Abstract
The X-ray crystal structures of homoprotocatechuate 2,3-dioxygenases isolated from Arthrobacter globiformis and Brevibacterium fuscum have been determined to high resolution. These enzymes exhibit 83% sequence identity, yet their activities depend on different transition metals, Mn2+ and Fe2+, respectively. The structures allow the origins of metal ion selectivity and aspects of the molecular mechanism to be examined in detail. The homotetrameric enzymes belong to the type I family of extradiol dioxygenases (vicinal oxygen chelate superfamily); each monomer has four βαβββ modules forming two structurally homologous N-terminal and C-terminal barrel-shaped domains. The active-site metal is located in the C-terminal barrel and is ligated by two equatorial ligands, H214NE1 and E267OE1; one axial ligand, H155NE1; and two to three water molecules. The first and second coordination spheres of these enzymes are virtually identical (root mean square difference over all atoms, 0.19 Å), suggesting that the metal selectivity must be due to changes at a significant distance from the metal and/or changes that occur during folding. The substrate (2,3-dihydroxyphenylacetate [HPCA]) chelates the metal asymmetrically at sites trans to the two imidazole ligands and interacts with a unique, mobile C-terminal loop. The loop closes over the bound substrate, presumably to seal the active site as the oxygen activation process commences. An “open” coordination site trans to E267 is the likely binding site for O2. The geometry of the enzyme-substrate complexes suggests that if a transiently formed metal-superoxide complex attacks the substrate without dissociation from the metal, it must do so at the C-3 position. Second-sphere active-site residues that are positioned to interact with the HPCA and/or bound O2 during catalysis are identified and discussed in the context of current mechanistic hypotheses.
Bacterial ring-cleaving dioxygenases are critical enzymes in the catabolism of aromatic compounds that enter the environment from a myriad of sources (9, 17, 18, 44). The substrates for most of these enzymes are ortho- or para-dihydroxylated aromatics and molecular oxygen. Both atoms of oxygen from O2 are incorporated into the product as the ring is cleaved. The resulting aliphatic products are further metabolized to intermediates of core metabolic cycles through well-established pathways (18, 55, 74).
Dioxygenases that act on ortho-dihydroxylated aromatic compounds are divided into two classes, termed intradiol and extradiol, which differ in their mode of ring cleavage and the oxidation state of the active-site metal (44, 63). Intradiol dioxygenases utilize Fe3+ and cleave the bond between the carbons bearing the two hydroxyls; extradiol dioxygenases utilize Fe2+ or, rarely, Mn2+ and cleave one of the carbon-carbon bonds adjacent to the ortho-hydroxyl substituents. Although intradiol and extradiol dioxygenases oxidize an overlapping set of substrates, they are not structurally related and are proposed to have fundamentally different mechanisms (44, 45, 61, 62). Briefly, the mechanism of the intradiol dioxygenases is thought to involve activation of the aromatic substrate for electrophilic attack by molecular oxygen. In contrast, in the extradiol dioxygenase mechanism, the substrate activates the metal center for dioxygen binding and subsequent nucleophilic attack on the aromatic substrate.
On the basis of sequence alignments, the extradiol group of dioxygenases has been divided into three families (20, 22, 73, 80). As recently reviewed by Vaillancourt et al. (80), type I includes the 2,3-dihydroxybiphenyl 1,2-dioxygenases, such as the single-domain enzyme from Rhodococcus globerulus (5) and the two-domain enzymes from Pseudomonas putida KF715 (29), Pseudomonas sp. strain KKS102 (BPHC_PS102) (39, 68), Pseudomonas cepacia LB400 (BPHC_LB400) (27, 32), and Bacillus sp. strain JF8 (BPHC_JF8) (28). Type I also includes most of the catechol 2,3-dioxygenases (2,3-CTDs), such as the xylE gene product from the TOL plasmid of P. putida mt-2 (51). A consensus sequence for the metal binding region of type I enzymes has been defined (7, 22). The type II extradiol dioxygenases include such enzymes as protocatechuate 4,5-dioxygenase (LigAB) from Pseudomonas paucimobilis (53), which has two different types of subunits, and 2,3-CTD from Alcaligenes eutrophus JMP222 (38). The type III extradiol dioxygenases include gentistate dioxygenase from Haloferax sp. strain D1227 (24), human homogentisate dioxygenase (77), and 1-hydroxy-2-naphthoate dioxygenase from Nocardioides sp. strain KP7 (35). The type III enzymes have been identified as part of the cupin superfamily (20, 21).
Extradiol dioxygenases of each type have no intense visible chromophore or electron paramagnetic resonance (EPR) spectrum, which has slowed mechanistic and structural investigations. However, early studies using type I and II enzymes showed that NO, used as an O2 mimic, binds to the active-site Fe2+ to convert it to an EPR-active form (2, 44). Hyperfine interactions with 17O-labeled solvents and substrates, detected as a broadening of this EPR spectrum, showed that at least three sites in the iron coordination can be occupied by exogenous molecules (1, 3). Solvent bound initially to the iron is displaced by the substrate as both hydroxyl groups bind. Later, magnetic circular dichroism-circular dichroism (MCD/CD) (47) and extended X-ray absorption fine structure (EXAFS) (71) studies showed that the Fe2+ ions in both the resting enzyme and the substrate complex are pentacoordinate in the absence of NO, suggesting that ligands (probably the solvents) are displaced as the substrate binds and that there is an open site that could be occupied by O2 (or NO) as part of the activation mechanism. This would bring the substrate and oxygen into close spatial and electronic proximity in a prescribed orientation that could be controlled by the enzyme. Similar mechanistic strategies (12) have recently been invoked for a wide range of nonheme iron-containing enzymes of the “2-His, 1-carboxylate facial triad” group (30) which all share the property that one face of the active-site Fe2+ coordination is bound by solvents which can be displaced by substrates as part of the catalytic cycle. The crystal structures (27, 68) of 2,3-dihydroxybiphenyl 1,2-dioxygenase bore out all of the conclusions deduced from the earlier spectroscopic studies of related extradiol dioxygenases. One important finding from the EXAFS and EPR studies of 2,3-CTD was that the substrate chelates the iron asymmetrically, probably due to the deprotonation of only one hydroxyl function (71). This could serve to direct the position of oxygen attack. Also, it may differentiate extradiol from intradiol ring cleavage mechanisms, because the substrates bind as dianionic chelate complexes in the latter enzyme class (56).
Insight into the structural details of the extradiol dioxygenases has recently been gained through additional X-ray crystallographic studies. The majority of extradiol enzymes that have been structurally characterized belong to type I. Type I enzymes can be further subdivided based on their preference for substrates that have one or two aromatic rings. The crystal structure of at least one member of each subclass has been solved: P. putida mt-2 2,3-CTD (40), which catalyzes the cleavage of monocyclic aromatics, and BPHC_LB400 (27) and BPHC_PS102 (68), which catalyze the cleavage of bicyclic aromatics. These structures demonstrate that these enzymes have a common fold, though different oligomeric structures. The different folds found in the type II extradiol dioxygenase LigAB (76) and the type III extradiol dioxygenase homogentisate dioxygenase (77) suggest that these types arose independently of the type I enzymes. However, all three types appear to share similar active-site features and probably similar catalytic mechanisms.
Although the spectroscopic and crystallographic structural studies conducted thus far are generally in agreement, several questions remain that can potentially be addressed through structural studies of additional extradiol enzymes. Among these are the origin of the mechanistically relevant asymmetry in substrate binding in the active site, the identities and roles of the second-sphere active-site residues that may participate in catalysis, a rationale for the strict specificity of the ring cleavage site for extradiol and intradiol dioxygenases, and the origin of the remarkable ability of the homologous extradiol dioxygenases to select between iron and manganese as an active-site metal. In this study, we examine these aspects of extradiol dioxygenase catalysis by using two enzymes that both catalyze the oxygen-dependent extradiol cleavage of 3,4-dihydroxyphenylacetate (also known as homoprotocatechuate [HPCA]) to produce 5-carboxymethyl-2-hydroxymuconic semialdehyde. Interestingly, homoprotocatechuate 2,3-dioxygenase (2,3-HPCD) from Arthrobacter globiformis (MndD) utilizes Mn2+ (7, 86), whereas the enzyme isolated from Brevibacterium fuscum (Bf 2,3-HPCD) utilizes Fe2+ (48, 84). Primary sequence analysis indicates that MndD has 14 of the 18 amino acid residues previously identified as conserved in the type I family (7, 32). Despite this, MndD was found to have on average only 23% sequence identity with the type I family (7). In contrast, the sequence identity between MndD and Bf 2,3-HPCD is 82%. In comparison, the Mn2+-dependent enzyme BPHC_JP8 has less than 25% sequence identity with other 2,3-dihydroxybiphenyl 2,3-dioxygenases (28) and only 30% sequence identity with MndD. A multiple sequence alignment of MndD and Bf 2,3-HPCD with several type I extradiol dioxygenases indicates low but significant sequence homology, without the need for large gaps or insertions. However, MndD and Bf 2,3-HPCD contain an additional 20 to 30 residues at the C terminus beyond those present in the majority of type I extradiol dioxygenases. Here we report the refined structures of MndD, Bf 2,3-HPCD, and their substrate complexes. Active-site residues and their possible roles are identified through comparison of these structures with those of other extradiol dioxygenases. Although some insight is gained into the origin of metal ion specificity, the true basis of this important process is shown not to lie in the final folded active-site region, which is nearly identical in the two 2,3-HPCD enzymes.
MATERIALS AND METHODS
Enzyme preparation.
Homogenous enzyme samples were obtained by using previously published procedures for the expression and purification of Bf 2,3-HPCD and MndD from the respective recombinant hosts (7, 84, 86). Enzymatic activity was monitored spectrophotometrically (41). The average preparations of Bf 2,3-HPCD and MndD used in crystallization had specific activities of 10 and 20 U/mg, respectively. Protein solutions were concentrated to 100 mg/ml and stored at 18°C under anaerobic conditions or flash frozen and stored at −80°C.
Prior to some crystallization trials, the proteins were modified proteolytically. Bovine pancreas trypsin (Sigma-Aldrich, St. Louis, Mo.) was added (final concentration, 2 mg/ml) to a concentrated solution (75 to 100 mg/ml) of Bf 2,3-HPCD or MndD. This mixture was then incubated at 18°C under an anaerobic hood for several days. Proteolysis was monitored by native polyacrylamide gel electrophoresis (PAGE) until the desired degree of cleavage was achieved (see Results for details).
Crystallization.
Bf 2,3-HPCD crystallized in two forms. The P3221 form grew as hexagonal rods (0.15 by 0.15 by 0.5 mm) in hanging drops at 18°C when equal volumes of protein (20 mg/ml) were combined with a solution containing 1.5 M (NH4)2SO4 and 100 mM morpholinepropanesulfonic acid (MOPS) (pH 7.5). The I41 form grew as bricks (0.4 by 0.4 by 0.8 mm) at 18°C. Here 10 μl of proteolytically cleaved Bf 2,3-HPCD (100 mg/ml) was combined with 90 μl of a stock solution consisting of 10 to 15% polyethylene glycol 5000 monomethyl ether (MME), 0.2 M magnesium acetate, and 100 mM MOPS (pH 7.5) in a 96-well cell culture plate.
Thin plates of MndD (dimensions, 0.1 by 0.5 by 0.5 mm) grew from batch crystallization experiments with incubation at 18°C. Typically 10 μl of proteolytically cleaved MndD (100 mg/ml) was combined with 90 μl of a stock solution consisting of 6 to 10% polyethylene glycol 8000, 0.2 M magnesium acetate, and 100 mM sodium cacodylate at pH 6.5 in a 96-well cell culture plate. These crystals were found to belong to the monoclinic space group C2 with one dimer per asymmetric unit.
Data collection.
A number of the original data sets were collected at room temperature on a Siemens HI-STAR area detector and processed by using XENGEN (33). X rays were generated by a Rigaku RU200 X-ray generator equipped with a copper anode operating at 45 kV and 200 mA. These included the MndD room temperature data set (Native 1), the Bf 2,3-HPCD trigonal data set (Native 3), and all of the Bf 2,3-HPCD tetragonal data sets used in heavy atom phasing.
High-resolution, low-temperature data sets of Bf 2,3-HPCD (Native 2) and MndD (Native 2) were collected at the Argonne National Laboratory at beamline 19-ID. Data were produced by using monochromatic radiation and were collected with a 3-by-3 charge-coupled device detector. Diffraction intensities were indexed with DENZO and scaled with SCALEPACK (58). Data were collected at −80°C by using a mother liquor containing 20% glycerol.
Crystalline enzyme-substrate (ES) complexes were prepared and mounted under anaerobic conditions under an argon atmosphere in a glove box (Coy Systems). Crystals were incubated for 1 h in anaerobic solutions containing the substrate (1 mM HPCA) and their respective mother liquors and were then mounted in quartz capillaries. A layer of mother liquor and then a layer of mineral oil were added to the top of the capillaries prior to their removal from the anaerobic chamber. The capillaries were then sealed with low-melt glue to maintain anaerobic conditions during the experiment. Diffraction patterns for the anaerobic ES complexes were measured at room temperature on an R-AXIS IV++ imaging plate detector and were processed by using CRYSTALCLEAR (Molecular Structure Corporation) (59). X rays were generated by utilizing a Rigaku RU200 X-ray generator operating at 45 kV and 100 mA, equipped with a copper anode and confocal MaxFlux Blue focusing optics (Molecular Structure Corporation).
Crystals of the ES complex remained colorless during incubation with the substrate and subsequent data collection, indicating that anaerobic conditions were maintained. After data collection, the capillaries were opened to expose the crystals to O2. The crystals immediately turned an intense yellow, the color of the product of the extradiol dioxygenase reaction. This suggested that both MndD and Bf 2,3-HPCD are active within the crystalline lattice and that the metal remained in its active state.
Phasing.
Molecular replacement attempts using the structural homologues 2,3-CTD (Protein Data Bank [PDB] identifier, 1MPY) and BPHC_LB400 (PDB identifier, 1HAN) were unsuccessful. Therefore, phases for the Bf 2,3-HPCD I41 crystal form were obtained by multiple isomorphous replacement (MIR) utilizing K2HgI4, UO2(CH3COO)2, and an iodinated substrate analog (3-iodo, 4-hydroxyphenylacetate) to produce derivatives. Heavy atoms were located by difference Patterson and difference Fourier analysis using the CCP4 program suite (15); heavy atom positions were refined by using the MLPHARE program (57). Data and phasing statistics are shown in Table 1. The K2HgI4 derivative had excellent phasing statistics, especially at low resolution, producing Fourier maps with clear boundaries for solvent and protein. Although UO2(CH3COO)2 and the iodinated substrate analog were very weak derivatives, they facilitated the identification of the noncrystallographic symmetry operators. The MIR phases were refined and extended to 2.5 Å by solvent flattening, histogram matching, and fourfold noncrystallographic symmetry averaging using the DM program (16). A Fourier map based on the DM-refined MIR phases was sufficient to position 96% of the Bf 2,3-HPCD sequence.
TABLE 1.
Phasing data sets for Bf 2,3-HPCDa
| Parameter | Bf 2,3-HPCD (Native 1) | K2HgI4 | UO2(CH3COO)2 | 3-Iodo-4-hydroxy phenyl acetate |
|---|---|---|---|---|
| Space group | I41 | I41 | I41 | I41 |
| Cell axes (Å) | a = b = 157.2, c = 123.8 | a = b = 157.0, c = 123.2 | a = b = 157.3, c = 123.0 | a = b = 156.3, c = 124.6 |
| Data resolution (Å) | 30-2.5 | 30-3.0 | 30-3.2 | 30-2.8 |
| Unique reflections | 51,441 | 28,827 | 24,629 | 31,615 |
| Completeness (%) | 97.4 | 94.6 | 97.7 | 86.2 |
| Redundancy | 2.60 | 3.54 | 2.96 | 3.27 |
| 〈I/σI〉b | 14.0 (1.2) | 7.2 (1.0) | 6.99 (1.1) | 12.2 (1.4) |
| Rsym (%)b | 5.5 (29.15) | 9.0 (34.57) | 9.2 (30.6) | 6.1 (22.6) |
| Phasing resolution | 30-3.0 | 30-3.0 | 10-3.2 | 30-3.5 |
| No. of binding sites | 4 | 4 | 4 | |
| Phasing power (centric/acentric)c | 0.80/0.59 | 0.43/0.28 | 0.67/0.61 |
Figure of merit (3.0 Å) centric/acentric = 0.48/0.27; cosine of the error in the phase.
Data in parentheses are for the highest-resolution bin.
Phasing power = 〈FH〉/E, where FH is the RMS heavy-atom structure factor amplitude and E is the residual lack of closure error.
Refinement.
Five to ten percent of all data sets were permanently set aside for calculation of Rfree (10). All structural models were manipulated by using the O program (37) and were refined with CNS (11). Where applicable, strict noncrystallographic symmetry restraints were used in the refinement until the final few rounds, at which point they were completely released. Solvent molecules were included in the models when peaks larger than 4σ and 1σ were found in Fo-Fc and 2Fo-Fc maps, respectively, and when the solvent molecules formed good-geometry hydrogen bonds with the protein or other solvents. The van der Waals interaction energy was turned off for all ligands to the active-site metal so that the refinement of ligand-to-metal distances was driven purely by the electron density.
The Bf 2,3-HPCD trigonal crystal form and the MndD structure were solved by molecular replacement using the CNS and AMORE (52) programs, respectively, with a molecular replacement search model from a partially refined monomer of Bf 2,3-HPCD (I41, Native 2) that had been stripped of all heteroatoms. Unambiguous solutions were obtained in both cases and were subsequently submitted to simulated annealing torsional dynamics. This reduced the initial Rfactor from 34.2 to 26.8% for the Bf 2,3-HPCD trigonal crystal form and from 37 to 28.9% for MndD. A high-resolution, low-temperature MndD data set (MndD C2, Native 2) became available at a later date and was used to complete the refinement.
Sequence correction.
Two regions of the previously published MndD sequence (7) were discovered to be in error. The new sequences for residues 230 to 255 and 335 to 360 are DKMGALRISDRIERGPGRHGVSNAFY and GADGFSYTRKDETGEAAEGFKLGAQV, respectively. The resequenced portion from residue 230 to residue 255 is consistent with the electron density found for the MndD structure for this region. No electron density was available for the second resequenced region, because these residues were removed upon trypsin digestion. However, the new sequence is more consistent (i.e., homologous) with the sequence of the C terminus of Bf 2,3-HPCD.
Accession numbers.
The atomic coordinates for the structures of Bf 2,3-HPCD and MndD have been deposited in the Protein Data Bank. Accession numbers are given in Table 2.
TABLE 2.
Data collection and refinement statistics
| Parametera | Bf 2,3-HPCD
|
MndD
|
||||
|---|---|---|---|---|---|---|
| Native 2 | Complex | Native 3 | Native 1 | Native 2 | Complex | |
| Data collection statistics | ||||||
| Space group | I41 | I41 | P3221 | C2 | C2 | C2 |
| Temp (°C) | −80 | 18 | 18 | 18 | −80 | 18 |
| Cell axes (Å) | a = 157.1 | a = 157.0 | a = 118.9 | a = 138.4 | a = 137.8 | a = 139.4 |
| c = 121.3 | c = 122.3 | c = 110.3 | b = 59.2 | b = 59.0 | b = 59.3 | |
| c = 102.8 | c = 102.3 | c = 103.2 | ||||
| Cell angles (°) | β = 118.3 | β = 119.0 | β = 118.4 | |||
| Data resolution (Å) | 20-1.5 | 20-2.1 | 20-2.3 | 20-1.75 | 20-1.50 | 20-1.90 |
| Completeness (%) | 92.9 (79.5) | 97.5 (99.3) | 80.8 (42.2) | 88.7 (65.0) | 96.9 (93.4) | 96.7 (93.6) |
| Redundancy | 2.55 (1.51) | 2.2 (2.2) | 4.8 (2.18) | 2.6 (1.4) | 3.0 (2.8) | 3.80 (3.8) |
| I/σI> | 21.2 (8.7) | 10 (3.2) | 6.7 (1.0) | 12.2 (1.0) | 20.0 (5.0) | 15.5 (6.3) |
| Rsym (%) | 4.1 (12.7) | 5.1 (21.6) | 8.9 (32.3) | 5.4 (23.9) | 8.1 (27.1) | 5.9 (23.6) |
| Refinement statistics | ||||||
| Resolution range (Å) | 20-1.6 | 30-2.1 | 20-2.3 | 20-1.8 | 20-1.5 | 20-1.9 |
| Rfactor (%)a | 16.8 (18.7) | 17.0 (24.7) | 16.1 (24.7) | 15.6 (26.5) | 16.2 (18.6) | 16.6 (25.2) |
| Rfree (%)a | 19.5 (23.9) | 21.2 (28.2) | 20.8 (29.3) | 18.7 (29.8) | 18.5 (20.5) | 20.0 (29.6) |
| No. of non-hydrogen atoms | 11,517 | 10,883 | 5,957 | 5,547 | 5,868 | 5,538 |
| No. of solvent molecules | 1,105 | 373 | 193 | 351 | 665 | 316 |
| Average B (Å2) | ||||||
| Main chain | 15.98 | 37.7 | 14.7 | 12.6 | 8.6 | 21.9 |
| Side chain | 18.4 | 39.7 | 15.8 | 15.2 | 11.0 | 24.0 |
| Waters/Fe | 27.6/23.8 | 43.0/39.5 | 19.6/17.2 | 25.5/14.9 | 19.8/16.0 | 35.0/18.6 |
| Ligand | 46.1 | 29.3 | ||||
| Iron occupancy | 1.0 | 1.0 | 1.0 | 0.5 | 0.4 | 1.0 |
| RMSD | ||||||
| Bond (Å) | 0.023 | 0.016 | 0.013 | 0.020 | 0.022 | 0.019 |
| Angle (°) | 2.14 | 1.91 | 1.76 | 2.00 | 2.10 | 1.90 |
| PDB identifier | 1F1X | 1Q0C | 1O0O | 1F1R | 1F1U | 1F1V |
Data in parentheses are for the highest-resolution bin. RMSD, root mean square difference.
RESULTS
Protein analysis.
Initial attempts to obtain crystals of either Bf 2,3-HPCD or MndD suitable for structural analysis were unsuccessful, producing malformed crystal plates (MndD) or a triclinic crystal form [Bf 2,3-HPCD (Native 3)] that could not be reproduced in quantity. Native PAGE of purified Bf 2,3-HPCD or MndD showed a very diffuse band, suggesting that the enzymes were present in more than one form.
It was noted that a proteolytic fragment of MndD occasionally appeared during purification and that when this occurred, better crystals were obtained. Controlled digestion by trypsin of Bf 2,3-HPCD and MndD produced more-homogeneous enzymes as judged by native PAGE. Trypsin-treated Bf 2,3-HPCD grew in a new crystal form (I41). Both cleaved proteins were active and had no substantial change in Vmax or Km.
Overall model quality.
All models presented in this study were refined against their respective data sets to Rfactors less than 18.0% and Rfrees less than 22.0%. Structural analysis using the PROCHECK program (42) demonstrated that all models conformed to typical protein stereochemistry and exhibited >90% of their residues in the most favored or allowed regions of a Ramachandran plot. Refinement statistics for the various models are shown in Table 2.
In general, the first two to three residues of Bf 2,3-HPCD and MndD are disordered and therefore were not modeled. Also, the data sets typically showed no density beyond residue 322 or 323. The exception was the P3221 crystal form of Bf 2,3-HPCD, for which the data did yield sufficient electron density to model 37 of these C-terminal residues.
Oligomeric structure.
Bf 2,3-HPCD and MndD form very similar tetramers, each with 222 point symmetry and overall dimensions of approximately 75 by 75 by 110 Å. Within the tetramer, each monomer has distinct interactions with the other three monomers. The most extensive interaction is that between monomers A and B (or C and D), where a 3,369-Å2 surface area is buried upon formation of the dimer. The interaction of dimer AC with dimer BD is composed almost entirely of solvent-mediated polar interactions. In this interface there are three solvent pockets: one at the center of the A-B interface, one at the center of the C-D interface, and one around the 222 symmetry point. Conversely, the interactions between monomers A and C (or B and D) and between monomers A and D (or B and C) bury only 1,733 and 1,006 Å2, respectively. The interaction of the AB dimer with the CD dimer includes both hydrophobic and solvent-mediated polar interactions. There are no biochemical data to suggest that either enzyme exists as anything but tetramers in solution (48, 86). The active-site cavities are located approximately along the long edges of the rectangular tetramers. The active-site metal of monomer A is 31, 33, and 44 Å away from the corresponding metals of monomers B, C, and D, respectively. Biochemical studies of Bf 2,3-HPCD and MndD do not indicate any allostery between monomers (48, 86).
The C-terminal 43 residues of Bf 2,3-HPCD run transverse from the corners of the tetramer, down the long axis, and occupy space between the A-C and B-D monomer contacts. As stated earlier, these residues can be removed upon incubation of Bf 2,3-HPCD or MndD with trypsin. Inclusion of the C terminus, as seen in the P3221 crystal form of Bf 2,3-HPCD, increases the width of the tetramer to 80 Å. This small increase in volume is not consistent with the large decrease in volume apparent upon treatment with trypsin, as viewed by native PAGE, suggesting that the C termini are mobile and can take extended conformations.
Monomer structure.
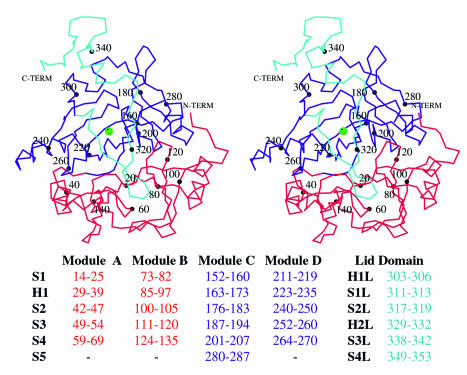
The monomer structures of Bf 2,3-HPCD and MndD are composed of two structurally homologous barrel-shaped domains: an N-terminal domain (residues 1 to 145), a C-terminal domain (residues 145 to 290), and a lid domain (residues 291 to 353) (Fig. 1). The substrate-binding determinants and the active-site metal are located within the C-terminal domain barrel. Each barrel can be further subdivided into two modules, each of which has the same βαβββ motif: modules A and B (N-terminal domain) and modules C and D (C-terminal domain). The four β strands of each module combine to form a mixed-topology β sheet with an amphipathic helix lying against the β sheet. The basic fold of the individual modules has been described for a number of other structures (4, 6, 25) including several that are not type I extradiol dioxygenases, i.e., human glyoxylase I (14), bleomycin resistance protein (19), and Pseudomonas fluorescens 4-hydroxyphenylpyruvate dioxygenase (69). This superfamily has been labeled the vicinal oxygen chelate superfamily (4).
FIG. 1.
Divergent stereo of a Cα trace of a monomer of Bf 2,3-HPCD. MndD and Bf 2,3-HPCD monomers are composed of an N-terminal domain (red), a C-terminal domain (blue), and a lid domain (cyan). The substrate binds to the iron (green sphere) in a pocket in the barrel of the C-terminal domain. Every 20th residue is labeled. Residues involved in the secondary structural elements are listed at the bottom.
In the construction of the two barrel domains in MndD and Bf 2,3-HPCD, the first strand from module A (or C) forms an antiparallel β duplex with the first strand of module B (or D). The two domains are coupled through the interactions of hydrophobic residues from the outer surface of each barrel to create a hydrophobic core. The C-terminal domain is unique in that after module D, the chain returns to module C and adds a fifth strand, to which the lid domain is attached. The lid domain contributes significantly to the shape of the active-site opening and is probably the principal determinant of the character of the outer substrate-binding pocket. The lid domain begins by forming one edge of the mouth of the active site, interacting with strands S2D, S4D, and S1D (see Fig. 1 for secondary-structure labeling). It then travels to the other side of the monomer, touches a portion of the opening to the N-terminal barrel, and then returns toward the opening of the C-terminal barrel. It is at this point in the sequence that the lid domain is susceptible to cleavage by trypsin. The trigonal form of Bf 2,3-HPCD demonstrates that the remaining residues of the lid can fold into a small helix (H2L) and two β strands (S3L and S4L) that interact with the loop between S3C and S4C and with the first portion of the lid domain. This portion of the lid domain covers the remaining opening to the active-site cavity, excluding all but the smallest molecules from the active-site cavity.
The monomer structures of Bf 2,3-HPCD and MndD are essentially identical (RMS difference over 319 structurally equivalent Cαs, 0.250 Å), as expected from their high sequence identity. MndD and Bf 2,3-HPCD also have high structural homology with the individual subunits of two other type I extradiol dioxygenases, 2,3-CTD (RMS difference over 285 Cαs, 1.65 Å) and BPHC_LB400 (RMS difference over 240 Cαs, 1.79 Å), despite sequence identities of <25%. BPHC_LB400 is a homooctamer, while the other proteins assemble as homotetramers with similar structural features. Additionally, 2,3-CTD and BPHC_LB400 do not have H2L, S3L, and S4L of the lid domain, because this region is absent in the these enzymes.
Active-site cavity.
We believe that the trypsin-cleaved structures are representative of the conformation the structure assumes before binding the substrate. Evidence for this will be presented in the following sections. The active-site metal is located approximately 15 to 20 Å within the C-terminal barrel. The outer rim of the active site has a very broad, flat opening constructed by residues 290 to 305 of the lid domain, the loop between strands S2C and S3C, and the loop between strands S4C and S1D. An approaching substrate would first encounter residues Y189, V287, R292, Y305, V300, G206, and G207. Farther in, the substrate would encounter a circular opening, approximately 8 to 10 Å in diameter, defined by the side chains of R293, I181, A190, W304, R243, and H213. The side chain of R293 is disordered in the substrate-free, trypsin-cleaved forms of MndD and Bf 2,3-HPCD. There is ample room for the substrate to enter if R293 and H2L of the lid domain move out of the way. Finally, upon interacting with the active-site metal, the substrate would settle in a pocket formed by the side chains of W192, H248, V250, S251, and V257.
Native metal coordination.
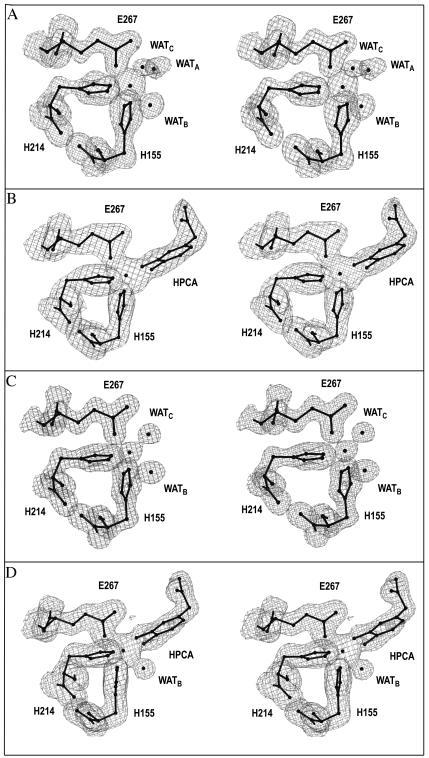
The electron densities for the ligands to the active-site metals of Bf 2,3-HPCD and MndD are shown in Fig. 2A and C, respectively. All the ligands to the metal are provided by the C-terminal domain and arise from β strands. The active-site metal is coordinated by two histidines, one glutamic acid, and two to three solvent molecules, depending on the crystal form, in octahedral geometry. This confirms the previous mutagenesis work, which showed that these three residues are critical to both activity and the ability to bind manganese in MndD (8). In our terminology we identify H214NE2 and E267OE1 as equatorial ligands and H155NE2 as an axial ligand. The three potentially open ligand positions (A, B, and C) are trans to H214NE2, E267OE1, and H155NE2, respectively. In two of the native structures (Bf 2,3-HPCD, Native 2, and MndD, Native 1), the three open positions are, in fact, occupied by solvent molecules. The solvent in site A appears to be the weakest ligand to the iron, because it typically has the highest B-factor and a significantly longer bond length, and it is absent in one of the native structures (Bf 2,3-HPCD, Native 3). At this resolution, changes in bond lengths of 0.25 to 0.3 Å are deemed significant. In the other sites, the average ligand distance is 2.1 to 2.2 Å depending on the native structure examined (Table 3). Having a weakly bound solvent or nothing in site A is consistent with the MCD/CD (47) and X-ray absorption spectroscopy (71) studies of 2,3-CTD, which show a pentacoordinate metal center with five N or O ligands at an average ligand-to-metal distance of 2.09 Å. It is assumed that, on average, one of the solvent molecules exists as a hydroxide, resulting in charge neutrality at the active site. The solvent molecule at site C has the shortest bond lengths, suggesting that it has the most hydroxide character. A superposition of the MndD and Bf 2,3-HPCD active sites does not indicate any discernible differences in the metal coordination geometry or the positions of critical residues (RMS deviation over 64 atoms, 0.12 Å).
FIG. 2.
2Fo-Fc electron density for the ligands to the iron in the native and complex structures of Bf 2,3-HPCD and MndD. Electron density is contoured at 1σ. (A) Bf 2,3-HPCD with no substrate (Native 2, I41); (B) Bf 2,3-HPCD soaked with 1 mM HPCA; (C) MndD with no substrate (Native 2, C2); (D) MndD soaked with 1 mM HPCA. This figure was produced by use of the modeling program O (37).
TABLE 3.
Ligand distances for Bf 2,3-HPCD and MndD
| Ligand bond | Ligand distance (Å) for:
|
|||||
|---|---|---|---|---|---|---|
| Bf 2,3-HPCD
|
MndD
|
|||||
| Native 2 | Native 3 | Complex | Native 1 | Native 2 | Complex | |
| Fe-H155NE2 | 2.2 | 2.2 | 2.2 | 2.2 | 2.2 | 2.1 |
| Fe-H214NE2 | 2.1 | 2.3 | 2.2 | 2.1 | 2.0 | 2.2 |
| Fe-E267OE1 | 2.1 | 1.9 | 2.0 | 2.0 | 2.1 | 2.0 |
| Fe-Site A | 2.5 (OH2) | 2.3 (O4) | 2.7 (OH2) | 2.1 (O4) | ||
| Fe-Site B | 2.3 (OH2) | 2.1 (OH2) | 2.2 (OH2) | 2.4 (OH2) | 2.3 (OH2) | |
| Fe-Site C | 2.2 (OH2) | 2.0 (O3) | 2.1 (OH2) | 2.2 (OH2) | 1.9 (O3) | |
Substrate binding.
Unambiguous electron density is seen for the substrate in the complexes of both Bf 2,3-HPCD and MndD (Fig. 2B and D). In the refined structure of the MndD ES complex, the Mn2+ is hexacoordinate with HPCAO3 in site C and HPCAO4 in site A. All of the ligands to the metal originate from the inner surface of the barrel in regular β-sheet secondary structure and therefore are relatively rigid. Additional hydrogen bonds between H155ND1 and D154OD2, and between H214ND1 and H213ND1, further restrict ligand movement. Binding of the substrate results in the dissociation of two of the solvent molecules, while a third solvent molecule remains bound to the metal at site B. Besides interacting with the metal, this solvent also forms hydrogen bonds with H200NE2 and the side chain of N154.
The Bf 2,3-HPCD-substrate complex demonstrates that Fe2+ ligation also occurs through both hydroxyls of the substrate. The most significant difference between the two complexes is the absence of the solvent molecule in site B, leading to a square pyramidal Fe2+ geometry leaving one open coordination site. In both complexes the binding is asymmetric, with HPCAO3 exhibiting the shorter bond to the iron (Table 3).
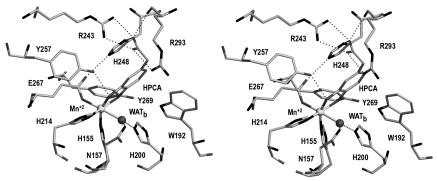
Several residues in the active-site cavity appear to be critical to the positioning of the substrate (Fig. 3). The substrate is wedged into a planar crevice. One wall is formed by Y257 and H248, both of which are conserved among all members of the type I extradiol dioxygenase family. Y257OH forms a hydrogen bond with HPCAO3, while the center of the imidazole ring of H248 stacks with HPCAC5 and HPCAC6. On the opposite side of the substrate, the side chain of W192 is approximately perpendicular to the plane of the substrate where W192NH interacts with the π-electron cloud of HPCA. The corresponding residue in 2,3-CTD and BPHC_LB400 is a phenylalanine, which is similarly oriented. At the bottom of the crevice, the edge of the aromatic ring (HPCAC5, HPCAC6) is within van der Waals contact with the side chains of V250 and S251 and with the backbone carbonyl of V250. The close proximity of these residues would severely restrict the abilities of MndD and Bf 2,3-HPCD to catalyze the cleavage of any substrates with bulky substituents at these positions. On the opposite side of the ring, HPCAC2 has more distant contacts; it is approximately 4.5 Å from the side chain of W304. W304 is therefore an important determinant of what substituents could be placed at C2 and still retain functional binding. In BPHC_LB400, the benzyl substituent at this position is accommodated by having a much abbreviated lid domain.
FIG. 3.
Divergent stereo diagram of the interaction of the substrate with residues of the active site in MndD. Important potential hydrogen bonds are shown as dotted lines. The active-site Mn2+ is shown as a light gray sphere, while a single equatorial solvent is shown as a dark sphere. This figure was prepared by use of SETOR (23).
The most interesting interactions are those with the acetate substituent of the substrate. The acetate group projects into a pocket of the active site lined by W304, R293, H248, and Y257. The guanidinium group of R243 is at the bottom of this pocket, where it makes a salt link with the carboxylate of the substrate. In addition, the carboxylate is within hydrogen bonding distance of H248ND1, R293O, and R293NH1. The binding of an acetate substituent in this pocket results in the partial ordering of R293, which in turn closes off most of the entrance to the active site. These specific interactions are likely crucial to the formation of a catalytically competent ES complex. Removing the carboxylate (4-methylcatechol) or the methylene group (3,4-dihydroxybenzoate) results in a 30- or 555-fold decrease in the relative activity of MndD, respectively (86).
Major movements upon substrate binding.
The binding of the substrate is facilitated by and/or facilitates the movement of several residues in the active-site cavity. The movements appear to propagate outward from several residues directly adjacent to the substrate.
In the native structure, a hydrogen bonding network links Y257OH with H248NE2 and H248ND1 with R293O. When a substrate binds, these interactions are disrupted, as also noted for the analogous residues in BPHC_LB400 (79). Y257OH forms a new hydrogen bond with HPCAO3. The plane of the imidazole ring of H248 slides approximately 1.4 Å along the plane of the substrate ring. The hydrogen bond of Y257OH with H248NE2 is broken, and H248NE2 forms a new hydrogen bond with E267OE2, one of the ligands to the metal. E267OE2 shifts slightly to optimize this interaction with H248NE2. The movement of H248 also breaks the hydrogen bond of H248ND1 with R293O. This interaction is replaced by one with the acetate group of HPCA, which forms a hydrogen bond with both H248ND1 and R293O. These movements permit the acetate group of the substrate to interact with the guanidinium group of R243. A smaller (0.6-Å) but similar shift of the homologous H241 of BPHC_LB400 occurs when a substrate binds that enzyme (79).
In the native structure, D294 forms a hydrogen bond with R243NE. Upon substrate binding, the conformation of D294 switches from gauche− (χ1 = −90°) to trans (χ1 = −168°), away from R243 and toward the 293-to-302 loop. This appears to be caused by the movement of R293 (due to the ordering of the side chain) and the loss of the hydrogen bond of R293O with H248ND1. In addition, the interaction of the acetate group of HPCA with R293 means that the electrostatic component of the interaction between R293 and D294 is no longer required. The ordering of the side chain of R293 causes the side chain of Y305 to move outward toward the solvent by approximately 1 to 2 Å. These displacements permit V300CB to move approximately 2.9 Å inward toward R243. There are also large changes in the positions of P299, V301, and P302, as this whole loop packs closer to the protein.
A second distinct set of displacements propagates from near the active-site metal to the interface between the A and B (or C and D) subunits. In the native structure, a solvent-filled pocket directly adjacent to the active-site metal is separated from a second solvent-filled pocket in the AB (or CD) dimer interface by the side chains of Y269 and D272. In the ES complex, Y269 and D272 have moved away from the dimer interface toward the substrate while maintaining the hydrogen bond of Y269OH with D272OE1. The side chain of Y269 fills a large portion of the solvent pocket adjacent to the iron, where Y269CE2 is only ∼3.6 Å from HPCAO4. These displacements cause the side chain of Q271, a residue in the AB (or CD) dimer interface, to change conformations (from a χ1 of −150° to a χ1 of −76°). The trigger for the second group of conformational changes is unclear, but they may be due to a change in the conformational stability in this area when solvents are expelled upon substrate binding.
A superposition of the Bf 2,3-HPCD and MndD substrate-bound structures indicates that all of the conformational changes upon substrate binding are conserved between the two structures. The most telling conformational changes are the ordering of the side chain of R293, the movement of the 293-to-302 loop (most specifically the side chain of V300), and the conformational change of Y269. The structures, therefore, suggest that the active sites of MndD and Bf 2,3-HPCD can be in an open or a closed conformation depending on the binding of the substrate.
P3221 crystal form.
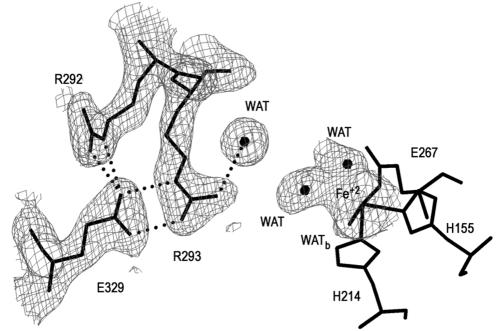
Analysis of native and cleaved Bf 2,3-HPCD and MndD by native PAGE and by their propensities to crystallize indicated that there was most likely a flexible amino or carboxy terminus on these proteins. Sequence alignment of several enzymes from the extradiol family with a diverse substrate range indicated that Bf 2,3-HPCD and MndD have unique carboxy termini. The function of these residues is unknown. We were able to obtain one data set from a crystal form of Bf 2,3-HPCD that did not require trypsin cleavage. Interestingly, 36 of the missing 42 C-terminal residues became visible in the electron density. The average B-factor for these residues is 25.5 Å2. As discussed earlier, the portion of the lid domain that remained after trypsin cleavage reduced the accessibility of the active site to small molecules. However, since the side chain of R293 was disordered, the mouth of the active site still permitted access to a molecule the size of HPCA. The structure of the P3221 crystal form demonstrates that a helix from the trypsin-sensitive portion of the lid domain is capable of covering the remaining opening of the active-site cavity. Figure 4 shows the electron density around R293 in the P3221 crystal form of Bf 2,3-HPCD. The size and shape of the electron density in the coordination sphere of the Fe2+ available to exogenous substrates suggested a partially occupied carboxylate, e.g., acetate. However, since acetate was not present in the crystallization solutions, we chose to model this density as two solvent molecules.
FIG. 4.
Electron density and residue interactions in the region surrounding the binding site of the acetate group of the substrate in the P3221 crystal form of Bf 2,3-HPCD. For purposes of clarity, ligands to the iron and their respective densities are not shown. Additional electron density at the iron is modeled by two solvent molecules. Dotted lines, potential hydrogen bonds. This figure was produced by use of the modeling program O (37).
Analysis of the P3221 crystal form indicates that the protein is in the closed conformation. R293 is well ordered, the 298-to-302 loop has moved toward the protein, and Y269 has rotated towards the metal ion. This suggests that exogenous ligand binding may trigger the flexible C terminus to cover the opening to the active site. The ordering of the side chain of R293 near R292 creates a latching interaction site with E329 (Fig. 4). The movement in the 298-to-303 loop is required to avoid a van der Waals clash between the side chains of V300 and I334 (d < 2.5 Å). Despite the fact that the sequence identity between Bf 2,3-HPCD and MndD drops below 60% in the C-terminal mobile fragment, residues that are involved in the interaction between the fragment and the protein are generally conserved.
DISCUSSION
In this study, we present the X-ray structures of two isofunctional extradiol dioxygenases that were isolated from different organisms and utilize different active-site metals. In agreement with the high sequence homology of the enzymes, the structures are remarkably similar, especially in the vicinity of the active site. The enzymes have much lower sequence homology to other structurally characterized extradiol dioxygenases, but the present structures show that the active-site metal ligand structure and key active-site amino acids are conserved. One novel feature of the new structures is the presence of a lid domain that apparently closes over the substrate as it binds. The detailed comparison of these structures with each other and with the other structurally characterized dioxygenases gives new insight into the roles of the active-site residues and the mechanism of specific ring cleavage by the extradiol dioxygenase family. These aspects of the study are discussed here.
Effect of the carboxy terminus on crystallization.
It was observed in this study that the space group and quality of the crystals obtained correlated with the presence or absence of the C-terminal fragment. The I41 space group of cleaved Bf 2,3-HPCD has one tetramer per asymmetric unit, with each monomer exhibiting unique contacts with crystallographically related molecules. In each case, if the flexible portion of the C terminus remained bound in the closed conformation, it would result in direct collisions with neighboring molecules and would prevent the formation of this crystal form. A similar comparison with the C2 crystal form of MndD suggested that the C-terminal fragment could fit comfortably within the solvent space of the crystal and not disrupt crystal contacts. However, the presence of the flexible C-terminal fragment would most likely interfere with the conformation of the immediately preceding residues P318 to V323, which form the most intimate crystal contacts. This postulate is consistent with the fact that disordered MndD crystals were obtained prior to treatment of the protein with trypsin. In the P3221 crystal form of Bf 2,3-HPCD, there are no specific contacts between the flexible C-terminal fragment and neighboring molecules in the lattice, indicating that order is not imposed by interactions with neighboring molecules. Thus, not only could a nearly complete lid domain be seen, but these observations also suggested that the lid domain has the probable function of closing over the active site after the substrate binds.
Metal ion specificities of MndD and Bf 2,3-HPCD.
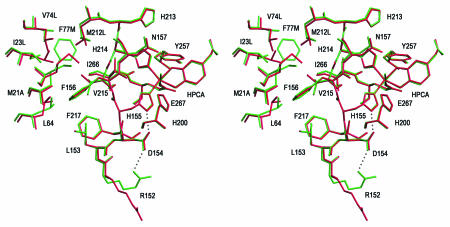
MndD and Bf 2,3-HPCD display a high degree of sequence identity, which fails to reveal any obvious basis for the observed metal ion specificity. The structures presented here permit an analysis of residues in and around the active site that might influence the binding and reactivity of a particular metal. Surprisingly, there are no significant differences in the positions of atoms within the first (H155, H214, and E267) or the second (H200, N157, H213, Y257, D154, and W192) sphere of the active-site metal. However, there are a number of amino acid differences at the interface between the two β barrels of the C-terminal domain (10 to 15 Å from the metal). The amino acid differences are complementary exchanges of hydrophobic residues so that the character of the interface is maintained. The reordering of this interface could have a long-range effect on the metal selectivity, but no clear rationale for this emerges. There are also a number of differences in the sequence or position of residues in the solvent pocket between the A and B (or C and D) subunits. One of these residues is R152, which in MndD forms a hydrogen bond with the side chain of D154OD1, while in Bf 2,3-HPCD it extends out into the cavity, forming hydrogen bonds with D272′O (symmetry-related monomer in tetramer), E57OE1, and several solvent molecules (Fig. 5). Since the side chain of D154 also forms a hydrogen bond to the imidazole of H155, a metal ligand, the position of R152 could have an indirect effect on the electronic character of the metal site. It is not clear if these differences are significant enough to have any impact on metal selectivity.
FIG. 5.
Divergent stereo diagram of the superposition of MndD (green) and Bf 2,3-HPCD (red) in an area around the active site. Sequence differences for Bf 2,3-HPCD are given after the sequence number. Significant differences occur only 10 to 15 Å away from the iron. This figure was prepared by use of SETOR.
A number of groups are studying a similar question of metal selectivity in superoxide dismutases (SODs). The structures of SODs that are dependent on mononuclear manganese or iron have been determined (46, 75). Residue differences in the second coordination sphere have been postulated to affect the redox “window” of the metal in these cases (82, 83). There has also been some success in altering the metal selectivity by mutating second-sphere residues (31, 67). However, these two SOD systems have significant differences in metal environments. In contrast to Bf 2,3-HPCD and MndD, the iron-dependent and manganese-dependent SODs have fewer residues in common (sequence identity, <50%), suggesting that more of the structure could be involved in tuning the specificity and properties of the metal center. Nevertheless, it is possible to exchange iron and manganese in the case of purified SOD, whereas no such exchange has been achieved with the 2,3-HPCDs studied here. Thus, it cannot be conclusively shown that Fe2+-substituted MndD or Mn2+-substituted Bf 2,3-HPCD is inactive. Given the functional and structural similarities of Bf 2,3-HPCD and MndD, it seems unlikely that a better pair of enzymes could be identified with which to address the question of metal ion specificity. The fact that little insight has been gained from the high-resolution structures of the enzymes suggests that the folded enzymes do not distinguish between the metals. It is possible that metal selection occurs during the folding process.
Enzyme mechanism.
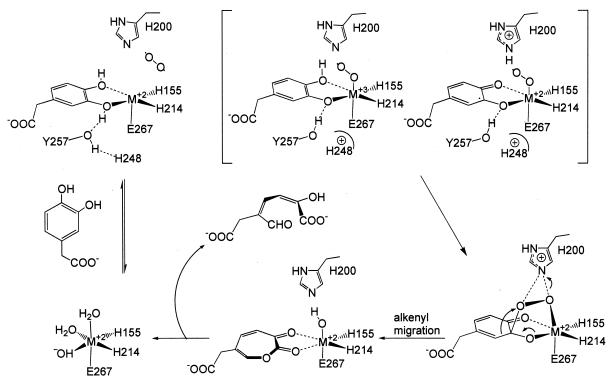
The high-resolution structures of the substrate-free and -bound forms of MndD and Bf 2,3-HPCD allow us to evaluate and expand the proposals that have been made for the mechanism of the extradiol dioxygenases (3, 13, 48, 71). Key questions relate to the mechanism of oxygen activation, the attack on the substrate, and the specificity of the ring cleavage site. A mechanistic proposal based on our structural insight is shown in Fig. 6.
FIG. 6.
Proposed enzyme mechanism. The typical geometry (E267OE1, H214NE1, O2, and HPCAO4 equatorial ligands) is rotated (H155NE1, H214NE1, HPCAO3, and HPCAO4 equatorial ligands) for clarity and for comparison to previously published mechanism diagrams (63, 71).
Metal site organization.
All of the current mechanistic proposals for the extradiol dioxygenases begin with a catecholic substrate and oxygen binding to Fe2+. The structures of MndD and Bf 2,3-HPCD show that the active-site metal (M2+) is ligated in adjacent positions on one side of the metal coordination sphere by three endogenous ligands (H155, H214, and E267) in what has been termed a 2-His-1-carboxylate facial triad motif (30). This motif, with two or three waters on the opposite face, allows the substrates to be organized and juxtaposed in the active site by simultaneously binding to the iron through displacement of the solvents. In both MndD and Bf 2,3-HPCD, the catecholic substrate binds with each of the ring hydroxyls trans to a histidyl ligand. Despite this apparent symmetry in the complex, the binding is markedly asymmetric, with the HPCAO3-M2+ forming a shorter bond, suggesting that only this hydroxyl function is deprotonated. As a result, the metal center is charge neutral, just as it is proposed to be in the substrate-free structure due to the presence of a hydroxide ligand. This displaceable M2+-bound hydroxide trans to H155NE2 in the active site of the enzyme as isolated probably acts as the general base that accepts the proton from HPCAO3 during the substrate-binding process. The negative charge on HPCAO3 may also be stabilized by hydrogen bonding from Y257OH (∼2.7 Å). A similar role has been proposed for the analogous tyrosine residues in BPHC_PS102 and BPHC_LB400 (66, 81). The notion of a monoanionic ES complex for extradiol dioxygenases has been suggested previously by X-ray absorption spectroscopy studies of 2,3-CTD (71), UV resonance Raman and crystallographic studies of BPHC_LB400 (79), and studies of 4-nitrocatechol binding to these enzymes (64, 78). The asymmetry of the complex offers a possible means by which the site of attack of activated oxygen on the aromatic ring and, from this, the regiospecificity of ring cleavage are determined.
Oxygen binding.
Studies utilizing NO as an oxygen analog showed that NO coordinates to the Fe2+ center of extradiol dioxygenases with an affinity that is dramatically enhanced by substrate binding (3). By extension, O2 is proposed to bind to the metal center in the solvent binding site (site C) that remains after the substrate is bound. This assignment is strongly supported by the crystal structure of BphC with NO bound in the analogous site (66). It may be necessary to stabilize the M2+-O2 bond to prevent adventitious oxidation of the metal center. One residue that that might carry out this function is H200, a conserved residue in the active sites of extradiol dioxygenases (27, 65, 68). The present structures show that H200NE2 is well positioned to interact with a metal-bound dioxygen and stabilize it, much like the function of the distal histidine in myoglobin and hemoglobin (49, 50, 54, 60, 70, 72, 85). H200 could also carry out a second function, since it is approximately 3.5 Å from HPCAO4. If this hydroxyl must be deprotonated as part of the oxygen insertion reaction of the reaction mechanism, H200 could serve as a general base for this reaction, as suggested for the equivalent reaction in BphC (68). A protonated H200 would also be more effective in stabilizing an M2+-O2 bond, so oxygen binding may promote removal of the second proton from HPCA. However, it must also be stated that the H200NE2 geometry is not ideal for the formation of a reasonable hydrogen bond with HPCAO4; reorganization would be required. Groce and Lipscomb have recently directly demonstrated the importance of H200 by generating the H200F mutant and showing that the rate of turnover is greatly decreased despite little effect on the substrate-binding reaction itself (26). This suggests that the primary effects of this residue are exerted on later steps in the reaction cycle, including oxygen binding, activation, and insertion. Interestingly, the H200F mutant also switches the ring cleavage specificity for some alternative substrates from extradiol to intradiol, suggesting that it has a profound effect on directing either the reactivity or the relative geometry of the substrate and oxygen in the active site.
Oxygen activation and attack on substrate.
It has been proposed that, subsequent to O2 binding, the metal center acts as a conduit for the flow of electron density from the electron-rich substrate to the bound dioxygen (3, 48), generating a semiquinone-M2+-superoxide intermediate. The oxygen activated in this way would then attack the adjacent substrate at C-2 or C-3 to form a peroxy intermediate. The proximity of Y257 to the axial binding site forces HPCAC3 closer to the putative oxygen binding site, as illustrated by the 15 to 20° deviation of HCPAO3 from octahedral geometry. The present structures show that the substrate would have to rotate about the HPCAC3/C5 axis in order to bring HPCAC2 close enough to form the M2+-O-O-HPCAC2 peroxy adduct favored by earlier mechanistic postulates (3, 48, 71). This rotation is unlikely because of the many interactions the acetate group makes with the protein and the potential for collisions between the substrate and W192. On the other hand, the structure shows that an M2+-O-O-HPCAC3 peroxy adduct can be formed easily without much distortion or dissociation of the incipient superoxide. A similar conclusion was reached for the BphC-substrate complex (66), suggesting that this may be a consistent aspect of the extradiol dioxygenase mechanism.
Ring-opening reaction and its regiospecificity.
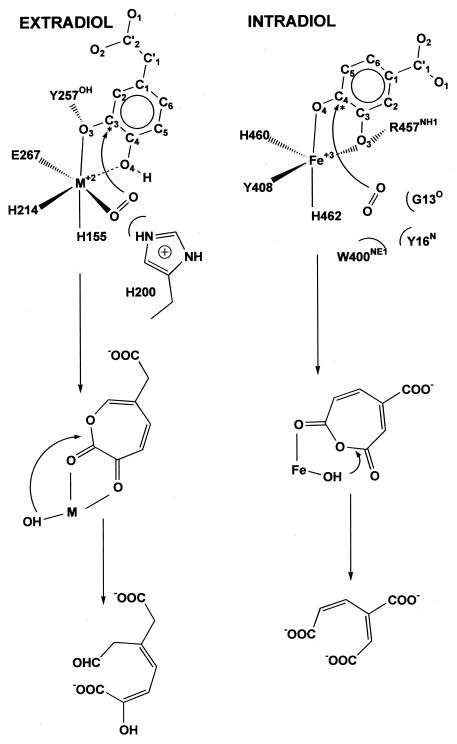
The ring-opening reaction is envisioned as a Criegee rearrangement in which the O-O bond is broken and one atom is inserted into the ring in a concerted process. This reaction would be promoted by removal of the proton from the second hydroxyl group of HPCA and proton donation to the peroxy intermediate undergoing O-O bond cleavage by one or more active-site acid or base catalysts as discussed above. Following the ring insertion chemistry, hydrolysis of the intermediate by the remaining oxygen atom from O2 bound to the metal center would complete the reaction. This sequence would ensure the overall dioxygenase stoichiometry observed for these enzymes. The proposed Criegee-type oxygen insertion reaction could occur in one of two ways, yielding either the C2-C3 lactone species that would lead to extradiol cleavage or the C3-C4 anhydride species that would yield the intradiol product after hydrolysis, as shown in Fig. 7.
FIG. 7.
Comparison of the binding characteristics of intradiol and extradiol dioxygenases. Numbering for extradiol enzymes is based on that of Bf 2,3-HPCD; numbering for intradiol dioxygenases is based on that of protocatechuate 3,4-dioxygenase from P. putida. Asterisks indicate the position at which oxygen is proposed to attack during the formation of the peroxy intermediate. The proposed location of oxygen is indicated.
What, then, are the factors that determine the regiospecificity of oxidative ring cleavage? Despite a common peroxo intermediate structure, the two enzyme classes differ in the type of metal center to which the peroxo intermediate is coordinated. For extradiol dioxygenases, the peroxo intermediate binds to an M2+ His2 Glu center, with the peroxo oxygen trans to the Glu residue. For intradiol enzymes, it binds to an Fe+3 His2 Tyr center, with the peroxo oxygen trans to a His residue. It is possible that these differing ligand environments are responsible for the different regiospecificities of ring cleavage. Support for this notion can be found in studies of biomimetic [Fe(L)catecholate] complexes where the site of oxidative cleavage can be changed by the nature and denticity of the ancillary ligand L as well as by the oxidation state of the metal center (34, 36, 43).
It has also been suggested that the precise stereochemistry of the metal-peroxo-substrate unit determines the sites of cleavage (13). In this proposal, the axial orientation of the peroxo-substrate bond (see Fig. 6) that results from the presence and relative positions of the three available binding sites on the metal, together with the ferrous oxidation state of the iron and the presence of a proton donor to promote the Criegee rearrangement, favors alkenyl group migration to form the C2-C3 lactone intermediate. The present structural studies show that each of these features is present in the active sites of the 2,3-HPCD enzymes. Mutational studies that examine the effects of different metal ligands and active-site residues are needed to test these mechanistic proposals. The structural studies described here provide a starting point for such mutational studies, which are currently under way.
Acknowledgments
This work was supported by grants GM46436 (to D.H.O.), GM24689 (to J.D.L.), and GM33162 (to L.Q.) from the National Institutes of Health and by a National Institutes of Health training grant (GM-07323, to M.W.V.). L.P.W. was supported by grant DE-FG02-01ER63268 from the Office of Science (BER), U.S. Department of Energy. We acknowledge the Minnesota Supercomputer Institute for providing computational resources. Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Energy Research, under contract W-31-109-ENG-38.
Special thanks go to Stephan L. Ginell, Norma E. C. Duke, Rongguang Zhang, and Andrzej Joachimiak, who assisted in the collection of data sets obtained on beamline ID-19 at the Argonne National Laboratory.
REFERENCES
- 1.Arciero, D. M., and J. D. Lipscomb. 1986. Binding of 17O-labeled substrate and inhibitors to protocatechuate 4,5-dioxygenase-nitrosyl complex. Evidence for direct substrate binding to the active site Fe2+ of extradiol dioxygenases. J. Biol. Chem. 261:2170-2178. [PubMed] [Google Scholar]
- 2.Arciero, D. M., J. D. Lipscomb, B. H. Huynh, T. A. Kent, and E. Münck. 1983. EPR and Mössbauer studies of protocatechuate 4,5-dioxygenase. Characterization of a new Fe2+ environment. J. Biol. Chem. 258:14981-14991. [PubMed] [Google Scholar]
- 3.Arciero, D. M., A. M. Orville, and J. D. Lipscomb. 1985. [17O]water and nitric oxide binding by protocatechuate 4,5-dioxygenase and catechol 2,3-dioxygenase. Evidence for binding of exogenous ligands to the active site Fe2+ of extradiol dioxygenases. J. Biol. Chem. 260:14035-14044. [PubMed] [Google Scholar]
- 4.Armstrong, R. N. 2000. Mechanistic diversity in a metalloenzyme superfamily. Biochemistry 39:13625-13632. [DOI] [PubMed] [Google Scholar]
- 5.Asturias, J. A., L. D. Eltis, M. Prucha, and K. N. Timmis. 1994. Analysis of three 2,3-dihydroxybiphenyl 1,2-dioxygenases found in Rhodococcus globerulus P6. Identification of a new family of extradiol dioxygenases. J. Biol. Chem. 269:7807-7815. [PubMed] [Google Scholar]
- 6.Bergdoll, M., L. D. Eltis, A. D. Cameron, P. Dumas, and J. T. Bolin. 1998. All in the family: structural and evolutionary relationships among three modular proteins with diverse functions and variable assembly. Protein Sci. 7:1661-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boldt, Y. R., M. J. Sadowsky, L. B. M. Ellis, L. Que, Jr., and L. P. Wackett. 1995. A manganese-dependent dioxygenase from Arthrobacter globiformis CM-2 belongs to the major extradiol dioxygenase family. J. Bacteriol. 177:1225-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boldt, Y. R., A. K. Whiting, M. L. Wagner, M. J. Sadowsky, L. Que, Jr., and L. P. Wackett. 1997. Manganese(II) active site mutants of 3,4-dihydroxyphenylacetate 2,3-dioxygenase from Arthrobacter globiformis strain CM-2. Biochemistry 36:2147-2153. [DOI] [PubMed] [Google Scholar]
- 9.Broderick, J. B. 1999. Catechol dioxygenases. Essays Biochem. 34:173-189. [DOI] [PubMed] [Google Scholar]
- 10.Brünger, A. T. 1992. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Acta Crystallogr. D 49:24-46. [DOI] [PubMed] [Google Scholar]
- 11.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 12.Bugg, T. D., J. Sanvoisin, and E. L. Spence. 1997. Exploring the catalytic mechanism of the extradiol catechol dioxygenases. Biochem. Soc. Trans. 25:81-85. [DOI] [PubMed] [Google Scholar]
- 13.Bugg, T. D. H., and G. Lin. 2001. Solving the riddle of the intradiol and extradiol catechol dioxygenases: how do enzymes control hydroperoxide rearrangements? Chem. Commun. 11:941-952. [Google Scholar]
- 14.Cameron, A. D., B. Olin, M. Ridderstrom, B. Mannervik, and T. A. Jones. 1997. Crystal structure of human glyoxalase I: evidence for gene duplication and 3D domain swapping. EMBO J. 16:3386-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 16.Cowtan, K. 1994. DM: an automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newsl. Protein Crystallogr. 31:34-38. [Google Scholar]
- 17.Dagley, S. 1984. Microbial degradation of aromatic compounds. Dev. Ind. Microbiol. 25:53-65. [Google Scholar]
- 18.Dagley, S. 1977. Microbial degradation of organic compounds in the biosphere. Surv. Prog. Chem. 8:121-170. [PubMed] [Google Scholar]
- 19.Dumas, P., M. Bergdoll, C. Cagnon, and J. M. Masson. 1994. Crystal structure and site-directed mutagenesis of a bleomycin resistance protein and their significance for drug sequestering. EMBO J. 13:2483-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunwell, J. M., A. Culham, C. E. Carter, C. R. Sos-Aguirre, and P. W. Goodenough. 2001. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26:740-746. [DOI] [PubMed] [Google Scholar]
- 21.Dunwell, J. M., S. Khuri, and P. J. Gane. 2000. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64:153-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eltis, L. D., and J. T. Bolin. 1996. Evolutionary relationships among extradiol dioxygenases. J. Bacteriol. 178:5930-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans, S. V. 1993. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph. 11:134-138, 127-128. [DOI] [PubMed] [Google Scholar]
- 24.Fu, W. J., and P. Oriel. 1998. Gentisate 1,2-dioxygenase from Haloferax sp. D1227. Extremophiles 2:439-446. [DOI] [PubMed] [Google Scholar]
- 25.Gerlt, J. A., and P. C. Babbitt. 2001. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu. Rev. Biochem. 70:209-246. [DOI] [PubMed] [Google Scholar]
- 26.Groce, S. L., and J. D. Lipscomb. 2003. Conversion of extradiol aromatic ring-cleaving homoprotocatechuate 2,3-dioxygenase into an intradiol cleaving enzyme. J. Am. Chem. Soc. 125:11780-11781. [DOI] [PubMed] [Google Scholar]
- 27.Han, S., L. D. Eltis, K. N. Timmis, S. W. Muchmore, and J. T. Bolin. 1995. Crystal structure of the biphenyl-cleaving extradiol dioxygenase from a PCB-degrading pseudomonad. Science 270:976-980. [DOI] [PubMed] [Google Scholar]
- 28.Hatta, T., G. Mukerjee-Dhar, J. Damborsky, H. Kiyohara, and K. Kimbara. 2003. Characterization of a novel thermostable Mn(II)-dependent 2,3-dihydroxybiphenyl 1,2-dioxygenase from a polychlorinated biphenyl- and naphthalene-degrading Bacillus sp. JF8. J. Biol. Chem. 278:21483-21492. [DOI] [PubMed] [Google Scholar]
- 29.Hayase, N., K. Taira, and K. Furukawa. 1990. Pseudomonas putida KF715 bphABCD operon encoding biphenyl and polychlorinated biphenyl degradation: cloning, analysis, and expression in soil bacteria. J. Bacteriol. 172:1160-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hegg, E. L., and L. Que, Jr. 1997. The 2-His-1-carboxylate facial triad: an emerging structural motif in mononuclear non-heme iron(II) enzymes. Eur. J. Biochem. 250:625-629. [DOI] [PubMed] [Google Scholar]
- 31.Hiraoka, B. Y., F. Yamakura, S. Sugio, and K. Nakayama. 2000. A change of the metal-specific activity of a cambialistic superoxide dismutase from Porphyromonas gingivalis by a double mutation of Gln-70 to Gly and Ala-142 to Gln. Biochem. J. 345:345-350. [PMC free article] [PubMed] [Google Scholar]
- 32.Hofer, B., L. D. Eltis, D. N. Dowling, and K. N. Timmis. 1993. Genetic analysis of a Pseudomonas locus encoding a pathway for biphenyl/polychlorinated biphenyl degradation. Gene 130:47-55. [DOI] [PubMed] [Google Scholar]
- 33.Howard, A. J., G. L. Gilliland, B. C. Finzel, T. L. Poulos, D. H. Ohlendorf, and F. R. Salemme. 1987. Use of an imaging proportional counter in macromolecular crystallography. J. Appl. Crystallogr. 20:383-387. [Google Scholar]
- 34.Ito, M., and L. J. Que. 1997. Biomimetic extradiol cleavage of catechols: insights into the enzyme mechanism. Angew. Chem. Int. Ed. Engl. 36:1342-1344. [Google Scholar]
- 35.Iwabuchi, T., and S. Harayama. 1998. Biochemical and molecular characterization of 1-hydroxy-2-naphthoate dioxygenase from Nocardioides sp. KP7. J. Biol. Chem. 273:8332-8336. [DOI] [PubMed] [Google Scholar]
- 36.Jo, D. H., Y. M. Chiou, and L. Que, Jr. 2001. Models for extradiol cleaving catechol dioxygenases: syntheses, structures, and reactivities of iron(II)-monoanionic catecholate complexes. Inorg. Chem. 40:3181-3190. [DOI] [PubMed] [Google Scholar]
- 37.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for finding protein models in electron density maps and the location of error in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 38.Kabisch, M., and P. Fortnagel. 1990. Nucleotide sequence of metapyrocatechase I (catechol 2,3-oxygenase I) gene mpcI from Alcaligenes eutrophus JMP222. Nucleic Acids Res. 18:3405-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimbara, K., T. Hashimoto, M. Fukuda, T. Koana, M. Takagi, M. Oishi, and K. Yano. 1989. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J. Bacteriol. 171:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kita, A., S.-I. Kita., I. Fujisawa, K. Inaka, T. Ishida, K. Horiike, M. Nozaki, and K. Miki. 1999. Crystal structure of catechol 2,3-dioxygenase from Pseudomonas putida mt-2, metapyrocatechase, an archetypical extradiol-cleaving dioxygenase. Structure 7:25-34. [DOI] [PubMed] [Google Scholar]
- 41.Kita, H. 1965. Crystallization and some properties of 3,4-dihydroxyphenylacetate 2,3-oxygenase from Pseudomonas ovalis. J. Biochem. 58:116-122. [DOI] [PubMed] [Google Scholar]
- 42.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 43.Lin, G., G. Reid, and T. D. Bugg. 2001. Extradiol oxidative cleavage of catechols by ferrous and ferric complexes of 1,4,7-triazacyclononane: insight into the mechanism of the extradiol catechol dioxygenases. J. Am. Chem. Soc. 123:5030-5039. [DOI] [PubMed] [Google Scholar]
- 44.Lipscomb, J. D., and A. M. Orville. 1992. Mechanistic aspects of dihydroxybenzoate dioxygenases. Met. Ions Biol. Syst. 28:243-298. [Google Scholar]
- 45.Lipscomb, J. D., J. W. Whittaker, D. M. Arciero, A. M. Orville, and S. A. Wolgel. 1988. Mechanisms of catecholic dioxygenases, p. 259-282. In S. R. Hagedorn, R. S. Hanson, and D. A. Kunz (ed.), Microbial metabolism and the carbon cycle. Harwood Academic Publishers, New York, N.Y.
- 46.Ludwig, M. L., A. L. Metzger, K. A. Pattridge, and W. C. Stallings. 1991. Manganese superoxide dismutase from Thermus thermophilus. A structural model refined at 1.8 Å resolution. J. Mol. Biol. 219:335-358. [DOI] [PubMed] [Google Scholar]
- 47.Mabrouk, P. A., A. M. Orville, J. D. Lipscomb, and E. I. Solomon. 1991. Variable-temperature variable-field magnetic circular dichroism studies of the Fe(II) active site in metapyrocatechase: implication for the molecular mechanism of extradiol dioxygenases. J. Am. Chem. Soc. 113:4053-4061. [Google Scholar]
- 48.Miller, M. A., and J. D. Lipscomb. 1996. Homoprotocatechuate 2,3-dioxygenase from Brevibacterium fuscum. A dioxygenase with catalase activity. J. Biol. Chem. 271:5524-5535. [DOI] [PubMed] [Google Scholar]
- 49.Mims, M. P., A. G. Porras, J. S. Olson, R. W. Noble, and J. A. Peterson. 1983. Ligand binding to heme proteins. An evaluation of distal effects. J. Biol. Chem. 258:14219-14232. [PubMed] [Google Scholar]
- 50.Nagai, K., B. Luisi, D. Shih, G. Miyazaki, K. Imai, C. Poyart, A. De Young, L. Kwiatkowsky, R. W. Noble, S. H. Lin, et al. 1987. Distal residues in the oxygen binding site of haemoglobin studied by protein engineering. Nature 329:858-860. [DOI] [PubMed] [Google Scholar]
- 51.Nakai, C., H. Kagamiyama, M. Nozaki, T. Nakazawa, S. Inouye, Y. Ebina, and A. Nakazawa. 1983. Complete nucleotide sequence of the metapyrocatechase gene on the TOL plasmid of Pseudomonas putida mt-2. J. Biol. Chem. 258:2923-2928. [PubMed] [Google Scholar]
- 52.Navaza, J. 1994. AMORE: an automated package for molecular replacement. Acta Crystallogr. A 50:157-163. [Google Scholar]
- 53.Noda, Y., S. Nishikawa, K. Shiozuka, H. Kadokura, H. Nakajima, K. Yoda, Y. Katayama, N. Morohoshi, T. Haraguchi, and M. Yamasaki. 1990. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J. Bacteriol. 172:2704-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olson, J. S., A. J. Mathews, R. J. Rohlfs, B. A. Springer, K. D. Egeberg, S. G. Sligar, J. Tame, J. P. Renaud, and K. Nagai. 1988. The role of the distal histidine in myoglobin and haemoglobin. Nature 336:265-266. [DOI] [PubMed] [Google Scholar]
- 55.Ornston, L. N., and R. Y. Stanier. 1966. The conversion of catechol and protocatechuate to β-ketoadipate by Pseudomonas putida. J. Biol. Chem. 241:3776-3786. [PubMed] [Google Scholar]
- 56.Orville, A. M., J. D. Lipscomb, and D. H. Ohlendorf. 1997. Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding. Biochemistry 36:10052-10066. [DOI] [PubMed] [Google Scholar]
- 57.Otwinowski, Z. 1991. Maximum likelihood refinement of heavy atom parameters, p. 80. In W. Wolf, P. R. Evans, and A. G. W. Leslie (ed.), Isomorphous replacement and anomalous scattering. Daresbury Laboratory, Warrington, United Kingdom.
- 58.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276A:307-326. [DOI] [PubMed] [Google Scholar]
- 59.Pflugrath, J. W. 1999. The finer things in X-ray diffraction data collection. Acta Crystallogr. D 55:1718-1725. [DOI] [PubMed] [Google Scholar]
- 60.Phillips, S. E. 1980. Structure and refinement of oxymyoglobin at 1.6 Å resolution. J. Mol. Biol. 142:531-554. [DOI] [PubMed] [Google Scholar]
- 61.Que, L., Jr. 1989. The catechol dioxygenases, p. 467-524. In T. M. Loehr (ed.), Iron carriers and iron proteins. VCH, New York, N.Y.
- 62.Que, L., Jr. 1993. Oxygen activation at nonheme iron centers, p. 347-393. In J. Reedijk (ed.), Bioinorganic catalysis. Marcel Dekker, New York, N.Y.
- 63.Que, L., Jr., and R. Y. N. Ho. 1996. Dioxgyen activation by enzymes with mononuclear non-heme iron active sites. Chem. Rev. 96:2607-2624. [DOI] [PubMed] [Google Scholar]
- 64.Reynolds, M. F., M. Costas, M. Ito, D.-H. Jo, A. A. Tipton, A. K. Whiting, and L. J. Que. 2003. Nitrocatechol as a probe of a Mn(II)-dependent extradiol-cleaving catechol dioxygenase (MndD): comparison with relevant Fe(II) and Mn(II) model complexes. J. Biol. Inorg. Chem. 8:263-272. [DOI] [PubMed] [Google Scholar]
- 65.Sato, H., T. Watanabe, Y. Murata, A. Ohtake, M. Nakamura, C. Aizawa, H. Saito, and N. Maehara. 1999. New exfoliative toxin produced by a plasmid-carrying strain of Staphylococcus hyicus. Infect. Immun. 67:4014-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sato, N., Y. Uragami, T. Nishizaki, Y. Takahashi, G. Sazaki, K. Sugimoto, T. Nonaka, E. Masai, M. Fukuda, and T. Senda. 2002. Crystal structures of the reaction intermediate and its homologue of an extradiol-cleaving catecholic dioxygenase. J. Mol. Biol. 321:621-636. [DOI] [PubMed] [Google Scholar]
- 67.Schwartz, A. L., E. Yikilmaz, C. K. Vance, S. Vathyam, R. L. Koder, and A. F. Miller. 2000. Mutational and spectroscopic studies of the significance of the active site glutamine to metal ion specificity in superoxide dismutase. J. Inorg. Biochem. 80:247-256. [DOI] [PubMed] [Google Scholar]
- 68.Senda, T., K. Sugiyama, H. Narita, T. Yamamoto, K. Kimbara, M. Fukuda, M. Sato, K. Yano, and Y. Mitsui. 1996. Three-dimensional structures of free form and two substrate complexes of an extradiol ring-cleavage type dioxygenase, the BphC enzyme from Pseudomonas sp. strain KKS102. J. Mol. Biol. 255:735-752. [DOI] [PubMed] [Google Scholar]
- 69.Serre, L., A. Sailland, D. Sy, P. Boudec, A. Rolland, E. Pebay-Peyroula, and C. Cohen-Addad. 1999. Crystal structure of Pseudomonas fluorescens 4-hydroxyphenylpyruvate dioxygenase: an enzyme involved in the tyrosine degradation pathway. Structure 7:977-988. [DOI] [PubMed] [Google Scholar]
- 70.Shaanan, B. 1983. Structure of human oxyhaemoglobin at 2.1 Å resolution. J. Mol. Biol. 171:31-59. [DOI] [PubMed] [Google Scholar]
- 71.Shu, L., Y. M. Chiou, A. M. Orville, M. A. Miller, J. D. Lipscomb, and L. Que, Jr. 1995. X-ray absorption spectroscopic studies of the Fe(II) active site of catechol 2,3-dioxygenase. Implications for the extradiol cleavage mechanism. Biochemistry 34:6649-6659. [DOI] [PubMed] [Google Scholar]
- 72.Sigfridsson, E., and U. Ryde. 1999. On the significance of hydrogen bonds for the discrimination between CO and O2 by myoglobin. J. Biol. Inorg. Chem. 4:99-110. [DOI] [PubMed] [Google Scholar]
- 73.Spence, E. L., M. Kawamukai, J. Sanvoisin, H. Braven, and T. D. Bugg. 1996. Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and biochemical properties of a third family of extradiol dioxygenases. J. Bacteriol. 178:5249-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stanier, R. Y., and L. N. Ornston. 1973. The β-ketoadipate pathway. Adv. Microb. Physiol. 9:89-151. [PubMed] [Google Scholar]
- 75.Stoddard, B. L., P. L. Howell, D. Ringe, and G. A. Petsko. 1990. The 2.1-Å resolution structure of iron superoxide dismutase from Pseudomonas ovalis. Biochemistry 29:8885-8893. [DOI] [PubMed] [Google Scholar]
- 76.Sugimoto, K., T. Senda, H. Aoshima, E. Masai, M. Fukuda, and Y. Mitsui. 1999. Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions. Structure 7:953-965. [DOI] [PubMed] [Google Scholar]
- 77.Titus, G. P., H. A. Mueller, J. Burgner, S. Rodriguez De Cordoba, M. A. Penalva, and D. E. Timm. 2000. Crystal structure of human homogentisate dioxygenase. Nat. Struct. Biol. 7:543-546. [DOI] [PubMed] [Google Scholar]
- 78.Tyson, C. A. 1975. 4-Nitrocatechol as a colorimetric probe for non-heme iron dioxygenases. J. Biol. Chem. 250:1765-1770. [PubMed] [Google Scholar]
- 79.Vaillancourt, F. H., C. J. Barbosa, T. G. Spiro, J. T. Bolin, M. W. Blades, R. F. B. Turner, and L. D. Eltis. 2002. Definitive evidence for monoanionic binding of 2,3-dihydroxybiphenyl to 2,3-dihydroxybiphenyl 1,2-dioxygenase from UV resonance Raman spectroscopy, UV/Vis absorption spectroscopy, and crystallography. J. Am. Chem. Soc. 124:2485-2496. [DOI] [PubMed] [Google Scholar]
- 80.Vaillancourt, F. H., J. T. Bolin, and L. D. Eltis. 2003. Ring-cleavage dioxygenases, p. 359-395. In J. L. Ramos (ed.), The pseudomonads, vol. III. Kluwer Academic/Plenum, New York, N.Y.
- 81.Vaillancourt, F. H., G. Labbe, N. M. Drouin, P. D. Fortin, and L. D. Eltis. 2002. The mechanism-based inactivation of 2,3-dihydroxybiphenyl 1,2-dioxygenase by catecholic substrates. J. Biol. Chem. 277:2019-2027. [DOI] [PubMed] [Google Scholar]
- 82.Vance, C. K., and A. F. Miller. 1998. Simple proposal that can explain the inactivity of metal-substituted superoxide dismutases. J. Am. Chem. Soc. 120:461-467. [Google Scholar]
- 83.Vance, C. K., and A. F. Miller. 1998. Spectroscopic comparisons of the pH dependencies of Fe-substituted (Mn)superoxide dismutase and Fe-superoxide dismutase. Biochemistry 37:5518-5527. [DOI] [PubMed] [Google Scholar]
- 84.Wang, Y. Z., and J. D. Lipscomb. 1997. Cloning, overexpression, and mutagenesis of the gene for homoprotocatechuate 2,3-dioxygenase from Brevibacterium fuscum. Protein Expr. Purif. 10:1-9. [DOI] [PubMed] [Google Scholar]
- 85.Weiss, J. J. 1964. Nature of the iron-oxygen bond in oxyhaemoglobin. Nature 203:182-183. [PubMed] [Google Scholar]
- 86.Whiting, A. K., Y. R. Boldt, M. P. Hendrich, L. P. Wackett, and L. Que, Jr. 1996. Manganese(II)-dependent extradiol-cleaving catechol dioxygenase from Arthrobacter globiformis CM-2. Biochemistry 35:160-170. [DOI] [PubMed] [Google Scholar]