Abstract
Genetic instability can be induced by unusual DNA structures and sequence repeats. We have previously demonstrated that a large palindrome in the mouse germ line derived from transgene integration is extremely unstable and undergoes stabilizing rearrangements at high frequency, often through deletions that produce asymmetry. We have now characterized other palindrome rearrangements that arise from complex homologous recombination events. The structure of the recombinants is consistent with homologous recombination occurring by a noncrossover gene conversion mechanism in which a break induced in the palindrome promotes homologous strand invasion and repair synthesis, similar to mitotic break repair events reported in mammalian cells. Some of the homologous recombination events led to expansion in the size of the palindromic locus, which in the extreme case more than doubled the number of repeats. These results may have implications for instability observed at naturally occurring palindromic or quasipalindromic sequences.
Keywords: palindrome, hairpin, sequence repeats, sister chromatid recombination
Perfect palindromes and inverted repeats separated by spacers, both of which have the potential to form secondary structures, manifest genetic instability in Escherichia coli (1), yeast (2), and in the mouse (3, 4). As a result, palindromes and inverted repeats are considered “at-risk motifs” for the genome (2). The degree of instability associated with palindromes and inverted repeats depends on several factors, such as the repeat length, the distance between repeats, and the homology shared by the repeats (1, 5). A history of inverted repeat instability has been suggested during the evolution of the human genome by the analysis of the distribution of Alu elements. The superabundance and dense clustering of Alu repeats predicts that nearby inverted Alu elements would be frequent in the genome. However, closely spaced highly homologous inverted Alu repeats are substantially underrepresented relative to direct repeats in regions able to be sequenced, as if they may have been removed from the genome (5, 6).
In E. coli, long palindromes confer inviability to replicons, presumably because of the induction of strand breaks that lead to degradation of the replicon (1). In eukaryotes, long palindromes can be propagated, but they nevertheless manifest instability. For example, in yeast, a 2-kb palindrome stimulates intra- and interchromosomal homologous recombination three and four orders of magnitude, respectively, and is also deleted at high frequency (7). A much smaller palindrome of 140 bp has been shown to create meiotic recombination hotspots by generating sites for double-strand breaks (DSBs) (8).
In an earlier report, we demonstrated that a long palindrome in the mouse germ line manifests a high degree of instability. The previously characterized events were primarily nonhomologous deletions at the center of symmetry, although we had also observed frequent gene conversion events within direct repeats contained in the palindrome by using a β-galactosidase sperm assay (4, 9). We now show that other outcomes of homologous recombination occur at high frequency at repeat sequences within or adjacent to the palindrome, approaching 50% in some mice. In particular, recombination events can result in repeat expansion, substantially increasing the size of the palindromic locus. The expansion events are most easily explained by a gene conversion mechanism that does not involve reciprocal exchange.
Materials and Methods
Transgenic Mice and Sperm Analysis.
The derivations of mouse lines 78 and 2275 have been previously described (4). Mouse lines were maintained by breeding with (C57BL/6 × CBA/Ca)F1 mice, and transgenic mice were identified by Southern blot analysis of tail-tip DNA by using the entire lacZ gene as probe.
Sperm preparations were similar to those previously described (4). Sperm were squeezed from the caudal epididymis of adult males into a PBS solution (PBS with 10 mM Hepes and 1% BSA). Sperm (2 × 106) were placed in 0.5 ml of PBS solution, washed twice, and then resuspended in 0.2 ml of PBS solution. For staining, 20 μl (2 × 105) of sperm was added to 30 μl of the PBS solution, and then 50 μl of 1 mM 5-chloromethylfluorescein di-β-d-galactopyranoside (CM-FDG; Molecular Probes) was added. After 60 sec, 1 ml of PBS solution was added. Sperm were analyzed on a FACScan (Becton Dickinson) after 30 min.
Results
Gene Conversion Within the Line 78 Palindromic Transgene.
We have previously introduced a gene conversion reporter substrate into the mouse germ line that, on integration, formed a large palindrome (4). The palindrome, designated 78 for the number of the founder mouse, consists of two complete copies of the reporter substrate (78 parent; Fig. 1A). Each copy is 7.65 kb and is comprised of two defective lacZ genes, lacZ4Δ, which contains a 4-bp deletion within an otherwise intact lacZ gene and inlacZ, which is a 1.8-kb internal lacZ fragment. In addition to creating a frameshift, the 4-bp deletion in lacZ4Δ disrupts a SacI site. A simple gene conversion (SGC) involving the lacZ4Δ and inlacZ genes can restore the sequence at the SacI site to create a lacZ+ gene (78 lacZ+; Fig. 1A). Because the promoter for the lacZ4Δ gene is derived from the spermatid-specific protamine 1 (Prm-1) gene, lacZ+ expression is detected postmeiotically by assaying β-galactosidase activity in spermatids or sperm.
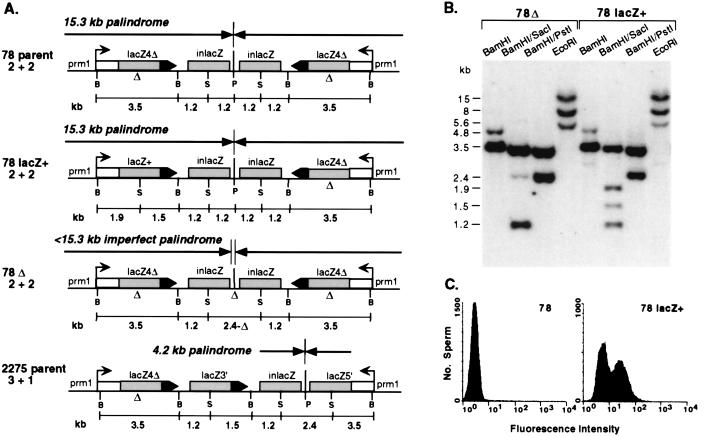
Figure 1.
(A) Structure of the transgene palindrome in mouse line 78 and derivative transgene structures. The line 78 palindrome consists of two copies of a lacZ gene conversion substrate that was injected into fertilized mouse eggs as a PstI (P) fragment. Each copy of the substrate consists of two defective lacZ genes, lacZ4Δ and inlacZ. The shaded regions denote the 1.8-kb homologous sequences. A SGC in which one of the lacZ4Δ genes is converted by one of the inlacZ repeats results in a lacZ+ gene. A common rearrangement in the line 78 palindrome is a central deletion designated 78Δ, which produces central asymmetry. Frequently, the central deletion encompasses the central PstI site but does not extend into the lacZ repeats, which are 270 bp away. One unique rearrangement, found in mouse 2275, is an inversion arising from recombination of one inlacZ repeat with an oppositely oriented lacZ4Δ gene. This rearrangement results in a smaller palindrome and a conversion of the (2 + 2) arrangement of repeats to a (3 + 1) arrangement. The promoter upstream of the outer lacZ genes is the mouse protamine 1 (Prm1) promoter, which is active postmeiotically in the male germ line (37). lacZ4Δ , lacZ gene with a 4-bp deletion (Δ) at a SacI site (S); inlacZ, internal lacZ fragment; lacZ3′, 3′ portion of lacZ; lacZ5′, 5′ portion of lacZ; B, BamHI. (B) Southern blot analysis of tail-tip DNA from mice with 78Δ and 78 lacZ+ transgene structures. The probe in this and subsequent blots is a lacZ gene fragment. (C) Flow cytometric analysis to detect β-galactosidase activity in sperm from mice with lacZ transgene structures 78 and 78 lacZ+. The assay is based on the expression of a recombined lacZ+ gene in spermatids, with retention of the expressed β-galactosidase in mature sperm (38). Caudal epididymal sperm were incubated with the fluorogenic β-galactoside, CM-FDG. For the line 78 mouse, a small number of sperm (≈0.5%) are detected to have fluorescence in the interval above the background sperm fluorescence. For the 78 lacZ+ mouse, ≈50% of the sperm are β-galactosidase positive. Although sharing of β-galactosidase occurs in the spermatid syncitia, a portion of the mature sperm in the caudal epididymus do not completely retain β-galactosidase activity (38).
The parental line 78 transgene locus was transmitted in a Mendelian fashion but rearranged at high frequency (4). Approximately 15–30% of progeny from mice with the palindrome had variant arrangements that frequently involved deletions at the center of symmetry at or adjacent to the central PstI site (78Δ; Fig. 1A). The central deletions in the 78Δ mice created an imperfect palindromic structure, i.e., an inverted repeat with a unique spacer sequence. Progeny from mice with the 78Δ structures stably transmitted the deletions, such that new transgene deletions or other rearrangements were found in <1% of progeny.
In addition to central deletions, parental line 78 mice were also found to undergo SGC within the transgene to create a lacZ+ gene (4). When sperm isolated from line 78 transgenic mice were incubated with a fluorogenic β-galactoside, ≈0.5% were β-galactosidase positive, as assayed by flow cytometry. The SGC events were substantially reduced in 78Δ mice relative to parental 78 mice (<0.02% of sperm), even when the lacZ repeats were intact (4). Thus, an imperfect palindrome is stabilized from further rearrangement, such that both SGC and nonhomologous deletions are significantly less frequent than with a perfect palindrome.
Previously, we had not reported obtaining progeny mice with a recombined transgene. However, more recently we obtained 78 lacZ+ recombinant mice in which one copy of the lacZ4Δ gene was converted to a lacZ+ gene to contain a SacI site at the former position of the 4-bp deletion (Fig. 1B). These SGC events could have presumably occurred between lacZ4Δ and inlacZ genes present on the same chromatids or sister chromatids, on either the same side or opposite sides of the center of symmetry. Flow cytometric analysis confirmed that recombinants identified by Southern blot analysis had β-galactosidase-positive sperm (Fig. 1C).
Other Homologous Recombination Events Stimulated by the Palindrome: Complex Gene Conversion (CGC) Events.
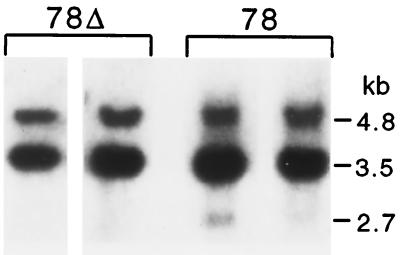
In addition to the SGC events, homologous recombination events were detected that did not restore lacZ function. These events were detected at less than single copy level within tail-tip DNA from most mice with otherwise parental 78 transgene structures (Fig. 2). Restriction analysis of DNA from line 78 mice by using BamHI digestion is expected to give two fragments of 4.8 and 3.5 kb. In addition to these two fragments, a weak 2.7-kb fragment was also consistently present at varying intensity (Fig. 2). A fragment of this size is expected to arise from recombination between lacZ4Δ and inlacZ genes, which would produce a lacZ3′ gene (see below). This 2.7-kb recombinant fragment was not detected in mice containing even a small deletion at the center of symmetry (78Δ; Fig. 2).
Figure 2.
Southern blot analysis of tail-tip DNA from mice with lacZ transgene structures 78Δ and 78. DNA from two different mice for each genotype was cleaved with BamHI. The 2.7-kb recombinant lacZ3′ fragment is visible in mice with the parental line 78 structure at varying subsingle copy intensities but not in mice with a central deletion because the transgene locus is stabilized.
To determine whether this recombinant fragment would be transmitted, tail-tip DNA from line 78 progeny was analyzed by Southern analysis. As reported previously, one mouse containing this fragment (number 2275) had an inversion within the transgene locus (4). The inversion maintained a palindromic structure, but the size of the palindrome was 4.2 kb, reduced from the parental line 78 palindrome of 15.3 kb (2275 parent; Fig. 1A). This inversion was apparently the result of crossing-over between lacZ4Δ and inlacZ genes located on opposite sides of the center of symmetry. The inversion created truncated lacZ genes, i.e., lacZ3′ which was on a 2.7 kb BamHI fragment and lacZ5′, and was accompanied by conversion of the 4-bp deletion in the recombining lacZ4Δ gene, such that both lacZ3′ and lacZ5′ were SacI+. As a result of the inversion, the four lacZ repeats in mouse line 2275 were arranged in a 3 + 1 configuration around the palindrome's center of symmetry, rather than the original 2 + 2 structure present in line 78 mice.
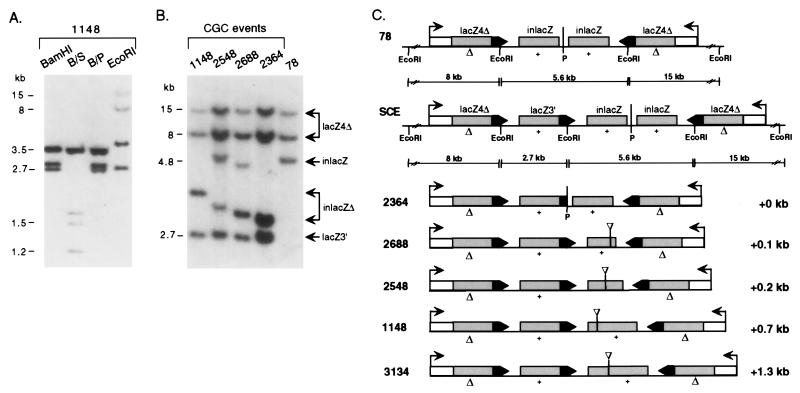
In addition to the inversion, more complex recombinant transgene structures were also obtained (Fig. 3). For example, mouse 1148 contained the 2.7-kb recombinant BamHI fragment, as well as the parental 3.5-kb fragment, but lost the 4.8-kb fragment that contained the inlacZ repeat and instead had a novel 2.8-kb fragment (Fig. 3A). EcoRI digestion gave a result similar to BamHI digestion. The EcoRI fragments containing the two outer lacZ4Δ were intact, but instead of the central inlacZ fragment, there was the recombinant 2.7-kb fragment and a novel fragment of 3.7 kb (Fig. 3B). Whereas mouse 2275 maintained the central PstI site in the palindrome, mouse 1148 did not, implying a disruption to the perfect symmetry of the palindrome.
Figure 3.
CGC transgene structures derived from the 78 line. (A) Southern blot analysis of tail-tip DNA from a mouse with the 1148 transgene structure. The 2.7-kb recombinant lacZ3′ fragment is visible as a single copy band in both BamHI (B) and EcoRI digestion because of the repeat structure of the locus. S, SacI; P, PstI. (B) Southern blot analysis of tail-tip DNA from mice with CGC transgene structures. Lanes are labeled with the number of the mouse from which the DNA was derived. Tail-tip DNA was cleaved with EcoRI (note: lanes for mice 2548 and 2364 were overloaded). (C) CGC transgene structures. Parental 78 and the SCE structures are shown with their EcoRI restriction maps. Although SCE expansion and contraction products have not been observed, the expansion product provides a framework with which to compare the CGC structures. To the right of the CGC transgene structures is the length each locus has expanded relative to the parental 78 locus. Δ, 4-bp deletion; +, SacI site.
Although mouse 1148 was the only mouse recovered with this exact transgene pattern, it was one of several mice obtained from different parents with related patterns. For example, mouse 2364 contained the 2.7-kb EcoRI fragment with the lacZ3′gene but had a different additional lacZ-hybridizing fragment of 2.9 kb (Fig. 3B). Mice 2548 and 2688 were similar in having the lacZ3′ fragment and a new lacZ fragment. They appeared, however, to be mosaics in which the rearranged structure was found in combination with the line 78 structure.
Deduced structures of these transgene loci are shown in Fig. 3C. In common with each mouse is a lacZ3′ gene (complete or partial, as for 2364) and an additional fragment containing deleted inlacZ genes. The 2.7-kb lacZ3′ fragment could be part of an expansion product derived from a sister chromatid exchange (SCE) between lacZ4Δ and inlacZ genes (Fig. 3C). However, the deleted inlacZ genes, present at the disrupted center of symmetry, as well as the lack of a reciprocal product (see below), imply that these structures arose from a more complex event than a SCE. We have, therefore, classified these recombination events as CGC events (see below). The CGC event often led to an overall increase in the size of the transgene locus, with mouse 3134 having expanded the locus by 1.3 kb.
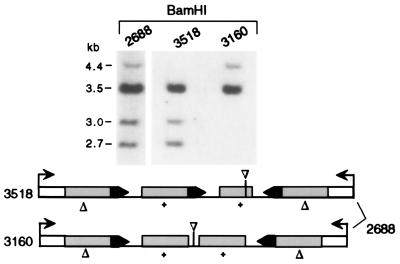
As the complex restriction pattern in CGC mice 2548 and 2688 suggested that they were mosaic for two transgene structures, these mice were mated to determine whether the transgene patterns would segregate. Mouse 2548 did not give rise to progeny. However, mouse 2688 gave rise to progeny with either of two transgene structures (Fig. 4), confirming that it was mosaic. Progeny mouse 3518, for example, had a CGC structure containing the 2.7-kb recombinant fragment, whereas mouse 3160 had fragments consistent with the line 78 structure. Further analysis with PstI indicated that the line 78 structure in mouse 3160 (and other progeny from 2688) was not intact but rather contained a deletion at the center of symmetry of ≈0.4 kb (data not shown). For mouse 2548, PstI analysis of its genomic DNA demonstrated that the center of symmetry was intact in the non-CGC transgene structure, indicating that it was parental 78. The similar intensity of the CGC and 78 transgene structures in mice 2688 and 2548 indicated that the CGC event giving rise to the 2.7-kb recombinant fragment occurred soon after fertilization, perhaps during the first cleavage divisions.
Figure 4.
Segregation of the CGC transgene structure from mosaic mouse 2688. Progeny contain either the CGC or the 78Δ structure (mice 3518 and 3160, respectively). Positions of the BamHI sites are shown in Fig. 1A.
Stabilization of the Transgene Locus in Mice Containing CGC Structures.
The CGC transgene structures contained central asymmetry, suggesting that they were stabilized from further rearrangement, as found for the 78Δ transgenes. To test this, mice 1148 and 2364 structures were bred (Table 1). Only two mice derived from line 1148 contained new rearrangements, and none of the mice from line 2364 were further rearranged. Combining the transgenic progeny from the two lines, therefore, 2% of the mice contained rearrangements, significantly less than the 15–30% of progeny in line 78.
Table 1.
Breeding analysis of mice containing CGC events
| Line | Number of mice
|
|||
|---|---|---|---|---|
| Progeny | Transgenic | Parental transgene | Rearranged transgene | |
| 1148 | 109 | 68 | 66 | 2 |
| 2364 | 41 | 19 | 19 | 0 |
Progeny from line 1148 are derived from 9 different parents, whereas those from line 2364 are derived from 3 different parents.
We also tested whether recombination leading to a lacZ+ gene was decreased in the CGC lines. Gene conversion in these lines involving lacZ4Δ, and a SacI + lacZ gene fragment would result in a lacZ+ gene, as with line 78, similar to the SGC described above for the parental transgene. In addition to SGC, the CGC structure in some cases permits alternative recombination events. For example, in line 1148, a lacZ+ gene could result from a crossover between the adjacent lacZ4Δ and lacZ3′ genes. Sperm from a number of line 1148 males were stained with a fluorescent β-galactoside and assayed by flow cytometry. Unlike with the line 78 mice, no positive sperm were detected above background levels (≈0.02%; data not shown), suggesting that the transgene in these mice was stabilized against further recombination.
Frequent Recombination Stimulated by a 4.2-kb Palindrome.
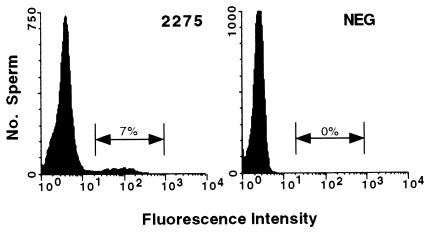
Unlike the transgenes in the CGC mice, the inversion in mouse 2275 maintained a perfect palindrome (Fig. 1A). Although smaller than the line 78 palindrome, retention of a palindromic structure suggested that it might still be unstable. To determine the frequency of homologous recombination within the inversion, mouse 2275 was bred, and sperm from progeny were incubated with a fluorogenic β-galactoside and assayed by flow cytometry. Results from one line 2275 male are shown in Fig. 5. A clear fluorescent population is seen, with ≈7% of sperm being β-galactosidase positive. In general, the number of positive sperm from line 2275 males was found to be between 2–3% but was found to be as high as 15% in one mouse (data not shown). Therefore, homologous recombination within the line 2275 transgene is roughly 5-fold higher than in the line 78 transgene.
Figure 5.
Flow cytometric analysis of sperm from a mouse with a 2275 transgene structure or a mouse without a transgene (NEG) incubated with the fluorogenic β-galactoside.
Repeat Expansion by Homologous Recombination Within the Palindrome.
In addition to SGC events, the structure of the line 2275 transgene permits alternative recombination events that would give rise to a lacZ+ gene, which could account for the higher frequency of β-galactosidase positive sperm than seen with the line 78 transgene. To determine the structure of the recombinant transgenes as well as other rearrangements, line 2275 males were mated. Although frequently rearranged, the transgene was transmitted at a Mendelian ratio (Table 2). Several different transgene structures were observed in progeny, including both central deletions, as previously reported (9), and homologous recombinants.
Table 2.
Breeding analysis of mice containing the 2275 inversion
| Parent | Number of mice
|
||||
|---|---|---|---|---|---|
| Progeny | Transgenic | Parental transgene | Transgene deletion | Homologous recombinants (expansion mice) | |
| 2275 | 36 | 16 | 7 | 6 | 3* |
| 2949 | 40 | 21 | 14 | 6 | 1* |
| 3308 | 16 | 9 | 1 | 3 | 5 (mice 3645, 3679, 3692) |
| 3368 | 25 | 8 | 2 | 3 | 3 (mice 3688, 3691) |
| Total: | 117 | 54 (46%) | 24 | 18 | 12 |
Data for mice 2275 and 2949 have been previously reported (9).
The transgene structures in these mice were not examined in as much detail as in subsequent mice, but more limited analysis suggested that each had undergone a transgene expansion.
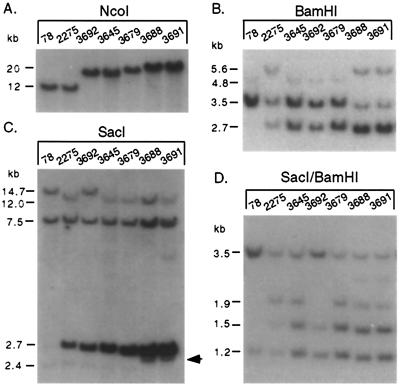
Of the homologous recombinants, a number of different structures, including those with a functional lacZ gene, were obtained. Strikingly, the majority of recombinants had undergone an expansion of the number of lacZ repeats, as determined by Southern blot analysis of tail-tip DNA (Table 2; Fig. 6). This expansion was evident from Southern analysis by both the increased size of the transgene locus and the increased lacZ hybridization signal. The increased size of the transgene locus was determined by using NcoI restriction digestion, as this enzyme cleaves only at the outermost lacZ repeats (Fig. 7). The transgene loci from lines 78 and 2275 gave NcoI fragments of 11.8 kb (Fig. 6A). In contrast, several of the recombinant mice exhibited significantly larger NcoI fragments (≥20 kb).
Figure 6.
Expansion CGC structures derived from the 2275 line. Southern blot analysis of tail-tip DNA is shown with the indicated restriction enzyme. Relative intensities of bands were determined by PhosphorImager (Molecular Dynamics) analysis. The arrow indicates the apparently identical 2.6-kb SacI fragment at the center of symmetry for the mouse pair 3688/3691. The 2.4-kb SacI fragment from the center of symmetry of the other mice is faint because of snapback of the inverted sequences before hybridization of the probe.
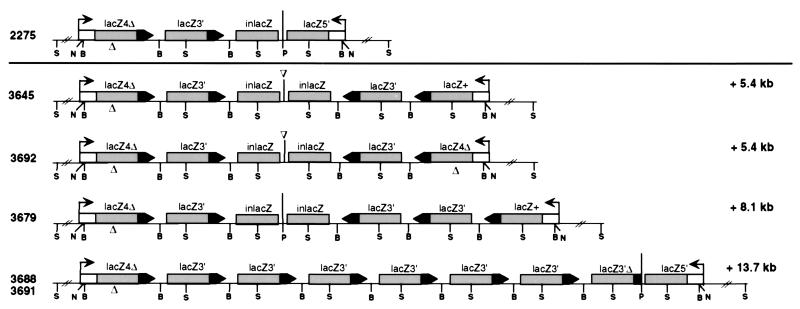
Figure 7.
Expansion CGC transgene structures derived from the 2275 line. The vertical bar indicates the center of symmetry. An unequivocal assignment of the arrangement of repeats cannot be made for all expansions. For mice 3645 and 3692, a (3 + 3) arrangement, as shown, or (4 + 2) arrangement, i.e., with both lacZ3′ genes on the same side, cannot be distinguished because the central PstI site is deleted. The (3 + 3) arrangement is diagrammed, because this structure can be obtained in one recombination event (see Fig. 8). Similarly for mouse 3679, it cannot be determined whether the lacZ+ gene is located on the left or right side of the center of symmetry. The latter arrangement is shown because recombination appears to have been initiated on the right side of the center of symmetry. S, SacI; N, NcoI; B, BamHI.
Further restriction analysis was performed to determine the expansion structure. With BamHI digestion, the intensity of the 2.7-kb fragment was clearly increased in the expansion mice relative to the 2275 parent (Fig. 6B). The intensity of the 3.5-kb BamHI fragment was also increased in three of the mice, and this increased intensity appeared to be associated with restoration of the 4.8-kb BamHI fragment that was found in line 78 mice. The remaining two expansion mice showed no increased intensity of the 3.5-kb BamHI fragment and maintained the 5.6-kb fragment found in the parental 2275 mice. With SacI digestion, the lacZ sequences in line 78 mice gave outer repeat fragments of ≈14.7 and 7.5 kb, as well as a central 2.4-kb band that hybridized poorly to the lacZ probe because of snapback of the palindromic sequence (Fig. 6C). In line 2275, the 14.7-kb fragment was cleaved into 12- and 2.7-kb fragments as a result of the inversion event, but the 7.5- and 2.4-kb bands remained intact. Progeny derived from line 2275 mice that expanded transgene locus gave the outer SacI fragments, either 14.7 or 12 kb and 7.5 kb, as well as the 2.7-kb band. In these mice, the 2.7-kb band was significantly more intense than that found with the 2275 transgene, as was also found in the BamHI digestion, indicating that the expansion occurred in the more centrally located portion of the locus rather than at the outer sequence. SacI/BamHI digestion confirmed that the 2.7-kb fragment was of the same origin in each of the mice, because it was always cleaved to fragments of 1.5 and 1.2 kb (Fig. 6D).
In most of the expansion mice, the central 2.4-kb SacI fragment was largely intact, although weakly hybridizing to the lacZ probe, as in 2275 mice. Nevertheless, for mice 3645 and 3692 the central PstI site was disrupted, indicating a small deletion at this central site (Fig. 7). Mice 3688 and 3691 were exceptions, in that the 2.4-kb SacI fragment was replaced with a 2.6-kb fragment (arrow, Fig. 6C). This fragment hybridized more strongly to the lacZ probe than did the 2.4-kb band found in the other mice, indicating that snapback of this fragment was less frequent. This lack of snapback was likely the result of significant asymmetry in the fragment. The PstI site was nevertheless retained in these mice.
Restriction mapping and quantitation of hybridization signals were used to infer the transgene structures in these mice (Fig. 7). The expansions ranged from the addition of two lacZ repeats to the addition of five repeats, the latter more than doubling the number present in the parental 2275 mouse. The amount of sequence added ranged from 5.4 to 13.7 kb. In mice 3645 and 3692, which had the addition of two repeats, it is ambiguous whether the repeat arrangement is (3 + 3), as shown, or (4 + 2), because the central PstI site was lost. The outer lacZ5′ gene fragment in both mice was transformed to a full-length lacZ gene, either a lacZ+ gene in mouse 3645 or a lacZ4Δ gene in mouse 3692. Mouse 3679 had three additional lacZ repeats (8.1 kb), to form either a (3 + 4) arrangement, as shown, or a (4 + 3) repeat arrangement. Like mouse 3645, the outer lacZ gene fragment was transformed to a full-length lacZ+ gene.
Especially striking were mice 3688 and 3691, in which there was an expansion to a total of nine repeats, with the additional five repeats located on one side of the palindrome, creating an (8 + 1) repeat arrangement. These mice, both of whom were derived from the same parent, appeared to have the same transgene structure.
A limited amount of breeding was undertaken with mice 3679 and 3692, such that 10 transgenic progeny were characterized for each. Rearranged transgenes were obtained from both mice, including a deletion, a possible further expansion (mouse 3679), and a gene conversion of a lacZ4Δ gene (mouse 3692) (data not shown). Although mouse 3679 may have maintained a perfect palindrome, mouse 3692 contained a slight disruption of the center of symmetry because the central PstI site was lost. Mice 3688 and 3691 also contained an imperfect palindrome. Although these mice were not mated, the transgene locus in mouse 3691 appeared unstable, because tail-tip DNA gave a faint novel fragment (Fig. 6C). Therefore, although asymmetry can stabilize a locus, a small amount of asymmetry as in mouse 3692 or large numbers of repeats may overcome the stabilization.
Discussion
We have demonstrated that a palindrome in the mouse germ line is extremely unstable and undergoes both homologous recombination and nonhomologous deletion at high frequency. Homologous recombination was previously reported to result in ≈0.5% lacZ+ sperm through a simple gene conversion mechanism involving repeats within the palindrome (4), and the predicted product was verified in this report by the recovery of mice containing the SGC transgene structure. More frequent complex homologous recombination products were also recovered in which the repeat configuration of the palindrome was altered. Some of these CGC events expanded the size of the palindromic locus. In the most extreme case, the expansion more than doubled the number of repeats, increasing the size of the locus by almost 14 kb.
CGC events were found to occur nearly as frequently as nonhomologous deletions in the 2275 inversion line. Of 54 transgenic progeny, 18 (33%) contained deletions, whereas 12 (22%) had CGC events, which included the expansions. In parental line 78, CGC events were also detected, although at a lower frequency than deletions (≤10% versus ≈30% deletions). The lower observed frequency for line 78 may be attributable to the symmetric arrangement of repeats, as some CGC events would be indistinguishable from the parental transgene or even from deletions.
Although studies have yet to be performed in tissues in mice, in cultured mammalian cells both homologous and nonhomologous mechanisms are used to repair DSBs (10). The homologous and nonhomologous rearrangements that occur at palindromes, therefore, may be frequent because strand breaks are frequently induced at these structures as a result of hairpin formation or cruciform extrusion. In the mouse, breaks are predicted to occur at the center of symmetry of a palindrome because of nicking at the hairpin tip (4, 9, 11, 12). A role for hairpin nicking is also suggested in E. coli, as the product of the SbcCD gene, which regulates palindrome stability, encodes a hairpin endonuclease (13). Breaks could also be introduced during replication as a result of the inability of DNA polymerase to proceed through a hairpin structure, as has been inferred from studies in yeast and E. coli (1, 2, 14). In this case, breaks would be offset from the center of symmetry, rather than involve sequences at the tip of the hairpin structure.
Unlike breaks at a hairpin base, breaks located at the tip of a hairpin have the potential to give rise to rearrangement products in which one-half of the palindrome remains intact. These products have previously been observed in the formation of nonhomologous deletions within the line 78 palindrome (4, 9). In the current study, one complete half of the palindrome was also retained in some CGC mice, as evidenced by the retention of sequences up to and including the central PstI site (e.g., mouse 2364, Fig. 3C; mouse pair 3688/3691), implying that strand breaks at the center of symmetry stimulates CGC events as well as deletions. The CGC events like deletions resulted in imperfect palindromes (i.e., inverted repeats with a central asymmetry) in most cases, which would have a decreased probability of forming secondary structure to promote further strand breakage. Consistent with this, we found that nonexpansion CGC products were significantly less likely than a perfect palindrome to undergo subsequent rearrangements (Table 1), as was also seen with imperfect palindromes formed by a deletion (4). We have recently noted, however, that an expanded transgene locus with a small central asymmetry is nevertheless prone to frequent rearrangements, suggesting that a large number of repeats may overcome the stabilization introduced by asymmetry.
Homologous recombination within the palindrome was not restricted to one developmental stage. CGC events giving rise to the products in mice 2688 and 2548 apparently occurred soon after fertilization, because the CGC product was present at a level similar to the nonrecombinant locus in tail tip DNA (Fig. 3C). The CGC event detected in mice 3688 and 3691 may have occurred somewhat later, because, although this event was transmitted twice, it was one of at least four transgene structures transmitted by the parent mouse (Table 2). Other CGC events occurred much later, possibly as late as meiosis, because they were found only once. An example is the unique rearrangement found in mouse 1148. This mouse was one of 51 transgenic progeny from a line 78 male (mouse 919), which transmitted a number of rearranged transgenes in addition to the one found in mouse 1148 (4). Previously, nonhomologous deletions within the palindrome were also found to have a broad timing during development (4), which again suggests that CGC events and deletions may be two alternative outcomes of break repair processes.
A number of homologous recombination mechanisms have been proposed in meiotic and mitotic cells, some of which involve reciprocal exchange (15). Palindrome recombination that occurs during early development (at least those recoverable events that occur between repeats on the same side of the center of symmetry) allows a determination of whether these events involved reciprocal exchange, because the reciprocal recombination product would be “captured” within the mouse. For an early SCE, therefore, an expansion product containing five lacZ repeats would be paired with a contraction containing three lacZ repeats. In the CGC mice in which the events occurred early in development (i.e., 2688 and 2548), reciprocal products were not observed. In mouse 2548, the CGC product was present with the parental 78 transgene, which has four lacZ repeats, and in mouse 2688, it was present with a 78Δ transgene, which also has four lacZ repeats. Also difficult to explain by a simple exchange mechanism is that CGC products often had complex structures. None of the CGC products derived from line 78 had the five complete lacZ repeats expected from an unequal SCE (Fig. 3C). Rather the two lacZ repeats located at the center of symmetry frequently contained deletions. The structure of the expansion in mouse 3679 would appear to be more consistent with an exchange mechanism, because each of the repeats is apparently intact (Fig. 7). However, to derive the 3679 structure by one event, repeats on opposite sides of the center of symmetry (i.e., the lacZ5′ and inlacZ repeats) on sister chromatids would have to undergo the exchange, an event that would have led to a nonrecoverable acentric/dicentric chromatid pair.
We infer, therefore, that CGC events generally involve a nonexchange gene conversion mechanism, similar to mechanisms proposed for homologous DSB repair in cultured mammalian cells (16–18) and in other organisms (19–21). As discussed above, these events would be initiated by strand breaks at or near the center of symmetry of the palindrome when a hairpin or cruciform is extruded. A broken end would invade a homologous repeat on the sister chromatid (or same chromatid) to initiate repair synthesis, and then the newly synthesized strand would rejoin the other end of the original broken chromatid.
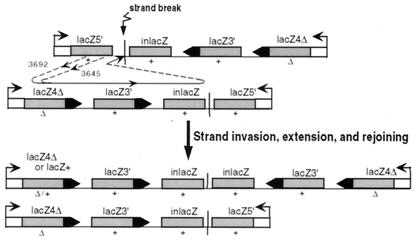
Application of this model to the repeat expansions in mice 3645 and 3692 is shown in Fig. 8. The broken end at the lacZ5′ gene invades the lacZ4Δ gene on the opposite side of the symmetry center on the sister chromatid. For mouse 3645, the invasion would occur after the SacI site of the invading lacZ5′ strand, whereas for mouse 3692, it would occur before the SacI site, creating lacZ+ and lacZ4Δ genes, respectively. Strand invasion would prime DNA synthesis that would extend through the lacZ3′ and inlacZ genes, possibly being impeded from further extension by a hairpin on the invaded strand. The new strand would then rejoin the original broken chromatid. Rejoining events at nonhomologous sequences could have occurred by end-joining, giving rise to a coupled homologous/nonhomologous break repair event, similar to those recently reported during interchromosomal recombination between nonpalindromic sequences (22).
Figure 8.
Proposed mechanism of repeat expansion at a palindrome in the mammalian germ line. As a consequence of secondary structure at the palindrome's center of symmetry, a strand break occurs. The broken strand invades a homologous lacZ repeat on the sister chromatid and primes DNA synthesis. The extended strand then rejoins the original chromatid, by nonhomologous mechanisms in some instances. The invading strand can be from either the lacZ5′ side of the palindrome (as shown) or the inlacZ side (not shown) and can invade the same (not shown) or opposite (as shown) side of the sister chromatid.
This replication-based recombination mechanism could explain the derivation of each of the CGC events, with variations in terms of which strand invades and what sequences are copied. For large repeat expansions (i.e., for mice 3688/3691), multiple rounds of strand invasion and extension from the sister chromatid could occur. Alternatively, a single invasion of a lacZ repeat on the same chromatid could occur, followed by extension that would continue into the newly synthesized DNA, as in rolling circle replication. For mice that contain truncated lacZ genes in the central repeats (e.g., 2688), synthesis may have stopped before reaching the center of symmetry, and/or the noninvading end may have been degraded before rejoining. During any of these repair events, however, the sequence being copied would be expected to remain unchanged.
Because we detected events during early divisions in the mouse, at least a fraction of the events we recovered were mitotic. The infrequent association with crossing-over, as well as the immutability of the sequence being copied, is a frequent outcome of mitotic homologous recombination in cultured mammalian cells (23). This lack of crossing-over contrasts with recombination during meiosis, in which events are frequently resolved as crossovers and in which sister chromatid events are disfavored relative to homologue recombination events, at least as described for yeast (24). Because the mice that we have bred are hemizygous for the palindrome, none of our recombination events involve the homologue. It would be interesting, however, to determine whether recombination leading to exchange events between homologues could be stimulated at meiosis by DSBs arising at palindromic sequences, and whether the DSBs would be Spo11-dependent (25, 26), as found in yeast for DSBs at a small palindrome (27).
The palindromic structure we have studied was fortuitously formed during the integration of a transgene and is therefore not a naturally occurring sequence in the mouse genome. Nevertheless, the instability observed here may shed light on genetic instability observed at natural repeats that have the potential to form hairpin structures. For example, hairpin structures have been predicted to form at the quasipalindromic triplet repeats associated with several genetic diseases (28–30). These repeats manifest a high degree of instability with dramatic expansions in the number of repeats, which has often been attributed to replication slippage (31). Because homologous recombination by gene conversion can lead to repeat expansion at a harpin-forming transgene, it is possible that gene conversion could contribute to triplet repeat expansion when hairpin structures are formed. Recent experiments with triplet repeats in plasmid substrates in E. coli have demonstrated expansions consistent with the gene conversion mechanism observed here (32, 33). As well, repeat expansion has been observed in yeast during DSB-induced gene conversion (34) [see Darlow and Leach (29) for a discussion of the potential roles of secondary structures and DNA replication and recombination in models of repeat tract expansion]. Palindrome-generated instability has also been implicated in the etiology of constitutional t(11;22) translocations, in which breakpoints occur at the center of symmetry of AT-rich palindromes (35, 36). In this case, however, end-joining events at the palindrome breaks, rather than gene conversion events, have been postulated to be involved.
Acknowledgments
We thank Susanna Lewis for comments on the manuscript. This work was supported by a grant from the National Institutes of Health to M. J. (HG01740).
Abbreviations
- DSB
double-strand break, SGC, simple gene conversion
- CGC
complex gene conversion
- SCE
sister chromatid exchange
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Leach D R. BioEssays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 2.Gordenin D A, Resnick M A. Mutat Res. 1998;400:45–58. doi: 10.1016/s0027-5107(98)00047-5. [DOI] [PubMed] [Google Scholar]
- 3.Collick A, Drew J, Penberth J, Bois P, Luckett J, Scaerou F, Jeffreys A, Reik W. EMBO J. 1996;15:1163–1171. [PMC free article] [PubMed] [Google Scholar]
- 4.Akgün E, Zahn J, Baumes S, Brown G, Liang F, Romanienko P J, Lewis S, Jasin M. Mol Cell Biol. 1997;17:5559–5570. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lobachev K S, Stenger J E, Kozyreva O G, Jurka J, Gordenin D A, Resnick M A. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stenger J E, Lobachev K S, Gordenin D, Darden T A, Jurka J, Resnick M A. Genome Res. 2001;11:12–27. doi: 10.1101/gr.158801. [DOI] [PubMed] [Google Scholar]
- 7.Lobachev K S, Shor B M, Tran H T, Taylor W, Keen J D, Resnick M A, Gordenin D A. Genetics. 1998;148:1507–1524. doi: 10.1093/genetics/148.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nag D K, Kurst A. Genetics. 1997;146:835–847. doi: 10.1093/genetics/146.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis S, Akgün E, Jasin M. Ann NY Acad Sci. 1999;870:45–57. doi: 10.1111/j.1749-6632.1999.tb08864.x. [DOI] [PubMed] [Google Scholar]
- 10.Liang F, Han M, Romanienko P J, Jasin M. Proc Natl Acad Sci USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis S. Proc Natl Acad Sci USA. 1994;91:1332–1336. doi: 10.1073/pnas.91.4.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis S M. Nucleic Acids Res. 1999;27:2521–2528. doi: 10.1093/nar/27.12.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connelly J C, Kirkham L A, Leach D R. Proc Natl Acad Sci USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cromie G A, Millar C B, Schmidt K H, Leach D R. Genetics. 2000;154:513–522. doi: 10.1093/genetics/154.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pâques F, Haber J E. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson C, Moynahan M E, Jasin M. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson R D, Jasin M. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belmaaza A, Chartrand P. Mutat Res. 1994;314:199–208. doi: 10.1016/0921-8777(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 19.Kogoma T. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- 20.Pâques F, Leung W, Haber J. Mol Cell Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gloor G B, Lankenau D H. Trends Genet. 1998;14:43–46. doi: 10.1016/s0168-9525(97)01365-6. [DOI] [PubMed] [Google Scholar]
- 22.Richardson C, Jasin M. Mol Cell Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jasin M. In: DNA Damage and Repair. Nickoloff J A, Hoekstra M F, editors. III. Totowa, NJ: Humana; 2001. pp. 207–235. [Google Scholar]
- 24.Kleckner N. Proc Natl Acad Sci USA. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baudat F, Manova K, Yuen J P, Jasin M, Keeney S. Mol Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 26.Romanienko P J, Camerini-Otero R D. Mol Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 27.Nasar F, Jankowski C, Nag D K. Mol Cell Biol. 2000;20:3449–3458. doi: 10.1128/mcb.20.10.3449-3458.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, Mariappan S V S, Catasti P, Ratliff R, Moyzis R K, Laayoun A, Smith S S, Bradbury E M, Gupta G. Proc Natl Acad Sci USA. 1995;92:5199–5203. doi: 10.1073/pnas.92.11.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darlow J M, Leach D R. Genetics. 1995;141:825–832. doi: 10.1093/genetics/141.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gacy A M, Goellner G, Juranic N, Macura S, McMurray C T. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 31.Bowater R P, Wells R D. Prog Nucleic Acid Res Mol Biol. 2000;66:159–202. doi: 10.1016/s0079-6603(00)66029-4. [DOI] [PubMed] [Google Scholar]
- 32.Jakupciak J P, Wells R D. J Biol Chem. 1999;274:23468–23479. doi: 10.1074/jbc.274.33.23468. [DOI] [PubMed] [Google Scholar]
- 33.Jakupciak J P, Wells R D. J Biol Chem. 2000;275:40003–40013. doi: 10.1074/jbc.M007153200. [DOI] [PubMed] [Google Scholar]
- 34.Richard G F, Goellner G M, McMurray C T, Haber J E. EMBO J. 2000;19:2381–2390. doi: 10.1093/emboj/19.10.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurahashi H, Shaikh T H, Hu P, Roe B A, Emanuel B S, Budarf M L. Hum Mol Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 36.Edelmann L, Spiteri E, Koren K, Pulijaal V, Bialer M G, Shanske A, Goldberg R, Morrow B E. Am J Hum Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peschon J J, Behringer R R, Palmiter R D, Brinster R L. Proc Natl Acad Sci USA. 1987;84:5316–5319. doi: 10.1073/pnas.84.15.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jasin M, Zalamea P. Proc Natl Acad Sci USA. 1992;89:10681–10685. doi: 10.1073/pnas.89.22.10681. [DOI] [PMC free article] [PubMed] [Google Scholar]