Abstract
The hyperthermophilic, facultatively heterotrophic crenarchaeum Thermoproteus tenax was analyzed using a low-coverage shotgun-sequencing approach. A total of 1.81 Mbp (representing 98.5% of the total genome), with an average gap size of 100 bp and 5.3-fold coverage, are reported, giving insights into the genome of T. tenax. Genome analysis and biochemical studies enabled us to reconstruct its central carbohydrate metabolism. T. tenax uses a variant of the reversible Embden-Meyerhof-Parnas (EMP) pathway and two different variants of the Entner-Doudoroff (ED) pathway (a nonphosphorylative variant and a semiphosphorylative variant) for carbohydrate catabolism. For the EMP pathway some new, unexpected enzymes were identified. The semiphosphorylative ED pathway, hitherto supposed to be active only in halophiles, is found in T. tenax. No evidence for a functional pentose phosphate pathway, which is essential for the generation of pentoses and NADPH for anabolic purposes in bacteria and eucarya, is found in T. tenax. Most genes involved in the reversible citric acid cycle were identified, suggesting the presence of a functional oxidative cycle under heterotrophic growth conditions and a reductive cycle for CO2 fixation under autotrophic growth conditions. Almost all genes necessary for glycogen and trehalose metabolism were identified in the T. tenax genome.
Archaea were recognized as a distinct phylogenetic group more than 25 years ago (75). Their importance as the third major evolutionary line is well established, but our knowledge of their physiological capabilities remains limited. Central metabolic pathways within these organisms are far from fully understood (43, 47, 72).
Thermoproteus tenax was the first hyperthermophilic archaeum described (76). It is able to grow chemolithoautotrophically on H2, CO2, and So as well as chemoorganoheterotrophically in the presence of So and various organic substrates such as glucose, starch, amylose, glycerate, glycerol, ethanol, and malate (12, 76). Physiological and biochemical studies revealed T. tenax as a physiologically versatile organism with numerous archaeon-specific metabolic capabilities, regulation, and thermoadaptive traits.
Comparative studies of carbohydrate metabolism in hyperthermophilic archaea indicate that sugars are generally metabolized by variants of the Entner-Doudoroff (ED) and Embden-Meyerhof-Parnas (EMP) pathways. The so-called nonphosphorylative ED pathway (phosphorylation takes place only at the stage of glycerate) is the only pathway that has been identified for sugar degradation in the aerobes Sulfolobus solfataricus (9) and Thermoplasma acidophilum (7), whereas the anaerobes Pyrococcus furiosus (27, 28, 31, 66, 69), Thermococcus spp. (31, 44, 50), Desulfurococcus amylolyticus (20), and Archaeoglobus fulgidus (33) use modified versions of the EMP pathway. In contrast to findings for other hyperthermophilic archaea, T. tenax uses both variants—the ED and EMP pathways—for glucose metabolism, as shown by in vitro studies identifying specific intermediates and enzyme activities in cell extracts (49, 55). The utilization of both pathways was furthermore shown by in vivo 13C-labeling experiments with growing cells (57) and with cell suspensions (50). Several enzymes and their coding genes involved in the degradative pathways, predominantly in the EMP variant, have already been characterized (4, 5, 48, 54, 56, 57), but the brunt of core carbon biochemistry in T. tenax is still unknown.
The main difference between the T. tenax EMP variant and the classical version of this pathway concerns (i) the presence of a hexokinase (HK) with no notable regulatory properties (11), (ii) the replacement of the antagonistic enzyme couple ATP-dependent PFK and fructose bisphosphatase by a bidirectional PPi-dependent enzyme (56), (iii) the presence of two different glyceraldehyde-3-phosphate (GAP) dehydrogenases (GAPDHs), differing in phosphate dependence, reversibility of the catalyzed reaction, and allosteric properties (4, 5, 21, 40) and (iv) a pyruvate kinase (PK) with reduced allosteric potential (48). Enzyme as well as transcript studies with T. tenax (late-log-phase cells) indicate that regulation takes place on the protein and gene levels. Under heterotrophic conditions, the catabolic flux seems to be forced by the preferred expression of the PPi-dependent phosphofructokinase (PPi-PFK) and fructose-bisphosphate aldolase (FBPA), which form an operon in T. tenax (fbp-pfp operon) (34, 54, 56), and of the PK (48). A strong influence on the catabolic flux is also exerted by the allosterically regulated nonphosphorylating GAPDH (GAPN), which catalyze the unidirectional oxidation of GAP to 3-phosphoglycerate. In the presence of activators such as glucose-1-phosphate, fructose-6-phosphate, AMP, and ADP, i.e., under conditions which are characterized by the availability of storage carbohydrates (e.g., glycogen) and/or a low-energy charge of the cell, the catabolic carbon flux is most effective. In contrast, the classical, phosphorylating NADP+-dependent GAPDH and phosphoglycerate kinase (PGK) are of predominant importance for anabolism rather than for catabolism, as indicated by the enzymatic features of the NADP+-dependent GAPDH, by higher transcript amounts of both genes (gap and pgk), and by higher-level enzyme activities in autotrophically grown cells (5). Phosphoenolpyruvate synthetase (PEPS) activity was detected only in extracts of cells grown under autotrophic conditions (53); thus, the PEPS seems to exert a key function in driving the carbon flux into the anabolic direction.
Enzymatic studies with cell extracts (49, 55) and analyses of characteristic intermediates (55) identified the nonphosphorylative version of the ED pathway, which had been described originally for S. solfataricus (9) and T. acidophilum (7). In contrast to findings for the haloarchaeal semiphosphorylative pathway (with phosphorylation at the level of 2-keto-3-deoxy-gluconate [KDG]), in this version of the ED pathway phosphorylation takes place only at the level of glycerate. Glyceraldehyde generated by the KDG aldolase (KDGA) is therefore the characteristic intermediate of the nonphosphorylative version. So far, only glucose dehydrogenase (GDH), the first enzyme of the nonphosphorylative ED pathway of T. tenax, has been purified and characterized (57).
Because of its essential function for providing pentoses and NADPH for anabolic purposes, the pentose phosphate pathway (PPP) has been assumed for all three domains of life. Genome analysis confirmed by biochemical studies indicates that glucose-6-phosphate dehydrogenase seems to be generally absent from methanoarchaea and hyperthermophilic archaea (51, 52); therefore, the conventional PPP does not seem to be operative in these organisms. Also, in halophiles—although activity of glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase was detected in Halococcus saccharolyticus (23) and gene homologs of these enzymes were found in the genome of Halobacterium sp. strain NRC1—PPP is not involved in sugar catabolism, as indicated by 13C nuclear magnetic resonance studies of H. saccharolyticus. Possibly, the fragmentary PPP serves just for the synthesis of pentoses in haloarchaea.
The catabolic fate of pyruvate was monitored in T. tenax by fermentation analyses and enzyme measurements in crude extracts, which showed that the final oxidation of carbohydrates occurs via an oxidative citric acid cycle (CAC) linked to the glycolytic pathways by a pyruvate synthase (POR) (pyruvate:ferredoxin oxidoreductase) (49).
No information is available yet about the reaction sequence of CO2 fixation in T. tenax. However, there is some evidence indicating that the CO2 fixation occurs via the reductive CAC, as demonstrated for T. neutrophilus, a close relative of T. tenax (1). Most of the enzymes of the CAC work reversibly and could therefore be engaged in both directions. Only three counteracting enzyme pairs are thought to determine the oxidative or reductive direction of the cycle: (i) citrate synthase/citrate lyase, (ii) 2-oxoglutarate dehydrogenase/2-oxoglutarate synthase (2-oxoglutarate-ferredoxin oxidoreductase [KOR]), and (iii) succinate dehydrogenase/fumarate reductase. Experimental data (49) give no support for the presence of alternative CO2 fixation mechanisms such as the Calvin cycle via ribulose-1,5-bisphosphate carboxylase or the reductive acetyl coenzyme A (acetyl-CoA) pathway via carbon monoxide dehydrogenase (Wood-Ljungdahl pathway).
Two additional carbohydrates—glycogen and trehalose—were identified in T. tenax, thus raising questions about their metabolism and function. Glycogen is a branched polymer of α-1,4-linked and α-1,6-linked glucosyl moieties and represents an osmotic inactive carbon storage compound in all three domains of life. Although the presence of glycogen in T. tenax and other archaea (members of Thermoproteales and Sulfolobales) was demonstrated previously (32), not much is known about glycogen metabolism in archaea. Trehalose, a glucose disaccharide [α-d-glucopyranosyl-(1,1)-α-d-glucopyranoside], is found in members of all three major phylogenetic domains. Trehalose plays an important role as a compatible solute, being involved in stress response to high-level osmolarity (Escherichia coli) and heat (Saccharomyces cerevisiae) (16, 39). Although trehalose has been identified in several archaea (e.g., T. acidophilum, Methanothermobacter thermautotrophicus, and T. tenax) (37, 39), the role of trehalose in archaea is still unknown. In T. tenax, trehalose was identified as the exclusive solute in concentrations of up to 0.3 μmol/mg of protein (37).
These insights into the physiological capabilities of T. tenax motivated us to study in more detail the biochemical and genetic mechanisms that select the various pathways, regulate the carbon flux through them, and govern the synthesis and breakdown of low- and high-molecular-mass carbohydrate compounds. For a better understanding of the central metabolism in T. tenax, as well as in archaea in general, we aimed at a representative genome sequence analysis of T. tenax to use the genetic information for reconstruction of the carbohydrate metabolism by assigning genes by sequence similarity and/or by the function of their recombinant gene products. So far, only four complete crenarchaeal genome sequences have been published: those for Aeropyrum pernix (26), Pyrobaculum aerophilum (13), and two species of Sulfolobus (25, 52). Here we report on the reconstruction of the T. tenax-specific EMP and ED pathways and the reversible CAC as well as on glycogen and trehalose metabolism.
MATERIALS AND METHODS
Strains and growth conditions.
Mass cultures of T. tenax Kra1 (DSM 2078) (12, 76) were grown under autotrophic conditions as described previously (5). E. coli strains DH5α (Invitrogen), BL21(DE3), and BL21-CodonPlus(DE3)-RIL (Stratagene) for use in cloning and expression studies were grown under standard conditions (46) following the instructions of the manufacturers.
Library construction, shotgun sequencing, and primer walking.
Genomic DNA was prepared from T. tenax cells as described previously (48). For construction of a whole-genome shotgun library, genomic DNA from T. tenax was sheared with a HydroShear (GeneMachines, San Carlos, Calif.) to yield fragments with estimated sizes of 2.5 to 5.0 kbp. The fragmented DNA was separated in a low-melting-point agarose gel, and the dominant fraction of the desired size range was isolated. The fragments were purified with a QIAquick gel extraction kit (Qiagen, Hilden, Germany) and cloned using a TOPO shotgun subcloning kit (Invitrogen Corp., Carlsbad, Calif.). Positive clones were identified by their ampicillin resistance in combination with their white colony phenotype in a X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) screening. The isolated vectors served as templates in sequencing reactions using the primers M13uni(-43) (5′-agggttttcccagtcacgacgtt-3′) and M13rev(-49) (5′-gagcggataacaatttcacacagg-3′). The sequence data were collected using an ABI Prism DNA System 377 apparatus and an ABI Prism 3700 DNA analyzer and processed and assembled with Phred/Phrap/Consed software (University of Washington). Gaps were closed where needed by direct sequencing using chromosomal DNA as a template.
Automated analysis and annotation using the MAGPIE system.
The genomic sequence was analyzed using Multipurpose Automated Genome Project Investigation Environment (MAGPIE) automated genome analysis and annotation software (15, 17). The function of open reading frames (ORFs) involved in metabolic pathways was verified manually, and gene ontology categories were attached to the functional assignments.
Cloning of genes and expression in E. coli.
To prove the assignment of gene homologs in the T. tenax genome, the respective genes were cloned into a pET vector system via PCR mutagenesis. Expression of the gene products in E. coli BL21(DE3) and BL21-CodonPlus(DE3)-RIL was performed following the instructions of the manufacturer (Stratagene). For expression, the glucose-6-phosphate isomerase (PGI) gene (pgi) of T. tenax was cloned into vector pET-11c through the use of the two new restriction sites (NdeI and BamHI) introduced by PCR mutagenesis with the following primer set: PGI-NdeI (5′-TCCGAAGATAGCTGCATATGATAG-3′ [sense]) (the mutations are shown in boldface characters and the resulting restriction sites are underlined) and PGI-BamHI (5′-TCGGCCAGGATAGGATCCTTGATC-3′ [antisense]). The following primer set—devised on the basis of N- and C-terminal partial sequence information—was designed to amplify the glycogen phosphorylase (glgP) gene in pET-24a: GLGP-AseI-f (5′-GCCCCCCCGATTAATGATAGTGT-3′ [sense]) and GLGP-HindIII-rev (5′-AGATCAAGCTTTTGGCCGAGCTCA-3′ [antisense]). Both enzymes were expressed in BL21-CodonPlus(DE3)-RIL.
Isolation of recombinant hyperthermophilic enzymes.
Recombinant E. coli cells were suspended in buffer (e.g., 100 mM HEPES-KOH [pH 7.5] or 100 mM TRIS-HCl [pH 7.0 at 70 to 90°C, depending on the temperature for heat precipitation]) containing 7.5 mM dithiothreitol (DTT) or 10 mM β-mercaptoethanol and passed three times through a French pressure cell at 150 MPa. Cell debris and unbroken cells were removed by centrifugation (85,000 × g for 30 min at 4°C). The resulting crude extract was heat precipitated at 70 to 90°C for 20 to 30 min, depending on the stability of the respective enzyme. The fraction remaining after heat precipitation was centrifuged again (20,000 × g for 20 min at 4°C) and dialyzed overnight at 4°C. In general, after heat treatment only a few faint contaminant E. coli proteins were detectable by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining.
For the recombinant PGI, 0.5 g of E. coli cells was resuspended in 3 ml of 100 mM TRIS-HCl (pH 7.0 at 90°C) containing 10 mM β-mercaptoethanol. The heat precipitation was performed for 20 min at 90°C, and the protein was dialyzed overnight at 4°C with 1.6 liters of 100 mM TRIS-HCl (pH 7.0 at 55°C) containing 10 mM β-mercaptoethanol. For the recombinant glycogen phosphorylase, 10 g of E. coli cells was resuspended in 30 ml of 100 mM HEPES-KOH (pH 7.5) containing 7.5 mM DTT and the heat precipitation procedure was performed for 30 min at 90°C. The extract was dialyzed overnight at 4°C with 2 liters of 50 mM HEPES-KOH (pH 7.5) containing 7.5 mM DTT, and the dialyzed fraction was applied on Q-Sepharose Fast-Flow (Amersham Pharmacia Biotech) equilibrated in the same buffer and eluted with a linear salt gradient of 0 to 600 mM KCl. Fractions containing the homogeneous enzyme solution were pooled, dialyzed, and concentrated.
Enzyme assays.
PGI activity was determined in the anabolic direction (glucose 6-phosphate formation) at 50°C in a coupled assay with glucose-6-phosphate dehydrogenase (EC 1.1.1.49) from baker's yeast as an auxiliary enzyme. The assay (total volume, 0.5 ml) was performed at 50°C in 100 mM TRIS-HCl (pH 7.0) in the presence of 1 mM NADP+, 1 U of glucose-6-phosphate dehydrogenase, 0.05 to 7 mM fructose 6-phosphate, and 4.5 μg of PGI after heat precipitation. Enzyme activity in the catabolic direction (fructose 6-phosphate formation) was assayed at 50°C in a coupled assay using ATP-dependent phosphofructokinase (EC 2.7.1.11; Bacillus stearothermophilus), fructose-1,6-bisphosphate aldolase (EC 4.1.2.13; rabbit muscle), triosephosphate isomerase (TIM) (EC 5.3.1.1; rabbit muscle), and glycerol-3-phosphate dehydrogenase (EC 1.1.1.8; rabbit muscle) as auxiliary enzymes. The assay (total volume, 0.5 ml) was performed at 50°C in 100 mM TRIS-HCl (pH 7.0) in the presence of 1 mM NADH, 3 mM ATP, 5 mM MgCl2, 2 U of phosphofructokinase, 1 U of fructose-1,6-bisphosphate aldolase, 50 U of TIM, 9 U of glycerol-3-phosphate dehydrogenase, 0.1 to 14 mM glucose 6-phosphate, and 0.9 μg of recombinant PGI after heat precipitation.
Glycogen phosphorylase activity was assayed in a coupled enzyme assay at 50°C with phosphoglucomutase (PGM) (EC 5.4.2.2; rabbit muscle) and glucose-6-phosphate dehydrogenase (EC 1.1.1.49; baker's yeast) as auxiliary enzymes. The standard assay (volume, 1 ml) was performed at 50°C in 100 mM TRIS-HCl (pH 7.5) containing 10 mM KH2PO4 (Pi), 1 mM NADP+, 10 mM MgCl2, 3 U of PGM, 3 U of glucose-6-phosphate dehydrogenase, 0.1% (wt/vol) glycogen (bovine liver), and 5 to 15 μg of protein. For the different enzyme assays, the change in absorbance was monitored at 366 nm (ɛ NADH 50°C = 3.36 mM−1 cm−1; ɛ NADPH 50°C = 3.43 mM−1 cm−1).
Nucleotide sequence accession numbers
Gene homologs assigned in this paper have been submitted to the EMBL database, and accession numbers are given in the tables and text.
RESULTS AND DISCUSSION
Genome sequence features.
The genome assembly used for the annotation of the genes described in this paper consists of 19,850 sequence reads assembled in 53 contigs more than 2 kbp in length; the largest contig is 110 kbp in length. Contigs smaller than 2 kbp were excluded from the analysis. The total length of the contigs adds up to 1.81 Mbp, which represents about 98.5% of the estimated genome size of 1.84 Mbp. The average gap size is estimated to be about 100 bp. With sequence coverage at 5.3-fold, the sequence quality of the trimmed contigs is approximately 15 errors in 10,000 bp. The average G+C content of the genome is calculated to be 54%. The individual contigs were subjected to an automated analysis with MAGPIE software. The ORFs were identified on the basis of stop codons and their upstream start codons. An ORF size of 100 amino acids was chosen as the minimum cutoff level. The automated analysis tool identified and annotated 3,913 ORFs, which were then surveyed for the presence of genes related to central carbohydrate metabolism. Given an average G+C level of 54%, this leads to an overprediction of ORFs; however, at least 2,000 to 2,500 of these should be coding ORFs (given the number of coding regions in other organisms of the same size).
The central carbohydrate metabolism of T. tenax. (i) Variant of the EMP pathway. (a) Assignment of genes involved in the reversible EMP variant.
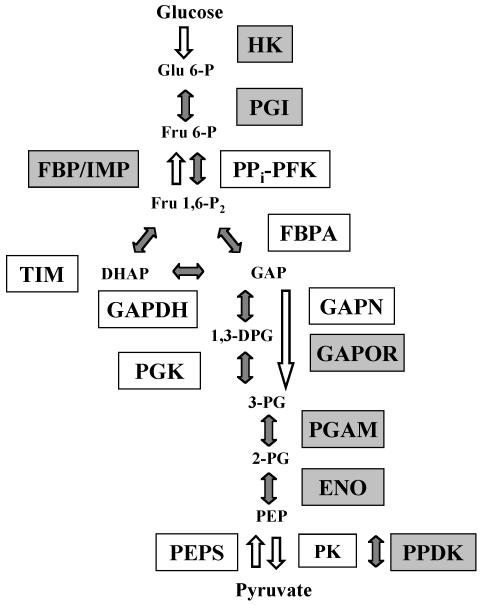
The following enzymes (and their coding genes) essential for a functional reversible EMP pathway have been identified and characterized to date: PPi-PFK (56), FBPA (34, 54), GAPDH (5), PGK (5), TIM (H. Walden, G. L. Taylor, E. Lorentzen, E. Pohl, H. Lilie, A. Schramm, T. Knura, K. Stubbe, B. Tjaden, and R. Hensel, submitted for publication), PK (48), and PEPS (63; B. Tjaden, C. Dörr, B. Siebers, and R. Hensel, unpublished data). As an additional constituent of the catabolic version of the pathway, the nonphosphorylating GAPN (4, 5, 21), and its coding gene were identified.
Genes still missing from the T. tenax genome for a functional EMP pathway were assigned (Table 1; Fig. 1). In the case of HK (11), enolase (ENO), PGI, and phosphoglycerate mutase (PGAM), the sequence similarity of the encoding genes allowed an unequivocal assignment. The HK was originally assigned as glucokinase, and only the characterization of the enzyme after expression of the respective gene in E. coli revealed that it is a true HK, catalyzing the phosphorylation of different sugars such as glucose, fructose, and mannose (11).
TABLE 1.
Identified genes coding for the enzymes of the variant of the EMP pathway in T. tenax
| Enzymea | EC no. | COG no.b | Gene | Accession no. | Evidence value and best hitc | Reference(s) |
|---|---|---|---|---|---|---|
| Hexokinase (HK) | 2.7.1.1 | 1940 | hxk | AJ510140 | 3e-99 P. aerophilum, glucokinase, (ROK family) | 11 |
| Glucose-6-phosphate isomerase (PGI) | 5.3.1.9 | 0166 | pgi | AJ621272 | le-93 P. aerophilum | |
| PPi-dependent phosphofructokinase (PPi-PFK) | 2.7.1.90 | 0205 | pfp | Y14655 | 0.0 T. tenax | 56 |
| Fructose-1,6-bisphosphatase type IV (FBP)/inositol-1-monophosphatase (IMP) | 3.1.3.11/ 3.1.3.25 | 0483 | fbpA/suhB | le-63 P. aerophilum, extragenic suppressor protein SuhB homolog | ||
| Fructose-bisphosphate aldolase (FBPA) | 4.1.2.13 | 1830 | fba | AJ310483 | 1.0e-148 T. tenax | 54 |
| Triosephosphate isomerase (TIM) | 5.3.1.1 | 0149 | tpi | AJ012066, AJ515539 | 1.0e-120 T. tenax | 74 |
| Glyceraldehyde-3-phosphate dehydrogenase (nonphosphorylating) (GAPN) | 1.2.1.9 | 1012 | gapN | Y10625 | 0.0 T. tenax | 4, 5, 21, 40 |
| Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) (GAPDH) | 1.2.1.13 | 0057 | gap | Y10126 | 1.0e-135 P. aerophilum | 5, 21 |
| Glyceraldehyde-3-phosphate oxidoreductase (ferrodoxin dependent) (GAPOR) | 2414 | gor | AJ621330 | 0.0 P. aerophilum, aldehyde ferredoxin oxidoreductase | ||
| 3-Phosphoglycerate kinase (PGK) | 2.7.2.3 | 0126 | pgk | Y10126 | 1.0e-124 P. aerophilum | 5 |
| Phosphoglycerate mutase (PGAM) | 5.4.2.1 | 3635 | gpmA | AJ6212333 | 1e-80 P. aerophilum, phosphonopyruvate decarboxylase | |
| Enolase (ENO) | 4.2.1.11 | 0148 | eno | AJ621325 | 1e-50 P. aerophilum | |
| Pyruvate kinase (PK) | 2.7.1.40 | 0469 | pyk | AF065890 | 0.0 T. tenax | 48 |
| Phosphoenolpyruvate synthetase (PEPS) | 2.7.9.2 | 0574 | pps | AJ515537 | 0.0 P. aerophilum | 63, Tjaden et al., unpublished |
| Pyruvate, phosphate dikinase (PPDK) | 2.7.9.1 | 0574 | ppdk | AJ515538 | 0.0 P. aerophilum | 63, Tjaden et al., unpublished |
Enzymes exclusively catalyzing the catabolic pathway are underlined; those operating exclusively in the anabolic direction are shown in bold. For enzymes that are biochemically characterized, the respective references are given. The abbreviations in parenthesis correspond to the respective enzymes shown in Fig. 1.
COG, Clusters of Orthologous Groups (http://www.ncbi.nlm.nih.gov/COG).
Evidence values represent the best Blastp hits and respective species; for homologs with functions deviating from those of the assigned T. tenax enzymes, the respective annotations are given.
FIG. 1.
Variant of the EMP pathway in T. tenax. The enzymes corresponding to homologs identified in this study are shown in grey-shaded boxes; enzymes identified and characterized previously are shown in open boxes. Open arrows indicate irreversible reactions. For abbreviations of enzyme names, see Table 1.
The PGI showed similarity only to an annotated homolog of P. aerophilum, and no similarity to the recently described unique PGIs of the cupin family identified in archaea (e.g., P. furiosus) (19, 71) and bacteria or to the classical PGIs found in bacteria and eucarya was observed. The pgi gene (897 bp) of T. tenax encodes a protein of 298 amino acids with a calculated molecular mass of 33.2 kDa. The heterologous expression of PGI resulted in an enzyme with an apparent molecular mass of 33 kDa (as detected by SDS-PAGE) which catalyzes the reversible glucose 6-phosphate/fructose 6-phosphate conversion. The enzyme follows Michaelis-Menten kinetics for glucose 6-phosphate in the catabolic direction and for fructose 6-phosphate in the anabolic direction. The Km values for glucose 6-phosphate and fructose 6-phophate were 2 mM and 0.17 mM, respectively; the Vmax value for the glucose 6-phosphate/fructose 6-phosphate isomerization was determined to be 170 U/mg of protein and for the reverse anabolic reaction was determined to be 44.5 U/mg of protein (assay temperature, 50°C). This third, new type of PGI differs significantly at the sequence level from members of the cupin family and classical PGIs, and characteristic sequence motifs identified for members of the two other families are absent (19, 71). BlastX searches revealed homologs of this new PGI family in archaea (P. aerophilum, T. acidophilum, T. volcanium, A. pernix, Ferroplasma acidarmanus, Sulfolobus tokodaii, and S. solfataricus) and extreme thermophilic bacteria (Anaerocellum thermophilum and Aquifex aeolicus). The gene coding for the putative PGAM (partial sequence information) showed high similarity to those coding for the recently characterized enzymes of Methanococcus jannaschii (AAB99632) (18, 68) and P. furiosus (AAL82083) (68).
T. tenax does not contain genetic information for glucose-6-phosphatase, suggesting either that glucose-6-phosphatase is absent or that T. tenax possesses a novel enzyme type that has not yet been recognized as such. In addition, gene homologs for ADP-dependent glucokinase or ADP-dependent phosphofructokinases present in other anaerobic, hyperthermophilic archaea (e.g., P. furiosus, A. fulgidus, and Thermococcus species) and for ATP-dependent PFK (Desulfurococcus species) are absent from the T. tenax genome, underlining the central function of the reversible nonallosteric PPi-PFK.
Surprisingly, three ORFs in the T. tenax genome showed convincing similarities to genes encoding enzymes with functions obviously equivalent to those of the assigned EMP enzymes fructose-1,6-bisphosphatase (FBP), ferredoxin-dependent GAP oxidoreductase (GAPOR), and pyruvate, phosphate dikinase (PPDK) (partial sequence information). The identified FBP homolog is a member of the recently described archaeal FBP (type IV) and related enzymes of the inositol-monophosphatase family (COG 0483) and shows 28% identity to the characterized FBP of P. furiosus (70) and 29% identity to the inositol-1-monophosphatase (IMP) of M. jannaschii (61). More recently a type V FBP (COG 1980) was detected in Thermococcus kodakaraensis, and homologs were identified in almost all archaeal genomes (42, 72). A type V FBP which is supposed to be the general archaeal gluconeogenetic FBP (72) is absent from T. tenax. Thus, biochemical studies of the T. tenax enzyme have to be awaited to elucidate a possible gluconeogenetic role in parallel to that of the bifunctional nonallosteric PPi-PFK. Ferredoxin-dependent GAPOR—like GAPN of T. tenax—catalyzes the nonphosphorylating, irreversible conversion of glyceraldehyde 3-phosphate to 3-phosphoglycerate and thus also fulfills a catabolic function. The conditions under which the expression of GAPOR is favored or discriminated against are yet unknown. The complete sequence information for the PPDK gene was obtained by the identification (via Southern hybridization) and sequencing of a genomic clone. The assignment of the PPDK homolog was confirmed by the functional properties of the encoded enzyme (63; Tjaden et al., unpublished data). The recombinant enzyme expressed in E. coli catalyzes the interconversion of PEP and pyruvate according to the following equation: ATP + Pi + pyruvate ↔ AMP + PPi + PEP. The reversibility of the PPDK reaction implies that the direction of conversion depends strongly on the ratio of the substrates and cosubstrates on both sides of the equation; therefore, the enzyme fulfills the functions of both the anabolic PEPS and the catabolic PK.
In summary, the genome data indicate that the variant of the EMP pathway in T. tenax is much more complex than previously suspected. In contrast to results seen with the classical pathway, no control point is present in the phosphorylating part of the EMP variant, as indicated by the presence of a HK with apparently reduced allosteric potential and a reversible nonallosteric PPi-dependent PFK. The major control point is shifted to the middle of the pathway at the level of GAP, where three different enzymes (GAPN, GAPDH, and GAPOR) are involved in the regulation of the carbohydrate flux. In addition, a second control point seems to emerge at the level of pyruvate/PEP conversion, which is catalyzed again by three different enzymes (PK, PEPS, and PPDK) (48, 63).
Other modifications of the EMP pathway are found in the anaerobic species P. furiosus (27, 28, 31, 66, 69), Thermococcus species (31, 44, 50), D. amylolyticus (20), and A. fulgidus (33). The glycolysis of D. amylolyticus differs from that of the classical version of the EMP pathway by the presence of a ferredoxin-dependent GAPOR replacing the “conventional” phosphate- and pyridine nucleotide-dependent GAPDH. A more extended variation is seen with P. furiosus and Thermococcus species (as well as with A. fulgidus), which additionally replace the classical ATP-dependent HK and phosphofructokinase with nonallosteric ADP-dependent glucokinases and phosphofructokinases. In addition, ADP-dependent phosphofructokinase activity was demonstrated in crude extracts from members of the orders Methanococcales (e.g., Methanococcus maripaludis) and Methanosarcinales (e.g., Methanosarcina mazei) (73) and a novel bifunctional ADP-dependent glucokinase/phosphofructokinase is described for M. jannaschii (45, 73). The absence of allosteric sugar kinases and the presence of GAPN and GAPOR enzyme activities in several archaea, as well as the identification of GAPN and GAPOR gene homologs—in addition to GAPDH—in many archaeal genomes (e.g., those of Pyrococcus spp.) (72), indicates that among all the different modifications of the EMP pathway in archaea, the regulation at the level of GAP might represent a unique feature of central carbon metabolism.
(b) Organization of the EMP genes.
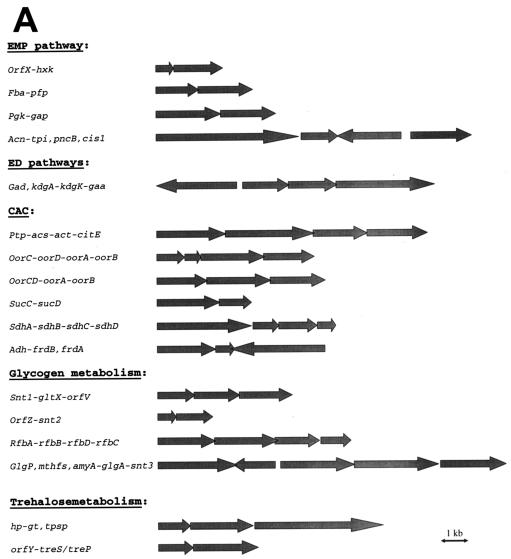
Analyses of gene organization and transcription studies reveal that several EMP genes are organized in operons, suggesting specific functional relationships between the genes or their products (Table 1) (see Fig. 3). Thus, as previously shown (54), the pfp gene coding for the PPi-dependent PFK, which catalyzes the reversible phosphorylation of fructose 6-phosphate, forms an operon with the fba gene encoding FBA (fba-pfp operon). The coordinated transcription of these genes in one operon seems meaningful for T. tenax, because both enzymes catalyze not only consecutive but also reversible reactions and are therefore of equivalent levels of importance for both directions of the pathway. Also, the phosphorylating NADP+-dependent GAPDH gene (gap) and the PGK gene (pgk) are clustered in an operon (pgk-gap operon) (5). In both operons, the two genes overlap by 4 bp (the coding regions overlap by 1 bp) and the formation of bicistronic as well as monocistronic mRNAs was demonstrated. Northern analyses reveal that the HK gene (hxk) is part of an operon (orfX-hxk operon) (11) and that, again, both genes overlap by 4 bp. The ORF located upstream (orfX) codes for a protein of unknown function, and only one homolog was identified in P. aerophilum (AE009933) (11).
FIG. 3.
(A) Organization of functionally related genes of the central carbohydrate metabolism in T. tenax. (Gene names are given in alphabetical order at the end of the legend; gene data are given in Table 1 to Table 6.) (B) Putative promoter structures of genes organized in operons. The nucleotide sequences (5′ end) and upstream regions of genes are shown. The putative crenarchaeal promoter sequences (3, 59, 60) with BRE sites [crenarchaeal consensus sequence (A/G)N(A/T)AA(A/T)] and the TATA box [crenarchaeal consensus sequence (C/T)TTTTAAA] are in light and dark grey boxes, respectively. Putative Shine-Dalgarno sequences (GAGG) (29) are underlined, the putative start codon is shown in boldface characters, and the stop codon of preceding genes is indicated with double underlining. The operon structures shown were confirmed for the EMP genes by Northern analysis. All other clusters shown represent putative operons. Gene names and corresponding enzyme names: acn, aconitase; adh, alcohol dehydrogenase; acs, acetyl-CoA synthetase; act, acetyl-CoA transferase; amyA, α-amylase; cis1, citrate synthase 1; citE, citrate lyase β chain; fba, FBPA; frdAB, fumarate reductase α and β chains; gaa, glucan-1,4-α-glucosidase; gad, GAD; gap, GAPDH; glgA, glycogen synthase; glgP, (glycogen) phosphorylase; gltX, glutamyl tRNA synthetase; gt, glycosyltransferase; hp, hypothetical protein, hxk, HK; kdgA, KDP(G)A; kdgK, KDGK; mthfs, 5-formyltetrahydrofolate cyclo-ligase; oorABCD, oxoacid:ferredoxin oxidoreductase α, β, γ, and δ chains; orfV, orfX, orfY, and orfZ, hypothetical proteins; pncB, nicotinate phosphoribosyltransferase; pfp, PPi-PFK; pgk, PGK; ptp, putative transport protein; rfbA, glucose-1-phosphate thymidylyltransferase; rfbB, dTDP-glucose-4,6-dehydratase; rfbC, dTDP-4-dehydrorhamnose-3,5-epimerase; rfbD, dTDP-4-dehydrorhamnose reductase; sdhABCD, succinate dehydrogenase α, β, γ, and δ chains; sucDC, succinyl-CoA synthetase α and β chains; snt, sugar phosphate nucleotidyltransferase; tpi, TIM; tpsp, TPSP; treS/treP, trehalose synthase or trehalose phosphorylase.
Remarkably, as shown by Northern analysis the TIM gene (tpi) forms an operon with the aconitase gene (acn), a constituent of the reversible CAC in T. tenax. The acn and tpi genes are separated by 40 bp. The tpi gene overlaps by 15 bp with a gene homolog coding for a putative nicotinate phosphoribosyl transferase (pncB gene; AJ515539), which is oriented in the opposite direction. Further downstream of the pncB gene (151 bp) is the gene coding for citrate synthase 1 (cis1) (63). This clustering of the tpi gene with CAC genes is rather unusual. In most archaea and bacteria analyzed so far, the tpi gene is adjacent to other EMP genes such as the gap, pgk, gpmA, and eno genes (63). The specific linkage of the tpi and acn genes in T. tenax may reflect the preferred functional relationship of both reversible pathways in this organism and hints at their strong coordination under anabolic and catabolic conditions.
(ii) Variants of the Entner-Doudoroff (ED) pathway. (a) As-signment of genes involved in the non- and semiphosphorylative ED pathways.
For reconstruction of the nonphosphorylative ED pathway of T. tenax, we screened for genes encoding the enzymes GDH, gluconolactonase, gluconate dehydratase (GAD), KDGA, aldehyde oxidoreductase/dehydrogenase, and glycerate kinase. Homologs of all these genes—with exception of a homolog of the gene encoding gluconolactonase—were found in the T. tenax genome sequence (Table 2; Fig. 2). The lack of the gluconolactonase gene homolog in T. tenax indicates that this enzyme (EC 3.1.1.17) is either absent or not related to known bacterial counterparts (e.g., Zymomonas mobilis). So far, archaeal gluconolactonase homologs were only identified in A. fulgidus and S. solfataricus (72). The assignment of the genes encoding GDH (gdh), GAD (gad), and KDGA (kdgA) was confirmed by identifying their encoded products. Thus, the gdh gene was unequivocally confirmed by identification of the N-terminal sequence of the GDH isolated from T. tenax cells (57) and of the activity of the recombinant protein expressed in E. coli. The GAD gene (gad) was recognized due to its neighborhood to the KDGA gene, and this assignment was confirmed by the enzymatic properties of the recombinant gene product (H. Ahmed, T. Ettema, B. Tjaden, J. Van der Oost, and B. Siebers, unpublished data). The gene showed significant similarity to those encoding members of the mandelate racemase/muconate-lactonizing enzyme family (ENO superfamily), which comprises numerous different enzyme activities (e.g., those of muconate cycloisomerase, galactonate dehydratase, putative isomerase, and the starvation-sensing protein RspA). The kdgA gene has been expressed in E. coli and biochemically characterized (Ahmed et al., unpublished data).
TABLE 2.
Identified genes coding for the enzymes of the semi- and nonphosphorylative ED pathways in T. tenax
| Enzymea | Candidate no. | EC no. | COG no.b | Gene | Accession no. | Evidence value and best hitc | Reference |
|---|---|---|---|---|---|---|---|
| Glucose dehydrogenase (GDH) | 1.1.1.47 | 1063 | gdh | AJ621346 | 2e-46 Haloferax mediterranei | 57 | |
| Gluconate dehydrogenase (GAD) | 4.2.1.39 | 4948 | gad | AJ621281 | 1.0e-139 S. tokodaii, 396- amino-acid hypothetical DgoA protein | Ahmed et al., unpublished | |
| 2-Keto-3-deoxy-(6-phospho-) gluconate aldolase [KD(P)GA] | 4.1.2.d | 0329 | kdgA | AJ621282 | 3e-68 S. solfataricus | Ahmed et al., unpublished | |
| 2-Keto-3-deoxy-gluconate kinase (KDGK) | 2.7.1.45 | 0524 | kdgK | AJ621283 | 1.0e-102 S. solfataricus, fructokinase | Ahmed et al., unpublished | |
| Aldehyde oxidoreductase, ferredoxin dependent (AOR) (3 candidates) | 1 | 1.2.7.d | 2414 | aor | AJ621293 | 0.0 P. aerophilum | |
| 2 | AJ621285 | 0.0 P. aerophilum | |||||
| 3 | AJ621317 | e-127 (N-terminal); e-107 (C-terminal) P. aerophilum | |||||
| Aldehyde dehydrogenase, NAD(P)+ dependent (ALDH) (2 candidates) | 1 2 | 1.2.1.d | 1012 | aldh | AJ621321AJ621277 | e-115 T. volcanium e-114 S. tokodaii | |
| Glycerate kinase (GK) | 2.7.1.31 | 2379 | garK | AJ621345 | 2e-56 A. pernix, hypothetical protein |
For enzymes that were biochemically characterized, the respective references are given. The abbreviations in parentheses correspond to the respective enzymes shown in Fig. 2.
For definition, see Table 1, footnote b.
For definition, see Table 1, footnote c. In cases in which two or more candidates were identified in the genome, the respective evidence values are given.
Final EC number has not been assigned.
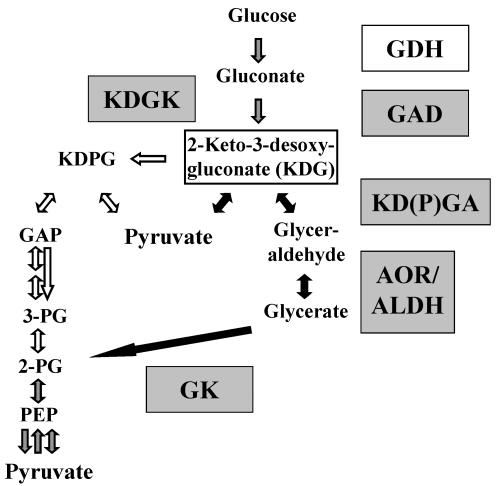
FIG. 2.
Variants of the ED pathway in T. tenax. The enzymes identified in the T. tenax genome are shown in grey-shaded boxes, and the previously characterized GDH is shown in an open box. The central intermediate KDG of both variants is boxed. Closed arrows indicate the reactions of the nonphosphorylative version of the ED pathway; open arrows indicate the reactions of the semiphosphorylative version. Grey-shaded arrows mark reactions involved in both versions. For abbreviations of enzyme names, see Table 2.
In contrast to the results seen with the S. solfataricus KDGA (6), which displays high-level sequence similarity (43% identity) to the sequence of the T. tenax enzyme, the latter is not specific for KDG but also uses 2-keto-3-deoxy-6-phospho-gluconate (KDPG) as a substrate and thus represents a 2-keto-3-deoxy-(6-phospho-)gluconate [KD(P)G] aldolase [KD(P)GA]. The assignment of the glycerate kinase gene is still unclear and was based only on sequence similarity to the putative genes of S. solfataricus, T. acidophilum, P. furiosus, and Thermotoga maritima (29, 26, 33, and 34% identity, respectively, by a Blast search). The gene encoding the enzyme which catalyzes the oxidation of glyceraldehyde to glycerate could not be assigned decisively. In the T. tenax genome, three ORFs (for candidate 3, only partial C- and N-terminal information is available) (Table 2) were found which show convincing similarities to genes encoding aldehyde:ferredoxin oxidoreductases; the sequences of two ORFs show high-level similarity to sequences annotated as genes coding for aldehyde dehydrogenases or for related enzymes which are—like GAPN—members of the aldehyde dehydrogenase superfamily. Since cell extracts of T. tenax are reported to catalyze the oxidation of glyceraldehyde with benzylviologen only but not with NADP+ or NAD+ as electron acceptors (49), we assume that ferredoxin-dependent oxidoreductases is responsible for oxidation of glyceraldehyde. A gene homolog for ferredoxin was identified in the T. tenax genome.
Surprisingly, an ORF with high-level similarity to genes encoding KDG kinases (KDGKs) and fructokinases was identified in the T. tenax genome. The close neighborhood to the kdgA gene suggests an involvement in the ED pathway (see below). The KDGK gene was expressed heterologously in E. coli and the corresponding enzyme activity has been confirmed, indicating that the semiphosphorylative ED pathway is also active in T. tenax (Ahmed et al., unpublished data). This finding is rather unexpected, since the semiphosphorylative ED pathway has been assumed to be characteristic of haloarchaea. The localization of a gene homolog coding for glucan-1,4-α-glucosidase (GAA) (gaa; see Table 5) in the close neighborhood of kdgK suggests that the ED variants are involved in the hydrolytic degradation of polysaccharides (e.g., glycogen) in T. tenax (see below). A homolog of the glucose-6-phosphate dehydrogenase gene of the kind typical of the classical ED pathway was not found in the T. tenax genome.
TABLE 5.
Identified genes coding for enzymes of the glycogen metabolism in T. tenax
| Enzymea | Candidate no. | EC no. | COG no.b | Gene | Accession no. | Evidence value and best hit |
|---|---|---|---|---|---|---|
| Phosphoglucomutase | 2.7.5.1 | 1109 | pgm | AJ621332 | 1.0e-154 P. aerophilum, phosphomannomutase | |
| Sugar phosphate nucleotidyl transferase (7 candidates) | 1 | 2.7.7.d | 1208 | snt | AJ621273 | 1.94e-91 P. aerohilum, sugar phosphate nucleotidyl transferase |
| 2 | AJ621322 | 1.6e-61 P. aerophilum, sugar phosphate nucleotidyl transferase | ||||
| 3 | AJ621298, AJ621299 | 2.5e-114 P. aerophilum, glucose-1-phosphate adenylyltransferase | ||||
| 4 | AJ621300 | 8.65e-104 P. aerophilum, sugar phosphate transferase | ||||
| 5 | AJ621349 | 2.96e-85 P. aerophilum, mannose-1-phosphate guanyltransferase | ||||
| 6 | AJ621350 | 1.0e-97 P. aerophilum, sugar phosphate nucleotidyl transferase | ||||
| 7 | 2.7.7.24 | 1209 | rfbA | AJ621292 | 1.6e-124 P. aerophilum, glucose-1-phosphate thymidylyltransferase | |
| Glycogen (starch) synthase | 2.4.1.11 | 0297 | glgA | AJ621297 | 2e-74 P. aerophilum, conserved hypothetical protein | |
| (Glucogen) phosphorylase | 2.4.1.1 | 0058 | glgP | AJ621294 | 0.0 P. aerophilum, conserved hypothetical protein | |
| α-Amylase | 3.2.1.1 | 1449 | amyA | AJ621296 | 0.0 P. aerophilum, conserved hypothetical protein | |
| Glucan 1,4-alpha-glucosidase (glucoamylase) | 3.2.1.3 | 3387 | gaa | AJ621284 | e-125 S. tokadaii, 599-amino-acid hypothetical protein | |
| α-Glucosidase (maltase) | 3.2.1.20 | 1501 | malZ | AJ621310 | 0.0 P. aerophilum |
The enzymes involved in glycogen degradation are underlined, and those involved in its biosynthesis are in bold.
For definition, see Table 1, footnote b.
For definition, see Table 1, footnote c. In cases in which two or more candidates were identified in the genome, the respective evidence values are given.
Final EC number has not been assigned.
Thus, the semiphosphorylative ED pathway seems to be present in this hyperthermophile probably in addition to the nonphosphorylative ED variant, and the KD(P)GA is the key activity in both variants. The organization of ED genes suggests a preference for the function of the ED variants in the hydrolytic degradation of glycogen.
(b) Organization of the ED genes.
Analysis of the organization of ED genes (Table 2; Fig. 2) revealed the presence of a gene cluster which comprises the genes coding for GAD (gad), KD(P)GA (kdgA), KDGK (kdgK), and GAA (gaa). The kdgA and kdgK genes as well as the kdgK and gaa genes overlap by 4 bp, giving good evidence that the three genes form an operon (Fig. 3B). The gene encoding the GAD is localized 68 bp downstream of the putative kdgA-kdgK-gaa operon and is oriented in the opposite direction (Fig. 3A).
(iii) Pentose metabolism. Assignment of genes involved in pentose metabolism.
T. tenax does not possess the complete gene set for oxidative degradation of carbohydrates via the PPP (Table 3). As seen with S. solfataricus (52), only the gene homologs for ribosephosphate isomerase and transketolase (partial sequence information for the N- and C-terminal sections) were found to date. Genes coding for the classic glucose-6-phosphate dehydrogenase, decarboxylating phosphogluconate dehydrogenase, ribulosephosphate-3-epimerase, and transaldolase seem to be absent. These findings might indicate that hyperthermophilic archaea either use different, nonhomologous enzymes (which show no sequence similarity to known counterparts) in the PPP or utilize a yet unknown, analogous pathway for carbohydrate catabolism. Thus, the biosynthesis of pentoses in hyperthermophilic archaea remains unclear. The presence of a nonoxidative PPP (as proposed for ribose biosynthesis in Methanococcus maripaludis) (67) seems to be improbable, since a gene homolog for the common ribulosephosphate-3-epimerase was not found in the T. tenax genome. However, two ORFs were detected in the T. tenax genome which showed similarity to genes coding for well-known enzymes of pentose metabolism such as ribokinase (sugar kinase of the ribokinase [pfkB] family) (ATP + d-ribose ↔ ADP + d-ribose 5-phosphate) and deoxyribose-phosphate aldolase (2-deoxy-d-ribose 5-phosphate ↔ d-glyceraldehyde 3-phosphate + acetaldehyde).
TABLE 3.
Identified genes coding for the enzymes of the pentose metabolism in T. tenax
| Enzyme | EC no. | COG no.a | Gene | Accession no. | Evidence value and best hitb |
|---|---|---|---|---|---|
| Ribosephosphate isomerase | 5.3.1.6 | 0120 | rpiA | AJ621331 | 5e-72 P. aerophilum |
| Transketolase | 2.2.1.1 | tktA | |||
| N-terminal section | 3959 | AJ621311 | 7e-94 P. aerophilum, N-terminal section | ||
| C-terminal section | 3958 | AJ621312 | 7e-70 P. aerophilum, C-terminal section | ||
| Deoxyribose-phosphate aldolase | 4.1.2.4 | 0274 | deoC | AJ621351 | 8e-51 M. thermautotrophicus |
| Ribokinase | 2.7.1.15 | 0524 | rbsK | AJ621324 | 1.0e-108 P. aerophilum, sugar kinase, possible phosphofructokinase |
(iv) The reversible CAC. (a) Assignment of genes involved in the oxidative and reductive CAC.
As shown in Table 4, not all enzymes necessary for the oxidative and reductive CAC were identified in the T. tenax genome. For some enzymes, not all expected subunits could be found or a confidential assignment is not possible because of intermediate levels of similarity to different enzymes. However, most of the enzymes could be assigned due to the high sequence similarity of their encoding genes to sequences of gene homologs (such as those encoding citrate synthase, aconitase, isocitrate dehydrogenase, succinyl-CoA synthetase, fumarase, and malate dehydrogenase) in other genomes. All CAC genes, with the sole exception of the citrate synthase-encoding gene, of which two copies (cis1 and cis2) were found, occurred only once in the genome. The cis1 gene was originally identified in the genomic clone harboring the genes encoding TIM and aconitase. The enzyme was expressed in E. coli and showed typical citrate synthase activity (63). The expression of cis2 is planned for the near future. Detailed enzymatic as well as transcription studies of both genes have to be completed before the physiological rationale behind the gene doubling can be resolved.
TABLE 4.
Identified genes coding for the enzymes of the citric acid cycle in T. tenax
| Enzymea | Candidate no. | EC no. | COGb no. | Gene | Accession no. | Evidence value and best hitc |
|---|---|---|---|---|---|---|
| Citrate synthase 1 | 4.1.3.7 | 0372 | cis1 | AJ515539 | 1.0e-126 P. aerophilum | |
| Citrate synthase 2 | 4.1.3.7 | 0372 | cis2 | AJ621309 | 0.0 P. aerophilum | |
| Citrate lyase | 4.1.3.6 | |||||
| β chain | 2301 | citE | AJ621302 | 3e-93 S. solfataricus | ||
| Aconitase | 4.2.1.3 | 1048 | acn | AJ515539 | 0.0 P. aerophilum | |
| Isocitrate dehydrogenase (NADP+) | 1.1.1.42 | 0538 | icd | AJ621308 | 0.0 P. aerophilum | |
| 2-Oxoglutarate dehydrogenase | ||||||
| Dihydrolipoamide dehydrogenase (E3 component) (2 candidates) | 1 | 1.8.1.4 | 1249 | lpd | AJ621347AJ621348 | 8e-85 T. acidophilum |
| 2 | 1.0e-172 P. aerophilum, mercuric reductase | |||||
| 2-Oxoglutarate synthase (2-oxoglutarate-ferredoxin oxidoreductase) (2 candidates) | 1.2.7.3 | |||||
| α chain | 1 | 0674 | oorA | AJ621327 | e-158 P. aerophilum, pyruvate ferredoxin oxidoreductase α subunit | |
| 2 | AJ621339 | 9e-94 P. aerophilum, pyruvate ferredoxin oxidoreductase α subunit | ||||
| β chain | 1 | 1013 | oorB | AJ621326 | e-142 P. aerophilum, pyruvate ferredoxin oxidoreductase β subunit | |
| 2 | AJ621340 | 3e-93 M. jannaschii, pyruvate ferredoxin oxidoreductase β subunit | ||||
| γ chain | 1 | 1014 | oorC | AJ621329 | 5e-69 P. aerophilum, pyruvate ferredoxin oxidoreductase γ subunit | |
| δ chain | 1 | 1144 | oorD | AJ621328 | 2e-42 P. aerophilum, pyruvate ferredoxin oxidoreductase δ subunit | |
| γ and δ chaind | 2 | oorCD | AJ621338 | 4e-40 M. thermautotrophicus, pyruvate oxidoreducatase γ subunit | ||
| Succinyl-CoA synthetase | 6.2.1.5 | |||||
| α chain | 0074 | sucD | AJ621271 | 2e-90 P. aerophilum | ||
| β chain | 0045 | sucC | AJ621270 | e-178 P. aerophilum | ||
| Succinate dehydrogenase/Fumarate reductase | 1.3.99.1 | |||||
| Flavoprotein subunit α (2 candidates) | 1 | 1053 | sdhA/frdA | AJ621269 | 0.0 P. aerophilum, succinate dehydrogenase flavoprotein subunit | |
| 2 | AJ621278 | 0.0 P. aerophilum, succinate dehydrogenase flavoprotein subunit | ||||
| Iron sulfur subunit β (2 candidates) | 1 | 0479 | sdhB/frdB | AJ621268 | 1.0e-112 P. aerophilum, succinate dehydrogenase iron-sulfur subunit | |
| 2 | AJ621279 | 1e-12 P. aerophilum, iron-sulfur protein, putative | ||||
| Cytochrome b subunit γ | 1 | sdhC/frdC | AJ621267 | 2e-29 P. aerophilum, succinate dehydrogenase cytochrome b subunit | ||
| Subunit δ | 1 | sdhD/frdD | AJ621266 | 3e-09 P. aerophilum, succinate dehydrogenase subunit D | ||
| Fumarase class II | 4.2.1.2 | fumC | AJ621286 | 1.0e-107 S. tokadaii | ||
| Fumarase class I N-terminal domain/tartrate dehydratase α subunit | 4.2.1.2/4.2.1.32 | 0114/1951 | fumA/ttdA | AJ621316 | 1.0e-122 P. aerophilum, fumarate hydratase class I α subunit | |
| Fumarase class I C-terminal domain/tartrate dehydratase β subunit | 0114/1838 | fumB/ttdB | AJ621315 | 8e-84 P. aerophilum, fumarate hydratase class I β subunit | ||
| Malate dehydrogenase | 1.1.1.37 | 0039 | mdh | AJ621301 | 1.0e-136 P. aerophilum |
Enzymes that are believed to exclusively catalyze the oxidative pathway are underlined; those operating exclusively in the reductive cycle are marked in bold.
For definition, see Table 1, footnote b.
For definition, see Table 1, footnote c. In cases in which two or more candidates were identified in the genome, the respective evidence values are given.
The ORF of candidate 2 codes for both the γ and δ subunits.
In addition to the gene encoding isocitrate dehydrogenase, a second gene homolog was identified which showed high similarity to genes encoding isocitrate dehydrogenases as well as 3-isopropylmalate dehydrogenases (leuB; AJ621334). However, due to its localization downstream of a putative leuA gene homolog, which codes for 2-isopropylmalate synthase (AJ621335), we assume that this homolog represents the leuB gene, which is involved in the leucine biosynthesis pathway (22). For the reversible fumarase (fumarate hydratase), three different ORFs were found in the T. tenax genome: one gene homolog (partial sequence information) with high similarity to genes encoding class II enzymes and two genes which show similarity either to the N-terminal and C-terminal domains, respectively, of genes encoding bacterial class I fumarases or alternatively to those encoding subunits α and β of tartrate dehydratases. Although both fumarase homologs have been annotated for many archaeal genomes, to our knowledge only the class II fumarase from S. solfataricus has been characterized (8).
The identification of the genes encoding enzymes determining the direction of the cycle—with the exception the gene encoding citrate synthase—reveals some problems. (i) For the citrate lyase, only a gene homolog for the β chain [citryl-CoA lyase subunit; (3S)-citryl-CoA ↔ acetyl-CoA + oxaloacetate] was found but homologs for subunits α [citrate-CoA transferase subunit; acetyl-CoA + citrate ↔ acetate + (3S)-citryl-CoA] and γ (acyl carrier protein subunit) are missing [Leuconostoc mesenteroides (αβγ)6 subunit composition] (2). To our knowledge nothing is known so far about the reaction mechanism of archaeal citrate lyases, although ATP citrate lyase activity was demonstrated in cell extracts of T. neutrophilus (1). The gene coding for the β subunit seems to form an operon with three gene homologs coding for a putative transport protein (ptp; AJ621305), putative acetyl-CoA synthetases (acetate-CoA ligase gene and acs; AJ621304), and acetyl-CoA transferases or carnitine dehydratases (act; AJ621303). Thus, acetate-CoA ligase (EC 6.2.1.1; ATP + acetate + CoA ↔ AMP + Pi + acetyl-CoA) might be responsible for the generation of acetyl-CoA, which is considered to be a free analog of the acetylated acyl carrier protein (γ subunit) (2) and the putative acetyl-CoA transferase might represent the citrate-CoA transferase subunit α. However, for a confidential assignment of citrate lyase experimental analyses have to be awaited. (ii) For the catabolic enzyme 2-oxoglutarate dehydrogenase, only the E3 component (dihydrolipoamide dehydrogenase; two candidates) of the multienzyme complex consisting of E1, dihydrolipoamide-S-succinyltransferase (E2), and E3 was found, which can be shared by different 2-oxo acid dehydrogenases and the glycine cleavage system (22, 74). (iii) KOR, which is thought to catalyze the reaction in the anabolic direction, could not be assigned unequivocally because of the high level of similarity between the different ferredoxin-dependent oxidoreductases. Two clusters of gene homologs were identified with similarity to genes encoding POR (pyruvate:ferredoxin oxidoreductase) and related 2-oxoacid synthases (2-oxoacid:ferredoxin oxidoreductases [OOR]). Both are organized in an operon which comprises genes encoding the α, β, γ, and δ subunits and thus—because of their four-subunit composition—might represent either POR or KOR (22, 35, 62). In addition, four additional gene clusters were identified which encode homologs for 2-oxoacid synthases (accession no. AJ621306, AJ621307, AJ621313, AJ621314, AJ621318, AJ621319, AJ621320, AJ621352, and AJ621353). (iv) For the succinate dehydrogenase (catabolic) and fumarate reductase (anabolic), two gene clusters were identified which allow no differentiation between the two enzymes due to their similarity. However, the genetic organization might give some evidence for the respective enzyme activities. We assume that the putative succinate dehydrogenase (candidate 1) is organized in an operon which encodes all four subunits (α, β, γ, and δ), whereas the fumarate reductase (candidate 2) is organized in a gene cluster with homologs coding for the flavoprotein subunit (chain α), the iron-sulfur protein (chain β; homology only to the N-terminal part), and a zinc-dependent class III alcohol dehydrogenase (AJ621280). The gene displacement in noncatalytic subunits of fumarate reductases has been previously described (22); thus, whereas the membrane-bound enzymes are composed of four or three subunits, the cytoplasmatic enzyme exhibits only one type of subunit.
In summary, all these findings suggest that the CAC in T. tenax is operative in the reductive and oxidative direction. The only missing enzyme is 2-oxoglutarate dehydrogenase. However, measurements in crude extracts demonstrate 2-oxoglutarate:benzylviologen oxidoreductase activity (49) and more recent reports about the POR in Clostridium thermoaceticum (14) demonstrate that the enzyme is active with ferredoxin as a cosubstrate in the oxidative as well as in the reductive reaction. Thus, 2-oxoglutarate oxidoreductase might be involved in both the oxidative and the reductive CAC in T. tenax.
No convincing candidates for the two key enzymes of the glyoxylate cycle—isocitrate lyase and malate synthase—were identified, suggesting that a functional glyoxylate cycle is absent from T. tenax. Surprisingly, putative gene homologs coding for the three subunits of the carbon monoxide dehydrogenase (medium, small, and large chains; AJ621345, AJ621344, and AJ621343, respectively) were identified in the T. tenax genome. All three subunits overlap by 4 bp and thus obviously form an operon. However, none of the other genes for a functional Wood-Ljungdahl pathway (e.g., from formylmethanofuran to methyl-tetrahydromethanopterin) were found, suggesting that a reductive acetyl-CoA pathway, which is described for several archaea (e.g., M. thermautotrophicus, Methanopyrus kandleri and in autotrophic species of Archaeoglobus [47]), is absent from T. tenax. However, the carbon monoxide dehydrogenase might be involved in coupling the POR reaction to CO oxidation via ferredoxin, as described for C. thermoaceticum (14).
(b) Organization of the CAC genes.
Obviously, several CAC genes are organized in operons or operon-like structures (Fig. 3; Table 4). Seven operons have been identified so far. The gene coding for the β subunit of citrate lyase (citE) seems to form an operon with the upstream-located gene homologs coding for a putative transport protein (ptp), acetyl-CoA synthetase (acs), and acetyl-CoA transferases (act) or carnitine dehydratases. The ptp and acs as well as the act and citE genes overlap by 4 bp and the acs and act genes overlap by 17 bp, forming the ptp-acs-act-citE operon and suggesting a functional connection of the genes. Further, the genes coding for the reversible enzymes TIM (tpi) and aconitase (acn) (see above) are linked in an operon (acn-tpi) (as confirmed by Northern analysis) (63), indicating a strong correlation between the reversible CAC and EMP pathways. The genes coding for the two putative candidates for KOR, e.g., oxoacid synthase (oxoacid:ferredoxin oxidoreductase; oor gene), are organized in operons which comprise all four subunits (α, β, γ, and δ). Whereas for candidate 1 each subunit is encoded by one gene (operon oorC-D-A-B), the second candidate comprises subunit γ and δ in one ORF (operon oorCD-A-B). All genes overlap by 4 bp. The fifth operon comprises two genes coding for α and β subunits of succinyl-CoA synthetase (sucC-D operon) which overlap by 4 bp. The sixth operon harbors four genes coding for the putative succinate dehydrogenase (sdh gene; candidate 1 Sdh/Frd). Whereas the downstream sdhA-located genes coding for the β, γ, and δ subunits overlap by 4 bp, the upstream-located sdhA gene coding for the α subunit is separated by 2 bp. The presence of putative promoter structures in front of sdhA only (Fig. 3B) indicates that all four genes form the sdhA-B-C-D operon. A second candidate, which encodes a putative fumarate reductase and comprises two genes with homology to sdhA/frdA and sdhB/frdB, was identified in the T. tenax genome. Here both ORFs overlap by 56 bp but are oriented in opposite directions, and the sdhB/frdB gene homolog overlaps by 4 bp with the adh gene which codes for a putative Zn-dependent alcohol dehydrogenase (adh-frdB operon).
(v) Glycogen metabolism. (a) Assignment of genes involved in glycogen metabolism.
By the use of the T. tenax genome, gene homologs for almost all enzymes necessary for catalyzing the synthesis and degradation of glycogen were found (Table 5). As determined on the basis of sequence similarity, seven genes are thought to encode sugar phosphate nucleotidyl transferases (snt) catalyzing the synthesis of the nucleotide-activated glucose. A possible role in glycogen synthesis is proposed for candidate 3 (snt3), which is found closely neighboring genes involved in glycogen metabolism (Fig. 3A). Because of the relatedness and numerous different annotations of gene homologs in the databases, a prediction of substrate specificity was not possible. Only for candidate 7, glucose-1-phosphate thymidylyltransferase (dTDP-glucose pyrophosphorylase [RfbA]; EC 2.7.7.24), can activity be suggested due to the closely neighboring genes coding for dTDP-glucose-4,6-dehydratase (RfbB; EC 4.2.1.46), dTDP-4-dehydrorhamnose reductase (RfbD; EC 1.1.1.133), and dTDP-4-dehydrorhamnose-3,5-epimerase (RfbC; EC 5.1.3.13). All four genes are involved in the biosynthesis of dTDP-rhamnose, which represents a precursor for the O-specific polysaccharide chain in the lipopolysaccharide of the outer membrane in most gram-negative bacteria (36), thus raising questions about their functions in archaea.
Only one gene was identified which shows convincing similarity to genes encoding glycogen synthase, which catalyzes the succeeding polymerization reaction. No gene homolog was found for the 1,4-α-glucan branching enzyme, which catalyzes the formation of α-1-6 bonds. For glycogen degradation, four different genes were identified by sequence similarity: the genes coding for glycogen phosphorylase, α-amylase, GAA, and α-glucosidase (maltase).
The glgP gene coding for glycogen phosphorylase was identified in T. tenax due to its similarity to genes encoding archaeal and bacterial glucan, glycogen, and maltodextrin phosphorylases. The glgP gene (1,458 bp), which codes for a protein of 485 amino acids, was expressed in E. coli. The recombinant protein was purified by heat precipitation and ion exchange chromatography and revealed a subunit molecular mass of 55 kDa (as determined by SDS-PAGE), a result in good agreement with the calculated molecular mass of 55.56 kDa. The enzyme catalyzes the Pi-dependent degradation of glycogen, forming glucose 1-phosphate. Glycogen phosphorylase follows Michaelis-Menten kinetics, with Km and Vmax values of 0.09 mg/ml and 0.9 U/mg of protein for glycogen and 0.09 mM and 0.9 U/mg of protein for Pi at 50°C. Thus, the glycogen phosphorylase seems to play a major role in the phosphorolytic degradation of glycogen.
Genes encoding debranching enzymes (such as pullulanases) seem to be missing from the T. tenax genome. Since most of the GAAs described are able to hydrolyze 1,6-α-bonds also, however, this enzyme might compensate for the missing activity of debranching enzyme. The gaa gene is found closely neighboring the genes coding for the key enzymes of the modified ED pathways, indicating the close functional relationship between hydrolytic degradation of polysaccharides and the ED variants. Finally, one gene homolog was found with similarity to genes encoding phosphomannomutase or PGM. Whether the respective gene product exhibits PGM activity and thus is engaged in the catabolic and anabolic directions remains to be analyzed.
In summary, most gene homologs necessary for glycogen metabolism were identified in the T. tenax genome, and the glycogen phosphorylase, which is involved in the phosphorolytic degradation of glycogen, is characterized. The only missing counterparts are enzymes that are involved in the formation (branching enzyme) and hydrolysis of 1,6-α bonds. Whether these enzymes show no similarity to known enzymes or are generally absent and their activity is taken over by glycogen synthase and GAA remains to be proven.
(b) Organization of genes involved in glycogen metabolism.
Three of the seven gene homologs coding for sugar phosphate nucleotidyl transferases are organized in operons (Fig. 3; Table 5). The gene of candidate 1 (snt1) forms an operon with the gltX gene (AJ621274 and AJ621275; coding for glutamyl tRNA synthetase) and orfV (AJ621276; coding for a hypothetical protein). snt1 and gltX overlap by 14 bp, and gltX and orfV overlap by 4 bp, forming the snt1-gltX-orfV operon. The gene of candidate 2 (snt2) overlaps by 4 bp with orfZ (AJ621323), which encodes a hypothetical protein in P. aerophilum (orfZ-snt2 operon). A functional connection is proposed for the putative glucose-1-phosphate thymidylyltransferase gene (rfbA; candidate 7) which forms an operon with the rfbB (4-bp overlap; dTDP-glucose-4,6-dehydratase; AJ621291), rfbD (10-bp overlap; dTDP-4-dehydrorhamnose reductase; AJ621290) and rfbC (3-bp gap; dTDP-4-dehydrorhamnose-3,5-epimerase; AJ621289) genes (rfbA-B-D-C operon). Four of the genes involved in the glycogen metabolism, the genes encoding glycogen phosphorylase (glgP), α-amylase (amyA), glycogen synthase (glgA), and sugar phosphate nucleotidyl transferases (candidate 3; snt3), are clustered in the genome of T. tenax. The glgA gene is located upstream of a gene homolog (mthfs; AJ621295) which encodes a putative 5-formyltetrahydrofolate cyclo-ligase and is oriented in the opposite direction. Both ORFs overlap by 22 bp. The amyA, glgA, and (separated by 44 bp) snt3 genes are localized downstream of the mthfs gene (75 bp) and in the same orientation as glgP. Since both amyA and glgA overlap by 1 bp (amyA-glgA operon), we assume that a functional linkage existed between these two genes at least, suggesting that the α-amylase is involved in glycogen metabolism. A similar combination of genes involved in glycogen degradation and synthesis has been previously reported for bacterial species (41). As described above, the gaa gene coding for GAA is organized in an operon with genes coding for the key enzymes of the ED pathway (see “Organization of the ED genes,” above).
(vi) Trehalose metabolism. (a) Assignment of genes involved in trehalose metabolism.
Sequence searches clearly indicated that trehalose is synthesized in T. tenax via the OtsA-OtsB pathway (Table 6). In this pathway, trehalose 6-phosphate is formed from glucose 6-phosphate and UDP-glucose by trehalose-6-phosphate synthase (TPS, or OtsA) and trehalose 6-phosphate is dephosphorylated by trehalose-6-phosphate phosphatase (TPP, or OtsB), as shown for E. coli and S. cerevisiae (16, 24). Strikingly, only one gene that was obviously a chimera composed of two domains, one coding for TPS and the other coding for TPP, was identified in T. tenax. Thus, the protein represents a TPS-phosphatase (TPSP). First studies of the recombinant TPSP demonstrate that the encoded enzyme exhibits both activities (M. Zaparty, unpublished data). Besides this key enzyme of trehalose synthesis, further enzymes involved in the trehalose metabolism were identified: PGM and (NTP) sugar phosphate nucleotidyltransferases, which are engaged in the glycogen metabolism, are also constituents of trehalose metabolism. Additionally, a gene homolog with similarity to either the gene encoding trehalose synthase (TreS; treS gene) or the gene encoding α,α-trehalose phosphorylase (TreP; treP gene) seems to be present in T. tenax. Whereas TreS catalyzes the conversion of the α-1,4 linkage in maltose to α-1,1 generating trehalose (specific for some bacteria, e.g., Thermus aquaticus and Pimelobacter) (65), TreP catalyzes the phosphorolytic degradation of trehalose, forming glucose and glucose 1-phosphate. So far, no biochemical information is available concerning whether the gene product represents an alternative pathway for trehalose synthesis or is involved in trehalose degradation.
TABLE 6.
Identified genes coding for enzymes of the trehalose metabolism in T. tenax
| Enzymea | Candidate no. | EC no. | COG no.b | Gene | Accession no. | Evidence value and best hitc |
|---|---|---|---|---|---|---|
| Phosphoglucomutase | 2.7.5.1 | 1109 | pgm | AJ621332 | 1.0e-154 P. aerophilum, phosphomannomutase | |
| Sugar phosphate nucleotidyl transferase (7 candidates) | 1 | 2.7.7.d | 1208 | snt | AJ621273 | 1.94e-91 P. aerophilum, sugar phosphate nucleotidyl transferase |
| 2 | AJ621322 | 1.6e-61 P. aerophilum, sugar phosphate nucleotidyl transferase | ||||
| 3 | AJ621298, AJ621299 | 2.5e-114 P. aerophilum, glucose-1-phosphate adenyl- yltransferase | ||||
| 4 | AJ621300 | 8.65e-104 P. aerophilum, sugar phosphate transferase | ||||
| 5 | AJ621349 | 2.96e-85 P. aerophilum, mannose-1-phosphate guanyl- transferase | ||||
| 6 | AJ621350 | 1.0e-97 P. aerophilum, sugar phosphate nucleotidyl transferase | ||||
| 7 | 2.7.7.24 | 1209 | rfbA | AJ621292 | 1.6e-124 P. aerophilum, glucose-1-phosphate thymidyl- yltransferase | |
| Trehalose-6-phosphate synthase-phosphatase | 0380 | tpsp | AJ621287 | 0.0 P. aerophilum, trehalose-6-phosphate synthase | ||
| 1877 | ||||||
| Trehalose synthase/α,α-Trehalose phosphorylase | 5.4.99.16/ 2.4.1.64 | 0438 | treS/treP | AJ621341 | 1.0e-176 P. aerophilum, trehalose synthase |
The enzymes involved in trehalose degradation are underlined, and those involved in its biosynthesis are in bold.
For definition, see Table 1, footnote b.
For definition, see Table 1, footnote c. In cases in which two or more candidates were identified in the genome, the respective evidence values are given.
Final EC number not yet assigned.
The bacterial and eucaryal OtsA-OtsB pathway has now been found in archaea, and the pathway in T. tenax is characterized by a bifunctional TPSP. So far, only the presence of the TreY-TreZ pathway has been described for archaea (e.g., some members of Sulfolobales), a pathway which is also present in some bacteria (e.g., Arthrobacter and Micrococcus spp.) (10, 30, 38). In this pathway, maltooligosyltrehalose synthase (TreY) converts the terminal α-1,4-linked residue of a glucose polymer in an α-1,1 linkage and trehalose hydrolase (TreZ) cleaves the disaccharide off. Gene homologs of treY and treZ gene were not identified in the T. tenax genome, indicating that this pathway is absent. To learn whether the third pathway for trehalose synthesis (the TreS pathway) is present will demand enzymatic characterization of the treP/treS gene product.
(b) Organization of genes involved in trehalose metabolism.
The organization of genes also involved in glycogen metabolism is discussed above (see also Table 5). The tpsp gene coding for the unusual TPSP is localized downstream of an ORF coding for a putative glycosyltransferase (gt); the latter overlaps by 4 bp with an upstream-located ORF (hp; AJ621288) which shows homology only to a gene coding for a hypothetical protein in P. aerophilum (Fig. 3; Table 6). Thus, at least the genes coding for the putative glycosyltransferase and the hypothetical protein seem to form an operon (hp-gt operon). Also, an ORF (orfY; AJ621342) (upstream of the treS/treP gene coding for the trehalose synthase or trehalose phosphorylase) was found with similarity to a gene encoding a hypothetical protein in P. aerophilum, S. solfataricus, and S. tokodaii. Both genes overlap by 4 bp and form the orfY-treS/treP operon (Fig. 3), but so far we have no indication about the function of the two genes.
Conclusions.
T. tenax genome data reveal the presence of pathways involved in central carbohydrate metabolism as well as in glycogen and trehalose turnover (Fig. 4). So far, T. tenax and S. solfataricus are the only facultative heterotrophic archaea sequenced. Thus, the presented analysis not only contributes to the diversity of carbon metabolism in archaea but also represents an important step towards understanding of archaeal metabolic networks, which allow adaptation to alternative growth conditions.
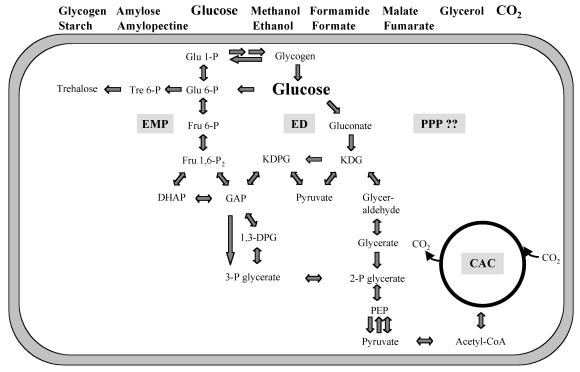
FIG. 4.
The central carbohydrate metabolism in T. tenax. A selection of growth substrates is given at the top; the labels of the different pathways are marked by grey-shaded boxes.
The identification of genes coding for HK, PGI, PPDK, GDH, GAD, KD(P)GA, KDGK, (glycogen) phosphorylase, and TPSP was confirmed by the functions of their recombinant gene products. In some cases, operon organization provides information about the function of genes, facilitating the reconstruction of pathways. Several genes organized in operons seem to overlap by 4 bp (the coding regions overlap by 1 bp) (Fig. 3), and coordinated transcription has been shown for three overlapping bigenic operons (orfX-hxk, fba-pfp, and pgk-gap) (5, 11, 54). For the acn-tpi operon, where the genes are separated by 40 bp, cotranscription was demonstrated by Northern analysis (63). Putative TATA boxes and B recognition element (BRE) sites were found only in front of the first gene of an operon (3, 59, 60) (except for the acn-tpi and rfbA-B-D-C operon), whereas putative Shine-Dalgarno sequences were often absent (29) (Fig. 3B). These results, as well as those determined for the transcription start sites of the single PK gene (pyk) (48) and the fpb-pfp operon (54), suggest also that in T. tenax, the translation of single genes and genes that are first in operons proceeds via leaderless transcripts (58, 64). The occurrence of monocistronic messages of the fba, pfp, pgk, gap, acn, and tpi genes, as well as the agreement of the N-terminal sequence of the PPi-PFK purified from T. tenax with the predicted translation start, suggests that posttranscriptional processing occurs (5, 11, 54).
The genome analysis strongly suggests the presence of three pathways for carbohydrate metabolism: (i) the EMP variant and (ii) the semiphosphorylative and (iii) the nonphosphorylative versions of the ED pathway. At the moment, we can only speculate about the physiological significance of the three different pathways. The organization of ED genes suggests that the ED variants are involved in hydrolytic degradation of glycogen, whereas the EMP variant is responsible for the phosphorolytic breakdown. The energy demand of the cell seems to have a strong influence on the selection of the different pathways. Whereas no ATP is generated by the ED variants, one ATP is gained using the EMP variant (taking into account that PPi, the phosphoryl donor of the phosphofructokinase, is a waste product of the cell). In T. tenax, GAP, the common intermediate of EMP pathway and semiphosphorylative ED version, can be converted into 3-phosphoglycerate in a one-step reaction—via GAPN or GAPOR—thus circumventing the formation of the extremely heat-labile intermediate 1,3-bisphosphoglycerate. Since the nonphosphorylative ED pathway avoids the most heat-labile intermediates (1,3-bisphosphoglycerate, GAP, and dihydroxyacetone phosphate), this pathway could be preferred at the upper temperature range of growth.
Further, the sequence data support the idea of the existence of a reversible CAC in T. tenax, indicating that—as already suggested—the organism uses the reaction cycle not only for catabolic purposes but also for CO2 fixation (1). Almost all gene homologs necessary for the glycogen metabolism (with the exception of the branching and debranching enzyme) were identified in the T. tenax genome. In addition, trehalose synthesis was shown to proceed via the OtsA-OtsB pathway in T. tenax. No evidence was found for a functional PPP, which has been thought to be essential for generation of pentoses and NADPH for anabolic purposes. Together with previous genome analyses of methanoarchaea and hyperthermophilic archaea, this finding suggests that the oxidative PPP is generally absent from archaea or that an analogous pathway exists with enzymes showing no significant similarity to known bacterial and eucaryal counterparts.
.
Acknowledgments
We thank Bettina Haberl (EPIDAUROS Biotechnologie AG) for assistance in the setup of sequencing reactions, Klaus Gellner (EPIDAUROS Biotechnologie AG) and Günter Raddatz (MPI für Entwicklungsbiologie, Tübingen) for bioinformatic support, and William Martin (Heinrich-Heine Universität Düsseldorf) for critical review of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft. The MAGPIE project is supported by Genome Prairie, Canada.
REFERENCES
- 1.Beh, M., G. Strauss, R. Huber, K.-O. Stetter, and G. Fuchs. 1993. Enzymes of the reductive citric acid cycle in the autotrophic eubacterium Aquifex pyrophilus and in the archaebacterium Thermoproteus neutrophilus. Arch. Microbiol. 160:306-311. [Google Scholar]
- 2.Bekal, S., J. van Beeumen, B. Samyn, D. Garmyn, S. Henini, C. Diviès, and H. Prévost. 1998. Purification of Leuconostoc mesenteroides citrate lyase and cloning and characterization of the citCDEFG gene cluster. J. Bacteriol. 180:647-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, S. D., and S. P. Jackson. 2001. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 4:208-213. [DOI] [PubMed] [Google Scholar]
- 4.Brunner, N. A., H. Brinkmann, B. Siebers, and R. Hensel. 1998. NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase from Thermoproteus tenax. The first identified archaeal member of the aldehyde dehydrogenase superfamily is a glycolytic enzyme with unusual regulatory properties. J. Biol. Chem. 273:6149-6156. [DOI] [PubMed] [Google Scholar]
- 5.Brunner, N. A., B. Siebers, and R. Hensel. 2001. Role of two different glyceraldehyde-3-phosphate dehydrogenases in controlling the reversible Embden-Meyerhof-Parnas pathway in Thermoproteus tenax: regulation on protein and transcript level. Extremophiles 5:101-109. [DOI] [PubMed] [Google Scholar]
- 6.Buchanan, C. L., H. Connaris, M. J. Danson, C. D. Reeve, and D. W. Hough. 1999. An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated substrates. Biochem. J. 343:563-570. [PMC free article] [PubMed] [Google Scholar]
- 7.Budgen, N., and M. J. Danson. 1986. Metabolism of glucose via a modified Entner-Doudoroff pathway in the thermoacidophilic archaebacterium Thermoplasma acidophilum. FEBS Lett. 196:207-210. [Google Scholar]
- 8.Colombo, S., M. Grisa, P. Tortora, and M. Vanoni. 1994. Molecular cloning, nucleotide sequence and expression of a Sulfolobus solfataricus gene encoding a class II fumarase. FEBS Lett. 340:93-98. [DOI] [PubMed] [Google Scholar]
- 9.De Rosa, M., A. Gambacorta, B. Nicolaus, P. Giardina, E. Poerio, and V. Buonocore. 1984. Glucose metabolism in the extreme thermoacidophilic archaebacterium Sulfolobus solfataricus. Biochem. J. 224:407-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Lernia, I., A. Morana, A. Ottombrino, S. Fusco, M. Rossi, and M. De Rosa. 1998. Enzymes from Sulfolobus shibatae for the production of trehalose and glucose from starch. Extremophiles 2:409-416. [DOI] [PubMed] [Google Scholar]
- 11.Dörr, Ch., M. Zaparty, B. Tjaden, H. Brinkmann, and B. Siebers. 2003. The hexokinase of the hyperthermophile Thermoproteus tenax: ATP-dependent hexokinases and ADP-dependent glucokinases, two alternatives for glucose phosphorylation in archaea. J. Biol. Chem. 278:18744-18753. [DOI] [PubMed] [Google Scholar]
- 12.Fischer, F., W. Zillig, K. O. Stetter, and G. Schreiber. 1983. Chemolithoautotrophic metabolism of anaerobic extremely thermophilic archaebacteria. Nature 301:511-513. [DOI] [PubMed] [Google Scholar]
- 13.Fitz-Gibbon, S. T., H. Ladner, U. J. Kim, K. O. Stetter, M. I. Simon, and J. H. Miller. 2002. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. USA 99:984-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furdui, C., and S. W. Ragsdale. 2000. The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J. Biol. Chem. 275:28494-28499. [DOI] [PubMed] [Google Scholar]
- 15.Gasterland, T., and C. W. Sensen. 1996. Fully automated genome analysis that reflects user needs and preferences. A detailed introduction to the MAGPIE system architecture. Biochimie 78:302-310. [DOI] [PubMed] [Google Scholar]
- 16.Giœver, H. M., O. B. Styrvold, I. Kaasen, and A. R. Strom. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J. Bacteriol. 170:2841-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon, P., T. Gaasterland, and C. W. Sensen. 2002. Genomic data representation through images: MAGPIE as an example, pp. 345-363. In C. W. Sensen (ed.), Essentials of genomics and bioinformatics. Wiley-VCH, Weinheim, Germany.
- 18.Graham, D. E., H. Xu, and R. H. White. 2002. A divergent archaeal member of the alkaline phosphatase binuclear metalloenzyme superfamily has phosphoglycerate mutase activity. FEBS Lett. 517:190-194. [DOI] [PubMed] [Google Scholar]
- 19.Hansen, T., M. Oehlmann, and P. Schönheit. 2001. Novel type of glucose-6-phosphate isomerase in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183:3428-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen, T., and P. Schönheit. 2000. Purification and properties of the first-identified, archaeal, ATP-dependent 6-phosphofructokinase, an extremely thermophilic non-allosteric enzyme, from the hyperthermophile Desulfurococcus amylolyticus. Arch. Microbiol. 173:103-109. [DOI] [PubMed] [Google Scholar]
- 21.Hensel, R., S. Laumann, J. Lang, H. Heumann, and F. Lottspeich. 1987. Characterization of two D-glyceraldehyde-3-phosphate dehydrogenases from the extremely thermophilic archaebacterium Thermoproteus tenax. Eur. J. Biochem. 170:325-333. [DOI] [PubMed] [Google Scholar]
- 22.Huynen, M. A., T. Dandekar, and P. Bork. 1999. Variation and evolution of the citric-acid cycle: a genomic perspective. Trends Microbiol. 7:281-291. [DOI] [PubMed] [Google Scholar]
- 23.Johnsen, U., M. Selig, K. B. Xavier, H. Santos, and P. Schönheit. 2001. Different glycolytic pathways for glucose and fructose in the halophilic archaeon Halococcus saccharolyticus. Arch. Microbiol. 175:52-61. [DOI] [PubMed] [Google Scholar]
- 24.Kaasen, I., J. McDougall, and A. R. Storm. 1994. Analysis of the otsBA operon for osmoregulatory trehalose synthesis in Escherichia coli and homology of the OtsA and OtsB proteins to the yeast trehalose-6-phosphate synthase/phosphatase complex. Gene 145:9-15. [DOI] [PubMed] [Google Scholar]
- 25.Kawarabayasi, Y., Y. Hino, H. Horikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Kato, T. Yoshizawa, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, S. Masuda, M. Yanagii, M. Nishimura, A. Yamagishi, T. Oshima, and H. Kikuchi. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Res. 8:123-140. [DOI] [PubMed] [Google Scholar]
- 26.Kawarabayasi, Y., Y. Hino, H. Horikawa, S. Yamazaki, Y. Haikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, H. Nakazawa, M. Takamiya, S. Masuda, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N, Kushida, A. Oguchi, K. Aoki, K. Kubota, Y. Nakamura, N. Nomura, Y. Sako, and H. Kikuchi. 1999. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6:83-101. [DOI] [PubMed] [Google Scholar]
- 27.Kengen, S. W., J. E. Tuininga, F. A. De Bok, A. J. Stams, and W. M. De Vos. 1995. Purification and characterization of a novel ADP-dependent glucokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 270:30453-30457. [DOI] [PubMed] [Google Scholar]
- 28.Kengen, S. W. M., F. A. M. de Bok, N.-D. van Loo, C. Dijkema, and A. J. M. Stams. 1994. Evidence for the operation of a novel Embden-Meyerhof pathway that involves ADP-dependent kinases during sugar fermentation by Pyrococcus furiosus. J. Biol. Chem. 269:17537-17541. [PubMed] [Google Scholar]
- 29.Kjems, J., H. Leffers, R. A. Garrett, G. Wich, W. Leinfelder, and A. Böck. 1987. Gene organization, transcription signals and processing of the single ribosomal RNA operon of the archaebacterium Thermoproteus tenax. Nucleic Acids Res. 15:4821-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi, K., M. Kato, Y. Miura, M. Kettoku, T. Komeda, and A. Iwamatsu. 1996. Gene analysis of trehalose-producing enzymes from hyperthermophilic archaea in Sulfolobales. Biosci. Biotechnol. Biochem. 60:1720-1723. [DOI] [PubMed] [Google Scholar]
- 31.Koga, S., I. Yoshioka, H. Sakuraba, M. Takahashi, S. Sakasegawa, S. Sakayu, and T. Ohshima. 2000. Biochemical characterization, cloning, and sequencing of ADP-dependent (AMP-forming) glucokinase from two hyperthermophilic archaea, Pyrococcus furiosus and Thermococcus litoralis. J. Biochem. 128:1079-1085. [DOI] [PubMed] [Google Scholar]
- 32.König, H., R. Skorko, W. Zillig, and W. D. Reiter. 1982. Glycogen in thermoacidophilic archaebacteria of the genera Sulfolobus, Thermoproteus, Desulfurococcus and Thermococcus. Arch. Microbiol. 132:297-303. [Google Scholar]
- 33.Labes, A., P. and Schönheit. 2001. Sugar utilization in the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus strain 7324: starch degradation to acetate and CO2 via a modified Embden-Meyerhof pathway and acetyl-CoA synthetase (ADP-forming). Arch. Microbiol. 176:329-338. [DOI] [PubMed] [Google Scholar]
- 34.Lorentzen, E., E. Pohl, P. Zwart, A. Stark, R. B. Russell, T. Knura, R. Hensel, and B. Siebers. 2003. Crystal structure of an archaeal class I aldolase and the evolution of (βα)8 barrel proteins. J. Biol. Chem. 278:47253-47260. [DOI] [PubMed] [Google Scholar]
- 35.Mai, X., and M. W. Adams. 1996. Characterization of a fourth type of 2-keto acid-oxidizing enzyme from a hyperthermophilic archaeon: 2-ketoglutarate ferredoxin oxidoreductase from Thermococcus litoralis. J. Bacteriol. 178:5890-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marolda, C. L., and M. A. Valvano. 1995. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J. Bacteriol. 177:5539-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martins, L. O., R. Huber, H. Huber, K. O. Stetter, M. S. Da Costa, and H. Santos. 1997. Organic solutes in hyperthermophilic archaea. Appl. Environ. Microbiol. 63:896-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maruta, K., H. Mitsuzumi, T. Nakada, M. Kubota, H. Chaen, S. Fukuda, T. Sugimoto, and M. Kurimoto. 1996. Cloning and sequencing of a cluster of genes encoding novel enzymes of trehalose biosynthesis from the thermophilic archaebacterium Sulfolobus acidocaldarius. Biochim. Biophys. Acta 1291:177-181. [DOI] [PubMed] [Google Scholar]
- 39.Nicolaus, B., A. Gambacorta, A. L. Basso, R. Riccio, M. De Rosa, and W. D. Grant. 1988. Trehalose in archaebacteria. Syst. Appl. Microbiol. 10:215-217. [Google Scholar]
- 40.Pohl, E., N. Brunner, M. Wilmanns, and R. Hensel. 2002. The crystal structure of the allosteric non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeum Thermoproteus tenax. J. Biol. Chem. 277:19938-19945. [DOI] [PubMed] [Google Scholar]
- 41.Preiss, J., and T. Romeo. 1994. Molecular biology and regulatory aspects of glycogen biosynthesis in bacteria. Prog. Nucleic Acids Res. Mol. Biol. 47:299-329. [DOI] [PubMed] [Google Scholar]
- 42.Rashid, N., H. Imanaka, T. Kanai, T. Fukui, H. Atomi, and T. Imanaka. 2002. A novel candidate for the true fructose-1,6-bisphosphatase in archaea. J. Biol. Chem. 277:30649-30655. [DOI] [PubMed] [Google Scholar]
- 43.Ronimus, R. S., and H. W. Morgan. Distribution and phylogenies of enzymes of the Embden-Meyerhof-Parnas pathway from archaea and hyperthermophilic bacteria support a gluconeogenetic origin of metabolism. Archaea, in press. [DOI] [PMC free article] [PubMed]
- 44.Ronimus, R. S., H. W. Morgan, and Y. H. R. Ding. 2001. Sequencing, expression, characterisation and phylogeny of the ADP-dependent phosphofructokinase from the hyperthermophilic, euryarchaeal Thermococcus zilligii. Biochim. Biophys. Acta 1527:384-391. [DOI] [PubMed] [Google Scholar]
- 45.Sakuraba, H., I. Yoshioka, S. Koga, M. Takahashi, Y. Kitahama, T. Satomura, R. Kawakami, and T. Ohshima. 2002. ADP-dependent glucokinase/phosphofructokinase, a novel bifunctional enzyme from the hyperthermophilic archaeon Methanococcus jannaschii. J. Biol. Chem. 277:12495-12498. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Schönheit, P., and T. Schäfer. 1995. Metabolism of hyperthermophiles. World J. Microbiol. Biotechnol. 11:26-57. [DOI] [PubMed] [Google Scholar]
- 48.Schramm, A., B. Siebers, B. Tjaden, H. Brinkmann, and R. Hensel. 2000. Pyruvate kinase of the hyperthermophilic crenarchaeote Thermoproteus tenax: physiological role and phylogenetic aspects. J. Bacteriol. 182:2001-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selig, M., and P. Schönheit. 1994. Oxidation of organic compounds to CO2 with sulphur or thiosulfate as electron acceptor in the anaerobic hyperthermophilic archaea Thermoproteus tenax and Pyrobaculum islandicum proceeds via the citric acid cycle. Arch. Microbiol. 162:286-294. [Google Scholar]
- 50.Selig, M., K. B. Xavier, H. Santos, and P. Schönheit. 1997. Comparative analysis of Embden-Meyerhof and Entner-Doudoroff glycolytic pathways in hyperthermophilic archaea and the bacterium Thermotoga. Arch. Microbiol. 167:217-232. [DOI] [PubMed] [Google Scholar]
- 51.Selkov, E., N. Maltsev, G. J. Olsen, R. Overbeek, and W. B. Whitman. 1997. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene 197:11-26. [DOI] [PubMed] [Google Scholar]
- 52.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siebers, B. 1995. Ph.D. thesis. University of Essen, Essen, Germany.
- 54.Siebers, B., H. Brinkmann, C. Dörr, B. Tjaden, H. Lilie, J. Van der Oost, and C. H. Verhees. 2001. Archaeal fructose-1, 6-bisphosphate aldolases constitute a new family of archaeal type class I aldolase. J. Biol. Chem. 276:28710-28718. [DOI] [PubMed] [Google Scholar]
- 55.Siebers, B., and R. Hensel. 1993. Glucose catabolism of the hyperthermophilic archaeum Thermoproteus tenax. FEMS Microbiol. Lett. 111:1-8. [Google Scholar]
- 56.Siebers, B., H.-P. Klenk, and R. Hensel. 1998. PPi-dependent phosphofructokinase from Thermoproteus tenax, an archaeal descendant of an ancient line in phosphofructokinase evolution. J. Bacteriol. 180:2137-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siebers, B., V. F. Wendisch, and R. Hensel. 1997. Carbohydrate metabolism in Thermoproteus tenax: in vivo utilization of the non-phosphorylative Entner-Doudoroff pathway and characterization of its first enzyme, glucose dehydrogenase. Arch. Microbiol. 168:120-127. [DOI] [PubMed] [Google Scholar]
- 58.Slupska, M. M., A. G. King, S. Fitz-Gibbon, J. Besemer, M. Borodovsky, and J. H. Miller. 2001. Leaderless transcripts of the crenarchaeal hyperthermophile Pyrobaculum aerophilum. J. Mol. Biol. 309:347-360. [DOI] [PubMed] [Google Scholar]
- 59.Soppa, J. 1999. Transcription initiation in archaea: facts, factors and future aspects. Mol. Microbiol. 31:1295-1305. [DOI] [PubMed] [Google Scholar]
- 60.Soppa, J. 1999. Normalized nucleotide frequencies allow the definition of archaeal promoter elements for different archaeal groups and reveal base-specific TFB contacts upstream of the TATA box. Mol. Microbiol. 31:1589-1592. [DOI] [PubMed] [Google Scholar]
- 61.Stec, B., H. Yang, K. A. Johnson, L. Chen, and M. F. Roberts. 2000. MJ0109 is an enzyme that is both an inositol monophosphatase and the “missing” archaeal fructose-1,6-bisphosphatase. Nat. Struct. Biol. 7:1046-1100. [DOI] [PubMed] [Google Scholar]
- 62.Tersteegen, A., D. Linder, R. K. Thauer, and R. Hedderich. 1997. Structures and functions of four anabolic 2-oxoacid oxidoreductases in Methanobacterium thermoautotrophicum. Eur. J. Biochem. 244:862-868. [DOI] [PubMed] [Google Scholar]
- 63.Tjaden, B. 2003. Ph. D.thesis. University of Duisburg-Essen, Essen, Germany.
- 64.Tolstrup, N., C. W. Sensen, R. A. Garrett, and I. G. Clausen. 2000. Two different and highly organized mechanisms of translation initiation in the archaeon Sulfolobus solfataricus. Extremophiles 4:175-179. [DOI] [PubMed] [Google Scholar]
- 65.Tsusaki, K., T. Nishimoto, T. Nakada, M. Kubota, H. Chaen, S. Fukuda, T. Sugimoto, and M. Kurimoto. 1997. Cloning and sequencing of trehalose synthase gene from Thermus aquaticus ATCC33923. Biochim. Biophys. Acta 1334:28-32. [DOI] [PubMed] [Google Scholar]
- 66.Tuininga, J. E., C. H. Verhees, J. Van der Oost, S. W. Kengen, A. J. Stams, and W. M. De Vos. 1999. Molecular and biochemical characterization of the ADP-dependent phosphofructokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 274:21023-21028. [DOI] [PubMed] [Google Scholar]
- 67.Tumbula, D. L., Q. Teng, M. G. Bartlett, and W. B. Whitman. 1997. Ribose biosynthesis and evidence for an alternative first step in the common aromatic amino acid pathway in Methanococcus maripaludis. J. Bacteriol. 179:6010-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van der Oost, J., M. A. Huynen, and C. H. Verhees. 2002. Molecular characterization of phosphoglycerate mutase in archaea. FEMS Microbiol. Lett. 212:111-120. [DOI] [PubMed] [Google Scholar]
- 69.Van der Oost, J., G. Schut, S. W. M. Kengen, W. R. Hagen, M. Thomm, and W. M. De Vos. 1998. The ferredoxin-dependent conversion of glyceraldehyde-3-phosphate in the hyperthermophilic archaeon Pyrococcus furiosus represents a novel site of glycolytic regulation. J. Biol. Chem. 273:28149-28154. [DOI] [PubMed] [Google Scholar]
- 70.Verhees, C. H., J. Akerboom, E. Schiltz, W. M. De Vos, and J. Van der Oost. 2002. Molecular and biochemical characterization of a distinct type of fructose-1,6-bisphosphatase from Pyrococcus furiosus. J. Bacteriol. 184:3401-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verhees, C. H., M. A. Huynen, D. E Ward, E. Schiltz, W. M. De Vos, and J. Van der Oost. 2001. The phosphoglucose isomerase from the hyperthermophilic archaeon Pyrococcus furiosus is a unique glycolytic enzyme that belongs to the cupin superfamily. J. Biol. Chem. 276:40926-40932. [DOI] [PubMed] [Google Scholar]
- 72.Verhees, C. H., S. W. M. Kengen, J. E. Tuininga, G. J. Schut, M. W. W. Adams, W. M. De Vos, and J. Van der Oost. 2003. The unique features of glycolytic pathways in archaea. Biochem. J. 375:231-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verhees, C. H., J. E. Tuininga, S. W. M. Kengen, A. J. Stams, and W. M. De Vos. 2001. ADP-dependent phosphofructokinases in mesophilic and thermophilic methanogenic archaea. J. Bacteriol. 183:7145-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wanner, C., and J. Soppa. 2002. Functional role for a 2-oxo acid dehydrogenase in the halophilic archaeon Haloferax volcanii. J. Bacteriol. 184:3114-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woese, C. R., and G. F. Fox. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA 74:5088-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zillig, W., K. O. Stetter, W. Schäfer, D. Janekovic, S. Wunderl, I. Holz, and P. Palm. 1981. Thermoproteales: a novel type of extremely thermoacidophilic anaerobic archaebacteria isolated from Icelandic solfatares. Zentbl. Bakteriol. Hyg. 1 Abt. Orig. C 2:205-227. [Google Scholar]