Abstract
Stem cells hold promise to revolutionize modern medicine by development of new therapies, disease models and drug screening systems. Standard cell culture systems have limited biological relevance because they do not recapitulate the complex 3-dimensional interactions and biophysical cues that characterize the in vivo environment. In this review, we discuss the current advances in engineering stem cell environments using novel biomaterials and bioreactor technologies. We also reflect on the challenges the field is currently facing with regard to translation of stem cell based therapies into the clinic.
Keywords: stem cells, bioengineering, bioreactors, biomaterials, in vitro models, models of disease, regenerative medicine, translation, high-throughput screening, personalized medicine
1. Introduction
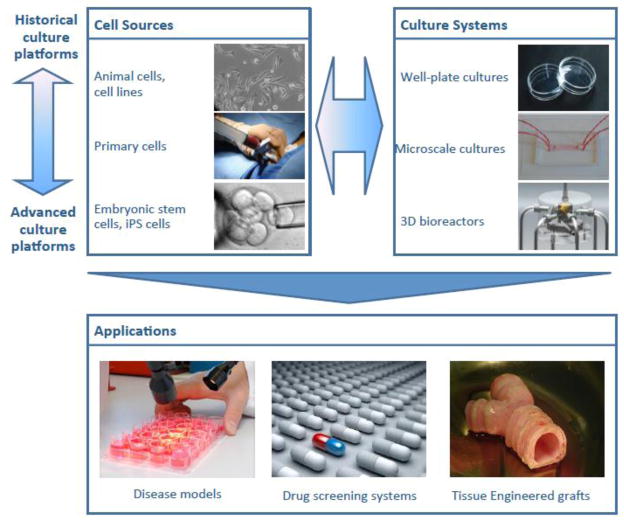
Stem cells provide enormous opportunities for improving human medicine, through the development of tissue replacement therapies, human in vitro models of disease, screening of therapeutic and toxic effects of chemical libraries, and “personalized” medicine. Furthermore, recent advances in stem cell biology, biomaterials, genetic engineering and biomedical engineering have allowed unprecedented ability to create controlled environments and ask specific biological questions. The progression from historical culture plates with animal cells and immortalized cell lines towards embryonic stem cells (ES) and induced pluripotent stem cells (iPS) in 3-dimensional (3D) bioreactors is truly paving the way for new applications in tissue engineering and regenerative medicine, the study of disease, and drug screening (Figure 1). Here we review advances in engineering stem cell environments using dynamic bioreactor systems, and discuss the importance of these novel tools to stem cell research as well as the applications of stem cells in pre-clinical and clinical settings.
Figure 1. Development of culture systems.
The progression from traditional cultures with animal cells and cell lines towards scaffold-bioreactor systems with human adult, embryonic and iPS cells. The new tissue engineering technologies are paving the way to the new generation of in vitro disease models, drug screening systems, and tissue-engineered implantable grafts.
2. Limitations of current stem cell research models
Ever since the time of Galen, the famous physician who reportedly dissected pigs and goats, researchers have sought experimental models of human biology. More recently, the Petri dish, invented at the end of the 19th century, has proven invaluable for experiments in cellular biology. And in fact, standard Petri dish cultures are still widely used: adherent cells are grown on synthetic surfaces (i.e. tissue culture plastic), basement membrane or extracellular matrix protein coatings (i.e. laminin, vitronectin, collagen), or feeder cells (i.e. mouse embryonic fibroblasts), and are bathed in culture medium containing appropriate nutrients and signaling molecules. Changing of cell culture medium is conducted batch-wise, resulting in the variation of medium composition over time.
In Petri dishes, the cells are essentially cultured in two dimensions. Stem cells generally grow in dense colonies with defined borders, which expand in size and merge with other colonies in the culture dish (Takahashi et al. 2007; Thomson et al. 1998). At confluence, cells are passaged for further expansion, or subjected to differentiation protocols. While this culture format recapitulates some aspects of tissues that are essentially two-dimensional (2D), such as skin or bladder, it falls short of providing environments experienced by most cells in the organism. In particular, Petri dish culture lacks the 3D cell-cell and cell-matrix interactions, provision of spatial and temporal gradients of biochemical and physical signals, and systemic regulation including cross-talk between different organ systems (Kaplan et al. 2005; Vunjak-Novakovic et al. 2005). Findings obtained in Petri dish cultures are therefore not always predictable of whole tissues and organs, and are difficult to translate into the in vivo settings of pre-clinical studies in animals, and clinical trials in human subjects.
In contrast to the controlled environments of cell culture systems, animal models allow assessment of stem cell developmental potential within whole organisms, and are therefore invaluable for studies of development, disease pathogenesis and toxicity testing (Cheshier et al. 1999; Sacco et al. 2010; Wobus and Loser 2011). After the discovery of mouse ES cells and the completion of human genome sequencing, creation of mice with specific gene knockouts and gene reporters has enabled the study of gene function during development, and cell lineage tracking experiments (Lloyd 2011). Furthermore, specific rodent strains with compromised immune systems have been developed that allow us to study the function of human cells in vivo without immune rejection (i.e. humanized mice) (Shultz et al. 2011).
However, despite these advantages, animal models present several limitations when used in disease modeling and toxicological studies. First, very few animal models faithfully reproduce human pathophysiology. Therefore it is important that all disease models - whether surgically or pharmacologically induced or genetic, are clearly defined with regards to the pathology that is being modeled, and to how it relates to the human condition. Second, there are important interspecies differences in pharmaco-toxicological effects between experimental animals and humans (Wobus and Loser 2011), which are only exacerbated when human cells are transplanted into immune-suppressed hosts, potentially also affecting physiological healing responses (Goldring et al. 2011). In this respect, progress in preparation of iPSc from large animals, such as pigs, would advance transplantation studies (Montserrat et al. 2011). Finally, for studies of transplanted cells, in vivo models offer less control over the cell microenvironment, and are challenging for on-line monitoring of the outcomes, compared to the in vitro systems, which are better defined and better controlled.
A critical application highlighting the importance of developing better in vitro systems to model human biology and physiology is that of drug development. A number of high-profile drugs have been recently withdrawn from the market, most commonly due to the cardiotoxicity, neurotoxicity and hepatotoxicity that were not observed until clinical trials (Report 2011). These negative side effects were not detected because of the limited functional capacity and genetic diversity of current research models, resulting in drugs that pass animal studies but fail in human studies. Part of the problem is that, in the conventional target-centric drug discovery model, compounds are typically not tested in a patient population until Phase II clinical trials have been carried out, thus contributing to a high drug attrition rate (Kola and Landis 2004, Kola 2008).
Further complicating the issue is that even compounds that have been found to be safe and efficacious in animal models can fail spectacularly in clinical trials, adding expense and risk. The cost of bringing a new drug to the clinic has been estimated at $1.2 billion, up from a 2003 average of $802 million (DiMasi et al. 2003), and pharmaceutical companies face their largest losses when drugs fail at the late stage of trials, such as Phase III and the post-marketing stage (Report 2011). In addition, some potential targets may be excluded from further development due to false negative results during in vitro testing.
The shift from using animal cells and immortalized cell lines to human stem cells and their differentiated progeny, and from simple Petri dish culture to more sophisticated environments capturing critical aspects of the in vivo system, could be hugely beneficial for drug development. Bioreactor-based screening technologies could improve prediction of in vivo outcomes in pre-clinical and clinical studies. Tissue models using cells from multiple patients could allow study of large and diverse genetic and disease pools, help overcome many of the current limitations of clinical trials (Li 2004). and help advance the state of the art from that of treatments designed for an “average” patient to a personalized medicine approach (Pouton and Haynes 2005, 2007; Rolletschek and Wobus 2009).
3. Stem cell cultivation in scaffold-bioreactor systems
Our understanding of stem cell biology has been greatly expanded by the development of novel culture environments using biomaterials and bioreactors that recapitulate selected aspects of native tissue (Gerecht et al. 2007; Gilbert et al. 2010; McBeath et al. 2004; Steiner et al. 2010). The importance of inductive signals present in 3D tissue settings has been recognized long ago, and simple techniques such as cultivation of mesenchymal stem cell (MSC) pellets, human embryonic stem cell (hESC) aggregates (embryoid bodies, or EBs) or neurospheres have been used to differentiate cells (Itskovitz-Eldor et al. 2000; Johnstone et al. 1998; Reynolds and Weiss 1992). However, these cultures present limitations in terms of size (mostly <0.5 mm), control over cell populations and biophysical signals (mechanical, electrical, hydrodynamic), and the lack of extracellular matrix.
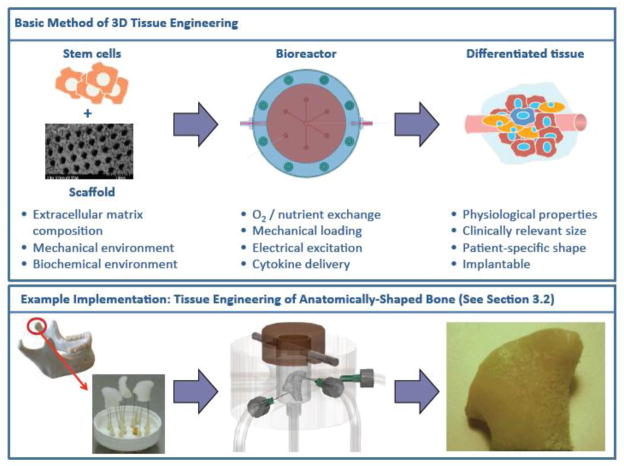
In the 1990s, it was proposed that 3D human tissue substitutes could be engineered in vitro by cultivation of cells on scaffolding materials, which act as temporary biodegradable templates for tissue development, in bioreactor systems providing environmental control and biochemical and biophysical signaling (Langer and Vacanti 1993) (see Figure 2). This approach was dubbed the “biomimetic paradigm” because it was based upon the idea that these scaffolds in combination with bioreactors would recapitulate the relevant developmental and morphological in vivo events in vitro. During native tissue development, cells are exposed to a myriad of signals including multiple cell types, extracellular matrix, cytokines, and physical factors, all of which are dynamic in nature. And in fact, since this time, by following the biomimetic paradigm, many biomaterial scaffolds and in vitro systems have been designed to recapitulate key aspects of this dynamic environment and support cell growth, matrix synthesis and tissue maturation.
Figure 2. 3D tissue engineering.
The so-called “biomimetic paradigm of tissue engineering” includes the cultivation of stem cells on scaffolds within bioreactors that provide in-vivo-like environments and produce differentiated, mature tissues. Engineering of anatomically-shaped bone, corresponding to case study #2 in Section 3.2, is included in the lower panel.
Developments in the field of biomaterials have made it possible to create 3D scaffolds with defined structural features (shape, porosity, pore size, surface roughness), mechanical properties, composition and degradation rate (Kraehenbuehl et al. 2011; Lutolf and Hubbel 2005). A variety of native, synthetic and composite materials is now available in geometries including fibers, meshes, sheets, sponges, hydrogels and combinations (Bhumiratana et al. 2011; Gerecht et al. 2007; Hofmann et al. 2007; Tran et al. 2010). Biodegradable materials allow encapsulation and sustained release of inductive biochemical signals (Ferreira et al. 2007; Son et al. 2011; Wang et al. 2009), and tethering of specific attachment groups (Hanjaya-Putra et al. 2011), to further tailor the cellular microenvironment and direct the cell function.
In order to induce cell growth in the third dimension and to support tissue development, it is critical to provide mass transport to and from all cells using dynamic culture systems such as bioreactors. In static cultures, mass transport is based on diffusion, and generally limits tissue development to thicknesses less than 0.2 mm due to drops in oxygen tension and increased concentrations of toxic metabolites (e.g., acidification). In bioreactors, stirring, perfusion, and dynamic loading have been applied to provide convective transport and allow tissue development on a millimeter to centimeter scale (Chahine et al. 2009; Grayson et al. 2010; Grayson et al. 2011; Hofmann et al. 2007; Marolt et al. 2006).
The direction and rate of medium flow and the viscosity of medium can be tailored to achieve mechanical stimulation by shear stress (Grayson et al. 2010; Grayson et al. 2011; Sikavitsas et al. 2003). Other examples of biophysical stimuli provided by bioreactors include compressive and tensile loading, electrical stimulation, medium flow resulting from stirring or perfusion (Baker et al. 2011; Bian et al. 2012; Maidhof et al. 2011), alone or in combinations. Finally, bioreactors allow online control and monitoring of temperature, pH, oxygen, medium concentrations of nutrients in order to meet the requirements of tissue under study.
3.1. Case study 1: Large-scale bioreactor cultivation of pluripotent stem cells
To realize the promise of pluripotent cells for drug discovery and cell therapy, scale up and automation of the manual culture process are necessary, as they assure robust and reproducible production of large numbers of relevant cells. In recent years, suspensionculture bioreactors have been investigated for the production of human pluripotent stem cells and their differentiated derivatives in clinically-relevant amounts, which range from several tens of millions to a few billion cells (Kehoe et al. 2010). These bioreactors offer advantages over monolayer culture, including control of the microenvironment, higher cell densities per volume, low contamination risk, application of scaffolds for encapsulation of anchorage-dependent cells, and tailoring of the hydrodynamic environment to study the effects on self-renewal and differentiation of stem cells.
Agitation is essential for ensuring that all cells are exposed to the culture medium of the same composition, keeping the aggregates or microcarriers suspended, and controlling the aggregate size (Kehoe et al. 2010). In static culture, cell aggregates can form large agglomerates with substantial necrotic areas in their centers, resulting from limited nutrient and oxygen transport (Gerecht-Nir et al. 2004). In stirred suspension bioreactors, impeller-associated hydrodynamic shear can regulate the size of ESC aggregates, prevent agglomeration and increase the final cell numbers. However, high levels of shear stress (>15 dynes/cm2) may cause damage to the cells. For hESCs sensitive to shear stress, slow turning lateral vessels and high-aspect rotating vessel bioreactors have been tested, as they contain a low shear environment (Gerecht-Nir et al. 2004). Hydrodynamic shear forces can also be applied in a controlled fashion to guide cell differentiation into specific cell lineages (Sargent et al. 2010).
In initial studies, growth and differentiation of mouse embryonic stem cells (mESCs) were evaluated in suspension bioreactor cultures of cell aggregates, microcarriers or hydrogels (Kehoe et al. 2010). The microcarrier and monolayer cultures showed similar cell doubling times (14–17 hours), and similar expression of pluripotency markers after 15 days of culture (Fok and Zandstra 2005). mESC aggregates were directly induced into EBs capable of multilineage differentiation by changing the medium composition. Stir-related shear stress reduced colony agglomeration, and regulated the size of aggregates. Uniform growth of mESC aggregates was achieved when the agitation rate was increased from 60 to 100 rpm.
Studies of mESCs in orbital suspension cultures showed that the hydrodynamic environment effectively regulated not only the size and yield of EBs from mESCs, but also the organization and phenotypes of differentiating cells (Sargent et al. 2010). To prevent agglomeration, mESCs have been cultured in hydrogel microcapsules in stirred suspension bioreactors, and differentiated into hematopoietic progenitors (Dang et al. 2004). More recently, gelatine microcarriers were used for mESC differentiation into the chondrogenic and osteogenic lineages. Differentiated cells formed mineralized tissue in vitro and in vivo in both ectopic and orthotopic locations. Interestingly, cultivation of mESC aggregates in the absence of microcarriers resulted in the formation of muscle tissue after ectopic transplantation (Alfred et al. 2011).
In one of the first studies of hESCs, differentiating EBs in rotating bioreactors that are characterized by laminar flow pattern with mild mixing, reached three times higher cell densities (36 × 106 cells/ml) than the static dish cultures (13 × 106 cells/ml) after 4 weeks of culture (Gerecht-Nir et al. 2004). The cells remained viable and gave rise to lineages of all three germ layers, suggesting that the bioreactor microenvironment did not alter early developmental events, and could be used to scale-up production of differentiated cells. Subsequent studies included the scale-up of mesoderm and cardiac cell differentiation from hESCs, using microprinting to prepare cell aggregates of uniform size that were then cultured in suspension bioreactors (Niebruegge et al. 2009).
Overall, the culture in suspension systems with differing agitation forces and vessel geometries (rotating dish, roller bottle, spinner bioreactors) significantly improved cell growth compared to static culture. In addition, cultivation of uniform size hESC aggregates under hypoxic conditions in spinner bioreactors further improved the yield of differentiating cells (reaching densities of 6 × 105 cells/ml after 16 days of culture) (Niebruegge et al. 2009).
In an alternate approach, encapsulation of mESCs and hESCs in poly-L-lysine coated alginate capsules, and liquefaction of the capsule cores was used to prepare cell aggregates of uniform sizes (Jing et al. 2010). Subsequent cultivation in suspension bioreactors allowed directed cell differentiation into cardiac lineage with higher efficiency than in static culture. In contrast, cultivation of hESCs on microcarriers in suspension bioreactors allowed monolayer-like cell growth, important for directed differentiation into definitive endoderm lineage (Lock and Tzanakakis 2009). Optimization of the agitation rate (to 45 rpm) resulted in up to 45-fold increase in the number of hESCs (~1.8 × 106 cells/ml) expressing pluripotent markers after 8 days of culture. Subsequent change from expansion to differentiation medium resulted in a population expressing definitive endoderm markers. Efficiency of differentiated cells production per unit of surface area was similar to that of static cultures, but the use of microcarriers bioreactors allowed more surface per unit volume.
These studies illustrate diverse effects of scaffolds and bioreactors on mESC and hESC fate, and the potential to develop bioprocesses where cell expansion, differentiation and storage could be integrated within a single culture system (Nie et al. 2009). It has been more challenging to scale up continuous expansion of undifferentiated hESCs compared to mESCs, due to their propensity to differentiate in the 3D environment, and low survival after dissociation into single cells. Biomaterials supporting undifferentiated hESC growth, such as hyaluronic acid hydrogel (Gerecht et al. 2007) and molecules enhancing cell survival, such as Rho-associated kinase (ROCK) inhibitor (Watanabe et al. 2007), could be implemented in such culture protocols.
In one example, Krawetz and colleagues reported continuous expansion of hESCs in stirred suspension bioreactors using ROCK inhibitor and rapamycin to promote cell survival and undifferentiated growth (Krawetz et al. 2010). Components tested to promote suspension cultures of undifferentiated hESCs included neural sphere culture medium, serum replacements, beta-D xylopiranose, extracellular matrix components (fibronectin, laminin), activin A, and basic fibroblast growth factor (Steiner et al. 2010).
3.2. Case study 2: Engineering of functional bone tissue from human stem cells
The shortage of donor tissues and organs suitable for transplantation, and the inherent limitations of artificial prosthetic devices make the development of stem-cell based biological tissue substitutes one of the most active areas of regenerative medicine research. One example is the development of functional bone tissue substitutes for surgical treatment of large bone injuries (Fröhlich et al. 2008).
Many studies have focused on cell lines and animal cells as models for bone tissue regeneration, and the signaling factors and molecular pathways involved in this process. However the development of therapies requires that the findings are tested with human cells. The treatment of smaller defects, such as augmentation of the jaw bones prior to dental implant placement, has been enhanced using culture-expanded primary cells and biomaterials (Schimming and Schmelzeisen 2004). Regeneration of extensive defects (on the centimeter-scale) necessitates large quantities of functional reparative cells (~ 108 per cm3 of bone), within a template with capacity to guide and undergo regeneration. Stem cells have the ability to self-renew, thereby allowing expansion to large numbers, and to give rise to specialized lineage progenitors, and therefore represent an excellent cell source for bone tissue engineering.
Adult human Mesenchymal Stem Cells (hMSCs), hESCs and iPSCs have all been shown to give rise to bone-forming cells. Research has mostly been performed using hMSCs from bone marrow, as these cells give rise to bone and cartilage during development, and have been shown to promote bone healing clinically in autologous transplants (Salter et al. 2012).
Bioreactor cultivation of human stem cells in an osteogenic scaffold supported cell survival, differentiation, maturation and deposition of bone matrix, while restricting the development of unwanted lineages (i.e. adipose), and facilitating a continued remodeling and vascularization following transplantation (Marolt et al. 2012). The design of scaffold-bioreactor systems is guided by the size and shape of the defect to be repaired and the need for establishing certain mechanical properties and ability for integration. A stepwise approach is often used to select and optimize culture parameters (Grayson et al. 2010; Grayson et al. 2008, 2011; Marolt et al. 2006). For instance, soluble factors to achieve osteogenesis of bone marrow stromal cells (BMSCs) have been extensively studied, and frequently involve a serum-based medium supplemented with a cocktail of osteogenic factors (Hofmann et al. 2007; Marolt et al. 2006; Sikavitsas et al. 2003). A variety of native and synthetic scaffolds has been found to influence bone formation via size, pore architecture (Hofmann et al. 2007), surface roughness (Dalby et al. 2007), cell attachment sites (Shin et al. 2004) and the matierial’s biomechanics (Engler et al. 2006).
Among the bioreactors for bone tissue engineering, those providing medium perfusion through the forming tissue have been demonstrated to best allow the scale-up of bone culture to clinical sizes (several centimeters) (Grayson et al. 2010; Martin and Vermette 2005, Salter et al. 2012). The interstitial flow through the scaffold is critical for cell survival and metabolic activity (Grayson et al. 2010; Grayson et al. 2008, 2011). Perfusion also causes shear stress on the cultured cells, stimulating bone development (Grayson et al. 2011; Sikavitsas et al. 2003).
Cultivation of bone substitutes under perfusion conditions requires a mechanical integrity and porosity of the scaffolds to allow fluid flow, with tight control over the perfusion rate and regime (continuous, intermittent etc), oxygen tension, and additional biophysical signals (e.g., mechanical loading). Our group has achieved reproducible formation of compact bone grafts (4 mm diameter × 4 mm thick discs) from bone marrow derived hMSCs cultured on decellularized bone scaffolds (Grayson et al. 2008; Grayson et al. 2011). Increasing the medium flow velocity from 80 to 1800 μm/s, corresponding to the initial shear stresses of 0.6÷20 mPa, significantly affected bone formation, with the flow velocity between 400÷800 μm/s yielding the best overall outcomes (cell numbers, cell connectivity and bone matrix formation).
This simple and robust scaffold-bioreactor model was easily translated to adult human adipose-derived stem cells (Fröhlich et al. 2010) and hESC-derived mesenchymal progenitors (Marolt et al. 2012), as well as to the cultivation of bone on synthetic silk-hydroxyapatite scaffolds (Bhumiratana et al. 2011). Composite scaffolds containing 4.6% hydroxyapatite resulted in the formation of bone constructs with the Young’s equilibrium modulus of 1.5 MPa. A similar perfusion culture approach was used to engineer a 1.5 cm condyle-shaped bone substitute, using anatomically shaped scaffolds and bioreactors, demonstrating the feasibility to prepare clinically relevant bone substitutes (Grayson et al. 2010).
The ability of cultured bone substitutes to survive in vivo is limited by the rate of vascularization, and many investigations are currently focusing on vascularized bone substitutes. Strategies include co-cultivation of various adult stem cells and adult vascular (progenitor) cells, as well as induction of pluripotent stem cells into bone-forming and vascular cells. The challenges include designing scaffold-bioreactor environments that would support development of specific tissues in specific scaffold regions (Correia et al. 2011). For engineering of vascularized bone from iPSCs (which can be used as a single source to derive cells of both bone and vascular lineages), the challenge is in efficient differentiation into the specific respective lineages, and in the restriction of unwanted developmental potential of the cells.
Differentiation of iPSCs generally results in mixed cell populations, potentially harboring residual pluripotent cells that could lead to development of tumors or unwanted tissues. Our strategy has been to first derive and expand specific lineage progenitors (for 5 passages), and then to perform the bone substitute engineering (5-week bioreactor cultivation). We have recently shown that ESCs can be induced into mesenchymal-like progenitors with osteogenic potential similar to that of bone marrow derived hMSCs. Importantly, cultivation in decellularized bone scaffolds in perfusion bioreactors resulted in development of homogenous bone tissue (cylindrical plugs, 4 mm in diameter and thickness) without the presence of other lineages, that remained stable and matured in vivo, and did not result in uncontrolled cell growth during 8 weeks following transplantation (Marolt et al. 2012).
4. Miniature bioreactors for precise, systematic studies of stem cell environments
Many aspects of the native tissue and stem cell niche develop under tight spatial and temporal control, with synergistic effects between various factors. These types of systems typically require tedious combinatorial experimentation, providing strong rationale for tight control over parameter variation and better mimicry of the complex in vivo tissue milieu in a high-throughput and scalable manner. Miniaturized versions of 3D bioreactor systems, known as micro-bioreactors, represent an important step towards accurate, multifactorial control of cultured cells and tissues. Because they reduce the amounts of cells and materials required for experimentation, they increase the number of experimental conditions and replicates that may be studied in parallel, making them ideally suited for high-throughput, combinatorial studies (Beebe et al. 2002; Squires and Quake 2005; Toner and Irimia 2005).
Furthermore, the short transport distances in micro-bioreactors allow for even more precise, “in vivo-like” control of certain environmental cues. For example, flow in micro-bioreactors is laminar: inertial forces are dominated by viscous forces, and transport occurs by molecular diffusion or by convective regime of well-defined hydrodynamic profile. Laminar flow allows for precise calculation of mass transport parameters and the establishment of flow profiles as a function of the system geometry, flow rate and fluid properties. Since the laminar streamlines remain constant over time and mixing occurs by diffusion, precise dynamic perturbations localized in space and time may be applied (Aasen et al. 2008; Chung et al. 2009; Cimetta et al. 2010; Cimetta et al. 2009; Diao et al. 2006; Jeon et al. 2000; Toetsch et al. 2009). Small transport distances are also key for enabling fast responses to environmental stimuli and their manipulation and control (Cimetta et al. 2009). In summary, micro-bioreactor systems are ideally suited for stem cell research due to their unique characteristics:
Down-scalable/parallelizable. Miniaturization leads to significant reductions in the required amounts of cells and materials for each experimental condition, thus allowing cutting costs and facilitating the increase in test numbers necessary for high-throughput, multi-factorial, quantitative studies. When experiments are massively parallelized, small but significant differences may be detected in biological responses that might otherwise go unnoticed in standard cell culture. In addition, parallelization also enables the screening of large numbers of variables, thus further reducing variability between separate experimental runs. Finally, significant multi-factorial characteristics may be attained by experimental designs in which cell density, cell types, substrate mechanics and biophysical stimuli may be independently and/or simultaneously controlled.
“In-vivo-like” fidelity. Tight spatial and temporal control of the delivery of regulatory factors result in a closer resemblance to the in vivo environments. The precise control over transport phenomena and delivery of exogenous stimuli allows the creation of compartmentalized concentration patterns (such as gradients) and of the physiological patterns of electrical and mechanical stimulation. In addition, it is possible to incorporate multiple tissue types on a single platform, either as 2D or pseudo-3D culture.
Key to micro-bioreactor fabrication are micro fabrication techniques, which have been used for applications ranging from micropatterning and microelectronics, to fabrication of micro electro-mechanical systems (MEMS) (McCreedy 2000; McDonald et al. 2000; Richards-Grayson et al. 2004). Typically, polymer-based micromachining is used for obtaining molds with high aspect-ratio structures, while photoresist-based photolithography allows fabrication of microscaled features with ~1 μm precision. Polydimethylsiloxane (PDMS) micromolding enables the production of polymeric devices from these molds with high accuracy and yield (Kane et al. 1999; Whitesides et al. 2001; Xia and Whitesides 1998). Many types of micro-bioreactor systems have employed these techniques to facilitate drug discovery (Dittrich and Manz 2006; Wen and Yang 2008), stem cell and medical research (Beebe et al. 2002).
4.1. Case study 3: Micro-bioreactors for high-throughput screening of environmental factors
One set of challenges in miniaturizing stem cell culture systems is to adapt the complex microenvironment that regulates stem cell fates to down-scaled in vitro models. Niche factors are multiple and diverse: for example, biochemical signaling cues and soluble factors from neighboring cells or elsewhere, must be recapitulated along with their associated mass transport characteristics. Moreover, the physiological or pathological cell milieu includes factors such as extracellular matrix elasticity and geometry, mechanical and electrical signals. For the niche effects to be investigated in a combinatorial fashion with physiological relevance, the key relevant factors must all be recapitulated on a microscale with high accuracy.
Certain niche factors, such as microflows are ideally suited for “in-vivo-like” control of transport and convective flow profiles. Figallo et al, for example, have developed a simple device comprised of an array of culture wells to enable systematic and precise variation of mass transport and hydrodynamic shear (Figallo et al. 2007). Using this microfluidic platform, hESCs were systematically studied for their cardiovascular differentiation potential, and it was demonstrated that higher percentages of cells expressing vascular-specific markers was inversely proportional to cell density (increasing from 2% to 23% as cell density decreased from 1 × 104 to 2 × 102 cells/cm2). This technology is compliant with standard imaging formats, and quantitative image processing.
Cimetta et al have developed a microfluidic device generating stable concentration gradients for long-term cell culture, and applied this method for studying Wnt3a regulation of β-catenin signaling (Cimetta et al. 2010). More recently, they designed a new microfluidic bioreactor providing space-resolved gradients of multiple molecular factors in 3D cell culture settings apt for studies of complex signaling pathways in human pluripotent stem cells. A single microbioreactor yields up to 120 data points, corresponding to 15 replicates of a gradient with 8 concentration levels (Cimetta et al. 2012).
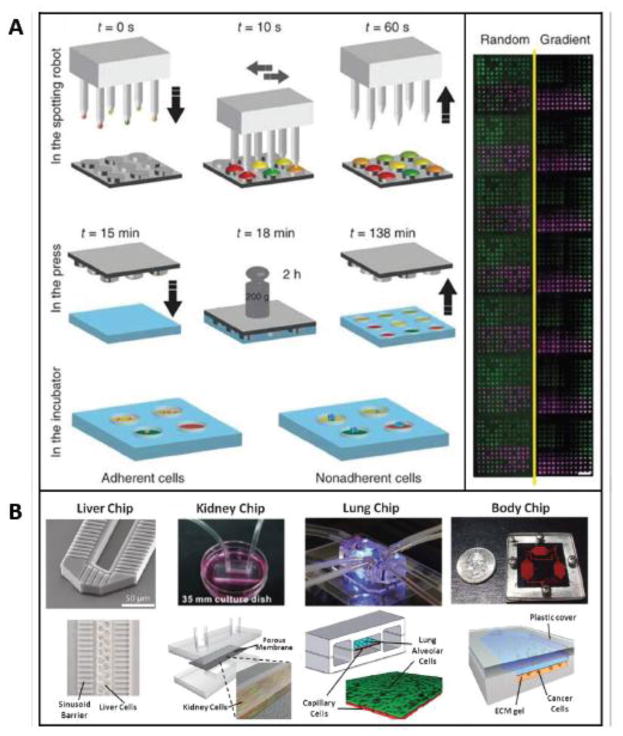
Other groups have focused on other factors in the stem cell niche, such as biochemical and extracellular matrix components, in a parallelized, high-throughput manner. Gobaa et al, for example, have developed a micro-platform that simultaneously probes the role of biochemical and biophysical niche factors on stem cell fate (Figure 3). The device is comprised as an array of soft hydrogel microwells with tunable stiffness (shear modulus of 1–50 kPa) which can be independently functionalized with combinations of proteins spotted by robotic technology (Gobaa et al. 2011). The authors addressed three representative stem cell questions to demonstrate the utility of their device: (i) the influence of cell density on adipogenic differentiation of hMSCs, (ii) the effects of substrate stiffness on osteogenic differentiation and (iii) the self-renewal of non-adherent mouse neural stem cells (NSCs). With this high-throughput hydrogel microwells system, features such as cell density, substrate mechanics and protein incorporation could be tested, and more than 2,000 experiments with several adherent and non-adherent cell types can be performed on a single glass slide. Using their method, they showed it is possible to probe the effects of key micro-environmental perturbations on the fate of single cells in high throughput.
Figure 3. Miniature bioreactors.
Miniature bioreactors have facilitated the precise, systematic studies of stem cell environments via (A) lab-on-chip devices mimicking organs and organ interactions (outlined in Section 4.2), and (B) high-throughput screening of environmental parameters (outlined in Section 4.1).
One of the challenges in high-throughput screening of soluble factors is to create devices that take advantage of microfluidic phenomena and exhibit the combinatorial diversities achieved by microarrays. Robotic fluid handling and microspotting techniques, for example, facilitate high throughput studies of the influence of different immobilized factors in microarrys, but require time-dependent stimulation to be applied simultaneously. Microfluidic platforms that facilitate studies of diffusible growth factors with spatial and temporal control have historically been difficult to scale up into larger, individually addressable culture chambers. Gomez-Sjoberg et al address these issues with their versatile, fully automated microfluidic cell culture system that creates arbitrary media formulations in independent cell culture chambers (Gomez-Sjoberg et al. 2007). Their microfluidic device is comprised of 96 rectangular culture chambers customizable in terms of cell seeding density, medium composition, and feeding schedule, and each chamber is imaged with time-lapsed microscopy, in long-term (ie > 7 days) experiments.
Certain biophysical cues, such as microflows, are facile to incorporate into micro-bioreactors, in contrast to other biophysical cues (e.g. mechanical stretch, electrical stimulation etc) that may require additional components and innovative solutions. Kensah and colleagues have constructed a novel scaled-down bioreactor for culturing cardiomyocytes that incorporates the application of mechanical stretch (Kensah et al. 2011). When exposed to cyclic longitudinal stretch during culture via a linear motor, cardiac tissues exhibited hypertrophy, and increased systolic force (~1.42 mN vs. ~0.96 mN in controls). Tandon and colleagues have microscale cell culture system with an interdigitated microarray of excimer-laser-ablated indium tin oxide electrodes for electrical stimulation of cultured cells (Tandon et al. 2010). They constructed systems in a range of geometries for cultivations of cardiomyocytes and human stem cells. Over 6 days of culture, electrical stimulation (2 ms duration, 1 Hz, 180 μm wide electrodes, 200 μm spacing), enhanced proliferation, elongation and alignment, and numbers of gap junctions in both cell types.
Another set of challenges with high throughput screening systems is to automate the cell culture and associated assessments. Lecault et al, for example, have developed a microfluidic platform containing thousands of nanoliter-scale chambers with the capability to perform live-cell imaging, in situ immunostaining, recovery of viable cells, and automated medium exchange (Lecault et al. 2011). Ye et al have developed a microfluidic platform which is able to rapidly extract multi-parametric measurements of plasma membrane permeability, nuclear size, mitochondrial transmembrane potential and intracellular redox states in anti-cancer drug-induced apoptosis of human liver carcinoma cells (Ye et al. 2007).
Parallelizable, high-throughput platforms of this kind will likely help characterize stem cells in terms of their self-renewal and differentiation, proliferative capacity and migration in response to combinations of stimuli. Going forward, better control over stem cell niche conditions such as cell density, biophysical cues, while allowing for increased multiplexing abilities, individual addressability, and online assay methods will undoubtedly contribute to higher quality, quantitative data relevant to toxicology, developmental biology, and beyond.
4.2. Case study 4: “Body on a chip” devices for drug efficacy/toxicity studies
One of the limitations of single cell cultures (whether of conventional type or bioreactor-based) is that, at best, they only capture the behavior of isolated, single tissues. A particularly important need for in vitro modeling of multi-organ systems is in the prediction of toxicity: in vitro approaches have hitherto only met with limited success, mainly because of the complexity of the in vivo toxic responses. In the animal or human body, there are multiple organs that are interconnected by the systemic circulation, leading to multi-organ interactions and multi-organ metabolism. In the body, cross-talk occurs through soluble agents: metabolites generated in the liver, for example, can influence the gene expression in other cell types and vice versa (Bhatia et al. 1999; Khetani et al. 2004; Takayama G et al. 2007). Drug screening based on simple culture systems is able to predict many cases of primary drug toxicity, but remains limited in predicting indirect toxicity (eg cardiotoxic effects of breakdown products of a drug metabolized in the liver) and of the pharmacokinetic bioavailability of drugs. Affordable in vitro models that mimic the in vivo multi-organ human system (so-called “Body-on-a-Chip” systems) are therefore of prime importance for drug discovery and chemical screening. Microscale systems are particularly well suited to the task because of their ability to recapitulate the cellular microenvironment.
Research towards developing “Body-on-a-Chip” systems has been spurred by the success of the development of microscale “Organs-on-Chips” that recapitulate the structural tissue arrangements and functional complexity for each component organ, and efforts in bioprinting, which may be employed for direct 3D cell writing with precise control (Chang et al. 2008). A variety of microscale organomimetic cell culture systems have been developed, such as liver, blood vessels, lung, kidney or brain on a chip, (Figure 3, reviewed in Huh et al. 2012). Notable examples include microscale cultures of cardiomyocytes from human stem cells (Kattman et al. 2006; Zhang et al. 2009), and human liver constructs that retain species-specific drug responses such as induction of P450s, drug metabolism, drug-induced hepatotoxicity and drug-drug interactions (Khetani and Bhatia 2008; Wang et al. 2010). These constructs compartmentalize into functional zones upon exposure to oxygen tension mimicking that along the hepatic sinusoid (Allen and Bhatia 2003; Allen et al. 2005). Even a human gut-on-chip device with intestinal flora and cyclic strain that mimics peristalsis has been developed (Kim et al. 2012).
Several groups have tackled the challenge of culturing multiple organs in a single culture platform, for studying systemic effects of drugs, or cell-cell communication via metabolites. Li et al, for example, have developed a simplified “well within a well” system in which various cell types are cultured in isolation from one another but connected via medium (Li 2008; Li et al. 2004), thus reproducing some organ interactions. When the chemotherapeutic agent tamoxifen was tested in this system against 6 cell types (liver, kidney, lung, central nervous system, blood vessels, and breast cancer) at increasing doses, cell survival values were obtained for each cell type, although rigorous characterization of multi-organ interactions were not performed.
Ma et al have developed a microfluidic device for testing metabolism-dependent drug toxicity. This device also contains two separate chamber arrays, a PDMS quartz layer containing human liver microsomes, and another for cell culture (Ma et al. 2009). Drug-containing solutions can be introduced continuously into the top layer, diffuse through the liver microsomes, and drugs and liver metabolites diffuse to the cell culture chamber. When tested with doses of Acetaminophen above saturation of the detoxification pathway that converts Acetaminophen to non-toxic conjugates, cytotoxic effects were increased, and subsequently reduced when administered below saturation doses. When tested for drug interactions with Acetaminophen, experimental results again aligned with expectations of cytotoxicity. Although the multi-tissue in vitro systems attempt to go beyond merely testing direct effects of drugs, towards reproducing some metabolism-dependent actions of the drug, one potentially important aspect which is lacking in these systems is their ability to mimic the dynamics of multi-organ interactions, and especially the physiologically realistic exchange of metabolites.
Several groups have attempted to address these issues. Shuler and colleagues have developed an integrated microscale cell culture analogue (called the μCCA) with several interconnected compartments linked by circulating culture medium (serving as a blood surrogate) for liver, lung, and fat cells (Ghanem and Shuler 2000, 2000; Sin et al. 2004; Viravaidya and Shuler 2004; Viravaidya et al. 2004). Interestingly, the first prototype of their device was made using two milk dilution bottles and a spinner flask (Sweeney et al. 1995)! The system was designed using simple physiologically based pharmacokinetic models, and used to analyze metabolic products of xenobiotics (in particular, naphthalene) and multidrug resistance with chemotherapy (Tatosian and Shuler 2009), using μL volumes. Two main problems with the device are the fluid links between the compartments that do not model certain nonlinear parameters such as organ volume, and the compartmentalized nature of tissues connected via systemic circulation, which includes some clearing of wastes.
Ahluwalia and colleagues have attempted to better mimic physiological cell-cell interactions. The chambers in their “multicompartment connected culture bioreactor” (MCB) system differ in dimensions, fabrication method, design principles, and applications, in order to mimic physiological cell-cell interactions with much more fidelity than previous systems (Vozzi et al. 2009; Vozzi et al. 2011). In particular, their device was designed with the goal to mathematically correlate nonlinear quantities (organ volume, blood flow, blood retention time, and metabolic rate) and conserve not only kinetic but also metabolic, volumetric, and exchange rate relationships between cells. The system was tested with murine hepatocytes and human umbilical vein endothelial cells, while monitoring albumin, urea, lactate and viability in a connected culture and monoculture in the bioreactor. When the two cell types were connected, increases in endothelial cell viability, hepatic glucose synthesis and albumin and urea production were observed, along with the decrease in lactate production. Although they only studied two cell types for a short period of time, their results showed potential for enhancing cell function, in particular using heterotypic signals from endothelial cells in cultures of hepatocytes.
Zhang, Zhao and colleagues have attempted to link together multiple cell types while maintaining compartmentalization of tissues within a single device (Zhang et al. 2009). Culture medium was supplied through channels to liver, kidney, and adipose chambers and optimized for the four tissue types by supplementation od localized controlled release of organ-specific growth factors (e.g. TGF-β1). Such a technique could, in principle, be applied to deliver a particular biochemical component to only specific cell types without compromising the functions of other cell types and potentially extend culture times.
Going forward, challenges with “human-on-chip” technologies revolve around achieving better predictability of in vivo conditions through more authentic representation of cellular behavior, and to construct systems in which conditions are parallelized, in order to achieve higher numbers of test conditions (currently this is an area of future work for the field). By producing consistent, realistic results, these devices will be more trusted as models capable of predicting hitherto unknown reactions and pathways in drug metabolism, for healthy and diseased tissues, and for personalized medicine applications. Ongoing developments of optical and electrical techniques compatible with multiplexing, could further facilitate these efforts. Finally, as the in vitro results become more predictable, systems that are easy to operate, more portable, and are less dependent on large equipment will become more and more important.
Translation of stem cell and drug discovery into clinical practice will continue to critically depend on preclinical studies of drug toxicity and disease. As engineered human tissues are being considered for in vitro applications in drug toxicity and disease models, advances in connecting different systems to model cross-talk and physiological interaction between organ systems will become of increasing importance. At this point, multi-tissue platforms that can truly simulate human multi-tissue interactions with spatial-temporal control represent a formidable goal. Fortunately, as tissue engineering approaches continue to make strides towards authentically reproducing the actual milieu of development, regeneration, and disease, the goal of truly developing integrated human physiology in vitro via controllable, modular, dynamic, interconnected human tissues, is becoming more attainable. As body-on-chip devices improve the links between in vitro and in vivo studies, we may hope that the demand for animal studies will be reduced, clinical trials will become more effective, and we may begin to enable a more personalized approach to evaluation of drug regimens.
4.3. Case study 5: Integration of advanced models with novel stem cell sources for studying human disease
Integration of advanced microbioreactors and bioengineered environments with novel stem cell sources can open new perspectives in understanding the fundamental biology of disease, towards more personalized approaches to studying the mechanisms of disease, and to develop treatment therapies (Bhadriraju and Chen 2002). In vitro models of primary cells or established cell lines are widely used as cellular screening models in toxicology (Rolletschek et al. 2004). Human cardiomyocytes derived from hESCs were used for more reliable cardiac safety pharmacology assays (Braam et al. 2010; Davis et al. 2011). In this study, patch clamp analyses and multichannel electrode arrays (MEA, increasing the numbers of test conditions and/or replicates of the system) allowed generation of field potential duration (FPD) values following exposure to different drugs. A dose-response effect on the prolongation of the FPD values was recorded and comparable to the known drug effects on QT intervals (15÷50% increases in QT time were measured for various drugs at therapeutic concentrations, and was further prolonged at higher concentrations). This method proved to be reliable for preclinical evaluation of new drugs and thus might be used in replacement of the limited, currently used assays.
Since the onset of hESC biology, researchers started focusing on modeling human disease “in a dish” and trying to elucidate their mechanisms. Successful attempts saw the production of hESC lines for cystic fibrosis (Mateizel et al. 2006; Pickering et al. 2005), Huntington’s disease (Bradley et al. 2011; Mateizel et al. 2006), and Fragile X syndrome (Eiges et al. 2007). However, the methods used to obtain genetically modified hESC are still inefficient (Giudice and Trounson 2008) thus failing to provide a solid source for predictive disease models that can be experimentally manipulated in vitro. iPS technology can offer an alternative after proving the feasibility of deriving patient-specific pluripotent stem cells, which can be used for a multiplicity of purposes, spanning from drug screenings, to cell replacement therapies and tissue engineering, and to disease modeling studies (Colman 2008).
Recently, two independent groups described the derivation of iPS cells from terminally differentiated mouse cells in stirred suspension cultures (Fluri et al. 2012; Shafa et al. 2012). These reprogrammed cells successfully responded to the “score-criteria” for iPSCs; the implementation of these novel strategies would thus enable standardized, scalable production and differentiation of cells to relevant phenotypes.
A series of diseases have already been successfully modeled in vitro via reprogramming of patient cells to iPS cells followed by differentiation into disease-relevant cells and tissues: neurodegenerative diseases, haematopoietic disorders, metabolic conditions and cardiovascular pathologies (Chamberlain et al. 2008; Ebert et al. 2009; Nishikawa et al. 2008; Park et al. 2008; Unternaehrer and Daley 2011). Patient-specific iPS cells were derived from Rett syndrome patients (Ananiev et al. 2011), successfully differentiated into neurons and used to better interpret the disease. Also, iPSCs were generated from patients with Amyotrophic lateral sclerosis (ALS), and were successfully directed to differentiate into motor neurons, the cell type destroyed in ALS (Dimos et al. 2008). Lee and Studer (Lee and Studer 2011) modeled familial dysautonomia (FD) and demonstrated a partial rescue of few disease-specific phenotypes in FD-iPS-derived via kinetin treatment. These FD-iPSCs could be a fundamental tool for high throughput screens of candidate drugs to reverse in vitro phenotypes and for mechanistic studies aimed at understanding disease pathogenesis.
It can be argued that the dynamics of disease progression in diseases such as Alzheimer’s or Parkinson’s develop in dramatically different scales in the patient and “in the dish”. However, researchers are now starting to adopt strategies to accelerate the onset of the pathological phenotypes in culture. Along this line, primary fibroblasts from patients with Parkinson’s (Soldner et al. 2009) and familial Alzheimer’s disease were successfully reprogrammed, differentiated into neurons and thoroughly characterized (Israel et al. 2012). In this case, iPSCs were successfully used to observe early onstage of Alzheimer’s-related phenotypes, even though it can take decades for overt disease to manifest in patients.
5. Current challenges
Advanced tissue culture platforms, recapitulating the in vivo environment in a controllable manner, have provided great tools to aid researchers in the transition from cell studies to animal models, human clinical trials and into the clinic. Advanced 3D and micro-bioreactor systems each offer unique advantages and present unique limitations associated with their respective size, providing unique engineering tradeoffs and design considerations for in vitro control of the stem cell niche (Figure 4). The challenges that remain to be resolved as the field progresses may be categorized into two main themes: (i) to establish conditions that are predictive of cell behavior in vivo, and (ii) to provide bioreactors beyond the laboratory bench.
Figure 4. Advanced culture platforms.
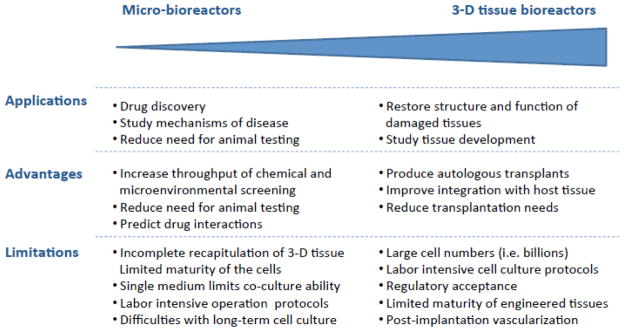
Micro-bioreactors and 3-D scaffold/bioreactor systems offer unique advantages and limitations associated with their respective size scale. These characteristics provide engineering tradeoffs and design considerations for in vitro control of the stem cell niche.
5.1. Producing conditions more predictive of cell behavior in vivo
Although bioreactors do outperform standard culture systems in their ability to provide matrix, molecular, and biophysical factors to cultured cells, current systems still lag behind the nature’s ability to deliver highly coordinated sequences of regulatory factors at the level of the cell, which are necessary to regulate cell function in a developing and adult organism.
For reasons that are still not fully understood, researchers have achieved more success with engineering mature tissues of certain types (such as cardiac and osteochondral) than other types (such as pancreatic and hepatic). Even with certain tissue types that grow reasonably well in bioreactor culture (such as bone and cartilage), challenges remain with respect to culturing multiple cell types on a single platform, whether in a 3D or micro-bioreactor configuration.
This issue is further complicated when considering in vitro models of disease: although many disease models have been developed using iPSCs, reproducible and consistent results between clones and lines have not yet been achieved. As for hESC lines, challenges remain in generating consistent total numbers of specific cell types, but the issue is further complicated by analysis of whether these cells undergo longitudinal changes that define the disease process. And although it may be argued that it is disease progression that may provide the most reliable assay with regard to compounds that could slow or prevent disease-specific cell death, until appropriately mature phenotypes are achieved consistently in culture, the field will face challenges with adapting engineered tissues to drug screening, disease models or implantable tissue grafts.
Another related challenge is that some known biophysical cues, such as molecular gradients, although relatively easy to apply in microfluidic conditions, are difficult to produce in the larger-scale 3D culture bioreactors, without applying significant shear stresses or using excessive amounts of expensive culture medium or scarce cells. Others, such as mechanical stimuli, extracellular matrix components, or even long-term cell culture conditions, may be easier to recapitulate in a 3D setting than within a micro-bioreactor.
Perhaps some of these challenges will be ameliorated through future high-throughput combinatorial studies performed in micro-bioreactors, which may shed more light on how tissues emerge from coordinated sequences of cell proliferation, differentiation, and functional assembly that are orchestrated by factors originating from the surrounding cells, matrix, and the external environment. In addition, future research on regeneration of adult tissues will likely help understand how cells respond to the entire milieu of injury or disease, and incorporate the most critical biophysical and/or molecular cues into the repertoire of current bioreactor systems. Finally, improvements in data acquisition and analysis techniques will aid the field in keeping up with an ever-expanding data set and with incorporating knowledge gleaned from basic research into even more advanced cell culture platforms.
5.2. Providing bioreactors beyond the laboratory bench
Challenges also remain in the area of bioreactor fabrication, operation, and medical regulation (Figure 4). Many of the bioreactor systems implemented up to date are at the development stage, and have been tested in stem cell research studies but not in basic biological studies. Closer collaboration between fields is expected to widen the opportunities for applying and optimizing bioreactor systems, and help disseminate user-friendly devices that can be easily and robustly operated. In the meantime, because bioreactors are often custom-designed, and because cells are highly sensitive to changes in their environment, even minute changes in molecular and biophysical cues may have implications on the reproducibility of the bioreactor regulation of cell survival, behavior, and differentiation.
A significant challenge is in the scale-up, standardization, and automation of cell culture protocols and bioreactor platforms. Automation will need to be applied wherever possible throughout the production pipeline, in order to derive the appropriate numbers of required cells, during culture when cells are matured (ie changing media etc), and coupled with quality control.
Furthermore, as bioreactor technology systems support engineering 3D tissue grafts for implantation, there will be further challenges to face related to the complexity of regulatory issues, and the design constraints of surgical techniques and the operating theater. For tissue-engineered constructs to be viable and clinically useful, for example, bioreactor design and protocols will need to be compatible with the transportation of biologically stable tissue engineered implants. In the meantime, in vitro applications for engineered tissues, such as experimental models of development and disease and chemical screening – will likely accelerate clinical acceptance of engineered tissues for implantation.
Acknowledgments
Studies described in this review have been supported by NIH (HL076485, DE016525, EB002520, AR061988 and RR026244 to GVN), New York State (C026449) and New York Stem Cell Foundation (Druckenmiller Fellowship to EC, independent investigator award to DM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nature Biotechnology. 2008;26(11):1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Alfred R, Taiani JT, Krawetz RJ, Yamashita A, Rancourt DE, Kallos MS. Large-scale production of murine embryonic stem cell-derived osteoblasts and chondrocytes on microcarriers in serum-free media. Biomaterials. 2011;32(26):6006–6016. doi: 10.1016/j.biomaterials.2011.04.015. [DOI] [PubMed] [Google Scholar]
- Allen JW, Bhatia SN. Formation of steady-state oxygen gradients in vitro: application to liver zonation. Biotechnol Bioeng. 2003;82(3):253–262. doi: 10.1002/bit.10569. [DOI] [PubMed] [Google Scholar]
- Allen JW, Khetani SR, Bhatia SN. In vitro zonation and toxicity in a hepatocyte bioreactor. Toxicol Sci. 2005;84(1):110–119. doi: 10.1093/toxsci/kfi052. [DOI] [PubMed] [Google Scholar]
- Ananiev G, Williams E, Li H, Chang Q. Isogenic pairs of wild type and mutant iPSc lines from Rett syndrome patients as in vitro disease model. PLoS ONE. 2011;6(9):e25255. doi: 10.1371/journal.pone.0025255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BM, Shah RP, Huang AH, Mauck RL. Dynamic tensile loading improves the functional properties of mesenchymal stem cell-laden nanofiber-based fibrocartilage. Tissue Engineering Part A. 2011;17(9–10):1445–1455. doi: 10.1089/ten.tea.2010.0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe DJ, Mensing GA, Walker GM. Physics and applications of microfluidics in biology. Annu Rev Biomed Eng. 2002;4:261–286. doi: 10.1146/annurev.bioeng.4.112601.125916. [DOI] [PubMed] [Google Scholar]
- Bhadriraju K, Chen CS. Engineering cellular microenvironments to improve cell-based drug testing. Drug Discovery Today. 2002;7(11):612–620. doi: 10.1016/s1359-6446(02)02273-0. [DOI] [PubMed] [Google Scholar]
- Bhatia SN, Balis UJ, Yarmush ML, Toner M. Effect of cell-cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999;13(14):1883–1900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- Bhumiratana S, Grayson WL, Castaneda A, Rockwood DN, Gil ES, Kaplan DL, et al. Nucleation and growth of mineralized bone matrix on silk-hydroxyapatite composite scaffolds. Biomaterials. 2011;32(11):2812–2820. doi: 10.1016/j.biomaterials.2010.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian L, Zhai DY, Zhang EC, Mauck RL, Burdick JA. Dynamic Compressive Loading Enhances Cartilage Matrix Synthesis and Distribution and Suppresses Hypertrophy in hMSC-Laden Hyaluronic Acid Hydrogels. tissue. Engineering Part A. 2012;18(7–8):715–724. doi: 10.1089/ten.tea.2011.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam SR, Tertoolen L, Van de Stolpe A, Meyer T, Passier R, Mummery CL. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Research. 2010;4(2):107–116. doi: 10.1016/j.scr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Bradley CK, Scott HA, Chami O, Peura TT, Dumevska B, Schmidt U, et al. Derivation of Huntington’s disease-affected human embryonic stem cell lines. Stem Cells and Development. 2011;20(3):495–502. doi: 10.1089/scd.2010.0120. [DOI] [PubMed] [Google Scholar]
- Chahine NO, Albro MB, Lima EG, Wei VI, Dubois CR, Hung CT, et al. Effect of dynamic loading on the transport of solutes into agarose hydrogels. Biophysical Journal. 2009;97(4):968–975. doi: 10.1016/j.bpj.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, Li XJ, Lalande M. Induced pluripotent stem (iPS) cells as in vitro models of human neurogenetic disorders. Neurogenetics. 2008;9(4):227–235. doi: 10.1007/s10048-008-0147-z. [DOI] [PubMed] [Google Scholar]
- Chang R, Nam J, Sun W. Direct cell writing of 3D microorgan for in vitro pharmacokinetic model. Tissue Engineering Part C Methods. 2008;14(2):157–166. doi: 10.1089/ten.tec.2007.0392. [DOI] [PubMed] [Google Scholar]
- Cheshier SH, Morrison SJ, Liao X, WIL In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. PNAS. 1999;96(6):3120–3125. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Sudo R, Mack PJ, Wan CR, Vickerman V, Kamm RD. Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab on a Chip. 2009;9:269–275. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- Cimetta E, Cannizzaro C, James R, Biechele T, Moon RT, Elvassore N, et al. Microfluidic device generating stable concentration gradients for long term cell culture: application to Wnt3a regulation of b-catenin signaling. Lab on a Chip. 2010;10:3277–3283. doi: 10.1039/c0lc00033g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimetta E, Sirabella D, Yeager K, Davidson K, Simon J, Moon RT, Vunjak-Novakovic Gordana. Microfluidic bioreactor for dynamic regulation of early mesodermal commitment in human pluripotent stem cells. Lab on a Chip. 2012 doi: 10.1039/C2LC40836H. Accepted Manuscript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimetta E, Figallo E, Cannizzaro C, Elvassore N, Vunjak-Novakovic G. Micro-bioreactor arrays for controlling cellular environments: Design principles for human embryonic stem cell applications. Methods. 2009;47:81–89. doi: 10.1016/j.ymeth.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman A. Induced pluripotent stem cells and human disease. Cell Stem Cell. 2008;3(3):236–237. doi: 10.1016/j.stem.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Correia C, Grayson WL, Park M, Hutton D, Zhou B, Guo XE, et al. In vitro model of vascularized bone: synergizing vascular development and osteogenesis. PloS ONE. 2011;6(12):e28352. doi: 10.1371/journal.pone.0028352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, et al. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nature Materials. 2007;6(12):997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- Dang SM, Gerecht-Nir S, Chen J, Itskovitz-Eldo rJ, Zandstra PW. Controlled, scalable embryonic stem cell differentiation culture. Stem Cells. 2004;22(3):275–282. doi: 10.1634/stemcells.22-3-275. [DOI] [PubMed] [Google Scholar]
- Davis RP, Van den Berg CW, Casini S, Braam SR, Mummery CL. Pluripotent stem cell models of cardiac disease and their implication for drug discovery and development. Trends in Molecular Medicine. 2011;17(9):475–484. doi: 10.1016/j.molmed.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Diao J, Young L, Kim S, Fogarty EA, Heilman SM, Zhou P, et al. A three-channel microfluidic device for generating static linear gradients and its application to the quantitative analysis of bacterial chemotaxis. Lab on a Chip. 2006;6:381–388. doi: 10.1039/b511958h. [DOI] [PubMed] [Google Scholar]
- DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. J Health Econ. 2003;22(2):151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321(5893):1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Dittrich PS, Manz A. Lab-on-a-chip: microfluidics in drug discovery. Nature Reviews - Drug Discovery. 2006;5:210–218. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- Ebert AD, Yu JY, Rose FF, Mattis VB, Lorson CL, Thomson JA, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457(7227):277–U271. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1(5):568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Ferreira LS, Gerecht S, Fuller J, Shieh HF, Vunjak-Novakovic G, Langer R. Bioactive hydrogel scaffolds for controllable vascular differentiation of human embryonic stem cells. Biomaterials. 2007;28(17):2706–2717. doi: 10.1016/j.biomaterials.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figallo E, Cannizzaro C, Gerecht S, Burdick JA, Langer R, Elvassore N, et al. Micro-bioreactor array for controlling cellular microenvironments. Lab on a Chip. 2007;7:710–719. doi: 10.1039/b700063d. [DOI] [PubMed] [Google Scholar]
- Fluri DA, Tonge PD, Song H, Baptista RP, Shakiba N, Shukla S, et al. Derivation, expansion and differentiation of induced pluripotent stem cells in continuous suspension cultures. Nature Methods. 2012;9(5):509–516. doi: 10.1038/nmeth.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok EY, Zandstra PW. Shear-controlled single-step mouse embryonic stem cell expansion and embryoid body-based differentiation. Stem Cells. 2005;23(9):1333–1342. doi: 10.1634/stemcells.2005-0112. [DOI] [PubMed] [Google Scholar]
- Fröhlich M, Grayson WL, Marolt D, Gimble JM, Kregar-Velikonja N, Vunjak-Novakovic G. Bone grafts engineered from human adipose-derived stem cells in perfusion bioreactor culture tissue. Engineering Part A. 2010;16(1):179–189. doi: 10.1089/ten.tea.2009.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich M, Grayson WL, Wan LQ, Marolt D, Drobnic M, Vunjak-Novakovic G. Tissue engineered bone grafts: biological requirements, tissue culture and clinical relevance. Current Stem Cell Research & Therapy. 2008;3(4):254–264. doi: 10.2174/157488808786733962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecht-Nir S, Cohen S, Itskovitz-Eldor J. Bioreactor cultivation enhances the efficiency of human embryoid body (hEB) formation and differentiation. Biotechnology And Bioengineering. 2004;86(5):493–502. doi: 10.1002/bit.20045. [DOI] [PubMed] [Google Scholar]
- Gerecht S, Burdick JA, Ferreira LS, Townsend SA, Langer R, Vunjak-Novakovic G. Hyaluronic acid hydrogel for controlled self-renewal and differentiation of human embryonic stem cells. PNAS. 2007;104(27):11298–11303. doi: 10.1073/pnas.0703723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem A, Shuler ML. Characterization of a perfusion reactor utilizing mammalian cells on microcarrier beads. Biotechnol Prog. 2000;16(3):471–479. doi: 10.1021/bp000047o. [DOI] [PubMed] [Google Scholar]
- Ghanem A, Shuler ML. Combining cell culture analogue reactor designs and PBPK models to probe mechanisms of naphthalene toxicity. Biotechnol Prog. 2000;16(3):334–345. doi: 10.1021/bp9901522. [DOI] [PubMed] [Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KE, Sacco A, Leonardi NA, Kraft P, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329(5995):1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice A, Trounson A. Genetic modification of human embryonic stem cells for derivation of target cells. Cell Stem Cell. 2008;2(5):422–433. doi: 10.1016/j.stem.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Gobaa S, Hoehnel S, Roccio M, Negro A, Kobel S, Lutolf MP. Artificial niche microarrays for probing single stem cell fate in high throughput. Nature Methods. 2011;8(11):949–955. doi: 10.1038/nmeth.1732. [DOI] [PubMed] [Google Scholar]
- Goldring CE, Duffy PA, Benvenisty N, Andrews PW, Ben-David U, Eakins R, et al. Assessing the safety of stem cell therapeutics. Cell Stem Cell. 2011;8(6):618–628. doi: 10.1016/j.stem.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Gomez-Sjoberg R, Leyrat AA, Pirone DM, Chen CS, Quake SR. Versatile, fully automated, microfluidic cell culture system. Anal Chem. 2007;79(22):8557–8563. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- Grayson WL, Bhumiratana S, Cannizzaro C, Chao PH, Lennon DP, Caplan AI, et al. Effects of initial seeding density and fluid perfusion rate on formation of tissue-engineered bone. Tissue Engineering Part A. 2008;14(11):1809–1820. doi: 10.1089/ten.tea.2007.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson WL, Fröhlich M, Yeager K, Bhumiratana S, Chan ME, Cannizzaro C, et al. Engineering anatomically shaped human bone grafts. Proc Nat Acad Sci U S A. 2010;107(8):3299–3304. doi: 10.1073/pnas.0905439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson WL, Marolt D, Bhumiratana S, Fröhlich M, Guo XE, Vunjak-Novakovic G. Engineering custom-designed osteochondral tissue grafts. Trends in Biotechnology. 2008;26(4):181–189. doi: 10.1016/j.tibtech.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson WL, Marolt D, Bhumiratana S, Fröhlich M, Guo XE, Vunjak-Novakovic G. Optimizing the medium perfusion rate in bone tissue engineering bioreactors. Biotechnology And Bioengineering. 2011;108(5):1159–1170. doi: 10.1002/bit.23024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanjaya-Putra D, Bose V, Shen YI, Yee J, Khetan S, Fox-Talbot K, et al. Controlled activation of morphogenesis to generate a functional human microvasculature in a synthetic matrix. Blood. 2011;118(3):804–815. doi: 10.1182/blood-2010-12-327338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Hagenmüller H, Koch AM, Müller R, Vunjak-Novakovic G, Kaplan DL, et al. Control of in vitro tissue-engineered bone-like structures using human mesenchymal stem cells and porous silk scaffolds. Biomaterials. 2007;28(6):1152–1162. doi: 10.1016/j.biomaterials.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Huh D, Torisawa YS, Hamilton GA, Kim HJ, Ingber DE. Microengineered physiological biomimicry: Organs-on-Chips. Lab On A Chip. 2012;12(12):2156–2164. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- Israel M, Yuan S, Bardy C, Reyna S, Mu Y, Herrera C, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482(7384):216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, et al. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Molecular Medicine. 2000;6(2):88–95. [PMC free article] [PubMed] [Google Scholar]
- Jeon NL, Dertinger SKW, Chiu DT, Choi IS, Stroock AD, Whitesides GM. Generation of Solution and Surface Gradients Using Microfluidic Systems. Langmuir. 2000;16:8311–8316. [Google Scholar]
- Jing D, Parikh A, Tzanakakis ES. Cardiac cell generation from encapsulated embryonic stem cells in static and scalable culture systems. Cell Transplantation. 2010;19(11):1397–1412. doi: 10.3727/096368910X513955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Experimental Cell Research. 1998;238(1):265–272. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Patterning proteins and cells using soft lithography. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- Kaplan D, Moon RT, Vunjak-Novakovic G. It takes a village to grow a tissue. Nature Biotechnology. 2005;23(10):1237–1239. doi: 10.1038/nbt1005-1237. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11(5):723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kehoe DE, Jing D, Lock LT, Tzanakakis ES. Scalable stirred-suspension bioreactor culture of human pluripotent stem cells. Tissue Engineering Part A. 2010;16(2):405–421. doi: 10.1089/ten.tea.2009.0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensah G, Gruh I, Viering J, Schumann H, Dahlmann J, Meyer H, et al. A novel miniaturized multimodal bioreactor for continuous in situ assessment of bioartificial cardiac tissue during stimulation and maturation. Tissue Eng Part C Methods. 2011;17(4):463–473. doi: 10.1089/ten.tec.2010.0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26(1):120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- Khetani SR, Szulgit G, Del Rio JA, Barlow C, Bhatia SN. Exploring interactions between rat hepatocytes and nonparenchymal cells using gene expression profiling. Hepatology. 2004;40(3):545–554. doi: 10.1002/hep.20351. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab On A Chip. 2012;12(12):2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- Kola I. The State of Innovation in Drug Development. Clin Pharmacol Ther. 2008;83(2):227–230. doi: 10.1038/sj.clpt.6100479. [DOI] [PubMed] [Google Scholar]
- Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3(8):711–716. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- Kraehenbuehl TP, Langer R, Ferreira LS. Three-dimensional biomaterials for the study of human pluripotent stem cells. Nature Methods. 2011;8(9):731–736. doi: 10.1038/nmeth.1671. [DOI] [PubMed] [Google Scholar]
- Krawetz R, Taiani JT, Liu S, Meng G, Li X, Kallos MS, et al. Large-scale expansion of pluripotent human embryonic stem cells in stirred-suspension bioreactors. Tissue Engineering Part C Methods. 2010;16(4):573–582. doi: 10.1089/ten.TEC.2009.0228. [DOI] [PubMed] [Google Scholar]
- Langer R, Vacanti JP. Tissue Engineering. Science. 1993;260(5110):920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- Lecault V, Vaninsberghe M, Sekulovic S, Knapp DJ, Wohrer S, Bowden W, et al. High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nature Methods. 2011;8(7):581–586. doi: 10.1038/nmeth.1614. [DOI] [PubMed] [Google Scholar]
- Lee G, Studer L. Modelling familial dysautonomia in human induced pluripotent stem cells. Philos Trans R Soc Lond B Biol Sci. 2011;366(1575):2286–2296. doi: 10.1098/rstb.2011.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AP. Accurate prediction of human drug toxicity: a major challenge in drug development. Chemico-Biological Interactions. 2004;150(1):3–7. doi: 10.1016/j.cbi.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Li AP. In vitro evaluation of human xenobiotic toxicity: scientific concepts and the novel integrated discrete multiple cell co-culture (IdMOC) technology. ALTEX. 2008;25(1):43–49. doi: 10.14573/altex.2008.1.43. [DOI] [PubMed] [Google Scholar]
- Li AP, Bode C, Sakai Y. A novel in vitro system, the integrated discrete multiple organ cell culture (IdMOC) system, for the evaluation of human drug toxicity: comparative cytotoxicity of tamoxifen towards normal human cells from five major organs and MCF-7 adenocarcinoma breast cancer cells. Chem Biol Interact. 2004;150(1):129–136. doi: 10.1016/j.cbi.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Lloyd KC. A knockout mouse resource for the biomedical research community. Ann N Y Acad Sci. 2011;1245:24–26. doi: 10.1111/j.1749-6632.2011.06311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock LT, Tzanakakis ES. Expansion and differentiation of human embryonic stem cells to endoderm progeny in a microcarrier stirred-suspension culture. Tissue Engineering Part A. 2009;15(8):2051–2063. doi: 10.1089/ten.tea.2008.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf MP, Hubbel JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nature Biotechnology. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- Ma B, Zhang G, Qin J, Lin B. Characterization of drug metabolites and cytotoxicity assay simultaneously using an integrated microfluidic device. Lab On A Chip. 2009;9(2):232–238. doi: 10.1039/b809117j. [DOI] [PubMed] [Google Scholar]
- Maidhof R, Tandon N, Lee EJ, Luo J, Duan Y, Yeager K, et al. Biomimetic perfusion and electrical stimulation applied in concert improved the assembly of engineered cardiac tissue. Journal of Tissue Engineering and Regenerative Medicine. 2011 doi: 10.1002/term.525. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marolt D, Augst A, Freed LE, Vepari C, Fajardo R, Patel N, et al. Bone and cartilage tissue constructs grown using human bone marrow stromal cells, silk scaffolds and rotating bioreactors. Biomaterials. 2006;27(36):6138–6149. doi: 10.1016/j.biomaterials.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Marolt D, I, Campos M, Bhumiratana S, Koren A, Petridis P, Zhang G, et al. Engineering bone tissue from human embryonic stem cells. Proc Natl Acad Sci U S A. 2012;109(22):8705–8709. doi: 10.1073/pnas.1201830109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin Y, Vermette P. Bioreactors for tissue mass culture: Design, characterization, and recent advances. Biomaterials. 2005;26(35):7481–7503. doi: 10.1016/j.biomaterials.2005.05.057. [DOI] [PubMed] [Google Scholar]
- Mateizel I, De Temmerman N, Ullmann U, Cauffman G, Sermon K, Van de Velde H, et al. Derivation of human embryonic stem cell lines from embryos obtained after IVF and after PGD for monogenic disorders. Human Reproduction. 2006;21(2):503–511. doi: 10.1093/humrep/dei345. [DOI] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Developmental Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- McCreedy T. Fabrication techniques and materials commonly used for the production of microreactors and micro total analytical systems. Trends in analytical chemistry. 2000;19(6):396–401. [Google Scholar]
- McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu HK, Schueller OJA, et al. Fabrication of microfluidic systems in poly(dimethylsiloxane) Electrophoresis. 2000;21(1):27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Montserrat N, De Oñate L, Garreta E, González F, Adamo A, Eguizábal C, et al. Generation of feeder free pig induced pluripotent stem cells without Pou5f1. Cell Transplantation. 2011 doi: 10.3727/096368911X601019. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Nie Y, Bergendahl V, Hei DJ, Jones JM, Palecek SP. Scalable culture and cryopreservation of human embryonic stem cells on microcarriers. Biotechnology Progress. 2009;25(1):20–31. doi: 10.1002/btpr.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebruegge S, Bauwens CL, Peerani R, Thavandiran N, Masse S, Sevaptisidis E, et al. Generation of human embryonic stem cell-derived mesoderm and cardiac cells using size-specified aggregates in an oxygen-controlled bioreactor. Biotechnology And Bioengineering. 2009;102(2):493–507. doi: 10.1002/bit.22065. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Goldstein RA, Nierras CR. The promise of human induced pluripotent stem cells for research and therapy. Nature Reviews Molecular Cell Biology. 2008;9(9):725–729. doi: 10.1038/nrm2466. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134(5):877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering SJ, Minger SL, Patel M, Taylor H, Black C, Burns CJ, et al. Generation of a human embryonic stem cell line encoding the cystic fibrosis mutation deltaF508, using preimplantation genetic diagnosis. Reproductive Biomedicine Online. 2005;10(3):390–397. doi: 10.1016/s1472-6483(10)61801-9. [DOI] [PubMed] [Google Scholar]
- Pouton CW, Haynes JM. Pharmaceutical applications of embryonic stem cells. Advanced Drug Delivery Reviews. 2005;57(13):1918–1934. doi: 10.1016/j.addr.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Pouton CW, Haynes JM. Embryonic stem cells as a source of models for drug discovery. Nature Review Drug Discovery. 2007;6(8):605–116. doi: 10.1038/nrd2194. [DOI] [PubMed] [Google Scholar]
- Report. Top R&D Drug Failures - Toxicity and Serious Adverse Events in Late Stage Drug Development are the Major Causes of Drug Failure. Global Business Intelligence Research; 2011. [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255(5052):1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Richards-Grayson AC, Shawgo RS, Johnoson AM, Flynn NT, Li Y, Cima MJ, et al. A BioMEMS Review: MEMS Technology for Physiologically Integrated Devices. Proceedings of the IEEE. 2004;92(1):6–21. [Google Scholar]
- Rolletschek A, Blyszczuk P, Wobus AM. Embryonic stem cell-derived cardiac, neuronal and pancreatic cells as model systems to study toxicological effects. Toxicology Letters. 2004;149:361–369. doi: 10.1016/j.toxlet.2003.12.064. [DOI] [PubMed] [Google Scholar]
- Rolletschek A, Wobus AM. Induced human pluripotent stem cells: promises and open questions. Biological Chemistry. 2009;390(9):845–849. doi: 10.1515/BC.2009.103. [DOI] [PubMed] [Google Scholar]
- Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, et al. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143(7):1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter E, Goh B, Hung B, Hutton D, Ghone N, Grayson WL. Bone tissue engineering bioreactors: a role in the clinic? Tissue Eng Part B Rev. 2012;18(1):62–75. doi: 10.1089/ten.TEB.2011.0209. [DOI] [PubMed] [Google Scholar]
- Salter E, Goh B, Hung B, Hutton D, Ghone N, Grayson WL. Bone tissue engineering bioreactors: a role in the clinic? Tissue Engineering Part B Reviews. 2012;18(1):62–75. doi: 10.1089/ten.TEB.2011.0209. [DOI] [PubMed] [Google Scholar]
- Sargent CY, Berguig GY, Kinney MA, Hiatt LA, Carpenedo RL, Berson RE, et al. Hydrodynamic modulation of embryonic stem cell differentiation by rotary orbital suspension culture. Biotechnology And Bioengineering. 2010;105(3):611–626. doi: 10.1002/bit.22578. [DOI] [PubMed] [Google Scholar]
- Schimming R, Schmelzeisen R. Tissue-engineered bone for maxillary sinus augmentation. J Oral Maxillofac Surg. 2004;62(6):724–729. doi: 10.1016/j.joms.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Shafa M, Day B, Yamashita A, Meng G, Liu S, Krawetz R, et al. Derivation of iPSCs in stirred suspension bioreactors. Nature Methods. 2012;9:465–466. doi: 10.1038/nmeth.1973. [DOI] [PubMed] [Google Scholar]
- Shin H, Zygourakis K, Farach-Carson MC, Yaszemski MJ, Mikos AG. Modulation of differentiation and mineralization of marrow stromal cells cultured on biomimetic hydrogels modified with Arg-Gly-Asp containing peptides. Journal of Biomedical Materials Research Part A. 2004;69(3):535–543. doi: 10.1002/jbm.a.30027. [DOI] [PubMed] [Google Scholar]
- Shultz LD, Brehm MA, Bavari S, Greiner DL. Humanized mice as a preclinical tool for infectious disease and biomedical research. Ann N Y Acad Sci. 2011;1245:50–54. doi: 10.1111/j.1749-6632.2011.06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikavitsas VI, Bancroft GN, Holtorf HL, Jansen JA, Mikos AG. Mineralized matrix deposition by marrow stromal osteoblasts in 3D perfusion culture increases with increasing fluid shear forces. PNAS. 2003;100(25):14683–14688. doi: 10.1073/pnas.2434367100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin A, Chin KC, Jamil MF, Kostov Y, Rao G, Shuler ML. The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol Prog. 2004;20(1):338–345. doi: 10.1021/bp034077d. [DOI] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell G, Cook E, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;6(136):964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son JS, Kim SG, Oh JS, Appleford M, Oh S, Ong JL, et al. Hydroxyapatite/polylactide biphasic combination scaffold loaded with dexamethasone for bone regeneration. Journal of Biomedical Materials Research Part A. 2011;99(4):638–647. doi: 10.1002/jbm.a.33223. [DOI] [PubMed] [Google Scholar]
- Squires TM, Quake SR. Microfluidics: Fluid physics at the nanoliter scale. Reviews of Modern Physics. 2005;77:977–1016. [Google Scholar]
- Steiner D, Khaner H, Cohen M, Even-Ram S, Gil Y, Itsykson P, et al. Derivation, propagation and controlled differentiation of human embryonic stem cells in suspension. Nature Biotechnology. 2010;28(4):361–364. doi: 10.1038/nbt.1616. [DOI] [PubMed] [Google Scholar]
- Sweeney LM, Shuler ML, Babish JG, Ghanem A. A cell culture analogue of rodent physiology: Application to naphthalene toxicology. Toxicol In Vitro. 1995;9(3):307–316. doi: 10.1016/0887-2333(95)00007-u. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takayama G, Taniguchi A, OT Identification of differentially expressed genes in hepatocyte/endothelial cell co-culture system. Tissue Engineering. 2007;13:159–166. doi: 10.1089/ten.2006.0143. [DOI] [PubMed] [Google Scholar]
- Tandon N, Marsano A, Maidhof R, Numata K, Montouri-Sorrentino C, Cannizzaro C, et al. Surface-patterned electrode bioreactor for electrical stimulation. Lab On A Chip. 2010;10(6):692–700. doi: 10.1039/b917743d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatosian DA, Shuler ML. A novel system for evaluation of drug mixtures for potential efficacy in treating multidrug resistant cancers. Biotechnol Bioeng. 2009;103(1):187–198. doi: 10.1002/bit.22219. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Toetsch S, Olwell P, Prina-Mello A, Volkov Y. The evolution of chemotaxis assays from static models to physiologically relevant platforms. Integrative Biology. 2009;(1):170–181. doi: 10.1039/b814567a. [DOI] [PubMed] [Google Scholar]
- Toner M, Irimia D. Blood-on-a-Chip. Annual Review of Biomedical Engineering. 2005;7:77–103. doi: 10.1146/annurev.bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran RT, Thevenot P, Zhang Y, Gyawali D, Tang L, Yang J. Scaffold Sheet Design Strategy for Soft Tissue Engineering. Nature Materials. 2010;3(2):1375–1389. doi: 10.3390/ma3021375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unternaehrer JJ, Daley GQ. Induced pluripotent stem cells for modelling human diseases. Philosophical Transactions of the Royal Society B. 2011;366:2274–2285. doi: 10.1098/rstb.2011.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viravaidya K, Shuler ML. Incorporation of 3T3-L1 cells to mimic bioaccumulation in a microscale cell culture analog device for toxicity studies. Biotechnol Prog. 2004;20(2):590–597. doi: 10.1021/bp034238d. [DOI] [PubMed] [Google Scholar]
- Viravaidya K, Sin A, Shuler ML. Development of a microscale cell culture analog to probe naphthalene toxicity. Biotechnol Prog. 2004;20(1):316–323. doi: 10.1021/bp0341996. [DOI] [PubMed] [Google Scholar]
- Vozzi F, Heinrich JM, Bader A, Ahluwalia AD. Connected culture of murine hepatocytes and HUVEC in a multicompartmental bioreactor. Tissue Eng Part A. 2009;15(6):1291–1299. doi: 10.1089/ten.tea.2008.0066. [DOI] [PubMed] [Google Scholar]
- Vozzi F, Mazzei D, Vinci B, Vozzi G, Sbrana T, Ricotti L, et al. A flexible bioreactor system for constructing in vitro tissue and organ models. Biotechnol Bioeng. 2011;108(9):2129–2140. doi: 10.1002/bit.23164. [DOI] [PubMed] [Google Scholar]
- Vunjak-Novakovic G, Meinel L, Altman G, Kaplan D. Bioreactor cultivation of osteochondral grafts. Orthodontics and Craniofacial Research. 2005;8(3):209–218. doi: 10.1111/j.1601-6343.2005.00334.x. [DOI] [PubMed] [Google Scholar]
- Wang F, Li Z, Tamama K, Sen CK, Guan J. Fabrication and characterization of prosurvival growth factor releasing, anisotropic scaffolds for enhanced mesenchymal stem cell survival/growth and orientation. Biomacromolecules. 2009;10(9):2609–2618. doi: 10.1021/bm900541u. [DOI] [PubMed] [Google Scholar]
- Wang WWW, Khetani SR, Krzyzewski S, Duignan DB, Obach RS. Assessment of a Micropatterned Hepatocyte Coculture System to Generate Major Human Excretory and Circulating Drug Metabolites. Drug Metabolism and Disposition. 2010;38(10):1900–1905. doi: 10.1124/dmd.110.034876. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature Biotechnology. 2007;25(6):681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Wen Y, Yang ST. The future of microfluidic assays in drug development. Expert Opinion on Drug Discovery. 2008;3(10):1237–1253. doi: 10.1517/17460441.3.10.1237. [DOI] [PubMed] [Google Scholar]
- Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Soft lithography in biology and biochemistry. Annual Review of Biomedical Engineering. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- Wobus AM, Loser P. Present state and future perspectives of using pluripotent stem cells in toxicology research. Archives of Toxicology. 2011;85(2):79–117. doi: 10.1007/s00204-010-0641-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Whitesides GM. Soft Lithography. Annual Review of Materials Science. 1998;28:153–184. [Google Scholar]
- Ye N, Qin J, Shi W, Liu X, Lin B. Cell-based high content screening using an integrated microfluidic device. Lab On A Chip. 2007;7(12):1696–1704. doi: 10.1039/b711513j. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhao Z, Abdul Rahim NA, van Noort D, Yu H. Towards a human-on-chip: Culturing multiple cell types on a chip with compartmentalized microenvironments. Lab on a Chip. 2009;9(22):3185–3192. doi: 10.1039/b915147h. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104(4):e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]