SYNOPSIS
Activity of the aminoglycoside phosphotransferase APH(3’)-Ia leads to resistance to aminoglycoside antibiotics in pathogenic Gram-negative bacteria, and contributes to the clinical obsolescence of this class of antibiotics. One strategy to rescue compromised antibiotics such as aminoglycosides is targeting the enzymes that confer resistance with small molecules. Previously we demonstrated that eukaryotic protein kinase (ePK) inhibitors could inhibit APH enzymes, due to the structural similarity between these two enzyme families. However, limited structural information of enzyme-inhibitor complexes hindered interpretation of the results. As well, cross-reactivity of compounds between APHs and ePKs represents an obstacle to their use as aminoglycoside adjuvants to rescue aminoglycoside antibiotic activity. Here, we structurally and functionally characterize inhibition of APH(3’)-Ia by three diverse chemical scaffolds – anthrapyrazolone, 4-anilinoquinazoline and pyrazolopyrimidine (PP) – and reveal distinctions in the binding mode of anthrapyrazolone and PP compounds to APH(3’)-Ia versus ePKs. Using this observation, we identify PP-derivatives that select against ePKs, attenuate APH(3’)-Ia activity and rescue aminoglycoside antibiotic activity against a resistant E. coli strain. The structures presented here and these inhibition studies provide an important opportunity for structure-based design of compounds to target aminoglycoside phosphotransferases for inhibition, potentially overcoming this form of antibiotic resistance.
Keywords: antibiotic resistance, aminoglycoside phosphotransferase APH(3’)-Ia, protein kinase inhibitor, structure-based drug design, crystal structure
INTRODUCTION
Antibiotic resistance is a serious global public health problem [1]. Resistance is an issue with all classes of currently used antibiotics, resulting in the emergence of pathogens untreatable using the available therapeutic arsenal. The genetic source of the problem is the “antibiotic resistome”, the reservoir of bona fide resistance genes plus genes capable of evolving into resistance-conferring function through antibiotic selection [2].
Within this context of need for new antimicrobials, industry is increasingly retreating from this space with a resultant atrophy of discovery pipeline output [3]. New approaches to circumvent difficulties in antimicrobial discovery include “repurposing” of compounds not previously used as antimicrobials [4]. Similarly, non-antimicrobials have been shown to potentiate the activity of antibiotics, although their cellular targets may not be known [5,6]. In this approach, ideally the adjuvant compound would be targeted against the resistance mechanism, freeing the established antimicrobial to impact its cellular target.
Enzyme-mediated antibiotic resistance is especially amenable to this combinatory approach, as exemplified by the clinical application of β-lactam-β-lactamase inhibitor combinations [7]. Fitting this model, aminoglycoside antibiotics offer a class of potent antimicrobials that have lost clinical efficacy as a result of enzyme-catalyzed modification [8]. Importantly, aminoglycosides are one of the few antibiotic classes that show good efficacy against Gram-negative pathogens, which can cause infections that are especially in need of new therapeutic options [9]. We are interested in investigating compounds targeted against aminoglycoside-modifying enzymes, which include aminoglycoside N-acetyltransferases (AACs), O-nucleotidyltransferases (ANTs) or O-phosphotransferases (APHs), also known as antibiotic kinases (AKs).
AK enzymes are one of the most common sources of aminoglycoside antibiotic resistance. They catalyze the transfer of the γ-phosphate group from ATP or GTP [10-12] in a regiospecific manner to the antibiotic substrates and thereby inactivate the drug. AK enzymes vary significantly in sequence, in substrate profile and in the modification site on the antibiotic substrate. Many studies have focused on detailed molecular characterization of AK enzymes and their interactions with aminoglycoside and nucleotide substrates [13-22]. These structural analyses demonstrated that despite sequence variation, AK enzymes adopt a common eukaryotic protein kinase (ePK)-like fold [22]. These efforts also demonstrated that the antibiotic binding site contains a high degree of functional and structural diversity, consistent with the chemical diversity of aminoglycoside substrates. In contrast to diversity in the structure of the antibiotic binding site, the nucleotide triphosphate (NTP) binding site has a higher degree of structural conservation. This site contains structural similarity with ePKs by virtue of its location at the interface between the N- and C-terminal lobes of the bilobal fold, the NTP contacting both lobes, the presence of a short inter-domain linker sequence (also known as the hinge) and the conservation of critical residues.
The NTP binding site of the ePK catalytic domain is a well-characterized drug target [23]. The similarity between the NTP binding site of ePKs and AKs, along with extensive libraries of small molecule ePK inhibitors (PKIs), prompted us to test the inhibition potential of PKIs against AKs [24]. This produced a matrix of inhibitory activity of 80 chemically diverse PKIs against 14 representative AKs; the compounds spanned 5 orders of magnitude in affinity for APHs and the study found broad and narrow spectrum inhibitors [24]. These findings confirmed that the NTP binding site of AKs can be exploited for inhibition and also demonstrated that PKIs are able to select for structural differences in AK enzymes. Missing from that work was significant structural analysis of the many enzyme-inhibitor pairs to rationalize the patterns of selectivity; we determined the structure of only one enzyme-general inhibitor pair (APH(2”)-IVa and quercetin).
One of the AK enzymes for which multiple, diverse and specific inhibitors were identified is APH(3’)-Ia. The gene encoding this enzyme (aphA1) was originally found on the transposable element Tn903 in E. coli [25] and is now widely distributed across Gram-negative bacterial pathogens responsible for clinical antibiotic resistance outbreaks (reviewed in [26]). The enzyme has high catalytic efficiency and activity against a broad spectrum of antibiotics [26,27]. Furthermore, APH(3’)-Ia demonstrates plasticity for its nucleotide substrate and can utilize both GTP and ATP as a phosphate donor [27].
In this current work, we present the 3D structure of APH(3’)-Ia and examine the structural basis of inhibition by three distinct PKI scaffolds. This analysis reveals the specific features of the enzyme-inhibitor interface that can be exploitable for the development of AK-specific inhibitors. Guided by these findings, we further studied APH(3’)-Ia inhibition by the pyrazolopyrimidine (PP) scaffold, identifying variants that are inactive against ePKs. We show that these PP derivatives are capable of attenuating APH(3’)-Ia activity in vitro and efficiently rescue aminoglycoside antibiotic action against an aminoglycoside-resistant E. coli strain. These results strengthen the possibility of repurposing PKI molecules and combining them with aminoglycosides as a strategy to overcome this type of antibiotic resistance.
EXPERIMENTAL
Protein expression and purification
APH(3’)-Ia purified as described previously for APH(4)-Ia [14].
Crystallization and structure determination
APH(3’)-Ia•Ca2+•ATP complex crystals were grown at room temperature using hanging drop vapor diffusion by mixing protein at 14 mg/mL with reservoir solution containing 0.1 M calcium acetate, 20% PEG3350 and 2 mM ATP. Working inhibitor solutions were prepared by dissolving inhibitor stock solutions (in 100% DMSO) into the following buffer: 0.6 M NaCl, 20 mM sodium malonate pH 7, 2.5 mM MgCl2, 0.5 mM CaCl2, 0.5 mM TCEP, such that final DMSO concentration was between 2-5% and final inhibitor concentration was between 0.05 – 0.3 mM (final concentration of compounds could only be estimated as volume was adjusted to maintain solubility). Working inhibitor solutions were mixed with 0.5-2 mM kanamycin A in water, 4 – 8 mg of protein dissolved in the above buffer, and incubated 1.5 – 2 h at 4°C. The mixtures were concentrated to a final protein concentration not less than 15 mg/mL, and final inhibitor concentrations between 1 – 6 mM, then centrifuged to remove insoluble components. Hanging drops were set up at room temperature and reservoir solutions that resulted in ternary complex crystals each contained 0.1 M sodium acetate pH 4.5 plus the following: SP600125 - 8% PEG 3350, 0.2 M NDSB-221; Tyrphostin AG 1478 - 14% PEG 3350, 0.3 M NDSB-221; PP1 - 18% PEG 3350; PP2 - 14% PEG 3350; 1-NA-PP1 - 7% PEG 3350; 1-NM-PP1 - 8% PEG 3350. All crystals were cryoprotected with paratone oil prior to shipment for diffraction data collection.
X-ray diffraction data collection
Diffraction data for APH(3’)-Ia•ATP complex was collected at 100 K, selenomethionine peak absorption wavelength for (0.97940 Å), at beamline 19-ID at the Structural Biology Centre, Advanced Photon Source. Diffraction data for each ternary complex were collected at 100 K, selenomethionine peak absorption wavelength (0.97856 Å), at beamlines 21-ID-F or 21-ID-G at Life Sciences Collaborative Access Team, Advanced Photon Source. All diffraction data was reduced with HKL-3000 [28], except for APH(3’)-Ia•kanamycin•1-NA-PP1 and 1-NM-PP1 ternary complexes, which were reduced with XDS [29] and Scala [30].
Structure Determination and Refinement
The structure of APH(3’)-Ia•Ca2+•ATP complex was determined by SAD using HKL-3000. Matthew's coefficient calculation suggested three copies in the asymmetric unit, and 21 total selenomethionine sites; 18 were located. Initial model building and refinement was performed with ARP/wARP [31] and Refmac [32], with later stages of refinement with PHENIX [33]. TLS parameterization groups were residues 1-24, 25-103, 104-271 for each chain, as determined by the TLSMD server [34]. ATP, Ca2+, and solvent molecules were built into positive Fo-Fc density in the NTP and aminoglycoside-binding sites after protein was fully built. All ternary complex structures were determined by Molecular Replacement with PHENIX, using a single chain of enzyme from APH(3’)-Ia•Ca2+•ATP complex. Refinement for PP1, PP2, AG 1478, 1-NA-PP1 and 1-NM-PP1 complexes was performed with PHENIX; PHENIX and then autoBUSTER [35] were utilized for SP600125. TLS parameterization was added immediately after MR. Atomic displacement parameters were refined as follows: anisotropic for protein and kanamycin atoms for PP1, PP2, 1-NA-PP1 and 1-NM-PP1 ternary complexes, isotropic for inhibitor atoms; isotropic for all atoms of ATP, SP600125 and AG 1478 complexes. Coot [36] was utilized for all electron density visualization. The positions of all inhibitors were verified by simulated annealing omit maps: coordinates of the inhibitors plus all atoms within 5 Å were removed, followed by simulated annealing using PHENIX and model building into residual positive Fo-Fc density. Occupancies of PP1, PP2, AG 1478, 1-NA-PP1 and 1-NM-PP1 molecules were refined while occupancies of SP600125 molecules were fixed at 1.0. Geometry of all structures was verified using PHENIX and the RCSB PDB Validation Server. Ramachandran statistics were verified: all residues fit favoured regions.
Structural analysis
Structure alignments were performed with PyMOL (http://www.pymol.org) or Coot. Structure images produced with PyMOL. The following structures of ePK-anthrapyrazolone complexes were used for comparisons: 1UKI, 1PMV and 2ZMD. Structures of ePK-4-anilinoquinazoline complexes used for comparisons were: 3NYV, 1D18, 1D19, 1M17, 1XKK, and 2ITY. The following structures of ePK-PP complexes were used for comparisons: 1QCF, 2IVV, 2V4L, 3EN4, 3EN5, 3EN6, 3EN7, 3ENE, 2WEI, 3I7B, 3I7C, 3EL8, 1QPE, 2ZV9, 3GEQ, 3NCG, 3MWU, 3N51, 3MA6, 3NZS, 3NZU, 2WXK, 2WXM. Modeling of 1-NA-PP1/1-NM-PP1 into ePK nucleotide-binding sites performed by manually aligning the purine cores of these compounds with those of the pyrazolopyrimidine inhibitors in the structures of H. sapiens Hck bound to PP1, PDB accession 1QCF [37] and H. sapiens PI3K bound to compound S2, PDB accession 3ENE [38], which were bound to the ePK enzymes in the “C3 up” conformation, followed by flipping 1-NA-PP1/1-NM-PP1 180° about the plane of the purine core such that they conform to the “C3 down” conformation in the ePK active sites. Amino acids from the ePK enzymes that overlapped in space with 1-NA-PP1/1-NM-PP1 were considered to be sterically-clashing residues. The following structures of PP compounds with large C3 substituents bound to protozoan kinases were used for comparisons: 2WEI, 3I7B, 3I7C, 3MA6, 3MWU, 3N51, and 3NCG. Shape complementarity was calculated using Sc [39]. Buried surface area calculated by PDBe PISA server [40].
Chemical synthesis
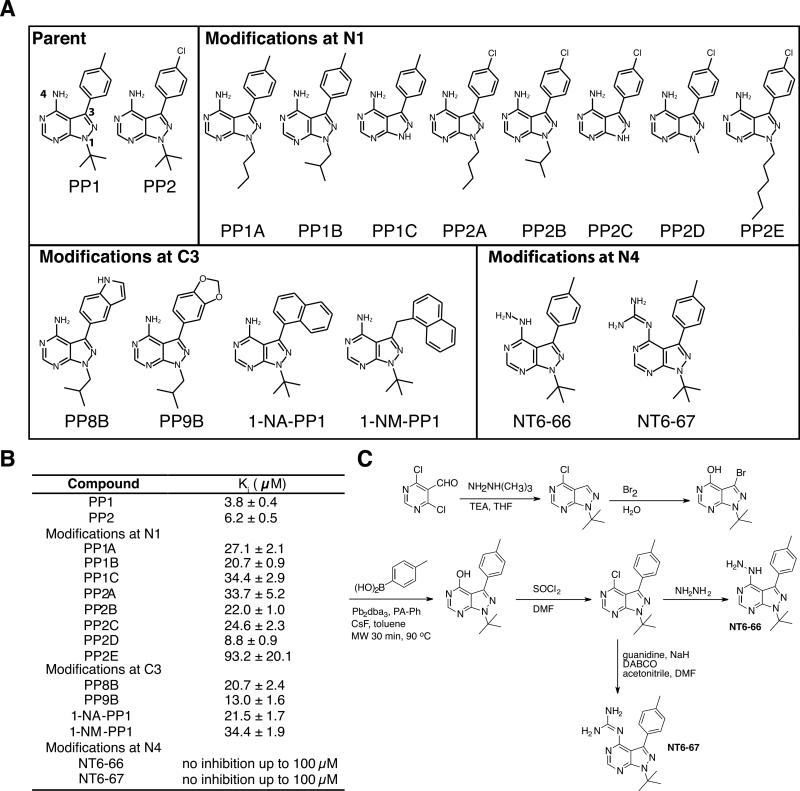
Compounds NT6-66 and NT6-67 were synthesized according to the scheme in Figure 5C and characterized by NMR and mass spectrometry in reference [41].
Figure 5. Structure-activity-relationship study of pyrazolopyrimidine inhibitors of APH(3’)-Ia.
A) Chemical structures of parent compounds and derivatives grouped by modification site. B) Measured inhibitory constants (Ki) of compounds against APH(3’)-Ia. C) Chemical synthesis scheme for compounds NT6-66 and NT6-67. Method adapted from Todorovic, N., Awuah, E., Shakya, T. & Wright, G. Microwave-assisted synthesis of N1-and C3-substituted pyrazolo [3, 4-d] pyrimidine libraries. Tetrahedron Letters (2011).
Antibiotic rescue factor determinations
Growth curves were measured with E. coli ΔtolC ΔbamB containing pET22-aphA1 or pGDP4-aac(3)-Ia (pBR322 backbone with lacI promoter) set up according to the CLSI broth microdilution method (CLSI document M7-A5). Inhibitors were dissolved in DMSO, diluted and added at a final concentration of 5% (v/v) while aminoglycoside concentration was held constant at ¼ MIC (32 μg/ml). Plates were incubated at 37°C with shaking in a Tecan Sunrise (Switzerland) plate reader over a 24 h period. Growth was monitored every 15 minutes by OD600 nm measurements and corrected for background (no growth control). Antibiotic rescue factors were calculated with kanamycin at ¼ MIC at 20 h incubation using the formula [24]:
A = Growth control, B = No aminoglycoside control, C = No APH(3’)-Ia inhibitor control, D = Growth in presence of aminoglycoside and APH(3’)-Ia inhibitor.
Inhibition kinetics
The in vitro activity of APH(3’)-Ia was monitored by coupling the release of GDP with pyruvate kinase/lactate dehydrogenase (PK/LDH) as previously described [24]. IC50 values were determined in duplicate and inhibitors added at a final concentration of 2% DMSO (v/v). Kis were determined from calculated IC50 values [42]. All inhibitors were competitive, as indicated by reversible inhibition behavior (dose dependence, slope factor ~1) as previously shown for PP1, PP2, SP600125, AG 1478, and further identified for 1-NA-PP1 and 1-NM-PP1.
Accession numbers
Coordinates and structure factors have been deposited in the Protein Data Bank with the following accession numbers: 4E7J (APH(3’)-Ia•Ca2+•ATP complex), 4FEU (APH(3’)-Ia•kanamycin•SP600125 complex), 4FEV (APH(3’)-Ia•kanamycin•PP1 complex); 4FEW APH(3’)-Ia•kanamycin•PP2 complex); 4FEX (APH(3’)-Ia•kanamycin•Tyrphostin AG 1478 complex); 4GKH APH(3’)-Ia•kanamycin•1-NA-PP1 complex); 4GKI (APH(3’)-Ia•kanamycin•1-NM-PP1 complex).
RESULTS AND DISCUSSION
Structural analysis of the APH(3’)-Ia • ATP complex
In order to gain a molecular understanding of the active site of APH(3’)-Ia, we first determined the structure of this enzyme in complex with a nucleotide triphosphate. In our activity assays, the kcat for modification of kanamycin by APH(3’)-Ia using GTP is 10-fold higher than with ATP (unpublished results), indicating GTP is a superior substrate. However, we could obtain crystals only of the APH(3’)-Ia•ATP complex, and this structure was determined by single anomalous dispersion (SAD) phasing using crystals grown from selenomethionine-derivatized protein (Table 1).
Table 1.
X-ray diffraction data collection and refinement statistics
| Ligands | Ca2+, ATP | Kanamycin, SP600125 | Kanamycin, PP1 | Kanamycin, PP2 | Kanamycin, Tyrphostin AG 1478 | Kanamycin, 1-NA-PP1 | Kanamycin, 1-NM-PP1 |
|---|---|---|---|---|---|---|---|
| PDB Code | 4EJ7 | 4FEU | 4FEV | 4FEW | 4FEX | 4GKH | 4GKI |
| Data collection | |||||||
| Space group Cell dimensions | C2221 | P1 | P1 | P1 | P1 | P1 | P1 |
| a, b, c, Å | 85.4, 152.5, 165.6 | 57.8, 94.2, 96.8 | 57.6, 94.2, 96.9 | 57.3, 93.6, 96.4 | 57.7, 93.7, 96.3 | 97.2, 97.3, 112.7 | 96.8, 97.1, 112.1 |
| α, β, γ, ° | 90, 90, 90 | 61.2, 73.1, 87.4 | 61.1, 73.1, 87.4 | 61.2, 73.2, 87.4 | 118.8, 103.6, 93.4 | 103.0, 106.2, 112.7 | 103.2, 106.2, 112.7 |
| Resolution, Å | 43.7 – 2.29 | 20.0 – 2.37 | 20.0 – 1.89 | 24.00 – 1.98 | 20.0 – 2.71 | 80.87 – 1.86 | 99.63 – 1.88 |
| Rsyma | 0.090 (0.530)b | 0.072 (0.423) | 0.047 (0.337) | 0.068 (0.405) | 0.061 (0.562) | 0.061 (0.401) | 0.062 (0.372) |
| I / σ(I) | 12.55 (1.50) | 16.35 (2.00) | 27.48 (3.02) | 19.26 (2.15) | 19.39 (2.04) | 11.1 (2.8) | 7.6 (2.1) |
| Completeness, % | 91.9 (69.4) | 98.4 (98.4) | 97.8 (96.5) | 98.1 (96.7) | 98.9 (98.5) | 97.2 (96.5) | 97.6 (96.5) |
| Redundancy | 5.7 (5.8) | 2.7 (2.7) | 3.1 (3.1) | 3.1 (3.1) | 3.9 (3.9) | 3.9 (3.9) | 2.3 (2.3) |
| Refinement | |||||||
| Resolution, Å | 43.67 – 2.40 | 20.0 – 2.37 | 19.97 – 1.89 | 23.90 – 1.98 | 19.9 – 2.69 | 54.78 – 1.86 | 54.38 – 1.88 |
| No. of reflections: working, test | 71660, 3148 | 67481, 3406 | 128187, 1972 | 111054, 5461 | 45056, 1939 | 275305, 13290 | 257334, 12522 |
| R-factor/free R-factorc | 21.5/25.2 (32.0/34.5) | 16.0/21.5 (18.8/21.8) | 15.4/20.4 (20.3/27.1) | 15.7/21.6 (25.1/34.0) | 18.4/24.0 (30.4/37.8) | 16.4/21.6 (25.8/28.9) | 16.6/21.6 (28.5/33.5) |
| No. of refined atoms, molecules | |||||||
| Protein | 6402, 3 | 12085, 6 | 12750 | 12422 | 10409, 5 | 25109, 12 | 25260, 12 |
| Kanamycin | 105 | 198, 6 | 198, 6 | 198, 6 | 165, 5 | 396, 12 | 396, 12 |
| Inhibitor | N/A | 51, 3 | 146, 6 | 147, 6 | 88, 4 | 288, 12 | 300, 12 |
| Solvent | 7 | 24 | 11 | 46 | 7 | 181 | 169 |
| Water | 302 | 685 | 1412 | 1190 | 178 | 2868 | 3307 |
| B-factors | |||||||
| Protein | 43.7 | 56.4 | 24.8 | 35.9 | 54.0 | 48.5 | 37.0 |
| Kanamycin | 41.1 | 48.7 | 17.3 | 26.6 | 43.8 | 38.6 | 26.7 |
| Ca2+, ATP | 46.7, 40.4 | N/A | N/A | N/A | N/A | N/A | N/A |
| Inhibitor | N/A | 52.0 | 28.0 | 36.7 | 66.9 | 52.1 | 48.3 |
| Solvent | 55.8 | 80.7 | 17.3 | 57.4 | 63.4 | 70.4 | 65.5 |
| Water | 41.3 | 48.4 | 29.2 | 35.7 | 42.9 | 49.5 | 37.7 |
| R.m.s.d. | |||||||
| Bond lengths, Å | 0.009 | 0.010 | 0.007 | 0.009 | 0.010 | 0.009 | 0.008 |
| Bond angles, ° | 0.694 | 1.07 | 1.13 | 1.14 | 1.30 | 1.173 | 1.159 |
Rsym = ΣhΣi|Ii(h) - 〈I(h)〉/ΣhΣiIi(h), where Ii(h) and 〈I(h)〉 are the ith and mean measurement of the intensity of reflection h.
Figures in parentheses indicate the values for the outer shells of the data.
R = Σ|Fpobs – Fpcalc|/ΣFpobs, where Fpobs and Fpcalc are the observed and calculated structure factor amplitudes, respectively.
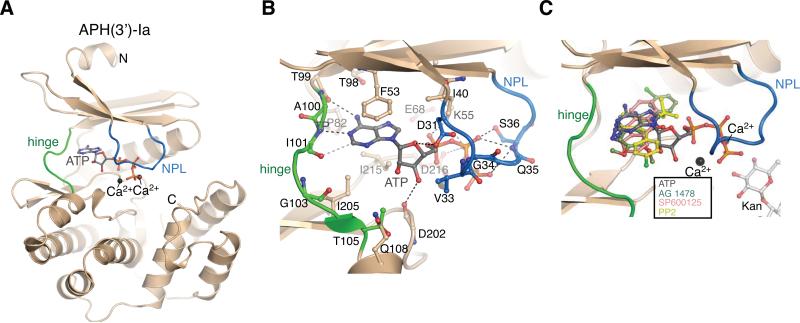
In this structure the APH(3’)-Ia protein adopted the typical bi-lobe ePK-like fold that is distinctive of all previously characterized AKs [13-22]. The nucleotide-binding sites located at the hinge connecting the two lobes contained unambiguous additional density corresponding to the ATP molecule (Figure 1A). The three ATP molecules in the asymmetric unit were identical in conformation except for a slight rotation of the terminal phosphate about the Pγ-O3B bond. Each ATP molecule adopted a bidentate chelating conformation with respect to two ions, which were modeled as Ca2+ due to the prevalence of these ions in the crystallization buffer, the magnitude of the electron density peaks and the enzyme-ion atomic bond distances. The position of the Ca2+ions in the APH(3’)-Ia•ATP complex corresponded to that of the two Mg2+ ions in the APH(3’)-IIIa-AMPPNP-Mg2+ complex [17,21]. The adenine moiety of ATP occupied a hydrophobic pocket formed by Phe53, Ile205 and Ile215 and establishes a number of hydrogen bonds with backbone atoms of the hinge, specifically between N6 and the carbonyl oxygen of Thr99, between N1 and the amide nitrogen of Ile101, and a weaker bond between the C2-H and the carbonyl oxygen of Ile101 (Figure 1B). Overall, the binding mode of ATP to APH(3’)-Ia was comparable with those of adenosine-based nucleoside-phosphates or non-hydrolyzable analogs in complex with other APH structures. Given that GTP is also a substrate of this enzyme and that the adenine and guanine bases have been shown to form distinct interactions with the hinge region of one APH enzyme [12], it should be noted that the APH(3’)-Ia•ATP complex structure presented may not represent the binding conformation of the guanine-containing nucleotide triphosphate.
Figure 1. Overview of APH(3’)-Ia structures.
A) Structure of the APH(3’)-Ia•Ca2+•ATP complex. APH(3’)-Ia hinge region is coloured green, nucleotide-positioning loop is coloured blue, Ca2+ ions shown in black spheres. N- and C-termini are labeled. B) Interactions between APH(3’)-Ia and ATP. C) Superposition of APH(3’)-Ia•ATP, APH(3’)-Ia•kanamycin•SP600125, APH(3’)-Ia•kanamycin•PP2 (representative of APH(3’)-Ia•kanamycin•PP1), APH(3’)-Ia•kanamycin•AG 1478 complexes. As hinge region of APH(3’)-Ia is nearly identical across all inhibitor complexes, only one copy of the enzyme is shown.
APH(3’)-Ia-kanamycin-inhibitor ternary complex structures
To characterize inhibition of APH(3’)-Ia at the molecular level, we selected PP1, PP2, SP600125 and tyrphostin AG 1478, representing three distinct chemical scaffolds, from the broad range of APH(3’)-Ia inhibitors [24]. These compounds were initially designed as ATP-competitive inhibitors, but these are all also GTP-competitive inhibitors of APH(3’)-Ia (unpublished results). We determined crystal structures of the APH(3’)-Ia•kanamycin•SP600125, APH(3’)-Ia•kanamycin•PP1, APH(3’)-Ia•kanamycin•PP2 and APH(3’)-Ia•kanamycin•tyrphostin AG 1478 ternary complexes to 2.37 Å, 2.69 Å, 1.89 Å and 1.97 Å, respectively (Figure 1C, Figure 2). All four ternary complex structures were solved by Molecular Replacement (MR) using the APH(3’)-Ia•ATP structure as a model. In all four inhibitor complex structures, the conformation of the APH(3’)-Ia hinge was highly conserved and nearly identical to that observed in the APH(3’)-Ia•ATP complex, indicating this is a stable interaction site. In contrast, the position of the nucleotide-positioning loop (NPL) was variable or disordered in some copies of the enzyme in the crystals. A similar phenomenon has been previously described for other structurally characterized members of the APH(3’) subgroup of enzymes and was attributed to structural flexibility of the NPL [43].
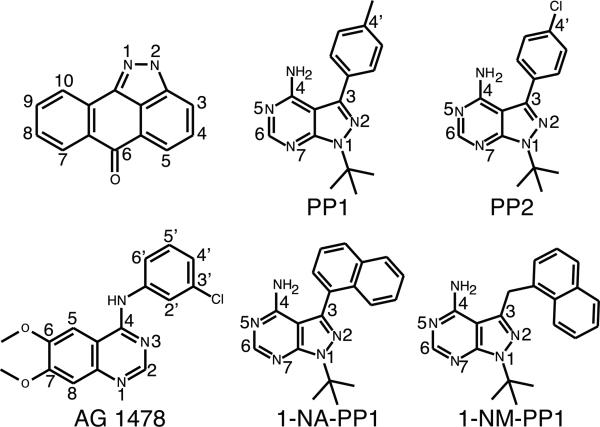
Figure 2.
Chemical structures of protein kinase inhibitors analyzed in this study
In all APH(3’)-Ia ternary complex structures, electron density corresponding to the inhibitor molecules was found at the ATP-binding site near the hinge (Figure 1C). We conducted a detailed analysis of the APH(3’)-Ia interactions with these four inhibitors and compared their interactions with those of ePKs.
First observation of a distinct binding mode of a PKI to an AK: inhibition of APH(3’)-Ia by the anthrapyrazolone SP600125
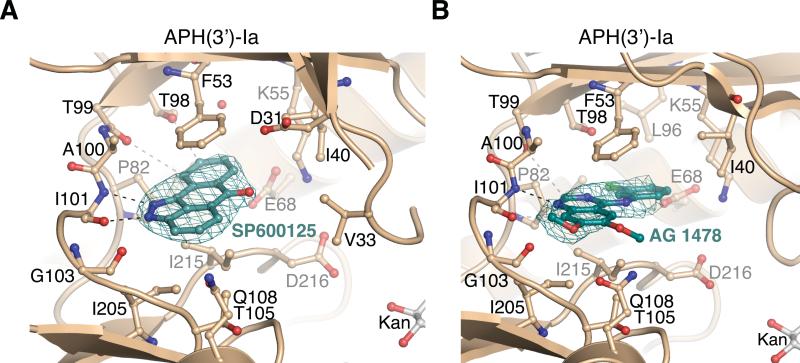
The SP600125 compound has been extensively used as an inhibitor of the Jun N-terminal kinase (JNK) family and the serine/threonine kinase Mps1 [44-46]. SP600125 was identified to be a potent inhibitor of APH(3’)-Ia and a general inhibitor of the APH(3’), APH(2’’), APH(4) and APH(9) subfamilies [24]. In the APH(3’)-Ia•kanamycin•SP600125 complex structure, the inhibitor molecule wedged into a hydrophobic pocket formed by the sidechains of Phe53, Ile205 and Ile215 (Figure 3A). The planar nature of the compound enabled it to form a π-stacking interaction with Phe53. SP600125 formed two hydrogen bonds with backbone atoms in the hinge region, specifically between N1 and the Ile101 amide hydrogen, and between N2 and the carbonyl oxygen of Ile101. Additional bonds were formed between the C10-H and the carbonyl oxygen of Thr99, and the C9-H and the carbonyl oxygen of Thr98.
Figure 3. Structures of anthrapyrazolone and 4-anilinoquinazoline inhibitors bound to APH(3’)-Ia.
A) APH(3’)-Ia•kanamycin•SP600125 and B) APH(3’)-Ia•kanamycin•tyrphostin AG 1478 complexes. Electron density shown is simulated annealing Fo-Fc omit density at 2.5 and 2.0 σ for SP600125 and AG 1478, respectively.
As SP600125 is a well-studied ePK inhibitor, we compared the binding mode of this compound to APH(3’)-Ia and to ePKs (Supplementary Figure S1A). According to our analysis, SP600125 adopted a conformation in the APH(3’)-Ia active site that was rotated 180° relative to its orientation in the active site of all three structurally-characterized complexes of this compound with ePKs [44-46]. As a result, the inhibitor's C7-C10 ring was positioned to face deeper into the APH(3’)-Ia active site, while in its interactions with ePKs, the C3-C5 ring faces into the deepest region of the active site. Thus, the structure of APH(3’)-Ia in complex with SP600125 provided the first example of a distinct mode of binding of a PKI to an antibiotic resistance enzyme.
We could rationalize the broad inhibition profile of SP600125 against multiple APH subgroups this using the crystallographic data: SP600125 is anchored to the APH(3’)-Ia enzyme through hydrogen bonds with the main chain atoms of the hinge region and the only interactions involving sidechains were with highly conserved positions. In these regards, the interactions mediated by SP600125 resemble those we previously observed in the binding of another APH general inhibitor, quercetin, in binding to APH(2’’)-IVa [24]. Based on these structural observations, we suggest the anthrapyrazolone scaffold as a source of effective general inhibitors of APH enzymes, and could be useful against bacterial strains carrying multiple aph genes. While the SP600125 molecule has been shown to bind ePK enzymes such as JNK family kinases, its interaction mode with ePKs is significantly different than that revealed in the APH(3’)-Ia•kanamycin•SP600125 complex structure, therefore retaining the possibility for development of AK-specific inhibitors.
4-anilinoquinazoline tyrphostin AG 1478 occupies a deeper binding pocket in APH(3’)-Ia that is also found in ePKs
4-anilinoquinazoline-based inhibitors in general have been optimized to be specific and potent inhibitors of ePKs [47]. Our data demonstrated that AG 1478 shows inhibitory activity with high specificity for the APH(3’) subfamily [24].
According to the APH(3’)-Ia•kanamycin•tyrphostin AG 1478 structure, the inhibitor molecule was held in the NTP binding site through hydrophobic interactions with the sidechains of Phe53, Ile205 and Ile215 (Figure 3B). In addition, through its N1 atom AG 1478 formed a hydrogen bond with the amide of Ile101. The 7-methoxy group of AG 1478 was accommodated into a cleft formed by Pro102 and the 6-methoxy group occupied an area near the deoxyribose-binding site for ATP. The quinazoline core and the 4-N-(3’-chloroanilino) groups were roughly coplanar, with rotations of 4-9° about the N4-C1, bond among the four of the five observed copies of the molecule in the asymmetric unit of the crystal. Notably, the 4-N-(3’-chloroanilino) group of AG 1478 wedged into a deeper pocket formed by APH(3’)-Ia Ile40, Lys55, Glu68, Leu72, Leu81, Pro82, Leu96 and Val217. The C4’ and C5’ positions approach the Lys55-Glu68 salt bridge, which is conserved in both APH and ePK enzymes.
The identification of the deeper binding pocket exploited by the 4-anilinoquinazoline group of AG 1478 prompted us to compare the local structural features of various APH enzymes in this region. Interestingly, APH(2”) enzymes lack the depth in this pocket due to larger residues in this area - Met85 in APH(2”)-Ib and Phe95 in APH(2”)-IVa as compared to Thr99 in APH(3’)-Ia. Similarly, APH(9)-Ia lacks the depth in this pocket due to the presence of Tyr100, among other significant changes in the ATP-binding site [16]. In contrast, the equivalent residue in APH(4)-Ia is a serine, which as in APH(3’)-Ia results in a deeper binding pocket in this enzyme. These subfamily-specific structural features correlate well with the AG 1478 inhibition profile [24].
We compared the APH(3’)-Ia•kanamycin•AG 1478 structure with six ePK•4-anilinoquinazoline complex structures available in the PDB (Supplementary Figure S1B). The binding position and orientation of the 4-anilinoquinazoline core of AG 1478 to APH(3’)-Ia was similar to the binding mode of 4-anilinoquinazolines to diverse ePKs. The positioning of the 4-N-substituent ring into the deep pocket of the ATP-binding site is conserved between APH(3’)-Ia and ePKs. Notably, the 4-substituent ring of this class of compounds is an area of derivatization which provides for selectivity against ePKs by accessing structural diversity in this deep pocket among these enzymes [51]. Providing the molecular evidence of a similar orientation of AG 1478 in its inhibition of APH(3’)-Ia suggests that a similar strategy for optimization of 4-anilinoquinazolines may be applied for discovery of compounds of this class with increased potency and/or specificity against the APH(3’) subfamily.
Inhibition of APH(3’)-Ia by pyrazolopyrimidine-based compounds takes place through a binding mode distinct from that observed for ePKs
The pyrazolopyrimidines are a well-known scaffold active against human PKs [37,38,48,49] as well as against protozoan calcium-dependent protein kinase (CDPK1) (PDB accession 3MA6 and [50]). Our previous work showed that the PP1 and PP2 representatives of this class were specific inhibitors of the APH(3’) subfamily [24].
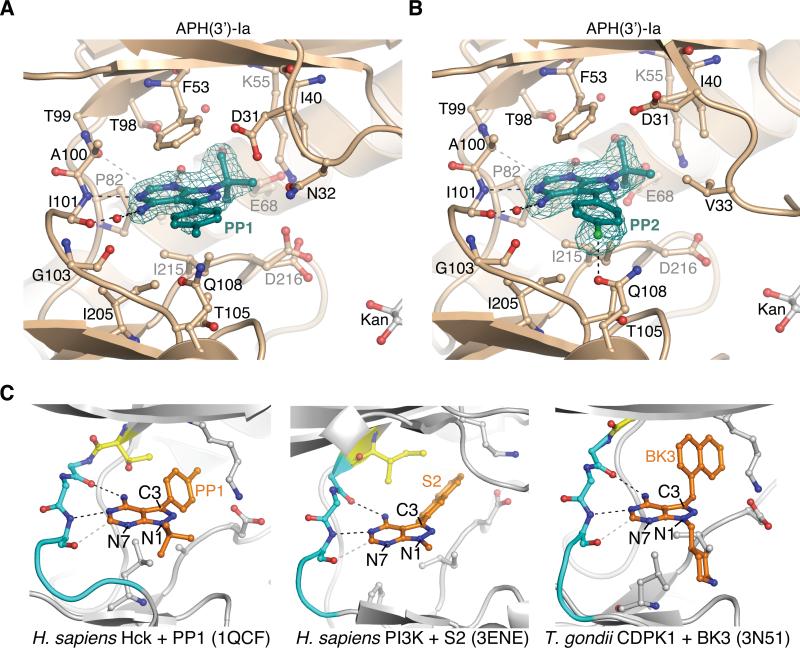
The orientation of PP1 and PP2 in the APH(3’)-Ia•kanamycin•PP1 and APH(3’)-Ia•kanamycin•PP2 ternary complex structures was such that the 3-(4’-methylphenyl or 4’-chlorophenyl) group points towards the C-terminal domain of the enzyme (Figure 4); we term this the C3-substituent “down” orientation. The PP compounds were anchored to APH(3’)-Ia primarily through hydrophobic interactions with the sidechains of Ile205, Ile215 and Phe53 and two hydrogen bonds to mainchain atoms in the hinge region: the N4 group with the carbonyl oxygen of Ile101 and N5 with the amide of Ile101. The larger residues in the deepest region of the binding pocket of APH(2’’) enzymes would obstruct the binding of PP1/PP2, and thus explain the subfamily specificity of these compounds.
Figure 4.
Binding mode of PP-based compounds to APH(3’)-Ia and comparison with PP-based compounds bound to ePKs
A) Structure of APH(3’)-Ia•kanamycin•PP1 complex. B) Structure of APH(3’)-Ia•kanamycin•PP2 complex. Electron density shown for both inhibitors is simulated annealing Fo-Fc omit density at 3.0 σ. C) Binding modes of PP-based inhibitors to ePKs. Left = PP1 bound to H. sapiens Hck (PDB accession 1QCF); middle = S2 bound to H. sapiens PI3K (PDB accession 3ENE); right = BK3 bound to T. gondii CDPK1 (PDB accession 3N51). Gatekeeper residues are coloured yellow, hinge residues cyan, also shown are sticks of conserved Lys-Asp/Glu salt bridge in active site.
To validate the interactions of PP compounds with APH(3’)-Ia, we investigated the effect of additional groups at the N1, C3 and N4 positions of the PP scaffold on in vitro inhibition of APH(3’)-Ia. PP variants with modifications at the N1 position ranging in size from isobutyl to hexyl [41] demonstrated 2- to 25-fold less inhibition of APH(3’)-Ia activity relative to PP1 (Figure 5). Modifications at C3 with large substituents including indole or naphthalen-1-ylmethyl also adversely affected APH(3’)-Ia activity (3- to 9-fold decrease relative to inhibition by PP1). Given the comparatively larger sizes of the tested modifications at C3, these results suggest APH(3’)-Ia is more sensitive to substitutions at N1 and are consistent with a C3 “down” binding orientation of PP compounds to the enzyme.
Next, we compared the molecular details of interactions of PP compounds and APH(3’)-Ia with those interactions formed between this class of compounds and ePKs. Through a search of the PDB, we identified 18 distinct complex structures between ePKs and PP-type compounds. Despite the diverse chemical nature of the PP compounds, the comparative analysis demonstrated a common binding position and orientation, with C3 substituents occupying deeper regions of the NTP sites (C3 “up”) (Figure 4C). This analysis showed that PP compounds adopt different binding orientation to APH(3’)-Ia and ePKs. Based on this analysis we hypothesized that PP compounds with large C3 substituents would clash with the enzyme if bound in this C3 “up” binding orientation; this is consistent with the data presented in Figure 5C and with a C3 “down” binding orientation instead.
Structural basis of inhibition of APH(3’)-Ia by C3-substituted PP compounds
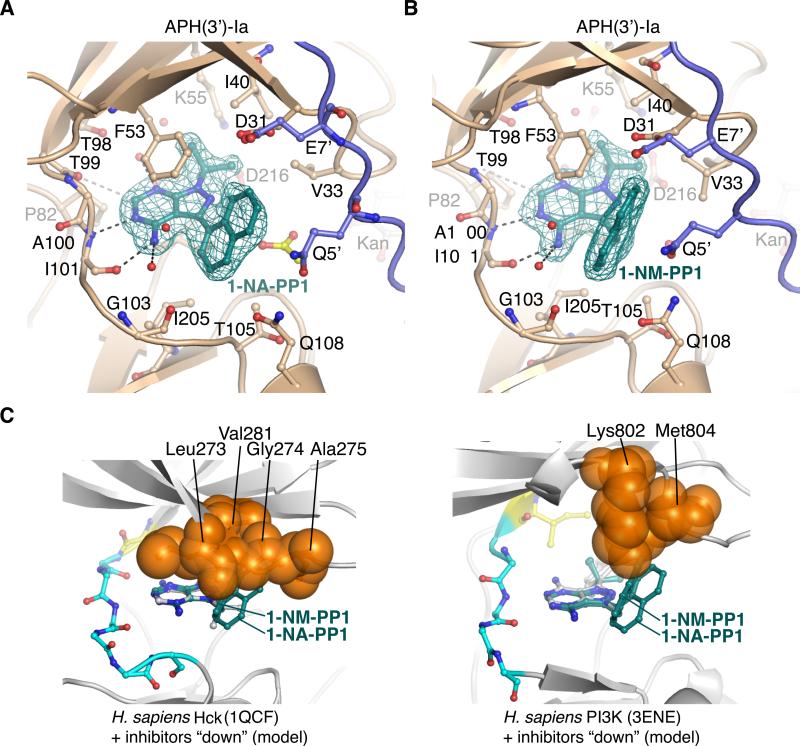
To validate this hypothesis, we determined the crystal structures of APH(3’)-Ia in complex with kanamycin and 1-NA-PP1 or 1-NM-PP1 - two PP variants with large aromatic substituents at the C3 position. These ternary complex structures were solved by MR to resolutions of 1.96 Å and 1.88 Å, respectively. In these structures, both compounds bind to APH(3’)-Ia in the C3 “down” binding orientation (Figure 6). The 3-napthalen-2-yl group of 1-NA-PP1 was rotated ~45° relative to the plane of the pyrazolopyrimidine which provides for substantial hydrophobic interaction with Phe53. Similarly, the napthalen-1-yl group of 1-NM-PP1 packed against one face of Phe53, with the additional methyl group linking the PP core and the napthalen-1-yl group facilitating a more acute packing angle against this residue. In addition, the 1-NA-PP1 and 1-NM-PP1 compounds formed hydrophobic interactions with the methyl group of Thr105 and the sidechains of Val33, Ile40, Ile205, and Ile215.
Figure 6. Structural analysis of C3-substituted compounds 1-NA-PP1 and 1-NM-PP1 bound to APH(3’)-Ia and modeling into ePKs.
A) Structure of APH(3’)-Ia•kanamycin•1-NA-PP1 complex. B) Structure of APH(3’)-Ia•kanamycin•1-NM-PP1 complex. Electron density shown for both inhibitors is simulated annealing Fo-Fc density at 3.0 σ. Symmetry-related protein chains contributing to interactions with the inhibitors are shown in purple. Acetate molecule shown in yellow. C) Modeling of 1-NA-PP1 and 1-NM-PP1 in “down” orientation into ATP-binding sites of H. sapiens Hck (PDB accession 1QCF, left) and PI3K (PDB accession 3ENE, right). Experimental structures of PP1 from the 1QCF and 3ENE structures are shown in grey sticks. Enzyme residues from ePKs expected to clash with 1-NA-PP1 and 1-NM-PP1 are shown as orange spheres.
In order to provide a quantitative comparison of APH(3’)-Ia interaction with PP compounds, we computed the shape complementarity between these inhibitors and the enzyme [39]. We calculated complementarity values of 0.740 and 0.761 (1.0 would indicate complete shape complementarity) for 1-NA-PP1 and 1-NM-PP1, respectively. In comparison, the corresponding values for PP1 and ATP are 0.691 and 0.754. Interestingly, the buried surface area between PP1 and the larger 1-NA-PP1 and 1-NMPP1 was nearly identical (432, 427, 422 Å2 buried, respectively). While overall the buried surface area of 1-NA-PP1, 1-NM-PP1 and the parent compound PP1 upon binding to APH(3’)-Ia were not significantly different, the increased shape complementarity and large aromatic substituents at the C3 atom of 1-NAPP1 and 1-NM-PP1 may provide for additional favorable hydrophobic packing interactions with APH(3’)-Ia, especially residue Phe53.
To the best of our knowledge, the complexes between PP compounds with large C3 substituents bound to CDPK1 from Toxoplasma gondii and Cryptosporidium parvum represent the only structurally-characterized examples of interactions of this type of PP derivative with a kinase catalytic domain [50]. Therefore, we compared the binding mode of these compounds observed in CDPK1 complexes with their binding to APH(3’)-Ia. In each of the seven retrieved structures, the inhibitors bound in the C3 “up” orientation to the protozoan kinases (Supplementary Figure S1C). In these complexes, the aromatic substituents at C3 were able to access the deeper pocket of the ATP-binding site due to the presence of smaller residues (glycines) in this area. This confirmed the prominent contrast with the C3 “down” orientation we observe for 1-NA-PP1 and 1-NM-PP1 bound to APH(3’)-Ia.
We modeled 1-NA-PP1 and 1-NM-PP1 into representative ePK active sites in the C3 “up” as well as in C3 “down” conformations (Figure 6C). This exercise demonstrates that 1-NA-PP1 and 1-NM-PP1 would clash with ePK residues in the N-terminal β-sheet or glycine-rich loop that form the “ceiling” of the NTP binding site. The ePKs analyzed appear to have a more constricted NTP binding site that would preclude the binding of 1-NA-PP1 and 1-NM-PP1 in the C3 “down” orientation.
Structure-selected APH(3’)-Ia inhibitors rescue aminoglycoside resistance
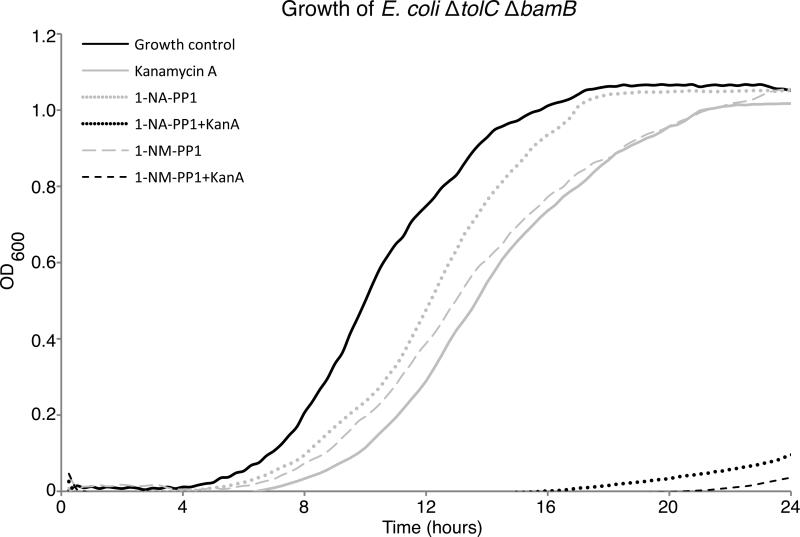
Having established the structural basis of APH(3’)-Ia inhibition by PKIs and identified the specific scaffolds with distinct binding to APH(3’)-Ia and ePKs, we then tested whether these compounds could re-sensitize kanamycin-resistant bacteria to the antibiotic. For these experiments a hyperpermeable efflux mutant strain of E. coli (ΔtolC ΔbamB) expressing aphA1 was grown in the presence of PKIs alone or in combination with kanamycin at one-quarter minimum inhibitory concentration (MIC) (Figure 7 and Supplementary Figure S2). The bacterial growth curves under these conditions were quantitatively compared using the antibiotic rescue factor (ARF), a measurement of the effectiveness of a compound for recovering antibiotic activity (a value of 1 would indicate complete restoration of antibiotic activity) [24]. All inhibitors had minimal impact on cell growth. The inhibitors AG1478 or SP600125 in combination with kanamycin demonstrated only modest antibiotic rescue activity. In contrast, PP1 or PP2 in combination with the antibiotic were able to decrease bacterial growth and displayed ARF values of 0.33 and 0.18, respectively. Most importantly, the additional of 1-NA-PP1 or 1-NM-PP1 in combination with the antibiotic lead to bacterial growth suppression with ARF values of 0.98 and 0.95, respectively, thus nearly completely re-sensitizing this antibiotic-resistant strain to kanamycin.
Figure 7. C3-substituted PP compounds rescue activity of kanamycin-resistant E. coli.
Shown are growth curves of E. coli ΔtolC ΔbamB expressing APH(3’)-Ia grown in the presence of 1-NAPP1 alone (64 μM, light grey circles), 1-NA-PP1 (64 μM) in combination with kanamycin at ¼ MIC (black circles), 1-NM-PP1 alone (32 μM, light grey dashed line) and 1-NM-PP1 (32 μM) in combination with kanamycin at ¼ MIC (dark grey dashed line). Also shown are normal growth control (solid black line) and kanamycin at ¼ MIC only (solid grey line). Inset: corresponding ARF [24] values.
To test for specificity, the pyrazolopyrimidines PP1, PP2, 1-NA-PP1 and 1-NM-PP1 were also tested against E. coli carrying a structurally unrelated aminoglycoside resistance gene, aminoglycoside acetyltransferase aac(3)-Ia, which we predicted would not show synergy with APH inhibitors. All pyrazolopyrimidines tested failed to re-sensitize this resistant strain to the aminoglycoside antibiotic gentamicin, consistent with our structural data that these compounds are specific to the APH(3’)-Ia enzyme (Supplementary Figure S3).
Antibiotic susceptibility tests with the hyperpermeable E. coli strain have demonstrated increased sensitivity to various antibiotics (unpublished data and also [52]). We have noticed this strain is particularly more sensitive to larger hydrophobic antibiotics (i.e. rifampin and erythromycin). Although 1-NA-PP1 and 1-NM-PP1 possess a marginally lower Ki compared to the parent compound PP1, their enhanced in vivo activity (in the absence of the major efflux component TolC) may be attributed to this increased sensitivity. Illustrating the importance of permeability, all pyrazolopyrimidines failed to resensitize wild type E. coli expressing aphA1 (Supplementary Figure S4). However, SP600125 and AG 1478 are also large hydrophobic compounds but do not rescue aminoglycoside resistance, therefore, compound size is not the only criterion for successful in vivo activity of PKIs; specific structural features shown by 1-NA-PP1 and 1-NM-PP1, particularly shape complementarity with the APH(3’)-Ia, are essential.
The compounds 1-NA-PP1 and 1-NM-PP1 were previously termed “bumped kinase inhibitors” (BKIs) and were shown to be inactive against wild-type mammalian kinases [53]. In these kinases, a “gatekeeper” residue, typically a methionine, phenylalanine or tyrosine residue, restricts the depth of the NTP binding site in ePKs and prevents such PP compounds with large C3 substituents from binding in the “up” orientation. Using the insight that 1-NA-PP1 and 1-NM-PP1 interact with APH(3’)-Ia in the C3 “down” orientation, modeling of these compounds into ePKs in the C3 “down” orientation suggested that these compounds would clash with residues in ePKs in the ribose-binding area of the binding site. This suggested that PP compounds with large C3 substituents are unable to bind ePKs in either the C3 “down” or in the C3 “up” orientation and are therefore ineffective against wild-type ePKs. Based on this analysis, our identification of the C3 “down” orientation of PP compounds to APH(3’)-Ia was an important advance in the identification of APH-specific inhibitors.
While the NTP binding site of APH(3’)-Ia shares enough structural features with ePK ATP-binding sites to enable the use of PKI molecules as initial leads for APH inhibitors, we demonstrated in this study that multiple classes of PKIs bind to this bacterial enzyme in a distinct orientation from their binding mode to ePKs. This observation allowed us to optimize compounds that select for binding to APH(3’)-Ia over ePKs. Keeping in mind the permeability barrier, which may be overcome with compound optimization, the APH inhibitor molecules studied here thus represent privileged chemical matter for design of the next generation of AK-specific inhibitors. We believe these findings provide a significant advance toward addressing antibiotic resistance by restoration of aminoglycoside antibiotic activity.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Rosa Di Leo for cloning; Kemin Tan and Zdzislaw Wawrzak for data collection and George Minasov for validation of structures.
FUNDING
The structures presented were solved by the Center for Structural Genomics of Infectious Diseases (CSGID, http://csgid.org); this project has been funded in whole or in part with U.S. Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272200700058C (2007 to September 27, 2012) and HHSN272201200026C (starting Sept. 1, 2012), the Canadian Institutes of Health Research (MT-13536 to G.D.W.), the Natural Sciences and Engineering Research Council of Canada (to A.C.) and the Canada Research Chairs program (to G.D.W.).
Abbreviations used
- 1-NA-PP1
(1-tert-butyl-3-naphthalen-2-yl-1H-pyrazolo[3,4-d]pyrimidin-4-amine
- 1-NM-PP1
1-(1-methylethyl)-3-(naphthalen-1-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine
- AG 1478
N-(3-chlorophenyl)-6,7-dimethoxyquinazolin-4-amine
- AMPPNP
adenosine 5’-(β,γ-imido)triphosphate
- AAC
aminoglycoside acetyltransferase
- ANT
aminoglycoside nucleotidyltransferase
- APH
aminoglycoside phosphotransferase
- AK
antibiotic kinase
- CDPK1
calcium-dependent protein kinase 1
- ePK
eukaryotic protein kinase
- JNK
Jun N-terminal kinase
- MIC
minimum inhibitory concentration
- NPL
nucleotide-positioning loop
- NTP
nucleotide triphosphate
- PKI
protein kinase inhibitor
- PP
pyrazolopyrimidine
- PP1
1-t-butyl-3-(4’-methylphenyl)pyrazolo[3,4-d]pyrimidin-4-amine
- PP2
1-t-butyl-3-(4’-chlorophenyl)pyrazolo[3,4-d]pyrimidin-4-amine
- SAD
single anomalous dispersion
- SP600125
anthra[1,9-cd]pyrazol-6(2H)-one
Footnotes
AUTHOR CONTRIBUTION
Peter Stogios determined the crystal structures, performed all structural analysis, interpreted the data and wrote the paper. Peter Spanogiannopoulos designed and performed enzyme kinetics and growth curve experiments, interpreted the data and wrote the paper. Elena Evdokimova performed crystallography. Olga Egorova performed protein purification and preliminary crystallography. Tushar Shakya designed and performed enzyme kinetics experiments. Nick Todorovic designed and performed chemical synthesis and wrote the paper. Alfredo Capretta interpreted the data and wrote the paper. Gerard Wright interpreted the data and wrote the paper. Alexei Savchenko interpreted the data and wrote the paper.
REFERENCES
- 1.Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century--a clinical super-challenge. N. Engl. J. Med. 2009;360:439–443. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- 2.Morar M, Wright GD. The genomic enzymology of antibiotic resistance. Annu. Rev. Genet. 2010;44:25–51. doi: 10.1146/annurev-genet-102209-163517. [DOI] [PubMed] [Google Scholar]
- 3.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 4.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089– 1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ejim L, Farha MA, Falconer SB, Wildenhain J, Coombes BK, Tyers M, Brown ED, Wright GD. Nat. Chem. Biol. Vol. 7. Nature Publishing Group; 2011. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. pp. 348–350. [DOI] [PubMed] [Google Scholar]
- 6.Kalan L, Wright GD. Antibiotic adjuvants: multicomponent anti-infective strategies. Expert Rev. Mol. Med. 2011;13:e5. doi: 10.1017/S1462399410001766. [DOI] [PubMed] [Google Scholar]
- 7.Drawz SM, Bonomo RA. Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright GD. Molecular mechanisms of antibiotic resistance. Chem. Commun. (Camb.) 2011;47:4055–4061. doi: 10.1039/c0cc05111j. [DOI] [PubMed] [Google Scholar]
- 9.Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 2011;35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 10.Shakya T, Wright GD. Nucleotide selectivity of antibiotic kinases. Antimicrob. Agents Chemother. 2010;54:1909–1913. doi: 10.1128/AAC.01570-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toth M, Chow JW, Mobashery S, Vakulenko SB. Source of phosphate in the enzymic reaction as a point of distinction among aminoglycoside 2“-phosphotransferases. J. Biol. Chem. 2009;284:6690–6696. doi: 10.1074/jbc.M808148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi K, Berghuis AM. Structural basis for dual nucleotide selectivity of aminoglycoside 2“-”phosphotransferase IVa provides insight on determinants of nucleotide specificity of aminoglycoside kinases. J. Biol. Chem. 2012;287:13094–13102. doi: 10.1074/jbc.M112.349670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fong DH, Xiong B, Hwang J, Berghuis AM. Crystal structures of two aminoglycoside kinases bound with a eukaryotic protein kinase inhibitor. PLoS ONE. 2011;6:e19589. doi: 10.1371/journal.pone.0019589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stogios PJ, Shakya T, Evdokimova E, Savchenko A, Wright GD. Structure and function of APH(4)-Ia, a hygromycin B resistance enzyme. J. Biol. Chem. 2011;286:1966–1975. doi: 10.1074/jbc.M110.194266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toth M, Frase H, Antunes NT, Smith CA, Vakulenko SB. Crystal structure and kinetic mechanism of aminoglycoside phosphotransferase-2“-”IVa. Prot. Sci. 2010;19:1565–1576. doi: 10.1002/pro.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fong DH, Lemke CT, Hwang J, Xiong B, Berghuis AM. Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila. J. Biol. Chem. 2010;285:9545–9555. doi: 10.1074/jbc.M109.038364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong DH, Berghuis AM. Structural basis of APH(3')-IIIa-mediated resistance to N1-substituted aminoglycoside antibiotics. Antimicrob. Agents Chemother. 2009;53:3049–3055. doi: 10.1128/AAC.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young PG, Walanj R, Lakshmi V, Byrnes LJ, Metcalf P, Baker EN, Vakulenko SB, Smith CA. The crystal structures of substrate and nucleotide complexes of Enterococcus faecium aminoglycoside-2”-phosphotransferase-IIa [APH(2”)-IIa] provide insights into substrate selectivity in the APH(2“”) subfamily. J. Bacteriol. 2009;191:4133–4143. doi: 10.1128/JB.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nurizzo D, Shewry SC, Perlin MH, Brown SA, Dholakia JN, Fuchs RL, Deva T, Baker EN, Smith CA. The crystal structure of aminoglycoside-3'-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance. J. Mol. Biol. 2003;327:491–506. doi: 10.1016/s0022-2836(03)00121-9. [DOI] [PubMed] [Google Scholar]
- 20.Fong DH, Berghuis AM. Substrate promiscuity of an aminoglycoside antibiotic resistance enzyme via target mimicry. EMBO J. 2002;21:2323–2331. doi: 10.1093/emboj/21.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burk DL, Hon WC, Leung AK, Berghuis AM. Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase. Biochemistry. 2001;40:8756–8764. doi: 10.1021/bi010504p. [DOI] [PubMed] [Google Scholar]
- 22.Hon WC, McKay GA, Thompson PR, Sweet RM, Yang DS, Wright GD, Berghuis AM. Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell. 1997;89:887–895. doi: 10.1016/s0092-8674(00)80274-3. [DOI] [PubMed] [Google Scholar]
- 23.Liao JJ-L. Molecular recognition of protein kinase binding pockets for design of potent and selective kinase inhibitors. J. Med. Chem. 2007;50:409–424. doi: 10.1021/jm0608107. [DOI] [PubMed] [Google Scholar]
- 24.Shakya T, Stogios PJ, Waglechner N, Evdokimova E, Ejim L, Blanchard JE, McArthur AG, Savchenko A, Wright GD. A small molecule discrimination map of the antibiotic resistance kinome. Chem. Biol. 2011;18:1591–1601. doi: 10.1016/j.chembiol.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 1981;147:217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- 26.Vakulenko SB, Mobashery S. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev., Am Soc Microbiol. 2003;16:430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siregar JJ, Miroshnikov K, Mobashery S. Purification, characterization, and investigation of the mechanism of aminoglycoside 3'-phosphotransferase type Ia. Biochemistry, American Chemical Society. 1995;34:12681–12688. doi: 10.1021/bi00039a026. [DOI] [PubMed] [Google Scholar]
- 28.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 29.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 31.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Painter J, Merritt EA. TLSMDweb server for the generation of multi-group TLS models. J. Appl. Crystallogr., International Union of Crystallography. 2006;39:109–111. [Google Scholar]
- 35.Bricogne G, Blanc E, Brandl M, Flensburg C. BUSTER-TNT 2.5. 1 and autoBUSTER 1.3. 1. Global Phasing Ltd; 2008. [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Schindler T, Sicheri F, Pico A, Gazit A, Levitzki A, Kuriyan J. Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Mol. Cell. 1999;3:639–648. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- 38.Apsel B, Blair JA, Gonzalez B, Nazif TM, Feldman ME, Aizenstein B, Hoffman R, Williams RL, Shokat KM, Knight ZA. Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat. Chem. Biol. 2008;4:691–699. doi: 10.1038/nchembio.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 40.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Todorovic N, Awuah E, Shakya T, Wright GD, Capretta A. Microwave-assisted synthesis of N1- and C3-substituted pyrazolo[3,4-d]pyrimidine libraries. Tetrahedron Lett. 2011;52:5761–5763. [Google Scholar]
- 42.Cer RZ, Mudunuri U, Stephens R, Lebeda FJ. IC50-to-Ki: a web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res. 2009;37:W441–5. doi: 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson PR, Boehr DD, Berghuis AM, Wright GD. Mechanism of aminoglycoside antibiotic kinase APH(3')-IIIa: role of the nucleotide positioning loop. Biochemistry. 2002;41:7001–7007. doi: 10.1021/bi0256680. [DOI] [PubMed] [Google Scholar]
- 44.Heo Y-S, Kim S-K, Seo CI, Kim YK, Sung B-J, Lee HS, Lee JI, Park S-Y, Kim JH, Hwang KY, et al. EMBO J. Vol. 23. Nature Publishing Group; 2004. Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125. pp. 2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scapin G, Patel SB, Lisnock J, Becker JW, LoGrasso PV. The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity. Chem. Biol. 2003;10:705–712. doi: 10.1016/s1074-5521(03)00159-5. [DOI] [PubMed] [Google Scholar]
- 46.Chu MLH, Chavas LMG, Douglas KT, Eyers PA, Tabernero L. Crystal Structure of the Catalytic Domain of the Mitotic Checkpoint Kinase Mps1 in Complex with SP600125. J. Biol. Chem. 2008;283:21495–21500. doi: 10.1074/jbc.M803026200. [DOI] [PubMed] [Google Scholar]
- 47.Levitzki A, Mishani E. Tyrphostins and other tyrosine kinase inhibitors. Annu. Rev. Biochem. 2006;75:93–109. doi: 10.1146/annurev.biochem.75.103004.142657. [DOI] [PubMed] [Google Scholar]
- 48.Knowles PP, Murray-Rust J, Kjaer S, Scott RP, Hanrahan S, Santoro M, Ibáñez CF, McDonald NQ. Structure and chemical inhibition of the RET tyrosine kinase domain. J. Biol. Chem. 2006;281:33577–33587. doi: 10.1074/jbc.M605604200. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Bishop A, Witucki L, Kraybill B, Shimizu E, Tsien J, Ubersax J, Blethrow J, Morgan DO, Shokat KM. Structural basis for selective inhibition of Src family kinases by PP1. Chem. Biol. 1999;6:671–678. doi: 10.1016/s1074-5521(99)80118-5. [DOI] [PubMed] [Google Scholar]
- 50.Ojo KK, Larson ET, Keyloun KR, Castaneda LJ, Derocher AE, Inampudi KK, Kim JE, Arakaki TL, Murphy RC, Zhang L, et al. Toxoplasma gondii calcium-dependent protein kinase 1 is a target for selective kinase inhibitors. Nat. Struct. Mol. Biol. 2010;17:602–607. doi: 10.1038/nsmb.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blencke S, Zech B, Engkvist O, Greff Z, Őrfi L, Horváth Z, Kéri G, Ullrich A, Daub H. Characterization of a Conserved Structural Determinant Controlling Protein Kinase Sensitivity to Selective Inhibitors. Chem. Biol. 2004;11:691–701. doi: 10.1016/j.chembiol.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 52.Liu A, Tran L, Becket E, Lee K, Chinn L, Park E, Tran K, Miller JH. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob. Agents Chemother. 2010;54:1393–1403. doi: 10.1128/AAC.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow, J., Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.