-4 and Mcsta have opposite effects on susceptibility with Mcsta decreasing and Mts-4 increasing susceptibility to mammary tumorigenesis suggesting that different genetic determinants underlie these QTLs effects on tumorigenesis. Altogether, the data suggest that distinct genetic determinants exist that confer susceptibility to irradiation-induced mammary tumorigenesis from those loci affecting chemically-induced and estrogen-induced mammary tumorigenesis.
As described by Cronkite et al 1960 [27], radiation-induced mammary tumor models exhibits both adenocarcinoma and fibroadenoma in Sprague Dawley rats. Notably, both adenocarcinoma and fibroadenoma were also detected in the F2[Dahl S x R] breast cancer cohort studied here concordant with the fact that both Dahl S and Dahl R rats were derived from Sprague Dawley rats selected for salt-sensitive and salt-resistant hypertension respectively. Given the same environmental insult, the spectrum of pathologies from malignant to benign, and the detection of multiple QTLs suggest that susceptibility to mammary tumorigenesis is a complex multifactorial event likely involving multiple genetic determinants and genetic modifiers. As spectrum of susceptibility, the data suggest that genetic analysis for sporadic breast cancer and fibroadenoma can be analyzed as one pathogenic event with a spectrum much like other diseases.
Although further studies are needed to identify causal genes in respective Mts-QTLs, the panel of candidate genes with reported gene expression changes or polymorphisms in breast cancer patients, Prlh for Mts-1, Anpep or Blm for Mts-2, Ece1 for Mts-3, and Smad2 for Mts-4_validate the hypotheses that these genes should be studied further in different experimental systems and in humans as potential susceptibility genes for mammary tumorigenesis. Although no statistically significant genetic interaction was detected among the QTLs, we note all Mts-QTLs 1–4 candidate genes are associated with key aspects of breast tumor progression and malignancy, which collectively could increase tumorigenesis susceptibility. Increased Prlh leading to higher prolactin levels is associated with increased risk for breast cancer, increased metastasis, disease progression, lower response to tamoxifen and worse clinical prognosis [41]. Furthermore, Prlh as a candidate gene for Mts-1 QTL is concordant with the observation that prolactin accelerated mammary tumorigenesis initiated by radiation [42]. Similarly, the candidate genes for Mts-2 QTL, Anpep or Blm are both implicated in breast cancer. Anpep is increased in breast cancer effusions [43], and its down regulation is associated with invasive colorectal cancer [30] and prostate cancer [31], while over expression of Anpep has been linked to Barrett’s adenocarcinomas [32]. Blm, the gene for Bloom syndrome, is a DNA repair gene which may play a role in breast cancer occurrence as its loss may contribute to somatic mutations and loss of heterozygosis, chromosomal instability, aneuploidy, and sensitivity to DNA damaging agents [44]. For Mts-3 QTL candidate gene, Ece1, cumulative studies point to its role in breast cancer invasiveness and more frequent recurrence [45]. Lastly, Mts-4 QTL candidate gene, Smad2, underlies Smad2-dependent epithelial mesenchymal transition of breast cancer cells [46], a key step in invasiveness and subsequent metastasis. Intriguingly, the deduced synergisms from these breast cancer roles for each Mts QTL candidate gene suggest the hypothesis that QTL-burden could increase risk, reiterating the need for further study.
Further inspection of the Rat Genome Database (RGD) reveals additional candidate genes within Mts-1, Mts-2, Mts-3 and Mts-4 QTLs that have been associated with different types of cancers (Table 2). However, more studies involving fine mapping through sub-strain construction will be necessary to identify the genes underlying these QTLs.
Table 2. Genes associated with cancer within mammary tumorigenesis susceptibility QTLs detected in F2 (Dahl S x R)-intercross female rats.
| Symbol | Description | Location (nt) | Association [ref] |
| Mts-1 (Chr9) | |||
| Ugt1a7c | UDP glucuronosyltransferase 1 family, polypeptide A7C | 87029500–87098362 | Pancreatic cancer [47] |
| Ugt1a1 | UDP glucuronosyltransferase 1 family, polypeptide A1 | 87091241–87098362 | Endometrial cancer [48], ovarian cancer [49] |
| Pam | Peptidylglycine alpha-amidating monooxygenase | 96893071–97047523 | Prostate cancer [50] |
| Ralbp1 | RalA binding protein 1 | 104617400–104653856 | Bladder cancer [51] |
| Mts-2 (Chr1) | |||
| Ntrk3 | Neurotrophic tyrosine kinase, receptor, type 3 | 133925530–134302139 | Pancreatic cancer [52] |
| Fzd4 | Frizzled family receptor 4 | 145953743–145957666 | Prostate cancer [53] |
| Mts-3 (Chr5) | |||
| Hdac1 | Histone deacetylase 1 | 148672515–148699810 | Breast cancer [54], cervical cancer [55],endometrial cancer [56], ovarian cancer [56] |
| Sfn | Stratifin | 151475395–151479996 | Breast cancer [57], endometrialcancer [58], prostate cancer (57) |
| Runx3 | Runt-related transcription factor 3 | 153950116–153973141 | Breast cancer [59], ovarian cancer [60],colorectal cancer [61] |
| Wnt4 | Wingless-type MMTV integration site family, member 4 | 156064371–156083198 | Endometrial cancer [62] |
| Sdhb | Succinate dehydrogenase complex, subunit B, iron sulfur(Ip) | 159818669–159839772 | Renal cancer [63] |
| Casp9 | Caspase 9, apoptosis-related cysteine peptidase | 160704234–160721796 | Breast cancer [64] |
| Tnfrsf1b | Tumor necrosis factor receptor superfamily, member 1b | 163666541–163697484 | Renal cancer [65] |
| Mts-4 (Chr18) | |||
| Smad4 | SMAD family member 4 | 70432705–70461541 | Ovarian cancer [66], endometrial cancer [67] |
| Smad7 | SMAD family member 7 | 72294803–72323354 | Endometrial cancer [68] |
Table legend: Genes and gene locations on rat chromosomes 1, 5, 9 and 18 regions as per RGD. nt, nucleotide; ref, reference.
In summary, the genome-wide scan for QTLs influencing breast cancer susceptibility identified genetic linkage to chromosomes 9, 1, 5 and 18 with radiation-induced mammary tumorigenesis in Dahl rats. The chromosomes 9, 5 and 18 QTLs were unique to this F2 (Dahl S x R)-intercross rat population suggesting that genetic mechanisms underlying radiation-induced mammary tumorigenesis differs substantially from mechanisms involved in chemically-induced and hormone-induced mammary tumorigenesis. Histopathology observations of a spectrum from poorly differentiated adenocarcinoma to benign fibroadenoma despite exposure to identical radiation dose in our F2[Dahl S x R]-intercross rat mammary tumor cohort suggest the importance of genetic susceptibility and/or gene modifiers in this gene-environment interaction paradigm.
Altogether, the detection of Mts- QTLs spanning fibroadenoma to poorly differentiated adenocarcinoma suggest the paradigm that genetic analysis of tumorigenesis-QTLs in humans could help elucidate breast cancer susceptibility genes which have remained elusive to date. Alternatively, the detection of candidate genes within Mts-QTLs associated with breast cancer in humans validates their analysis as causal susceptibility genes in human cohorts respectively. Elucidation of variants accounting for mammary tumorigenesis in Dahl rats will give insights into gene pathways important for gene-environment interactions, tumor initiation and progression.
Materials and Methods
Ethics Statement
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of Boston University School of Medicine (Permit Number: AN-13924).
Genetic Crosses
Dahl S/jrHsd and Dahl R/jrHsd rats were obtained from Harlan (Indianapolis, Indiana). Parental strains (Dahl R/jrHsd female x Dahl S/jrHsd male) were crossed to produce F1 progeny. The F2 subjects were derived from brother-to-sister mating of F1 hybrids to produce the F2 female (n = 150) segregating population.
Phenotypic Characterization and Genotyping
Animals were maintained on a LabDiet 5001 rodent chow (Harlan Teklad, Madison WI) containing 0.23% NaCl. We induced rat mammary gland tumors in 12 Dahl S/jrHsd, 12 Dahl R/jrHsd and 150 F2 female subjects as described [27] at 40 days of age via 127Cs-radiation. Rats were exposed to 400 r of whole body radiation. All subjects were sacrificed when tumor burden reached a total of 3 cm in diameter for animals harboring one or multiple tumors or at 12 months of age for those that did not develop tumors. Three out of 12 Dahl S/jrHsd, seven out 12 Dahl R/jrHsd and ninety eight out of 150 F2 female rats developed mammary tumors. Seven F2 female rat tumors were randomly selected for histological analysis (five were found to be carcinomas and two were fibroadenomas). Routine Hematoxylin and Eosin stained histological sections were obtained from 4% paraformaldehyde fixed tumor tissues and analyzed with a clinical tumor pathologist (Michael J O’Brien, MD, MPH) at Boston Medical Center. Tumor latency was computed from 40 days of age (at time of radiation). Tumor burden index was computed using the formula TBI = 1+[Tx2]+[TLH/TL] (T = # tumors; TLH = highest tumor latency; TL = tumor latency) as described [69]. We used 112 microsatellite markers informative for our F2 (Dahl S x R)-intercross spanning the whole genome (including all 20 autosomes plus the X chromosome) with an average marker density of 12.7 cM. Microsatellite markers were PCR genotyped and detected by 6% denaturing polyacrylamide gel electrophoresis.
Intercross Linkage Analysis
Phenotype distributions were analyzed for normality; data transformations were done when necessary and datasets that passed Kolmogorov-Smirnov normality testing (SigmaPlot11.2) were used for linkage analysis. QTL analysis was performed using the tumor burden index (TBI) as quantitative trait that was computed using the formula TBI = 1+[Tx2]+[TLH/TL] (T = # tumors; TLH = highest tumor latency; TL = tumor latency) as described [69]. Linkage maps, marker regression and composite interval mapping were done with the Map Manager QTXb19 (MMQTXb19) program for windows [70] which generates a likelihood ratio statistic (LRS) as a measure of the significance of a possible QTL. Genetic distances were calculated using Kosambi mapping function (genetic distances are expressed in centiMorgan, cM). Critical significance values (LRS values) for interval mapping were determined by a permutation test (2000 permutations at all loci tested) on our female cohort using Kosambi mapping function and an additive regression model. Values for suggestive linkage LRS = 6.0 (LOD 1.30) and for significant linkage LRS = 11.4 (LOD 2.48). LRS 4.6 delineates LOD 1-support interval. Confidence interval for a QTL location was estimated by bootstrap resampling method wherein histogram single peak delineates the QTL and peak widths define confidence interval for the QTL. Histograms which show more than one peak warn that the position for the QTL is not well defined or that there may be multiple linked QTLs (QTX Map Manager). We also performed interaction analysis using the Map Manager QTXb19 program applying a two-stage test paradigm for determination of interaction in which the pair of loci must pass two tests in order to be reported as having a significant interaction effect. First, the significance of the total effect of the two loci must be <0.00001 and second, the pairs of loci must exhibit a P value <0.01 for the interaction effect.
Funding Statement
The work was supported by grant HL58136 from the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bray F, McCarron P, Parkin DM (2004) The changing global patterns of female breast cancer incidence and mortality. Breast Cancer Res 6: 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Parkin DM (2001) Global cancer statistics in the year 2000. Lancet Oncol 2: 533–543. [DOI] [PubMed] [Google Scholar]
- 3. Shull JD (2007) The rat oncogenome: comparative genetics and genomics of rat models of mammary carcinogenesis. Breast Disease 28: 69–86. [DOI] [PubMed] [Google Scholar]
- 4. Szpirer C, Szpirer J (2007) Mammary cancer susceptibility: human genes and rodent models. Mamm Genome 18: 817–831. [DOI] [PubMed] [Google Scholar]
- 5. Easton DF (1999) How many more breast cancer predisposition genes are there? Breast Cancer Res 1: 14–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huggins C, Grand LC, Brillantes FP (1961) Mammary cancer induced by a single feeding of polynuclear hydrocarbons, and its suppression. Nature 189: 204–207. [DOI] [PubMed] [Google Scholar]
- 7. Gullino PM, Pettigrew HM, Grantham FH (1975) N-Nitrosomethylurea as mammary gland carcinogen in rats. J Natl Cancer Inst 54: 401–414. [PubMed] [Google Scholar]
- 8. Ito N, Hasegawa R, Sano M, Tamano S, Esumi H, et al. (1991) A new colon and mammary carcinogen in cooked food, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). Carcinogenesis 12: 1503–1506. [DOI] [PubMed] [Google Scholar]
- 9. Harvell DM, Strecker TE, Xie B, Buckles LK, Tochacek M, et al. (2001) Diet-gene interactions in estrogen-induced mammary carcinogenesis in the ACI rat. J Nutrition 131: 3087S–3091S. [DOI] [PubMed] [Google Scholar]
- 10. Gragtmans NJ, Myers DK, Johnson JR, Jones AR, Johnson LD (1984) Occurrence of mammary tumors in rats after exposure to tritium beta rays and 200-kVp X rays. Radiat Res 99: 636–650. [PubMed] [Google Scholar]
- 11. Jacrot M, Mouriquand J, Mouriquand C, Saez S (1979) Mammary carcinogenesis in Sprague-Dawley rats following 3 repeated exposures to 14.8 MeV neutrons and steroid receptor content of these tumor types. Cancer Lett 8: 147–153. [DOI] [PubMed] [Google Scholar]
- 12. Lemon HM, Kumar PF, Peterson C, Rodriguez-Sierra JF, Abbo KM (1989) Inhibition of radiogenic mammary carcinoma in rats by estriol or tamoxifen. Cancer 63: 1685–1692. [DOI] [PubMed] [Google Scholar]
- 13. Mandybur TI, Ormsby I, Samuels S, Mancardi GL (1985) Neural, pituitary and mammary tumors in Sprague-Dawley rats treated with X irradiation to the head and N-ethyl-N-nitrosourea (ENU) during the early postnatal period: a statistical study of tumor incidence and survival. Radiat Res 101: 460–472. [PubMed] [Google Scholar]
- 14. Shellabarger CJ, Cronkite EP, Bond VP, Lippincott SW (1957) The occurrence of mammary tumors in the rat after sublethal whole-body irradiation. Radiat Res 6: 501–512. [PubMed] [Google Scholar]
- 15. Welsch CW, Goodrich-Smith M, Brown CK, Miglorie N, Clifton KH (1981) Effect of an estrogen antagonist (tamoxifen) on the initiation and progression of gamma-irradiation-induced mammary tumors in female Sprague-Dawley rats. Eur J Cancer Clin Oncol 17: 1255–1258. [DOI] [PubMed] [Google Scholar]
- 16. Shepel LA, Lan H, Haag JD, Brasic GM, Gheen ME, et al. (1998) Genetic identification of multiple loci that control breast cancer susceptibility in the rat. Genetics 149: 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schaffer BS, Lachel CM, Pennington KL, Murrin CR, Strecker TE, et al. (2006) Genetic bases of estrogen-induced tumorigenesis in the rat: mapping of loci controlling susceptibility to mammary cancer in a Brown Norway x ACI intercross. Cancer Res 66: 7793–7800. [DOI] [PubMed] [Google Scholar]
- 18. Smith-Bindman R (2012) Environmental causes of breast cancer and radiation from medical imaging: findings from the Institute of Medicine report. Arch Intern Med 172: 1023–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Preston DL, Mattsson A, Holmberg E, Shore R, Hildreth NG, et al. (2002) Radiation effects on breast cancer risk: a pooled analysis of eight cohorts. Radiat Res 158: 220–235. [DOI] [PubMed] [Google Scholar]
- 20. Preston DL, Ron E, Tokuoka S, Funamoto S, Nishi N, et al. (2007) Solid cancer incidence in atomic bomb survivors: 1958–1998. Radiat Res 168: 1–64. [DOI] [PubMed] [Google Scholar]
- 21. Ronckers CM, Erdmann CA, Land CE (2004) Radiation and breast cancer: a review of current evidence. Breast Cancer Res 7: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thompson DE, Mabuchi K, Ron E, Soda M, Tokunaga M, et al. (1994) Cancer incidence in atomic bomb survivors. Part II: Solid tumors, 1958–1987. Radiat Res 137: S17–S67. [PubMed] [Google Scholar]
- 23. Dahl LK, Heine M, Tassinari L (1962) Effects of chronic salt ingestion evidence that genetic factors play an important role in susceptibility to experimental hypertension. J Exp Med 115: 1173–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Imaoka T, Nishimura M, Kakinuma S, Hatano Y, Ohmachi Y, et al. (2007) High relative biologic effectiveness of carbon ion radiation on induction of rat mammary carcinoma and its lack of H-ras and Tp53mutations. Int J Radiat Oncol Biol Phys 69: 194–203. [DOI] [PubMed] [Google Scholar]
- 25. Shellabarger CJ, Stone JP, Holtzman S (1978) Rat differences in mammary tumor induction with estrogen and neutron radiation. J Natl Cancer Inst 61: 1505–1508. [PubMed] [Google Scholar]
- 26. Vogel HH Jr, Turner JE (1982) Genetic component in rat mammary carcinogenesis. Radiat Res 89: 264–273. [PubMed] [Google Scholar]
- 27. Cronkite EP, Shellabarger CJ, Bond VP, Lippincott SW (1960) Studies on radiation-induced mammary gland neoplasia in the rat. I. The role of the ovary in the neoplastic response of the breast tissue to total- or partial-body X-irradiation. Radiat Res 12: 81–93. [PubMed] [Google Scholar]
- 28. Bernichtein S, Touraine P, Goffin V (2010) New concepts in prolactin biology. J Endocrinol 206: 1–11. [DOI] [PubMed] [Google Scholar]
- 29. Tran TH, Utama FE, Lin J, Yang N, Sjolund AB, et al. (2010) Prolactin inhibits BCL6 expression in breast cancer through a Stat5a-dependent mechanism. Cancer Res 70: 1711–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wiese AH, Auer J, Lassmann S, Nahrig J, Rosenberg R, et al. (2007) Identification of gene signatures for invasive colorectal tumor cells. Cancer Detect Prev 31: 282–295. [DOI] [PubMed] [Google Scholar]
- 31. Sorensen KD, Abildgaard MO, Haldrup C, Ulhoi BP, Kristensen H, et al. (2013) Prognostic significance of aberrantly silenced ANPEP expression in prostate cancer. Br J Cancer 108: 420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Razvi MH, Peng D, Dar AA, Powell SM, Frierson HF Jr, et al. (2007) Transcriptional oncogenomic hot spots in Barrett’s adenocarcinomas: serial analysis of gene expression. Genes Chromosomes Cancer 46: 914–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Broberg K, Huynh E, Schlawicke Engstrom K, Bjork J, Albin M, et al. (2009) Association between polymorphisms in RMI1, TOP3A, and BLM and risk of cancer, a case-control study. BMC Cancer 9: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ding SL, Yu JC, Chen ST, Hsu GC, Kuo SJ, et al. (2009) Genetic variants of BLM interact with RAD51 to increase breast cancer susceptibility. Carcinogenesis 30: 43–49. [DOI] [PubMed] [Google Scholar]
- 35. Grimshaw MJ (2005) Endothelins in breast tumour cell invasion. Cancer Lett 222: 129–138. [DOI] [PubMed] [Google Scholar]
- 36. Smollich M, Gotte M, Yip GW, Yong ES, Kersting C, et al. (2007) On the role of endothelin-converting enzyme-1 (ECE-1) and neprilysin in human breast cancer. Breast Cancer Res Treat 106: 361–369. [DOI] [PubMed] [Google Scholar]
- 37. Papageorgis P, Lambert AW, Ozturk S, Gao F, Pan H, et al. (2010) Smad signaling is required to maintain epigenetic silencing during breast cancer progression. Cancer Res 70: 968–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Petersen M, Pardali E, van der Horst G, Cheung H, van den Hoogen C, et al. (2010) Smad2 and Smad3 have opposing roles in breast cancer bone metastasis by differentially affecting tumor angiogenesis. Oncogene 29: 1351–1361. [DOI] [PubMed] [Google Scholar]
- 39. Gould KA, Tochacek M, Schaffer BS, Reindl TM, Murrin CR, et al. (2004) Genetic determination of susceptibility to estrogen-induced mammary cancer in the ACI rat: mapping of Emca1 and Emca2 to chromosomes 5 and 18. Genetics 168: 2113–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Quan X, Laes JF, Stieber D, Riviere M, Russo J, et al. (2006) Genetic identification of distinct loci controlling mammary tumor multiplicity, latency and aggressiveness in the rat. Mamm Genome 17: 310–321. [DOI] [PubMed] [Google Scholar]
- 41. Damiano JS, Wasserman E (2013) Molecular pathways: blockade of the PRLR signaling pathway as a novel antihormonal approach for the treatment of breast and prostate cancer. Clin Cancer Res 19: 1644–1650. [DOI] [PubMed] [Google Scholar]
- 42. Inano H, Suzuki K, Onoda M, Kobayashi H, Wakabayashi K (1999) Radiation-induced tumorigenesis of mammary glands in pituitary transplanted rats ovariectomized before onset of estrous cycle. Cancer Lett 138: 93–100. [DOI] [PubMed] [Google Scholar]
- 43. Stavnes HT, Nymoen DA, Langerod A, Holth A, Borresen Dale AL, et al. (2013) AZGP1 and SPDEF mRNA expression differentiates breast carcinoma from ovarian serous carcinoma. Virchows Arch 462: 163–173. [DOI] [PubMed] [Google Scholar]
- 44.Sassi A, Popielarski M, Synoqiec E, Morawiec Z, Wozniak K (2013) BLM and RAD51 genes polymorphism and susceptibility to breast cancer. Pathol Oncol Res DOI 10.1007/s12253-013-9602-8. [DOI] [PMC free article] [PubMed]
- 45. Smollich M, Gotte M, Yip GW, Yong ES, Kersting C, et al. (2007) On the role of endothelin-converting enzyme (ECE-1) and neprilysin in human breast cancer. Breast Cancer Res Treat 106: 361–369. [DOI] [PubMed] [Google Scholar]
- 46. Lv ZD, Kong B, Li JG, Qu HL, Wang XG, et al. (2013) Transforming growth factor-beta1 enhances the invasiveness of breast cancer cells by inducing a Smad2-dependent epithelial-to-mesenchymal transition. Oncol Rep 29: 219–225. [DOI] [PubMed] [Google Scholar]
- 47. Ockenga J, Vogel A, Teich N, Keim V, Manns MP, et al. (2003) UDP glucuronosyltransferase (UGT1A7) gene polymorphisms increase the risk of chronic pancreatitis and pancreatic cancer. Gastroenterology 124: 1802–1808. [DOI] [PubMed] [Google Scholar]
- 48. Deming SL, Zheng W, Xu WH, Cai Q, Ruan Z, et al. (2008) UGT1A1 genetic polymorphisms, endogenous estrogen exposure, soy food intake, and endometrial cancer risk. Cancer Epidemiol Biomarkers Prev 17: 563–570. [DOI] [PubMed] [Google Scholar]
- 49. Cecchin E, Russo A, Corona G, Campagnutta E, Martella L, et al. (2004) UGT1A1*28 polymorphism in ovarian cancer patients. Oncol Rep 12: 457–462. [PubMed] [Google Scholar]
- 50. Rocchi P, Boudouresque F, Zamora AJ, Muracciole X, Lechevallier E, et al. (2001) Expression of adrenomedullin and peptide amidation activity in human prostate cancer and in human prostate cancer cell lines. Cancer Res 61: 1196–1206. [PubMed] [Google Scholar]
- 51. Smith SC, Oxford G, Baras AS, Owens C, Havaleshko D, et al. (2007) Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res 13: 3803–3813. [DOI] [PubMed] [Google Scholar]
- 52. Sakamoto Y, Kitajima Y, Edakuni G, Sasatomi E, Mori M, et al. (2001) Expression of Trk tyrosine kinase receptor is a biologic marker for cell proliferation and perineural invasion of human pancreatic ductal adenocarcinoma. Oncol Rep 8: 477–484. [DOI] [PubMed] [Google Scholar]
- 53. Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, et al. (2007) Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell 12: 559–571. [DOI] [PubMed] [Google Scholar]
- 54. Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, et al. (2005) Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast. Breast Cancer Res Treat 94: 11–16. [DOI] [PubMed] [Google Scholar]
- 55. Lin Z, Bazzaro M, Wang MC, Chan KC, Peng S, et al. (2009) Combination of proteasome and HDAC inhibitors for uterine cervical cancer treatment. Clin Cancer Res 15: 570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weichert W, Denkert C, Noske A, Darb-Esfahani S, Dietel M, et al. (2008) Expression of class I histone deacetylases indicates poor prognosis in endometrioid subtypes of ovarian and endometrial carcinomas. Neoplasia 10: 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Horie-Inoue K, Inoue S (2006) Epigenetic and proteolytic inactivation of 14-3-3sigma in breast and prostate cancers. Semin Cancer Biol 16: 235–239. [DOI] [PubMed] [Google Scholar]
- 58. Nakayama H, Sano T, Motegi A, Oyama T, Nakajima T (2005) Increasing 14-3-3 sigma expression with declining estrogen receptor alpha and estrogen-responsive finger protein expression defines malignant progression of endometrial carcinoma. Pathol Int 55: 707–715. [DOI] [PubMed] [Google Scholar]
- 59. Subramanian MM, Chan JY, Soong R, Ito K, Ito Y, et al. (2009) RUNX3 inactivation by frequent promoter hypermethylation and protein mislocalization constitute an early event in breast cancer progression. Breast Cancer Res Treat 113: 113–121. [DOI] [PubMed] [Google Scholar]
- 60. Nevadunsky NS, Barbieri JS, Kwong J, Merritt MA, Welch WR, et al. (2009) RUNX3 protein is overexpressed in human epithelial ovarian cancer. Gynecol Oncol 112: 325–330. [DOI] [PubMed] [Google Scholar]
- 61. Imamura Y, Hibi K, Koike M, Fujiwara M, Kodera Y, et al. (2005) RUNX3 promoter region is specifically methylated in poorly-differentiated colorectal cancer. Anticancer Res 25: 2627–2630. [PubMed] [Google Scholar]
- 62. Bui TD, Zhang L, Rees MC, Bicknell R, Harris AL (1997) Expression and hormone regulation of Wnt2, 3, 4, 5a, 7a, 7b and 10b in normal human endometrium and endometrial carcinoma. Br J Cancer 75: 1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bardella C, Pollard PJ, Tomlinson I (2011) SDH mutations in cancer. Biochim Biophys Acta 1807: 1432–1443. [DOI] [PubMed] [Google Scholar]
- 64. Valladares A, Hernandez NG, Gomez FS, Curiel-Quezada E, Madrigal-Bujaidar E, et al. (2006) Genetic expression profiles and chromosomal alterations in sporadic breast cancer in Mexican women. Cancer Genet Cytogenet 170: 147–151. [DOI] [PubMed] [Google Scholar]
- 65. Al-Lamki RS, Sadler TJ, Wang J, Reid MJ, Warren AY, et al. (2010) Tumor necrosis factor receptor expression and signaling in renal cell carcinoma. Am J Pathol 177: 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Takakura S, Okamoto A, Saito M, Yasuhara T, Shinozaki H, et al. (1999) Allelic imbalance in chromosome band 18q21 and SMAD4 mutations in ovarian cancers. Genes Chromosomes Cancer 24: 264–271. [DOI] [PubMed] [Google Scholar]
- 67. Zhou Y, Kato H, Shan D, Minami R, Kitazawa S, et al. (1999) Involvement of mutations in the DPC4 promoter in endometrial carcinoma development. Mol Carcinog 25: 64–72. [DOI] [PubMed] [Google Scholar]
- 68. Dowdy SC, Mariani A, Reinholz MM, Keeney GL, Spelsberg TC, et al. (2005) Overexpression of the TGF-beta antagonist Smad7 in endometrial cancer. Gynecol Oncol 96: 368–373. [DOI] [PubMed] [Google Scholar]
- 69. Schmid MH, Bird P, Dummer R, Kempf W, Burg G (1999) Tumor burden index as a prognostic tool for cutaneous T-cell lymphoma. Arch Dermatol 135: 1204–1208. [DOI] [PubMed] [Google Scholar]
- 70. Manly KF, Cudmore RH Jr, Meer JM (2001) Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome 12: 930–932. [DOI] [PubMed] [Google Scholar]