Abstract
Sortases are transamidases that covalently link proteins to the peptidoglycan of gram-positive bacteria. The genome of the pathogenic bacterium Listeria monocytogenes encodes two sortases genes, srtA and srtB. The srtA gene product anchors internalin and some other LPXTG-containing proteins to the listerial surface. Here, we focus on the role of the second sortase, SrtB. Whereas SrtA acts on most of the proteins in the peptidoglycan fraction, SrtB appears to target minor amounts of surface polypeptides. We identified one of the SrtB-anchored proteins as the virulence factor SvpA, a surface-exposed protein which does not contain the LPXTG motif. Therefore, as in Staphylococcus aureus, the listerial SrtB represents a second class of sortase in L. monocytogenes, involved in the attachment of a subset of proteins to the cell wall, most likely by recognizing an NXZTN sorting motif. The ΔsrtB mutant strain does not have defects in bacterial entry, growth, or motility in tissue-cultured cells and does not show attenuated virulence in mice. SrtB-mediated anchoring could therefore be required to anchor surface proteins involved in the adaptation of this microorganism to different environmental conditions.
Listeria monocytogenes is a ubiquitous food-borne gram-positive bacterium, responsible for life-threatening infections in humans and animals (17). It is a facultatively intracellular pathogen that is able to enter and multiply in both professional (29) and nonprofessional phagocytes such as epithelial cells (19) or hepatocytes (47). After entry, bacteria rapidly lyse the phagosomal membranes and gain access to the cytosol, where they spread to adjacent cells by an actin-based motility process (for reviews, see references 11, 12, 25, and 46). The interaction of L. monocytogenes with host cells is a key event in the pathogenesis of listeriosis, a process that involves a number of surface proteins (see reference 10 for a review). Several distinct mechanisms of cell wall attachment and display of surface proteins have been described in gram-positive bacteria (10, 13, 37). Each mechanism is determined by specific structural motifs in the sequence of the protein. Until very recently, the only surface proteins known to be attached by a covalent linkage to the peptidoglycan (PG) were the LPXTG proteins, exemplified by protein A of Staphylococcus aureus and internalin (InlA) of L. monocytogenes. In this class, the C-terminal sorting signal is a conserved LPXTG motif that precedes a membrane-spanning hydrophobic domain and a tail that is mostly composed of positively charged residues (31, 44). The LPXTG sorting mechanism was elegantly deciphered in S. aureus. Its hydrophobic domains and charged tails retain target staphylococcal proteins in the membrane, allowing recognition and processing of the LPXTG motif by the membrane-anchored sortase, SrtA (33). The anchor structure of cell wall surface proteins in L. monocytogenes is also known: a C-terminal threonine is amide linked to the side chain amino group of m-diaminopimelic acid within cell wall peptides (14). Thus, in both organisms, the proteins are cleaved between the threonine and the glycine residue of the LPXTG motif and are amide linked to the PG, suggesting that this cell wall sorting mechanism is shared by all gram-positive bacteria.
Analyses of srtA deletion mutants demonstrated that SrtA of L. monocytogenes acts to process and anchor InlA and at least three other LPXTG proteins and is involved in listerial virulence (5, 20). In S. aureus, srtA mutants fail to anchor all of the LPXTG surface proteins, causing a reduction in the ability of such sortase mutants to establish animal infections (30). In the gram-positive human commensal organism Streptococcus gordonii, inactivation of srtA decreases the ability of the bacteria to colonize the oral mucosa in the mouse (6). More recently, genes encoding sortases were identified in Streptococcus suis (39), Streptococcus pyogenes (3), and the oral pathogens Streptococcus mutans (28) and Streptococcus pneumoniae (22, 24). Each of these species possess several paralogous copies of srtA. For example, S. suis possesses as many as five genes encoding sortases or sortase-like proteins (39). In this organism, SrtA is a major sortase that acts to anchor LPXTG proteins to the PG; the roles of the other four putative sortases remain unknown (38).
Bierne et al. recently identified another gene encoding a putative sortase in the genome of L. monocytogenes, designated srtB (5). Like SrtA, SrtB contains an N-terminal hydrophobic region, which could act as a signal peptide/transmembrane domain, and an essential cysteine residue within the catalytic TLXTC signature sequence, at the C terminus. However SrtB, which is only 23% identical to SrtA, contains different or additional amino acid structures relative to SrtA. SrtB orthologues also exist in the nonpathogenic species Listeria innocua, in S. aureus, and in a few other gram-positive bacteria whose genomes are available—S. pyogenes, Clostridium difficile, Bacillus anthracis, and Bacillus halodurans—suggesting that SrtB belongs to a second specific class of sortases (5, 34).
Very recently, staphylococcal SrtB has been shown to be specifically required to anchor the surface protein IsdC, which is not an LPXTG protein (34). In vitro studies suggest that SrtB recognizes and cleaves an NPQTN peptide motif. Here we studied the role of SrtB in L. monocytogenes by constructing ΔsrtB and ΔsrtA ΔsrtB knockout mutants. We show that SrtB has a “sorting” activity distinct from that of SrtA, and we identify SvpA, a virulence factor of L. monocytogenes (8), as a target of SrtB. The respective roles of SrtA and SrtB in surface protein anchoring and in the virulence of L. monocytogenes are discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, mammalian cells, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Plasmids were introduced into Listeria strains by electroporation. Listeria strains were grown at 37°C in brain-heart infusion (BHI) broth or agar (Difco), supplemented with 8 μg of erythromycin/ml, when carrying plasmids (18). RAW 264.7 mouse macrophages (ATTC TIB-71) were cultured in Dulbecco's modified Eagle medium (Gibco) supplemented with 2 mM glutamine, 1% nonessential amino acids (Gibco), and 10% fetal calf serum (Sera-Lab) that was decomplemented by heating for 30 min at 56°C. Cells were incubated at 37°C under 10% CO2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Property | Reference |
|---|---|---|
| Strains | ||
| BUG 1600 | L. monocytogenes EGD-e wild-type strain | 21 |
| BUG 1777 | Isogenic ΔsrtA mutant | 21 |
| BUG 1877 | Isogenic ΔsrtB mutant | This study |
| BUG 1878 | Isogenic ΔsrtA ΔsrtB mutant | This study |
| BUG 1988 | Isogenic ΔsrtA mutant carrying pP1srtB | This study |
| BUG 1989 | Isogenic ΔsrtB mutant carrying pP1srtB | This study |
| BUG 1990 | Isogenic ΔsrtA ΔsrtB mutant carrying pP1srtA | This study |
| BUG 1991 | Isogenic ΔsrtA ΔsrtB mutant carrying pP1srtB | This study |
| BUG 1992 | Isogenic ΔsvpA mutant | This study |
| Plasmids | ||
| pKSVΔsrtB | Thermosensitive plasmid pKSV7 carrying two coligated 500-bp fragments flanking srtB | This study |
| pau1A svpA | This study | |
| pP1srtA | Expressing srtA under the control of the constitutive promoter pProt | 5 |
| pP1srtB | Expressing srtB under the control of the constitutive promoter pProt | This study |
RNA extraction and reverse transcription-PCR (RT-PCR).
Cultures of L. monocytogenes (EGD-e) (at an optical density at 600 nm of 0.4) were centrifuged at 4,000 × g for 10 min, and the pellet was resuspended in 1 ml of Trizol (Life Technologies) and then transferred to a Bead Beater tube containing 500 μl of 0.1-mm mini glass beads (Polylabo). Bacteria were broken once for 30 s in the Bead Beater at maximum speed. The tubes were centrifuged (13,000 × g, 1 min), and the supernatants were transferred to new tubes containing 300 μl of chloroform:isoamyl alcohol. After 10 min of centrifugation at 13,000 × g, the aqueous phase was transferred to a tube containing 270 μl of isopropanol. Total RNA was then precipitated overnight at 4°C and washed with 1 ml of 75% ethanol. RNA was resuspended in 60 μl of diethyl pyrocarbonate-treated water. Contaminating DNA was removed by digestion with DNase I according to the manufacturer's instructions (Roche).
RT-PCR experiments were carried out with 1 μg of RNA and 2.5 pmol of specific reverse primers for each amplification. After denaturation at 65°C for 10 min, 12 μl of a mixture containing 2 μl of deoxynucleoside triphosphates (25 mM), 4 μl of 4× buffer, 2 μl of dithiothreitol, 1 μl of RNasin (Promega), and 1.5 μl of Superscript II (Invitrogen) was added. Samples were incubated for 60 min at 42°C, heated at 75°C for 15 min, and chilled on ice. Samples were diluted with 30 μl of H2O and stored at −20°C. PCR conditions were identical for all reactions. The 50-μl reaction mixtures consisted of 4 μl of the template, 10 pmol of each primer, 1 μl of deoxynucleoside triphosphates (10 mM), 5 μl of 5× buffer, and 0.5 μl of AmpliTaq DNA polymerase from Thermus aquaticus (Perkin-Elmer, Branchburg, N.J.) in a Gene Amp system 9600 thermal cycler (Perkin-Elmer).
The following pairs of primers were used to amplify the mRNAs: (i) for srtB, primer 1 (5′-TTT GGG GAA AAG TTT GAC CTT-3′) and primer 2 (5′-CTT GAA TAA CCA TAC GTC CTT-3′); (ii) for the cotranscription of srtB and lmo2180, primer 1 (5′-TTT GGG GAA AAG TTT GAC CTT-3′) and primer 2 (5′-CGC TTT CAA CTA ATC TAG CAC CCT-3′); (iii) for the cotranscription of lmo2182 and srtB, primer 1 (5′-GTG GCG ATT GTT CAT CAA AGT A-3′) and primer 2 (5′-CTT GAA TAA CCA TAC GTC CTT-3′); (iv) for the cotranscription of lmo2186 and svpA, primer 1 (5′-GAA TTC GGG CCT ATG GGT TGA AGG-3′) and primer 2 (5′-GGA TCC GAA AGA GTC ACA GGT GTT G-3′); and (v) for the cotranscription of svpA and lmo2184, primer 1 (5′-GGA CCA AAA TTG GCA AAA CCG-3′) and primer 2 (5′-TGA ACT AGA CGG AAT TCC AAC GAG-3′).
Chromosomal inactivation and cloning of srtB.
Two 500-bp fragments flanking srtB (lmo 2181; GenBank/EMBL accession no. AL591824) (21) were amplified by PCR from EGD-e chromosomal DNA with primers inside and outside the srtB locus. Primers for the 5′ fragment were SBS1 [5′-(CGCGGATCC)AATGAACTAATGGTGAAGGAATTG-3′] and SBS2 [5′-(AAAAGGCCT)CCCCAAAAACGATTTTATTTTCAC-3′]; primers for the 3′ fragment were SBS3 [5′-(AAAAGGCCT)TGAGAAAGCACCAAAATATAATAG-3′] and SBS4 [5′-(CCGGAATTC)AATCGATTCGCTTTCAACTAATC-3′]. After restriction of the amplified 5′ and 3′ fragments with BamHI and StuI and with StuI and EcoRI, respectively, the two fragments were coligated into the thermosensitive plasmid pKSV7 (16) digested with BamHI and EcoRI, yielding plasmid pKSVΔsrtB. This plasmid was then electroporated into wild-type L. monocytogenes (strain EGD-e), as well as into the previously constructed ΔsrtA mutant (5), to generate a single ΔsrtB mutant and a double ΔsrtA ΔsrtB mutant, respectively. Gene replacement was performed as described previously (16), resulting in a gene containing only the first eight codons and the stop codon of srtB. The resulting ΔsrtB mutants were verified by PCR analysis of chromosomal DNA using couples of internal or flanking primers SB1, SB4, SB10 (5′-GAATTTGCGCAAAATGTGGCG-3′), SB11 (5′-AAATCCGCAAACATGGATCCG-3′), and SB12 (5′-GGAGGAAGTGGCTAAATGTTC-3′). To complement the ΔsrtB mutation, srtB was PCR amplified from EGD-e chromosomal DNA by using primers 5′-(ACGAGCTC)ACGAAAGGAGTTTTAGAGAGT-3′ and 5′-(ACATGCATGC)CTATTATATTTTGGTGCTTTC-3′. The DNA fragment was digested with SacI and SphI and inserted into the SacI and SphI sites in the shuttle plasmid pP1 downstream from the constitutive promoter pProt of the protease gene from Streptococcus cremoris (15), yielding plasmid pP1srtB.
Construction of an svpA mutant.
The mutation in svpA, initially constructed in LO28, corresponds to the deletion of a 79-bp internal fragment of svpA and insertion of a promoterless aph3 gene conferring resistence to kanamycin. It was introduced into the chromosome of EGD-e by allelic replacement, using the recombinant integrative plasmid pAUL-A-svpAΩaphA3 described previously (8).
Immunofluorescence analyses.
For analysis of SvpA localization at the surfaces of bacteria grown in BHI, overnight cultures of L. monocytogenes were grown at 37°C and used immediately (stationary phase) or diluted 1:20 in BHI at 37°C for 3 h (exponential phase). Bacteria were washed twice in phosphate-buffered saline (PBS) and fixed in 3% paraformaldhehyde in PBS for 10 min. They were then labeled with an anti-SvpA polyclonal antibody (8) and an Alexa 488-conjugated secondary antibody (Molecular Probes). To study the distribution of surface proteins strongly bound to PG by using the anti-PG antibody, PBS-washed bacteria were boiled in 4% sodium dodecyl sulfate (SDS) for 30 min, further washed three times with PBS to remove traces of SDS, and processed for immunofluorescence analysis as described above for SvpA. The anti-PG antibody was used in this case at a 1:200 dilution. Preparations were observed with a Zeiss Axiovert 135 microscope. Image acquisition from the Zeiss microscope was carried out with a cooled charge-coupled device camera (Princeton), and the images processed with Metamorph software (Universal Imaging Corporation).
Fractionation experiments and Western blot analysis.
Fractions containing highly pure PG were obtained from the different L. monocytogenes strains grown overnight at 37°C in 500 ml of BHI medium, as previously described (5). This “PG fraction” contains proteins that associate with PG, withstanding extensive boiling in 4% SDS. The supernatant fractions were prepared from the same bacterial culture in which the PG fraction was obtained. After the first centrifugation step of the 500-ml culture, 10 ml of the supernatant was filtered with 0.22-μm-pore-size Millipore filters and the proteins were precipitated by a 10% trichloroacetic acid-acetone washing procedure as described previously (23). Equivalent amounts of supernatant fractions (1.5 ml of culture) were loaded for SDS-polyacrylamide gel electrophoresis (PAGE) analysis. The PG fraction was prepared 50-fold concentrated with respect to supernatant fractions. Membrane fractions and cytoplasmic fractions were prepared from bacteria grown overnight in 250 ml of BHI medium. Bacteria were collected by centrifugation (at 10,000 × g for 10 min at 4°C), washed in an equal volume of PBS (pH 7.4), and finally resuspended in 5 ml of PBS containing DNase I (30 μg/ml) and an appropriate volume of protease inhibitor cocktail (according to tables provided by Roche). After three freeze-thaw cycles, the bacterial suspension was passed through a French press (1,000 lb/in2) and centrifuged at low speed (3,000 × g for 10 min at 4°C) to remove unbroken cells. The supernatant of this centrifugation was subjected to high-speed centrifugation (at 40,000 × g for 30 min at 4°C) to separate the cytosol (supernatant) from the envelope material (pellet). The envelope material was further resuspended in 0.75 ml of PBS containing protease inhibitors. Both the cytosol and membrane fractions were stored at −20°C. Upon addition of an appropriate volume of Laemmli buffer, cytosol and membrane samples corresponding to 1.5 ml of the initial volume culture were used for SDS-PAGE analysis. Proteins were resolved by using the Tris-Tricine buffer system with 10% SDS-polyacrylamide gels (42) or, when specifically indicated, by Tris-glycine SDS (0.1%)-PAGE (7% polyacrylamide) using standard methods (1). Proteins were transferred to Immobilon-P membranes (polyvinylidene difluoride filter, catalog no. IPVH00010; Millipore). Membranes were probed with either monoclonal antibody (MAb) L7.7 (35) to detect InlA, an anti-P60 MAb (a gift from Andreas Bubert, University of Würzburg, Würzburg, Germany), the anti-InlB MAb B4-6 (9), a rabbit polyclonal antiserum (839) to detect proteins present in PG fractions (5), or anti-SvpA polyclonal antibodies (8).
In vitro invasion assays and intracellular multiplication assays.
Invasivity tests were performed as described previously (5). Intracellular multiplication assays in RAW 264.7 cells were performed as follows. Cells were infected at a multiplicity of infection of 1:1 and then left 15 min on ice to allow bacterial adherence (time zero). After 45 min of internalization at 37°C, noninvasive bacteria were killed by adding gentamicin at 10 μg/ml. The number of intracellular bacteria was estimated in cell lysates after 45 min (time, 90 min) and at selected intervals (from 90 min to 8 h postinfection).
Animal infections.
Animal experiments were performed according to the guidelines of the Institut National de la Santé et de la Recherche Médicale (INSERM) for laboratory animal husbandry. Specific-pathogen-free 6- to 8-week-old female Swiss mice (Janvier, Le Geneset St Isle, France) were used.
(i) Kinetics of bacterial growth in infected organs.
Bacteria were grown for 18 h in BHI broth, centrifuged, appropriately diluted in 0.15 M NaCl, and inoculated intravenously (i.v.) (0.5 ml) via the lateral tail vein with a dose of 105 bacteria per mouse. Groups of five mice were sacrificed 3 days after infection. Organs (spleens and livers) were aseptically removed and separately homogenized in 0.15 M NaCl. Bacterial counts in organ homogenates were determined on BHI agar plates, as described previously (2).
(ii) LD50.
The virulence of each mutant was estimated by the 50% lethal dose (LD50) by using the Probit method (2). Groups of five mice were challenged i.v. with various doses of bacteria, and mortality was monitored over a 14-day period.
RESULTS
The L. monocytogenes srtB locus.
Bierne et al. previously identified a gene encoding a second putative sortase, srtB (lmo2181) (5), at a distance of 1,300 kb from srtA in the Listeria genome (21). Comparison of the srtB region of L. monocytogenes to the S. aureus isd region, which contains the staphylococcal srtB gene and which is involved in iron acquisition (34), revealed similarities of genomic organization but not an exact conservation (Fig. 1A). The listerial srtB gene is present in a region apparently organized as a seven-gene operon, preceded by a putative promoter and flanked by transcriptional terminators. As in S. aureus, this gene cluster encodes a putative iron acquisition system and may be controlled by the iron regulator Fur, because a Fur box exists upstream of the promoter.
FIG. 1.
srtB loci in L. monocytogenes and S. aureus. (A) Genetic organization of the srtB locus in S. aureus (34) and in L. monocytogenes (21). ORFs are designated according to the L. monocytogenes genome nomenclature (lmo). The srtB gene corresponds to lmo2181. lmo2182, lmo2183, and lmo2184 are homologous to ABC iron transporter-encoding genes; svpA (lmo2185) and lmo2186 display homologies with isdC in S. aureus. lmo2179 and lmo2178 encode LPXTG proteins of unknown function that are unrelated to the staphylococcal IsdA and IsdB LPXTG proteins. Bent arrows indicate promoters; circles, putative transcription terminators. Fur boxes are indicated. Solid arrows flanking dotted lines, below the svpA locus of L. monocytogenes, indicate the positions of the primers used in the RT-PCR analysis (see panel D). The ΔsrtB and ΔsrtA ΔsrtB mutants were constructed from L. monocytogenes EGD-e by homologous recombination using a thermosensitive vector carrying two short, blunt-end-ligated PCR fragments (hatched rectangles), which were produced with primers SB1-SB2 and SB3-SB4. The in-frame deletion was confirmed by PCR analysis using primers flanking (SB1, SB4, SB10, SB12) (see panel E) or inside (SB11) srtB. (B) Schematic representation of IsdC from S. aureus and of Lmo2186 and SvpA from L. monocytogenes. The N-terminal peptide signal and the C-terminal hydrophobic domain and charged tail are shown. The percentage of identity (id) between the SvpA repeats and Lmo2186 is shown. (C) Sequence alignment of the C-terminal regions of IsdC, Lmo2186, and SvpA. Hydrophobic domains are italicized. The NPQTN cleavage motif in IsdC is boldfaced; putative SrtB cleavage motifs in SvpA and Lmo2186 are underlined. (D) Tris-acetate-EDTA-agarose gel electrophoresis of transcripts amplified by RT-PCR. Numbers (in kilobases) with arrows on the left correspond to sizes on the DNA ladder. Numbers above each lane correspond to those at the bottom of panel A: 1, srtB; 2, lmo2182 and srtB; 3, srtB and lmo2180; 4, svpA and lmo2184; 5, lmo2186 and svpA. (E) In-frame deletions in mutants were genetically verified by PCR analysis using primers flanking srtB (SB1-SB12) or srtA (SA5-SA7). DNA fragments were separated on an ethidium bromide-stained agarose gel.
The first gene of the operon, lmo2186, encodes a protein of 207 amino acids that displays 33% identity with IsdC, the only known target of the staphylococcal sortase SrtB. By contrast, the second gene, svpA (lmo2185), is not present in the isd operon and encodes a 569-amino-acid polypeptide previously identified as a listerial virulence factor (8). SvpA displays three domains that are homologous to regions within Lmo2186 and IsdC, and a proline-rich region possibly involved in protein-protein interactions (Fig. 1B). Both SvpA and Lmo2186 have an N-terminal signal peptide, a C-terminal hydrophobic domain, and a charged tail, resembling sorting signals, albeit without the LPXTG motif. However, an NAKTN motif in SvpA and an NKVTN or NPKSS motif in Lmo2186 may correspond to the NPQTN motif thought to be recognized by SrtB in IsdC (Fig. 1C).
Downstream from the svp genes are three genes homologous to components of a putative ferric hydroxamate ABC transporter (see reference 41 for a review). lmo2184 is predicted to encode a lipoprotein, lmo2183 is predicted to encode a permease, and lmo2182 is predicted to encode an ATP-binding protein. The first two share 49 and 38% identity, respectively, with the isdE and isdF genes in the isd locus of S. aureus, but the third gene has no homologue in S. aureus. The last gene of the locus, lmo2180, shares homology with the gene encoding the Gp46 protein of Listeria phage 2389 (GenBank/EMBL accession no. NC_003291), a protein of unknown function. The region upstream of this operon does not contain genes homologous to isdA and isdB in S. aureus but instead contains a short open reading frame (ORF) of unknown function and several ORFs homologous either to competence genes, including the negative regulator mecA, or to genes involved in amino acid transport and metabolism. Lastly, downstream of the operon are two genes encoding surface proteins with LPXTG motifs, but these display no homology with the LPXTG proteins IsdA and IsdB of S. aureus (32).
The organization of the isd locus of S. aureus is also partially conserved in the gram-positive extremophile B. halodurans as well as in B. anthracis (34, 36). A closer examination of the locus in B. halodurans revealed that the first two genes encode proteins of 221 and 1,071 residues, respectively, that share significant similarity with one another (35.1% identity), as in the L. monocytogenes srtB region. Moreover, the large protein shows homology to SvpA (up to 45% identity between residues 365 to 494 of SvpA and residues 385 to 511 encoded by the B. halodurans orf). Alignment of the C-terminal domains of the two proteins of B. halodurans indicates that the NPKTG motif of the short protein corresponds to an NSKTA motif in the large protein (sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org).
srtB is transcribed in bacteria grown in BHI at 37°C.
We analyzed the transcription of the srtB region by RT-PCR (Fig. 1D). In the wild-type strain, EGD-e, grown under laboratory conditions (see Materials and Methods), the srtB gene is transcribed, suggesting that it encodes a functional protein. The srtB gene is cotranscribed with the downstream gene lmo2180 and with the third gene of the putative ABC transporter (lmo2182). Finally, the first two genes of the locus, lmo2186 and svpA, and the first gene of the ABC transporter (lmo2184) are all cotranscribed. Taken together, these data suggest that this seven-gene cluster is a single transcriptional unit.
Inactivation of L. monocytogenes srtB in wild-type and ΔsrtA strains.
To analyze the function of the L. monocytogenes SrtB protein, we inactivated the srtB gene in strain EGD-e by introduction of an in-frame deletion, as described previously (5). We also generated the double ΔsrtA ΔsrtB mutant. PCR amplifications confirmed the deletion of srtB or srtA coding sequences in these strains (Fig. 1E and data not shown). The two strains displayed no growth defects in BHI medium, indicating that viability and cell division were not affected under this growth condition. To address a putative modification of the cell wall in the sortase mutants, we also tested their susceptibilities to penicillin and vancomycin, two antibiotics that alter cell wall synthesis, by using disk diffusion tests. We found no differences between the wild-type strain and the ΔsrtA, ΔsrtB, or ΔsrtA ΔsrtB mutant (data not shown). These results indicate that the sortase mutants have no obvious morphological cell wall alterations.
SrtA has previously been shown to target LPXTG proteins, including the invasion protein InlA (5). We therefore tested whether SrtB had an effect on InlA anchoring. The amounts of InlA in membranes and highly pure PG fractions from the wild-type, ΔsrtA, ΔsrtB, and ΔsrtA ΔsrtB strains, obtained by a fractionation procedure described previously (5), were determined by Western blotting. This set of samples was also probed with antibodies directed against p60 and InlB, two proteins known to be present in membrane fractions (10). As shown in Fig. 2A, InlA was present in the PG extracts of the wild-type and ΔsrtB strains, whereas, as expected, it was totally absent in cell walls from ΔsrtA and ΔsrtA ΔsrtB mutants. Membrane proteins did not appear in the PG fraction, indicating that there was no cross-contamination between membrane and PG fractions. This result demonstrates that the listerial SrtB protein is not involved in the anchoring of InlA to PG.
FIG. 2.
Roles of SrtA and SrtB in anchoring Listeria surface proteins to the PG. (A) Cultures of wild-type (wt), ΔsrtA, ΔsrtB, and ΔsrtA ΔsrtB EDGe strains were fractionated into membrane (M) and highly pure PG fractions, and proteins were detected by immunoblotting with anti-InlA, -p60, or -InlB antibodies. InlA is missorted in the ΔsrtA and ΔsrtA ΔsrtB mutants. Membrane-associated p60 and InlB proteins do not appear in PG fractions. (B) Immunofluorescence analysis of surface-bound proteins recognized by the polyclonal antiserum 839, which was raised against purified macromolecular L. monocytogenes PG. The labeling is mostly polar (arrows). A lack of SrtA notably reduces the amount and number of proteins that associate with PG, whereas a lack of both SrtA and SrtB fully suppresses the detection of these proteins. (C) Immunoblots of highly pure PG and supernatant (S) fractions, prepared from wild type, ΔsrtA, ΔsrtB, ΔsrtA ΔsrtB, ΔsrtA ΔsrtB/pP1srtA, and ΔsrtA ΔsrtB/pP1srtB EDG-e strains and probed with an anti-PG antiserum as previously described (5). The 60- to 70-kDa polypeptide is present in ΔsrtA and absent in ΔsrtA ΔsrtB strains. MW, molecular weight (in thousands).
SrtA anchors most of the PG-associated proteins.
To determine whether PG-associated proteins distinct from InlA were missorted at the surfaces of ΔsrtB mutant cells, we used an antiserum raised against proteins present in purified L. monocytogenes PG fractions (5) to analyze the surfaces of wild-type, ΔsrtA, ΔsrtB, and ΔsrtA ΔsrtB cells by immunofluorescence. In all cases, bacteria were boiled in the presence of 4% SDS prior to fixation and antibody labeling (see Materials and Methods). As shown in Fig. 2B, the anti-PG antiserum strongly labeled the wild-type and ΔsrtB strains, whereas it only weakly labeled the ΔsrtA mutant. No signal could be detected at the surface of the ΔsrtA ΔsrtB double mutant. These results suggested that SrtB anchors only a small subset of surface proteins, whereas SrtA sorts most of the proteins to the PG. These results were confirmed by Western blot analysis of PG and supernatant fractions. Indeed, a small amount of proteins recognized by the anti-PG antiserum was present in PG extracts prepared from the ΔsrtA mutant, in contrast to the amounts in PG extracts of wild-type and ΔsrtB strains (Fig. 2C). Interestingly, polypeptides remaining in the PG extracts from the ΔsrtA mutant, including a major protein of about 60 to 70 kDa, were absent in the ΔsrtA ΔsrtB strain extracts, suggesting that they could be targets of SrtB. No immunoreactive species were detected in PG extracts of the double ΔsrtA ΔsrtB mutant.
The ΔsrtA ΔsrtB strain was complemented for sortase expression with a plasmid expressing either the srtA gene (pP1srtA [5]) or the srtB gene (pP1srtB [this study]), placed under the control of a constitutive promoter. The antiserum specific for the listerial PG fraction recognized a pattern of several proteins of diverse apparent molecular weights in the PG fraction from the ΔsrtA ΔsrtB strain expressing SrtA, in contrast to the ΔsrtA ΔsrtB strain expressing SrtB, in which only a faint smear was detected (Fig. 2C).
Altogether, these results confirmed that SrtA is required for efficient sorting of most of the PG-bound proteins, and they demonstrated that a lack of SrtB affects the association with PG of only a relatively small number of protein species. In addition, the absence of proteins in PG extracts from the double ΔsrtA ΔsrtB mutant supports the notion that there is no other sortase activity in L. monocytogenes.
Inactivation of srtB abolishes association of SvpA with the bacterial surface.
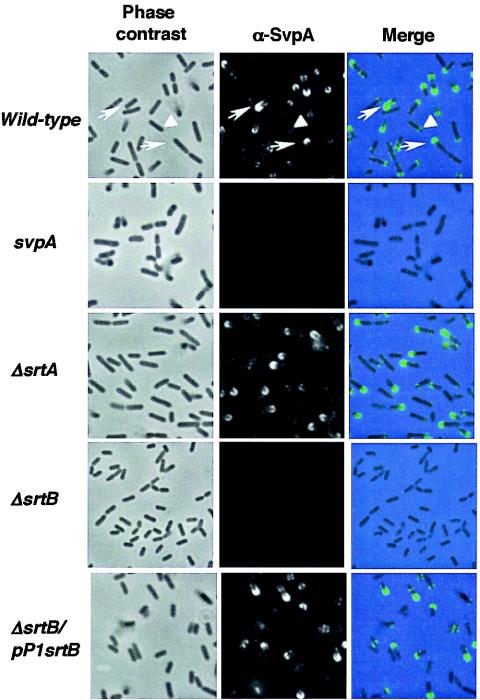
Because of the similarity between the region of srtB in L. monocytogenes and the isd locus in S. aureus, encoding both SrtB and its target, IsdC (Fig. 1A), it is possible that SvpA and Lmo2186, the first proteins encoded by the region of srtB, are substrates of the listerial SrtB. Moreover, SvpA and Lmo2186 have sequence similarities to IsdC, although they do not bear an NPQTN sorting motif. SvpA is located at the bacterial surface of L. monocytogenes strain L028 (8). Because a polyclonal anti-SvpA antibody was available, we focused our analysis on this protein. By immunofluorescence, we detected SvpA at the surface of the L. monocytogenes EGD-e wild-type strain, both in exponential phase (Fig. 3) and in stationary phase (data not shown). Notably, SvpA was detected either at one pole of the bacterium or laterally, and the intensity of labeling by the anti-SvpA antibody was heterogeneous in the bacterial population. To confirm the anti-SvpA labeling specificities, we generated an allelic svpA mutant of strain EGD-e (see Materials and Methods). As expected, the antibody did not label the surface of the svpA mutant strain. We addressed the role of the sortases SrtA and SrtB in SvpA anchoring. SvpA was no longer detected at the surface of the ΔsrtB mutant. In contrast, SvpA was present at the surface of the ΔsrtA mutant as well as at that of the ΔsrtB/pP1srtB complemented strain, demonstrating that expression of srtB in the ΔsrtB mutant restored surface anchoring (Fig. 3). These results indicate that expression of SrtB is required for the association of SvpA with the bacterial surface.
FIG. 3.
Display of SvpA on the surfaces of listeriae grown in BHI. The EDGe wild type, ΔsrtA, ΔsrtB, and svpA strains were analyzed by phase-contrast and immunofluorescence staining with the SvpA-specific polyclonal antibody. The merge image shows the phase contrast in blue and the SvpA labeling in green. SvpA is polarly (arrows) or laterally (arrowheads) localized at cell surfaces in the wild-type bacterial population. Inactivation of srtB abolished SvpA surface association, and complementation of the mutation restored it.
SrtB anchors SvpA to the cell wall.
To investigate whether SvpA is anchored to the PG, we studied the localization of SvpA in cytoplasmic, membrane, PG, and supernatant fractions. A major polypeptide of approximately 66 kDa, a molecular mass compatible with the predicted molecular mass of 63 kDa, was specifically detected in all fractions from the wild-type strain and was absent in the corresponding extracts from the svpA mutant (Fig. 4). This result indicates that SvpA is present in the cell wall. However, since the amount of the supernatant fraction loaded in gels was 50-fold diluted compared to that of PG fractions, the amount of SvpA found in PG extracts was small, representing only ∼20% of the whole expressed protein. Therefore, the bacterium retains only a small amount of SvpA at the surface, at least for bacteria grown in BHI in vitro. In addition to the 66-kDa protein, immunoreactive species of apparently lower molecular weight were also detected in PG fractions from SrtB-expressing bacteria (Fig. 5A). All of these anti-SvpA immunoreactive species were absent in extracts from the svpA mutant, suggesting that they correspond to proteolytic degradation products of SvpA.
FIG. 4.
Localization of SvpA in bacterial compartments. Cytoplasmic (C), membrane (M), PG, and supernatant (Sp) fractions of wild-type (wt) and svpA EGD-e strains were analyzed by Western blotting with an anti-SvpA polyclonal antibody. A major polypeptide of ∼66 kDa is specifically detected in extracts from the wild-type strain. The Sp fraction is 50-fold diluted with respect to the other fractions.
FIG. 5.
SrtB is required for SvpA anchoring to PG. (A) PG fractions (as in Fig. 2) of wild-type EDG-e, svpA, ΔsrtA, ΔsrtB, ΔsrtA ΔsrtB, ΔsrtA/pP1srtB, ΔsrtB/pP1srtB, and ΔsrtA ΔsrtB/pP1srtB strains were analyzed by Western blotting with anti-SvpA (top) or anti-InlA (bottom) antibodies by using the Tris-Tricine electrophoresis system to visualize proteins from high to low molecular weights. Inactivation of srtB abolishes the anchoring of SvpA to PG. MW, molecular weight (in thousands). (B) Supernatant fractions of the same strains were analyzed by Western blotting with the anti-SvpA antibody by using the Tris-glycine system to separate proteins in the 90- to 40-kDa range. The SvpA forms present in the samples of ΔsrtB and ΔsrtA ΔsrtB strains (labeled as SvpA*) run with higher mobility than those detected in all other strains (labeled as SvpA), which express SrtB. A 60-kDa band (indicated by the star) that appears in all strains, including the svpA mutant, is nonspecific.
We then tested whether the absence of sortases in L. monocytogenes affected the localization of SvpA in PG fractions. In agreement with the immunofluorescence experiments, PG extracts from the ΔsrtB mutant displayed undetectable levels of SvpA, whereas they contained InlA in amounts comparable to those in the wild-type strain (Fig. 5A). Conversely, PG extracts from the ΔsrtA mutant displayed SvpA but no InlA. PG extracts from the ΔsrtA ΔsrtB mutant displayed no detectable levels of either of the two proteins. These results indicate that the presence of SvpA in PG fractions is a SrtB-dependent process.
Complementation of ΔsrtB and ΔsrtA ΔsrtB strains with pP1srtB restored SvpA anchoring to the PG. However, amounts of the full-length protein in PG fractions from these complemented mutants were smaller than those in the wild-type or ΔsrtA strain. A similar effect was observed with the ΔsrtA strain expressing SrtB. This partial complementation may be due either to an overexpression of SrtB in these strains that would interfere with the normal anchoring process or to instability of SrtB.
SvpA species present in PG and supernatant fractions of strains that have a functional SrtB are probably linked to muropeptide fragments, as a result of muramidase digestion of the cell wall (for PG extracts) or of PG turnover (for supernatant extracts). In the absence of SrtB, the secreted SvpA should not be linked to PG residues. Therefore, one would expect to detect SvpA in the supernatants of ΔsrtB or ΔsrtA ΔsrtB mutants at a molecular weight lower than that detected in supernatants of wild-type and ΔsrtA strains. To address this hypothesis, we performed SvpA immunoblotting of supernatant fractions, using 7% acrylamide-Tris-glycine gels in order to optimize the separation of protein bands in the 90- to 40-kDa range (see Materials and Methods). Under these conditions, the SvpA forms present in the supernatant fractions of ΔsrtB and ΔsrtA ΔsrtB strains ran with a higher mobility than those detected in all other strains (Fig. 5B, SvpA*). This result strongly suggests that the SvpA species observed in the supernatant fractions of strains lacking SrtB are not bound to muropeptide molecules.
Overall, these results demonstrate that anchoring of SvpA to the bacterial cell wall is specifically mediated by SrtB.
SrtB-mediated SvpA anchoring is not required for the intracellular infectious process.
In order to determine whether inactivation of srtB could affect the cellular infectious process of L. monocytogenes, we first used the gentamicin survival assay (5) to investigate whether the ΔsrtB and ΔsrtA ΔsrtB mutants displayed any defect in the ability to invade epithelial cells (Caco-2 and Vero) as well as macrophages. Entry of the ΔsrtB mutant into all cell lines tested was not impaired and was similar to that of the wild-type strain (data not shown). In contrast, entry of the ΔsrtA ΔsrtB mutant was significantly decreased only in Caco-2 cells, to the level of entry of ΔsrtA and ΔinlA mutants. Therefore it is evident that inactivation of srtB in L. monocytogenes has no detectable effect on bacterial entry in the cellular systems tested.
Although inactivation of svpA did not affect the entry process, an svpA mutant of L. monocytogenes strain L028 was reported earlier to be more susceptible than wild-type bacteria to the bactericidal activity of phagosomes in bone marrow macrophages (8). We therefore compared the intracellular replication of isogenic EGD-e svpA, ΔsrtB, ΔsrtA, and ΔsrtA ΔsrtB mutants to that of wild-type EGD-e in RAW 264.7 macrophages during 8 h of infection (Fig. 6). Wild-type bacteria rapidly multiplied intracellularly, while svpA mutant bacteria were partially killed in the first 4 h of infection and surviving bacteria started to multiply thereafter, in agreement with previous observations (8). In contrast, intracellular growth of ΔsrtA, ΔsrtB, and ΔsrtA ΔsrtB mutants was identical to that of the wild-type strain. Similar results were obtained with epithelial cells (data not shown).
FIG. 6.
SrtB-mediated anchoring of SvpA is not required for bacterial growth in macrophages. RAW 264.7 macrophages were exposed to wild-type EGD-e bacteria (squares) or to the ΔsrtA (circles), ΔsrtB (triangles), or svpA (diamonds) mutant for 15 min at 4°C. After 45 min of internalization at 37°C, gentamicin was added to kill extracellular bacteria, and bacterial survival was monitored for 8 h postinfection. In contrast to that of the svpA mutant, the intracellular multiplication of the ΔsrtB mutant was not affected.
All the sortase mutants were also analyzed by immunofluorescence for actin tail formation in J774 and RAW 264.7 macrophages and in Vero cells. None of them displayed any obvious difference from the wild-type strain in the actin-based motility process (data not shown). These data indicate that inactivation of sortases does not affect the intracytosolic multiplication or motility of L. monocytogenes, at least in these cell types.
Effects of srtB inactivation on bacterial virulence.
ΔsrtA and svpA mutants have been shown to have impaired virulence in the mouse model of infection (5, 20). The role of SrtB in the virulence of L. monocytogenes was studied by determining the LD50s of the sortase mutants of EGD-e described in this work, after i.v. inoculation of Swiss mice. The LD50 of the ΔsrtB mutant was 104.5 per mouse, e.g., indistinguishable from that of the wild-type strain. In contrast, the LD50 of the ΔsrtA ΔsrtB double mutant, 105.75 per mouse, was ∼1.2 log units higher than that of the parental strain (and identical to that of the ΔsrtA single mutant), reflecting a moderate alteration of bacterial virulence. This result shows that the sortase encoded by srtB does not participate in the virulence of L. monocytogenes, at least in the mouse model of infection by the i.v. route.
The ability of the ΔsrtB and ΔsrtA ΔsrtB mutants to multiply in the target organs of infected mice was also evaluated. Mice were infected i.v. with 105 bacteria per mouse, and bacteria were recovered in the spleens 3 days after inoculation (the time corresponding to the peak of infection). Confirming earlier observations (20), the number of ΔsrtA mutant bacteria recovered per organ (107.22 ± 0.5 per mouse) was not significantly lower than the number recorded for the wild-type strain (107.3 ± 0.11 per mouse), and the counts recorded for the ΔsrtB mutant were very similar (107.25 ± 0.3 per mouse). With the ΔsrtA ΔsrtB mutant, the bacterial counts in the spleen were slightly lower (106.8 ± 0.09 per mouse) but not significantly different from those recorded for the two single mutants.
DISCUSSION
L. monocytogenes expresses two sortases that contribute differently to the anchoring of surface proteins. SrtA sorts most of the PG-associated proteins, including internalin and probably all the other LPXTG-containing proteins, while SrtB apparently targets only a few surface proteins, including the SvpA protein, which is not an LPXTG protein. Inactivation of srtB does not impair intracellular survival in vitro and does not attenuate bacterial virulence in the mouse, suggesting that the anchorage of SvpA to the cell surface does not play a role in bacterial survival inside cells.
Genes with homology to srtA occur in a variety of gram-positive bacteria, usually with more than one sortase-encoding gene homologue in each genome (40). However, genomes may contain several paralogous copies of srtA, encoding sortases with the same specificity (e.g., responsible for the attachment of LPXTG proteins), but also srt genes expressing sortases with different specificities (e.g., recognizing different target proteins).
Together with the identification of IsdC as a target of SrtB in S. aureus (34), our work indicates that class B sortases are responsible for the anchoring of a small subset of cell wall proteins in gram-positive bacteria. We show that the listerial SrtB mediates the sorting of at least one protein, SvpA, which does not bear the classical LPXTG motif. As in S. aureus, the gene encoding SrtB and the gene encoding its target, SvpA, are both part of the same locus. Amino acid sequence similarities exist between the products of the first two ORFs of the svp operon, SvpA and Lmo2186, and IsdC of the S. aureus isd locus. However, only three of the five residues constituting the S. aureus probable SrtB recognition motif are conserved in the C-terminal part of SvpA (residues N, T, and N [Fig. 1C]). As mentioned earlier, the genomic organization of the S. aureus isd locus is also partially conserved in B. halodurans and B. anthracis, and in these two organisms, the IsdC homologues contain putative SrtB recognition sequences (NPQTG and NPKTG, respectively). These observations support the hypothesis that the recognition motif of SrtB family members may be less stringent than that recognized by SrtA. This lower stringency may decrease the efficiency of SrtB enzymatic cleavage. The exact mechanism of SrtB-dependent cleavage in L. monocytogenes will require further biochemical characterization. Our results suggest that SrtB anchors a small number of proteins. We searched in the L. monocytogenes genome for proteins containing an NXXTX sequence upstream of a hydrophobic domain and a charged tail in the C-terminal part. Since only SvpA and Lmo2186 were found, it is tempting to speculate that SrtB may sort only these two proteins.
Immunofluorescence experiments showed that SvpA was detected at the bacterial surface in a SrtB-dependent manner. The anchoring of SvpA to PG was further studied by fractionation experiments and analyses of pure PG preparations of the different mutants constructed. These studies demonstrated that only a fraction of SvpA was anchored by SrtB to the PG. The fact that the extracellular species of SvpA displays a mobility similar to that of the species observed in the PG fraction argues for a release of SvpA from the cell wall as the result of PG turnover, as has been described for internalin (26).
What is the role of SrtB in the environmental adaptation of L. monocytogenes? During the course of our study, Schneewind and coworkers demonstrated that the S. aureus isd genes were regulated by iron (34) and encoded factors responsible for hemoglobin binding and the passage of heme iron to the cytoplasm (32), thus constituting a potential source of iron for this extracellular pathogen. The isd locus of S. aureus, initially described by Mazmanian et al. in 2002 (34), was also described in two other papers published the same year, but with different names (frp genes in reference 36; stbA, srH, and sir genes in reference 45). The genomic similarities between the region of srtB in L. monocytogenes and the S. aureus isd locus suggest that the svp-srtB region might also participate in iron utilization. However, the infectious processes of S. aureus and L. monocytogenes are distinct (extracellular versus intracellular), implying different requirements for iron, different sources of iron, and different mechanisms of iron acquisition. Although both pathogens may lyse erythrocytes, and therefore acquire hemoglobin, it is unlikely that this represents a significant source of iron for L. monocytogenes. Indeed, the life cycle of L. monocytogenes is predominantly intracellular during the whole infectious process in vivo, and bacteremia occurs only transiently, preceding the crossing of the blood-brain barrier (4). Addressing the role of the svp locus in iron acquisition may thus reveal unique features that are specific to Listeria.
We show here that inactivation of srtB does not alter the intracellular survival of L. monocytogenes in several cellular models and has no detectable effect on virulence in the mouse model. These results indicate that the SrtB-dependent anchorage of SvpA to the cell wall is not crucial for these processes. The svpA mutant of L. monocytogenes shows a severe defect in intracellular survival in bone marrow-derived macrophages and has reduced virulence in mice (8). It is possible that the reduced virulence of the svpA mutant might be due solely to its restricted growth capacity.
The facts that most SvpA is secreted in the medium and that the surface-bound fraction is not evenly distributed on the surface of the bacterium imply a complex temporal and spatial control of its expression and of its anchoring at the bacterial surface. The surface-anchored SvpA may be functional when the microorganism encounters particular extracellular conditions. It will therefore be interesting to study the expression of srtB and of the whole svp operon under different growth conditions, especially under iron-controlled conditions.
Several lines of evidence suggest that the regulation of the svpA region might be controlled not only by iron availability but also by stress response regulatory genes. Indeed, it was previously shown that expression of SvpA is reduced by MecA and the stress proteins ClpC and ClpP (7, 8). In Bacillus subtilis, the adaptor protein MecA, through interaction with ClpC, plays a dual role in the protein quality control network by rescuing or, together with ClpP, by degrading misfolded and/or aggregated proteins (43). It is of interest in this respect that the gene encoding MecA is located in the svp operon upstream region. Therefore, deciphering the links between this complex regulatory network, the srtB region, and the mechanisms of iron acquisition by L. monocytogenes will be an important issue.
In conclusion, SrtB represents a second class of sortase in L. monocytogenes, involved in the attachment of SvpA to the cell wall, most likely by recognizing an NXXTN sorting motif. L. monocytogenes is the second gram-positive organism, after S. aureus, and the first intracellular pathogen shown to express two different types of sorting enzymes, a property that may be shared by other gram-positive bacterial species. Inactivation of srtB does not affect listerial virulence in the mouse model, as assessed by LD50 or bacterial counts in the spleen after i.v. infection. In S. aureus, inactivation of srtB has no detectable effect on the establishment of infection, but it does have an effect on long-term persistence (34). At present, the possibility cannot be excluded that during a natural oral infection in humans, an active srtB allele might be required to promote full virulence. Future work will therefore be required to address the contributions of the different sortases in listeriosis by using animal models (27) that may be more relevant for human listeriosis than the mouse model. Lastly, Listeria exists ubiquitously in several ecological niches, and SrtB maybe required to anchor proteins involved in the adaptation of Listeria to particular environmental conditions.
Acknowledgments
We thank Nuria Goméz-López for technical assistance, Phillip Klebba for careful reading of the manuscript, and Elise Borezee and Patrick Berche for the kind gifts of plasmid pAUL-A-svpAΩaphA3 and the anti-SvpA antibody.
Work in the laboratory of A. Charbit was supported by CNRS, INSERM, and University Paris V. S. Newton was supported by a “Poste Orange” from INSERM and by grant 6074 from the Oklahoma Center for the Advancement of Science and Technology (OCAST). Work in the laboratory of F. Garcia-del Portillo is supported by the European Union (grant QLG2-CT-1999-00932). Work in the laboratory of P. Cossart is supported by the Ministère de l'Education Nationale et de la Recherche Scientifique et Technique and the Pasteur Institute. H. Bierne is on the staff of the Institut National de la Recherche Agronomique. P. Cossart is an international research investigator from the Howard Hughes Medical Institute.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl. 1990. Current protocols in molecular biology. Wiley Interscience, New York, N.Y.
- 2.Autret, N., I. Dubail, P. Trieu-Cuot, P. Berche, and A. Charbit. 2001. Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis. Infect. Immun. 69:2054-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, T. C., and J. R. Scott. 2002. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J. Bacteriol. 184:2181-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berche, P. 1995. Bacteremia is required for invasion of the murine central nervous system by Listeria monocytogenes. Microb. Pathog. 18:323-336. [DOI] [PubMed] [Google Scholar]
- 5.Bierne, H., S. K. Mazmanian, M. Trost, M. G. Pucciarelli, G. Liu, P. Dehoux, L. Jansch, F. G. Portillo, O. Schneewind, and P. Cossart. 2002. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 43:869-881. [DOI] [PubMed] [Google Scholar]
- 6.Bolken, T. C., C. A. Franke, K. F. Jones, G. O. Zeller, C. H. Jones, E. K. Dutton, and D. E. Hruby. 2001. Inactivation of the srtA gene in Streptococcus gordonii inhibits cell wall anchoring of surface proteins and decreases in vitro and in vivo adhesion. Infect. Immun. 69:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borezee, E., T. Msadek, L. Durant, and P. Berche. 2000. Identification in Listeria monocytogenes of MecA, a homologue of the Bacillus subtilis competence regulatory protein. J. Bacteriol. 182:5931-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borezee, E., E. Pellegrini, J. L. Beretti, and P. Berche. 2001. SvpA, a novel surface virulence-associated protein required for intracellular survival of Listeria monocytogenes. Microbiology 147:2913-2923. [DOI] [PubMed] [Google Scholar]
- 9.Braun, L., F. Nato, B. Payrastre, J. C. Mazie, and P. Cossart. 1999. The 213-amino-acid leucine-rich repeat region of the Listeria monocytogenes InlB protein is sufficient for entry into mammalian cells, stimulation of PI 3-kinase and membrane ruffling. Mol. Microbiol. 34:10-23. [DOI] [PubMed] [Google Scholar]
- 10.Cabanes, D., P. Dehoux, O. Dussurget, L. Frangeul, and P. Cossart. 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 10:238-245. [DOI] [PubMed] [Google Scholar]
- 11.Cossart, P. 2000. Actin-based motility of pathogens: the Arp2/3 complex is a central player. Cell Microbiol. 2:195-205. [DOI] [PubMed] [Google Scholar]
- 12.Cossart, P., and H. Bierne. 2001. The use of host cell machinery in the pathogenesis of Listeria monocytogenes. Curr. Opin. Immunol. 13:96-103. [DOI] [PubMed] [Google Scholar]
- 13.Cossart, P., and R. Jonquieres. 2000. Sortase, a universal target for therapeutic agents against gram-positive bacteria? Proc. Natl. Acad. Sci. USA 97:5013-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhar, G., K. F. Faull, and O. Schneewind. 2000. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry 39:3725-3733. [DOI] [PubMed] [Google Scholar]
- 15.Dramsi, S., I. Biswas, E. Maguin, L. Braun, P. Mastroeni, and P. Cossart. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Mol. Microbiol. 16:251-261. [DOI] [PubMed] [Google Scholar]
- 16.Dramsi, S., P. Dehoux, M. Lebrun, P. L. Goossens, and P. Cossart. 1997. Identification of four new members of the internalin multigene family of Listeria monocytogenes EGD. Infect. Immun. 65:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fortineau, N., P. Trieu-Cuot, O. Gaillot, E. Pellegrini, P. Berche, and J. L. Gaillard. 2000. Optimization of green fluorescent protein expression vectors for in vitro and in vivo detection of Listeria monocytogenes. Res. Microbiol. 151:353-360. [DOI] [PubMed] [Google Scholar]
- 19.Gaillard, J. L., P. Berche, J. Mounier, S. Richard, and P. Sansonetti. 1987. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect. Immun. 55:2822-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garandeau, C., H. Reglier-Poupet, I. Dubail, J. L. Beretti, P. Berche, and A. Charbit. 2002. The sortase SrtA of Listeria monocytogenes is involved in processing of internalin and in virulence. Infect. Immun. 70:1382-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 22.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 23.Kaniga, K., D. Trillinger, and J. E. Galan. 1995. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 177:7078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kharat, A. S., and A. Tomasz. 2003. Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro. Infect. Immun. 71:2758-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kocks, C., E. Gouin, M. Tabouret, P. Berche, H. Ohayon, and P. Cossart. 1992. Listeria monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68:521-531. [DOI] [PubMed] [Google Scholar]
- 26.Lebrun, M., J. Mengaud, H. Ohayon, F. Nato, and P. Cossart. 1996. Internalin must be on the bacterial surface to mediate entry of Listeria monocytogenes into epithelial cells. Mol. Microbiol. 21:579-592. [DOI] [PubMed] [Google Scholar]
- 27.Lecuit, M., and P. Cossart. 2002. Genetically-modified-animal models for human infections: the Listeria paradigm. Trends Mol. Med. 8:537-542. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S. F., and T. L. Boran. 2003. Roles of sortase in surface expression of the major protein adhesin P1, saliva-induced aggregation and adherence, and cariogenicity of Streptococcus mutans. Infect. Immun. 71:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackaness, G. B. 1962. Cellular resistance to infection. J. Exp. Med. 116:381-406. [PubMed] [Google Scholar]
- 30.Mazmanian, S. K., G. Liu, E. R. Jensen, E. Lenoy, and O. Schneewind. 2000. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. USA 97:5510-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 32.Mazmanian, S. K., E. P. Skaar, A. H. Gaspar, M. Humayun, P. Gornicki, J. Jelenska, A. Joachmiak, D. M. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906-909. [DOI] [PubMed] [Google Scholar]
- 33.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 34.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 99:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mengaud, J., M. Lecuit, M. Lebrun, F. Nato, J. C. Mazie, and P. Cossart. 1996. Antibodies to the leucine-rich repeat region of internalin block entry of Listeria monocytogenes into cells expressing E-cadherin. Infect. Immun. 64:5430-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrissey, J. A., A. Cockayne, J. Hammacott, K. Bishop, A. Denman-Johnson, P. J. Hill, and P. Williams. 2002. Conservation, surface exposure, and in vivo expression of the Frp family of iron-regulated cell wall proteins in Staphylococcus aureus. Infect. Immun. 70:2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osaki, M., D. Takamatsu, Y. Shimoji, and T. Sekizaki. 2003. Allelic variation in srtAs of Streptococcus suis strains. FEMS Microbiol. Lett. 219:195-201. [DOI] [PubMed] [Google Scholar]
- 39.Osaki, M., D. Takamatsu, Y. Shimoji, and T. Sekizaki. 2002. Characterization of Streptococcus suis genes encoding proteins homologous to sortase of gram-positive bacteria. J. Bacteriol. 184:971-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pallen, M. J., A. C. Lam, M. Antonio, and K. Dunbar. 2001. An embarrassment of sortases—a richness of substrates? Trends Microbiol. 9:97-101. [DOI] [PubMed] [Google Scholar]
- 41.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 42.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 43.Schlothauer, T., A. Mogk, D. A. Dougan, B. Bukau, and K. Turgay. 2003. MecA, an adaptor protein necessary for ClpC chaperone activity. Proc. Natl. Acad. Sci. USA 100:2306-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneewind, O., A. Fowler, and K. F. Faull. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103-106. [DOI] [PubMed] [Google Scholar]
- 45.Taylor, J. M., and D. E. Heinrichs. 2002. Transferrin binding in Staphylococcus aureus: involvement of a cell wall-anchored protein. Mol. Microbiol. 43:1603-1614. [DOI] [PubMed] [Google Scholar]
- 46.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, and B. Gonzalez-Zorn. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood, S., N. Maroushek, and C. J. Czuprynski. 1993. Multiplication of Listeria monocytogenes in a murine hepatocyte cell line. Infect. Immun. 61:3068-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]