SUMMARY
Electrocardiographic (ECG) measurements vary by ancestry. Genome-wide association studies (GWAS) have identified loci that contribute to ECG measurements; however most are performed in Europeans collected from population-based cohorts or surveys. The strongest associations reported are in NOS1AP with QT interval and SCN10A with PR and QRS durations. The extent to which these associations can be generalized to African Americans has yet to be determined. Using electronic medical records, PR and QT intervals, QRS duration, and heart rate were determined in 455 African Americans as part of the Vanderbilt Genome-Electronic Records Project and Northwestern University NUgene Project. We tested for an association between these ECG traits and >930K SNPs. We identified a total 36 novel associations with PR interval, QRS duration, QT interval, and heart rate at p< 1.0 ×10−6. Using published GWAS data, we compared our results with those previously identified in other populations. Five associations originally identified in other populations generalized with respect to statistical significance and direction of effect. A total of 43 associations have a consistent direction of effect with European and/or Asian populations. This work provides a catalogue of generalized versus non-generalized associations, a necessary step in prioritizing GWAS-identified regions for further fine-mapping in diverse populations.
Keywords: Electrocardiography, African Americans, GWAS, Generalization, Electronic Medical Records
INTRODUCTION
Candidate gene studies have identified several genes that affect cardiac depolarization and conduction measured by ECG traits. One set of candidates consists of genes that encode voltage gated potassium and sodium channels, which also play an important role in the pathophysiology of long QT syndrome (Lai et al. 1994; Splawski et al. 2002; Wang et al. 1996; Wang et al. 1995a; Wang et al. 1995b). These associations have been confirmed by molecular genetic studies, and, in the past five years, GWAS have also identified these associations with various ECG phenotypes in European and Asian-descent populations (Chambers et al. 2010; Newton-Cheh et al. 2009; Smith et al. 2009). These GWAS have not only implicated common variation in congenital long QT syndrome genes, but also have identified associations between genes not previously implicated in cardiac electrophysiology and ECG traits, such as NOS1AP with QT interval (Arking et al. 2006; Post et al. 2007). Genome-wide association studies for ECG traits have also revealed pleiotropy: for example, variants in the cardiac sodium channel gene SCN5A, has been associated with across multiple ECG traits (Holm et al. 2010; Jeff et al. 2011; Marroni et al. 2009; Newton-Cheh et al. 2007; Smith et al. 2009). Variants in SCN10A, a gene previously known to play a role in the action potentials in nociceptive nerve fibers (Abrahamsen et al. 2008; Akopian et al. 1999; Zimmermann et al. 2007), have been identified by GWAS for an association with PR and QRS interval (Chambers et al. 2010; Denny et al. 2010; Holm et al. 2010; Pfeufer et al. 2010; Sotoodehnia et al. 2010).
As of October 2011, there were 127 unique trait-SNP associations reported in the National Human Genome Research Institute’s (NHGRI) GWAS catalog for electrocardiographic (ECG) traits (http://www.genome.gov/gwastudies/). Most of these studies were limited to European (118 associations) or Asian (9 associations)-descent populations, and it is unclear if these GWAS-identified variants generalize across populations, particularly the extent to which these associations generalize to African Americans. There is considerable variability in ECG traits across populations. African Americans have a shorter QRS duration, QTc interval, and a longer PR interval compared to Europeans (Ramirez et al. 2011). From a genetic standpoint, there are also significant differences in allele frequencies and linkage disequilibrium patterns between Africans and Europeans. For example, rs7626962 (S1103Y) in SCN5A is rare in European and Asian-descent populations but is common in African Americans and has been linked to arrhythmia susceptibility (Jeff et al. 2011; Splawski et al. 2002). Expanding genetic association studies to diverse populations has been of recent interest to many investigators in the field. To date there has been one GWAS published for ECG traits in African Americans on PR interval and one fine-mapping study for QT interval in African Americans (Avery et al. 2012; Smith et al. 2011). Missing from the current literature is a comprehensive study of multiple ECG traits that explain the generalizability or lack thereof across populations. Furthermore, to date there are no GWAS reported for heart rate in African Americans.
Assessing the utility of electronic medical records (EMRs) systems coupled to DNA repositories as a tool for genome science is one of the primary objectives for the National Human Genome Research Institute’s electronic MEdical Records and GEnomics (eMERGE) Network (McCarty et al. 2011). Studies from eMERGE, including those on PR interval, have demonstrated that EMR-based genetic studies replicate existing findings and discover new ones (Crosslin et al. 2011; Denny et al. 2010; Denny et al. 2011; Turner et al. 2011). Here we performed a GWAS of ECG measurements to identify novel genetic associations and describe the extent to which previous associations generalize in African Americans within the eMERGE network. We also examine reasons for non-generalization of associations in our African American study population.
MATERIALS AND METHODS
Study population
African American subjects were collected from the Vanderbilt or Northwestern University biobanks. The Vanderbilt Genome-Electronic Records (VGER) accesses BioVU, the Vanderbilt biorepository of DNA extracted from blood collected for routine clinical care linked to de-identified electronic medical records (Roden et al. 2008). BioVU is a subset of the larger repository of de-identified EMRs known as the synthetic derivative. The Northwestern biobank, NUgene, combines DNA samples from consented participants with enrollment questionnaire and longitudinal data from the EMR (McCarty et al. 2011). Both biobanks were approved by Institutional Review Boards at their respective sites.
Study population demographics and characteristics are described in Table 1. Individuals with a normal ECG without evidence of cardiac disease before or within one month following the ECG, without concurrent use of medications that interfere with QRS duration, and who did not have abnormal electrolyte values at the time of the ECG were included. Using natural language processing combined with EMR (structured database queries) to query (specific to VGER), we excluded individuals with any of the following before or within one month of the ECG: indication of heart failure, arrhythmia, cardiomyopathy, cardiac conduction defect, or myocardial ischemia/infarct based on clinical notes, unstructured text, billing codes, and labs. VGER subjects were African American as indicated by observer reported ancestry, which is highly concordant with genetic ancestry (Dumitrescu et al. 2010). African American ancestry was self-reported for Northwestern subjects (McCarty et al. 2011), which is also known to be highly concordant with genetic ancestry. All ECGs had normal Bazett’s corrected QT intervals (<450ms), heart rates (between 50–100 bpm), and QRS duration (65–120 ms).
Table 1. Descriptive statistics of the study population.
Means and standard deviations were calculated in the final study population for all covariates and ECG traits unless otherwise noted.
| Study Population (n= 455) | ||
|---|---|---|
| Variable | Mean /% | SD |
| Female | 77% | -- |
| Age (yr) | 46 | 15 |
| PR duration (msec) | 159 | 21 |
| QRS duration (msec) | 82 | 8 |
| QTc duration (msec) | 410 | 21 |
| Heart Rate (msec) | 74 | 11 |
| On QT drug | 10% | -- |
| On PR drug | 21% | -- |
Genotyping
Genotyping for the eMERGE network was performed by the Center for Inherited Disease Research (CIDR) and the Broad Institute. All individuals that met the inclusion criteria (n = 501) were genotyped for >1.1 million SNPs using the Illumina 1M BeadChip at the Broad Institute. Data were cleaned by the eMERGE QC pipeline (Zuvich et al. 2011). Individuals with cryptic relatedness, ancestry inconsistent with observer- or self-reported ancestry, anomalous X-chromosome heterozygosity or poor genotyping efficiency were removed from further analysis (n = 46). All markers that were intensity only probes that had technical failure, minor allele frequency ≤ 0.05, genotyping efficiency <99%, discordant calls with duplicates, and Mendelian errors >0 were removed. Tests of association were performed with and without SNP that deviated from Hardy Weinberg Equilibrium (p-value <1.0 × 10 −4), and consistent findings were observed for both SNP sets (data not shown).
Statistical methods
Greater than 930K SNPs from the Illumina 1M BeadChip were tested for an association with PR interval, QRS duration, QTc interval, and heart rate using linear regression assuming an additive genetic model. Tests of association were performed unadjusted and adjusted for age, sex, PR/QT drug usage (for PR and QT interval only), and principal components (PCs; two to four PCs depending on the trait) using PLINK (Purcell et al. 2007). All results were plotted using Synthesis View, Haploview, or Locus Zoom (Barrett et al. 2005; Pendergrass et al. 2010; Pruim et al. 2010). Power calculations were performed using QUANTO (Gauderman & Morrison 2006) assuming genetic effect sizes based on the original published GWAS. The fixation index FST, a measure of population differentiation, was calculated using the Weir and Cockerham algorithm (Weir & Cockerham 1984). We calculated FST between European descent populations and our African American study population using the Platform for the Analysis, Translation, and Organization of large-scale data (PLATO) (Grady et al. 2010). Pair-wise linkage disequilibrium was calculated (r2) around the SCN5A/SCN10A and NOS1AP regions using the SeattleSNPs Genome Variation Server (gvs.gs.washington.edu/).
Generalization
Using the NHGRI GWAS catalog, we identified SNPs associated with PR interval, QTc interval, QRS duration, and heart rate at significance threshold of p<10−6 from European or Asian -descent populations. We then examined the significance level and direction of effect for the same SNP-trait pair in this African American dataset.
RESULTS
Discovery
None of the SNPs tested for an association met genome-wide significance (p<5.0×10−8) with any ECG trait. Although we did not detect associations at genome-wide significance, we were able to detect novel associations at p<10−6 (Table 2, Supplementary Fig. 1). Nine common variants were associated with increased heart rate, representing five independent disease loci. Three SNPs were in complete linkage disequilibrium with each other (r2= 1.0) in SERPINI1 on chromosome 3. Aside from SNPs associated with heart rate, nine associations at p<10−6 were observed for QT interval, QRS duration, and PR interval, separately (27 associations total). Collectively, these potentially novel associations represent 17 candidate genes throughout the genome associated with these traits. None of these associations have been reported by previously published GWAS for ECG traits in any population.
Table 2. Most significant genome-wide association study (GWAS) results in African Americans for heart rate, QT interval, QRS duration, and PR interval.
We performed single SNP tests of association between >930 SNPs and ECG traits using linear regression assuming an additive genetic model in African Americans (n=455). All tests of association were adjusted by age, sex, and principal components. Associations at p ≤ 10−6 are shown, and for each association, chromosome, rs number, nearest gene, coded allele, beta coefficient, and p-value are given.
| Heart Rate | |||||
|---|---|---|---|---|---|
| CHR | SNP | LOCATION | CODED ALLELE | BETA | P |
| 8 | rs1015003 | Intergenic | G | 3.94 | 1.21E-06 |
| 8 | rs6468401 | Intergenic | A | 3.93 | 1.55E-06 |
| 5 | rs816475 | Intergenic | T | 3.69 | 2.83E-06 |
| 3 | rs13090836 | SERPINI1 (intron) | T | 3.58 | 6.86E-06 |
| 3 | rs9815034 | SERPINI1 (intron) | A | 3.59 | 7.26E-06 |
| 3 | rs1473511 | SERPINI1 (intron) | T | 3.59 | 7.26E-06 |
| 12 | rs12824981 | TMEM132D (intron) | T | 4.56 | 8.90E-06 |
| 9 | rs35061590 | DAB2IP (intron) | T | 16.03 | 9.45E-06 |
| 9 | rs13290547 | DAB2IP (intron) | T | 16.03 | 9.45E-06 |
| QT Interval | |||||
| CHR | SNP | LOCATION | CODED ALLELE | BETA | P |
| 5 | rs6894385 | Intergenic | C | −9.92 | 1.03E-06 |
| 6 | rs9342616 | Intergenic | A | −6.92 | 1.04E-06 |
| 7 | rs12666280 | DPP6 (intron) | C | 8.93 | 1.66E-06 |
| 20 | rs237450 | Intergenic | A | 6.86 | 5.36E-06 |
| 4 | rs6819013 | Intergenic | A | 6.06 | 5.50E-06 |
| 4 | rs4698433 | FGFBP2 (near 5′ region) | T | 7.23 | 5.53E-06 |
| 4 | rs1483012 | LDB2 (intron) | G | 6.02 | 7.19E-06 |
| 16 | rs8045405 | Intergenic | G | −11.38 | 9.44E-06 |
| 15 | rs17237606 | UNC13C (intron) | G | −8.91 | 9.92E-06 |
| QRS Duration | |||||
| CHR | SNP | LOCATION | CODED ALLELE | BETA | P |
| 14 | rs1867082 | Intergenic | A | 2.82 | 1.46E-06 |
| 6 | rs504008 | NKAIN2 (intron) | C | 2.70 | 2.58E-06 |
| 5 | rs6861497 | Intergenic | A | 2.34 | 4.73E-06 |
| 11 | rs308309 | Intergenic | C | 4.39 | 4.95E-06 |
| 6 | rs12194062 | C6orf190 (intron) | T | 5.16 | 5.10E-06 |
| 4 | rs6820368 | ADH6 (intron) | C | 9.90 | 5.57E-06 |
| 12 | rs10784762 | Intergenic | T | −2.38 | 6.31E-06 |
| 16 | rs17444745 | Intergenic | A | −6.40 | 6.96E-06 |
| 1 | rs13375391 | HMCN1 (intron) | A | 5.82 | 9.37E-06 |
| PR Interval | |||||
| CHR | SNP | LOCATION | CODED ALLELE | BETA | P |
| 11 | rs1994318 | MICAL2 (intron) | A | −6.79 | 1.52E-06 |
| 6 | rs10447419 | Intergenic | A | −8.65 | 1.95E-06 |
| 10 | rs16926523 | MYO3A (intron) | A | 9.55 | 4.23E-06 |
| 2 | rs7604827 | VWC2L (intron) | C | 6.54 | 4.52E-06 |
| 3 | rs3733017 | BCL6 (intron) | G | −10.07 | 5.55E-06 |
| 15 | rs746265 | Intergenic | C | −6.27 | 7.92E-06 |
| 3 | rs1524976 | MAGI1 (intron) | A | 8.00 | 7.96E-06 |
| 1 | rs3103778 | MFSD2 (intron) | G | 6.30 | 8.67E-06 |
| 15 | rs12595668 | Intergenic | G | −6.26 | 9.01E-06 |
Generalization
In European-descent populations, 118 GWAS-identified SNPs have been reported for ECG traits in the NHGRI GWAS catalog as of October 2011. We examined the reported European-descent SNP-trait association to determine if the association “generalized” to African Americans. We considered an association generalized if the observed association was significant at a liberal significance threshold (p<0.05) and had a direction of effect consistent with the original study. Of the 118 SNPs identified in the NHGRI GWAS catalog, 92 were also directly genotyped in our study population: 26, 14, 19, and 33 SNPs were previously associated with heart rate, PR interval, QRS duration, and QT interval, respectively.
Overall, there were five SNP-trait associations that generalized to African descent populations with respect to both level of significance (p<0.05) and direction of effect: for heart rate rs4352210 (intergenic, β= −1.6, p= 0.03), for QTc interval, rs1112795 (SCN5A, β = − 4.77; p=0.05) and rs4725982 (KCNH2, β = 4.79; p=0.03), and for QRS duration, rs6795970 (SCN10A, β = 2.43; p=0.01) and rs11710077 (SCN5A, β= 2.42; p=0.004) (Table 3). No SNPs generalized for PR interval. There were six additional SNP-trait associations from the GWAS catalog that were significant (p-value <0.05) but trended in the opposite direction in African Americans compared with European-descent populations (after accounting for the coded allele): four for PR interval, three for heart rate, and one for QT interval (Supplementary Table 1).
Table 3. Association results for genome-wide association study (GWAS)-identified associations that generalize in African Americans.
We compared GWAS-identified association results originally identified in European or Asian-descent populations to 455 African Americans from VGER and NUgene in the eMERGE network. We declared a generalization as having a consistent direction of genetic effect (expressed as a beta coefficient) and p-value <0.05. Abbreviations: CAF= coded allele frequency.
| Trait | SNP | Coded Allele | Gene | Original Study | African Americans | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Population | P-value | Effect | CAF | P-value | Effect | CAF | ||||
| QT interval | rs11129795 | A | SCN5A | European | 5.0E-14 | −1.27 | 0.23 | 0.05 | −4.77 | 0.16 |
| QT interval | rs4725982 | T | KCNH2 | European and Asian-descent | 5.0E-16 | 1.58 | 0.22 | 0.03 | 4.79 | 0.23 |
| QRS duration | rs6795970 | A | SCN10A | European and Asian-descent | 4.0E-09 | 5.17 | 0.36 | 0.01 | 2.43 | 0.08 |
| QRS duration | rs11710077 | T | SCN5A | European | 1.0E-06 | 0.44 | 0.21 | 0.004 | 2.42 | 0.11 |
| Heart rate | rs4352210 | A | Intergenic | European | 2.00E-06 | −0.14 | 0.37 | 0.03 | −1.6 | 0.44 |
An additional 40 SNP-trait associations failed to achieve the liberal threshold of significance (p<0.05) in African Americans but did have consistent directions of effect compared with European-descent populations (Supplementary Table 2, Fig. 1). Based solely on direction of effect, approximately 50% and 52% of the associations tested for PR and QT intervals, respectively, have a consistent direction of effect in African Americans compared with European descent populations. For QRS duration, 37% of the associations tested generalized in African Americans while 44% generalized for heart rate. Collectively, 44 out of 92 (47%) SNP-trait associations had a consistent direction effect regardless of significance in African Americans compared to previously identified associations in European-descent populations. The remaining 48/92 (52%) SNP-trait associations identified in European-descent populations did not generalize to African Americans with respect to either level of significance or direction of effect.
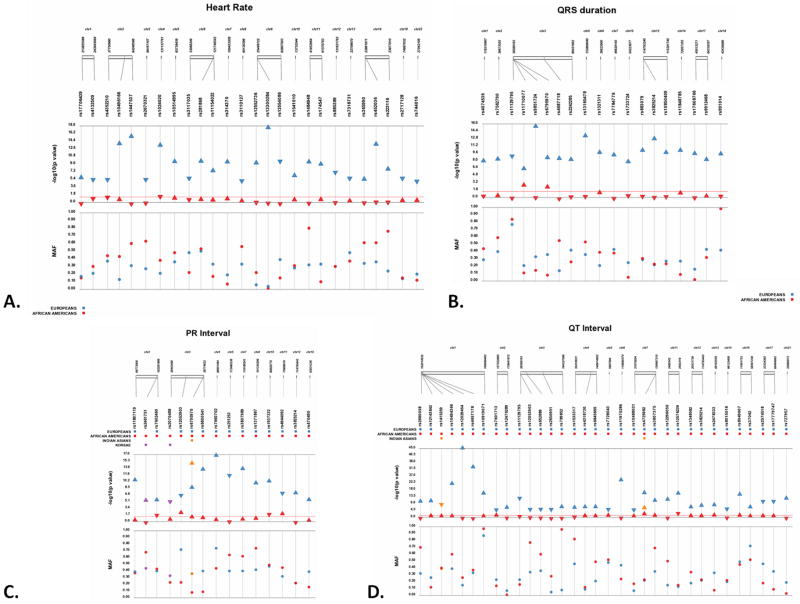
Figure 1. (A–D). SNP-trait association comparison between European or Asian -descent and African American populations across ECG traits.
Each SNP was tested for an association with each ECG trait assuming an additive genetic model adjusted for age and sex. P-values are −log10 transformed along the y-axis and corresponding location for each SNP is located on the x-axis. Each point represents a p-value for each population indicated by color (see legend). The direction of the arrows corresponds to the direction of the effect (measured by beta coefficient). The significance threshold is indicated by the red bar at p= 0.05. The bottom panel displays the minor allele frequency comparisons for both populations for each SNP.
There were ten SNPs reported in the NHGRI catalog that are associated with ECG traits in Asian -descent populations. Of these, six SNPs were also genotyped in our study population. None of these SNPs generalized in African Americans with respect to significance and direction of effect, although two of these associations had a consistent direction of effect compared with the original study population.
Power
SNP-trait associations that failed to achieve significance or failed to generalize to African Americans may represent lack of power or true differences in allelic architecture and/or linkage disequilibrium patterns between the two populations. To help distinguish between these two possibilities, we calculated the power to detect the mostly European-reported associations in this African American sample. Overall, 17 SNP-trait tests of association were adequately powered (>80%). Among the adequately powered tests of association, almost all (16/17) SNP-trait tests of association failed to generalize in African Americans with respect to level of significance anddirection of effect (Table 4).
Table 4. Association results for adequately powered genome-wide association study (GWAS)-identified SNPs that do not generalize in African Americans.
Using the effect size, mean, and standard deviations reported in the published study, we calculated our power to detect the same effect given our sample size and minor allele frequency in African Americans. CAF= coded allele frequency.
| SNP | Trait | Gene | Coded Allele | Original Study | African Americans | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CAF | P-value | Beta | CAF | P-value | Beta | Power | ||||
| rs10494366 | QT Interval | NOS1AP | G | 0.39 | 5.00E-22 | 12.2 | 0.60 | 0.88 | 0.28 | 0.99 |
| rs11129795 | QRS duration | SCN5A | G | 0.77 | 5.00E-10 | −8.24 | 0.84 | 0.73 | −0.25 | 0.99 |
| rs12296050 | QT Interval | KCNQ1 | T | 0.15 | 1.00E-11 | 13.16 | 0.50 | 0.52 | −1.20 | 0.99 |
| rs3807989 | PR interval | CAV1 | A | 0.40 | 7.00E-13 | 6.40 | 0.62 | 0.46 | 0.94 | 0.95 |
| rs3825214 | QRS duration | TBX5 | G | 0.22 | 3.00E-13 | 7.35 | 0.23 | 0.52 | −0.41 | 0.99 |
| rs3825214 | PR interval | TBX5 | G | 0.22 | 1.00E-07 | 5.88 | 0.22 | 0.79 | −0.40 | 0.81 |
| rs1321311 | QRS duration | CDKN1A | T | 0.21 | 3.00E-10 | 6.52 | 0.39 | 0.17 | 0.77 | 0.99 |
| rs174547 | Heart Rate | FADS1 | C | 0.33 | 2.00E-09 | 6.20 | 0.10 | 0.43 | 1.028 | 0.99 |
| rs223116 | Heart Rate | MYH7, NDNG | A | 0.24 | 3.00E-08 | 7.40 | 0.76 | 0.44 | −0.635 | 0.99 |
| rs2461751 | PR interval | Intergenic | G | 0.44 | 8.00E-06 | 4.54 | 0.68 | 0.83 | −0.30 | 0.80 |
| rs281868 | Heart Rate | SLC35F1 | G | 0.50 | 4.00E-10 | 6.30 | 0.53 | 0.41 | 0.64 | 0.99 |
| rs314370 | Heart Rate | SLC12A9 | C | 0.19 | 6.00E-10 | 7.60 | 0.07 | 0.28 | 1.58 | 0.99 |
| rs365990 | HR | MYH6 | G | 0.34 | 7.00E-06 | 5.25 | 0.61 | 0.61 | −0.41 | 0.99 |
| rs3807375 | QT Interval | KCNH2 | T | 0.35 | 5.00E-11 | 11.95 | 0.69 | 0.32 | 1.95 | 0.99 |
| rs452036 | Heart Rate | MYH6 | A | 0.36 | 4.00E-14 | 7.80 | 0.61 | 0.41 | −0.68 | 0.99 |
| rs7660702 | PR interval | ARHGAP24 | T | 0.74 | 3.00E-17 | 8.46 | 0.44 | 0.60 | 0.65 | 0.99 |
Allele Frequencies
We calculated the coded allele frequencies for previously identified SNPs (from European descent populations) in African Americans (Supplementary Table 3). To measure population differences, we calculated FST for all SNPs previously identified by GWAS in European descent populations that report sample sizes and minor allele frequencies (85 SNPs). There were 20 SNPs with an FST value >0.15 (ranging from 0.15 to 0.85), which is indicative of significant population differentiation at these loci. We observed the largest FST value (F= 0.85) for rs789852 located in the intron region of the TMEMFF gene located on chromosome 3. This SNP was previously associated with QT interval in Europeans (β= 0.25, p=7.0 × 10−7 ) (Marroni et al. 2009). This association did not generalize in African Americans and trended in the opposite direction compared to Europeans (β = −0.95, p =0.60).
DISCUSSION
Here we identified novel associations with heart rate, QT interval, QRS duration, and PR interval. We also characterized previously reported associations in our African American study population derived in a clinical based setting. To our knowledge we are the first to perform a genome-wide association study for heart rate in African Americans and to characterize GWAS-identified variants in European and Asian-descent populations to African Americans for heart rate, QT interval, and QRS duration.
Discovery
We have identified 36 novel associations in 17 candidate genes with the four ECG traits tested at p< 1.0 × 10 −6. While none of our associations met genome-wide significance, reporting these associations can provide insight to novel regions in the genome that effect ECG trait variability yet to be explored in African populations. Interestingly we were able to detect a novel region on chromosome 3 with three SNPs that are in high LD and are associated with increased heart rate in SERPINI1 (Supplementary Fig. 1C). The SERPINI1 gene encodes serine peptidase inhibitors secreted by axons in the brain and has been associated with familial encephalopathy (Davis et al. 1999).
We also identified a region on chromosome 9 within the DAB2IP gene encompassing two SNPs that were associated with increased heart rate. This gene encodes a GTPase-activating protein and is an inhibitor of cell growth and survival. DAB2IP has been identified by a GWAS for an association with prostate cancer in European and African descent populations (Gretarsdottir et al. 2010). Recently, a GWAS of individuals from Iceland and the Netherlands identified an association with DAB2IP with abdominal aortic aneurysms and other vascular diseases including early onset myocardial infarction (Gretarsdottir et al. 2010).
Generalization
Four of the five generalizations we identified in African Americans are located in voltage-gated channels (Table 3). For PR interval, we compared the SCN5A/SCN10 region association results in African Americans with previously identified European Americans (also ascertained from VGER; Supplementary Fig. 2)(Denny et al. 2010). Though not significant, the index SNP identified in Europeans was different compared to African Americans. This is likely due to differences in linkage disequilibrium patterns between the two populations (Supplementary Figs. 2 and 3).
For QRS duration, two of these variants are within the SCN5A/SCN10A region and generalize in African Americans (Table 3). SCN10A variant rs97595970 originally associated with PR interval has been previously reported to have pleiotropic effects with QRS duration (Chambers et al. 2010; Holm et al. 2010; Sotoodehnia et al. 2010). This association for QRS duration generalized to African Americans (β = 2.43, p= 0.01). There were a total of three associations that generalized in African Americans for QT interval, two of which are in voltage gated channels KCNH2 and SCN5A (Table 3). The third association, rs12143842, is located in NOS1AP, which has been consistently associated with prolonged QT interval (Arking et al. 2006). The effect of this variant accounts for more 1.5% of the trait variability in Europeans (Arking et al. 2006) and 1.3% in our African Americans. For PR interval, two previously identified associations in the SCN10A gene have consistent directions of effect with African Americans (Fig. 1, Supplementary Table 2). Non-synonymous variant rs6795970 (in the SCN10A gene) has been recently reported to have an association with increased PR interval in Asian and European-descent populations (Holm et al. 2010; Pfeufer et al. 2010). In African Americans the magnitude of this effect (β= 3.67) is consistent with Europeans (β= 5.17) (Fig. 1, Supplementary Table 2).
There were 48 associations that did not generalize to African Americans with respect to direction of effect and statistical significance. GWAS associations identified in European descent populations may not generalize in African Americans for several reasons. More often than not, this is due to discordant minor allele frequency differences and linkage disequilibrium (LD) patterns, both of which impact statistical power. On average for SNPs that do not generalize, the MAF in the original population was significantly different compared to the MAF in our study population (p ≤ 0.01). The MAF was lower in African Americans compared to the original study population for 60% of the SNPs that do not generalize (Fig. 1). Low minor allele frequency is directly correlated with power in genetic association studies. We also noticed this correlation in our analysis; 13/79 inadequately powered tests of association had a minor allele frequency < 0.09.
Replication of published GWAS in African Americans
As previously mentioned, there have been one recent GWAS and one fine-mapping study performed in African Americans for ECG traits (Avery et al. 2012; Smith et al. 2011). The first GWAS in African Americans for PR interval described an association between SCN5A intronic rs3922844 and increased PR interval (Smith et al. 2011). This association was also identified in an independent candidate gene study of SCN5A in African Americans (Jeff et al. 2011). In the present study, rs3922844 was associated with PR interval at p = 0.01 and a consistent direction of effect (β = 3.06) compared with the published reports (Supplementary Table 4). To date rs3922844 has only been identified in African descent populations and thus was not included in this generalization analysis. Although not statistically significant, we observed similar results for the other SNPs published by Smith et al (2011) (Supplementary Table 4). Despite the lack of power, these data collectively suggest and confirm previous reports that SCN5A has pleiotropic effects and is associated with multiple ECG traits in African Americans (Supplementary Table 4) (Jeff et al. 2011).
In the second study, GWAS regions identified for the QT interval in European and Asian populations were further fine-mapped in >8,000 African Americans (Avery et al. 2012). Approximately 40% of the associations reported in European or Asian populations generalized in the African American study population, which is consistent with this present study’s generalization results, despite differences in sample size. Three novel associations with QT interval specific to African Americans were also reported; of these, two of the reported SNPs were not on the Illumina 1M BeadChip used for this present study, and one SNP (rs12061601) did not replicate in our African American study population (Supplementary Table 5). The fine-mapping effort also highlighted differences in the index SNP associated with QT interval between African Americans and Europeans. We compared the index SNPs reported in African Americans from the fine-mapping study to our study population (Supplementary Table 5). Of the fifteen index SNPs reported, only six SNPs were directly genotyped in our study of which only two, rs1805120 and rs735951, trended towards significance (Supplementary Table 5).
The Effects of Linkage Disequilibrium
Linkage disequilibrium (LD) is often used in genetic association studies to select tagSNPs for genotyping that represent a region in the genome. While this method is cost effective, identifying an association with a tagSNP does not necessarily identify the true functional variant given that the un-assayed functional variant is most likely in LD with the assayed variant. TagSNPs are also population specific; therefore, associations identified in one population may not necessary generalize to another population if only the index variant is genotyped. Our observations in this African American population compared with European descent populations for ECG trait associations are consistent with known properties of tagSNPs. That is, there are differences in LD between African Americans and Europeans for known loci associated with PR interval, QRS duration, and QT interval. For the SCN10A/SCN5A and NOS1AP regions, we calculated pair-wise LD (r2) for each SNP pair. As expected, there is less linkage disequilibrium in African descent populations compared to European populations (Supplementary Figs. 3 and 4) (Rosenberg et al. 2010). This lack of LD could account for the non-generalization of associations in African Americans originally identified in European descent populations.
LD is often described as a “double-edged” sword: strong LD can be advantageous to identifying associations, but is it not useful when searching for the functional variants. Using populations with low amounts of LD can help fine map the functional variant (Rosenberg et al. 2010). Therefore, index associations that robustly associate with ECG traits in European descent populations but do not generalize to African descent populations could be prioritized for fine-mapping studies to identify the functional variant. Also, further large discovery and fine-mapping studies should be performed in African descent populations to identify additional ECG-trait associated loci not found in other populations.
Limitations and Strengths
There were several factors that limited our ability to detect novel associations at the GWAS level (at p< 1.0 × 10 −8) with ECG traits. Our study population compared to recently published GWAS for ECG traits is relatively small and underpowered. Specifically, we were underpowered (<80%) to detect effects that explain less than 10% of the trait variability at p<1.0 × 10 −8 even for common variants (MAF >0.05). Power also limited our ability to generalize European/Asian identified associations to African Americans. As previously mentioned power is directly correlated to allele frequencies. Most GWAS fixed-content products are biased to common variation and based on LD patterns of European populations (Spencer et al. 2009). Therefore, the SNPs previously identified may only be common in Europeans and may be rare in other populations such as African Americans, limiting our power to generalize these variants. It is important to note the power calculations we report in our generalization analysis were limited to the effect sizes detected in the original study. These effect sizes could be subject to the “winner’s curse” and may over estimate the true effect size (Xiao & Boehnke 2009; Zhong & Prentice 2010). Using these potentially overestimated effect sizes in our power calculations could consequently over estimate our power to detect these effects.
Another limitation to both the discovery and the generalization analyses was our liberal significance threshold. For discovery, a conservative Bonferroni threshold to correct for multiple testing is the generally accepted 5.0×10−8. Given our small sample size and this conservative threshold, we had sufficient power to detect large genetic effect sizes; however, these effects are not realistic given previously reported candidate gene and GWAS findings for these traits. To address this limitation, we arbitrarily choose a significance threshold of 1.0×10−6 to identify possible real associations that would be missed by the conservative significance threshold. For generalization, we declared a generalization as having a consistent direction of effect and meeting a liberal significance threshold of p<0.05. Given that the generalization analysis was limited to only a subset of loci that was previously identified, a genome-wide correction for multiple testing was not necessary.
Here we confirm the ability to use electronic medical records (EMRs) linked to DNA to identify genotype-phenotype relationships. We identified several potentially novel associations across the genome for heart rate, PR interval, QT interval, and QRS duration. Additionally, we generalized several associations identified by GWAS in European and Asian -descent populations to African Americans. Most importantly, we extensively characterized European and Asian identified associations in African Americans by reporting population specific information such as allele frequency and linkage disequilibrium differences, which may be important in both research and clinic-based settings.
We successfully assessed the generalizability of European identified genetic associations for ECG traits in African Americans derived from an electronic medical record. Additionally, we report novel associations for ECG traits in African Americans. Replication of these findings in an independent African American population is needed to confirm these associations. Indeed, in the future, replication may be possible within the eMERGE network given that the network has expanded with respect to study sites and sample size. Most of the associations identified in European and Asian -descent populations failed to generalize due to power (as a consequence of sample size and low minor allele frequencies). However, there were several adequately powered associations that did not generalize to African Americans. These data suggest that the functional variant has yet to be identified for these traits by GWAS and highlight the need for future fine mapping studies in a large African-descent population.
Supplementary Material
Supplementary Table 1. SNP-trait associations significant in African Americans (p<0.05) but trend in the opposite direction compared with previously identified association. We compared association results in African Americans to the association results from the original study accounting for the coded allele. This table only represents associations that are statistically significant in African Americans (p<0.05) but have discordant effect sizes (expressed as a beta coefficient) compared to the original study. Abbreviations: CAF= coded allele frequency.
Supplementary Figure 1. Genome-wide association results for heart rate in African Americans (n= 455). (A) Manhattan plot of GWAS results for heart rate. Association results are plotted for each maker on the x-axis across the genome, and the −log of the p-value is plotted on the y axis. (B)Locus Zoom Plot for the most significant association. We plotted the local association results around 100kb of the most significant SNP rs1015003 from our GWAS of heart rate. The −log of the p-value is plotted on the y-axis, and each SNP is plotted by location on the x-axis. Pairwise linkage disequilibrium with the index SNP, measured by r2, is indicated by color. (C) Locus Zoom Plot for the SERPINI1 region. We plotted the local association results around 100kb of the most significant SNP (rs13090836) region in the SERPINI1 region.
Supplementary Figure 2. Comparison of SCN5A/SCN10A Locus Zoom plots between African Americans (top panel) and European Americans from Denny et al (Denny et al. 2010) (bottom panel). Using locus specific plots, we compared the index SNP (indicated at the top of each plot) identified in African Americans to previously identified association results in European Americans from VGER.
Supplementary Table 2. Effect size and p-values for GWAS-identified associations with a consistent direction of effect in African Americans compared to European descent populations. We compared the association results from GWAS-identified variants to African American association results for SNPs that have a consistent direction of effect but did not meet our significance threshold (p<0.05) for all ECG traits.
Supplementary Table 3. Coded Allele Frequencies for GWAS-identified SNPs in African Americans compared to the original study population. We calculated allele frequencies for all GWAS-identified SNPs in African Americans after accounting for the coded allele.
Supplementary Table 4. Replication of previous associations for PR interval in VGER/NUgene African Americans. We tested previously reported associations with PR interval in African Americans from Smith et al. for replication in our study population (Smith et al. 2011). Using the same coded allele as the original study, we performed tests of association for all reported SNPs genotyped in our study population using an additive genetic model. Abbreviations: CAF= coded allele frequency
Supplementary Table 5. Replication of previous associations for QT interval in VGER/NUgene African Americans. We tested previously reported associations with QT interval in African Americans from Avery et al. for replication in our study population (Avery et al. 2012). Using the same coded allele as the original study, we performed tests of association for all reported SNPs that were also genotyped in our study population using an additive genetic model. Abbreviations: CAF= coded allele frequency, *= Novel association in African Americans for PR interval reported by Avery et al
Supplementary Figure 3. Linkage Disequilibrium plots around the SCN10A region in YRI HapMap samples compared to CEU samples. Pairwise linkage disequilibrium was calculated for all SNPs with a MAF of >0.05 within 100kb of the SCN10A gene using Seattle SNPs Genome Server (http://gvs.gs.washington.edu/GVS/index.jsp).
Supplementary Figure 4. Linkage Disequilibrium plots in the NOS1AP gene in YRI HapMap samples compared to CEU samples. Pairwise linkage disequilibrium was calculated for all SNPs with a MAF of >0.05 within the NOS1AP gene using Seattle SNPs Genome Server (http://gvs.gs.washington.edu/GVS/index.jsp).
Acknowledgments
We would like to thank the Vanderbilt University Center for Human Genetics Research, Computational Genomics Core (CGC) for their analytical and computational support. We also thank the team of clinicians and medical personnel for selecting study participants and collecting the data. This work was supported by NIH U01HG004609 (Northwestern University as part of eMERGE) and U01HG04603 (Vanderbilt University as part of eMERGE, also serving as the Administrative Coordinating Center). A portion of the dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center1s BioVU which is supported by institutional funding and by the Vanderbilt CTSA grant UL1 TR000445 from NCATS/NIH.
Footnotes
Additional supporting information for this article may be found in the online version of this article:
References
- Abrahamsen B, Zhao J, Asante CO, Cendan CM, Marsh S, Martinez-Barbera JP, Nassar MA, Dickenson AH, Wood JN. The cell and molecular basis of mechanical, cold, and inflammatory pain. Science. 2008;321:702–705. doi: 10.1126/science.1156916. [DOI] [PubMed] [Google Scholar]
- Akopian AN, Souslova V, England S, Okuse K, Ogata N, Ure J, Smith A, Kerr BJ, McMahon SB, Boyce S, Hill R, Stanfa LC, Dickenson AH, Wood JN. The tetrodotoxin-resistant sodium channel SNS has a specialized function in pain pathways. Nat Neurosci. 1999;2:541–548. doi: 10.1038/9195. [DOI] [PubMed] [Google Scholar]
- Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, West K, Kashuk C, Akyol M, Perz S, Jalilzadeh S, Illig T, Gieger C, Guo CY, Larson MG, Wichmann HE, Marban E, O’Donnell CJ, Hirschhorn JN, Kaab S, Spooner PM, Meitinger T, Chakravarti A. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- Avery CL, Sethupathy P, Buyske S, He Q, Lin DY, Arking DE, Carty CL, Duggan D, Fesinmeyer MD, Hindorff LA, Jeff JM, Klein L, Patton KK, Peters U, Shohet RV, Sotoodehnia N, Young AM, Kooperberg C, Haiman CA, Mohlke KL, Whitsel EA, North KE. Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans. PLoS Genet. 2012;8:e1002870. doi: 10.1371/journal.pgen.1002870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Chambers JC, Zhao J, Terracciano CM, Bezzina CR, Zhang W, Kaba R, Navaratnarajah M, Lotlikar A, Sehmi JS, Kooner MK, Deng G, Siedlecka U, Parasramka S, El-Hamamsy I, Wass MN, Dekker LR, de Jong JS, Sternberg MJ, McKenna W, Severs NJ, de SR, Wilde AA, Anand P, Yacoub M, Scott J, Elliott P, Wood JN, Kooner JS. Genetic variation in SCN10A influences cardiac conduction. Nat Genet. 2010;42:149–152. doi: 10.1038/ng.516. [DOI] [PubMed] [Google Scholar]
- Crosslin DR, McDavid A, Weston N, Nelson SC, Zheng X, Hart E, de AM, Kullo IJ, McCarty CA, Doheny KF, Pugh E, Kho A, Hayes MG, Pretel S, Saip A, Ritchie MD, Crawford DC, Crane PK, Newton K, Li R, Mirel DB, Crenshaw A, Larson EB, Carlson CS, Jarvik GP. Genetic variants associated with the white blood cell count in 13,923 subjects in the eMERGE Network. Hum Genet. 2012;131:639–52. doi: 10.1007/s00439-011-1103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Holohan PD, Shrimpton AE, Tatum AH, Daucher J, Collins GH, Todd R, Bradshaw C, Kent P, Feiglin D, Rosenbaum A, Yerby MS, Shaw CM, Lacbawan F, Lawrence DA. Familial encephalopathy with neuroserpin inclusion bodies. Am J Pathol. 1999;155:1901–1913. doi: 10.1016/S0002-9440(10)65510-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, Chai HS, Bastarache L, Zuvich R, Peissig P, Carrell D, Ramirez AH, Pathak J, Wilke RA, Rasmussen L, Wang X, Pacheco JA, Kho AN, Hayes MG, Weston N, Matsumoto M, Kopp PA, Newton KM, Jarvik GP, Li R, Manolio TA, Kullo IJ, Chute CG, Chisholm RL, Larson EB, McCarty CA, Masys DR, Roden DM, de AM. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011;89:529–542. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Ritchie MD, Crawford DC, Schildcrout JS, Ramirez AH, Pulley JM, Basford MA, Masys DR, Haines JL, Roden DM. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitrescu L, Ritchie MD, Brown-Gentry K, Pulley JM, Basford M, Denny JC, Oksenberg JR, Roden DM, Haines JL, Crawford DC. Assessing the accuracy of observer-reported ancestry in a biorepository linked to electronic medical records. Genet Med. 2010;12:648–50. doi: 10.1097/GIM.0b013e3181efe2df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauderman WJ, Morrison JM. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006 Ref Type: Computer Program. Web address: http://hydra.usc.edu/gxe/
- Grady BJ, Torstenson E, Dudek SM, Giles J, Sexton D, Ritchie MD. Finding unique filter sets in plato: a precursor to efficient interaction analysis in gwas data. Pac Symp Biocomput. 2010:315–326. [PMC free article] [PubMed] [Google Scholar]
- Gretarsdottir S, Baas AF, Thorleifsson G, Holm H, den HM, de Vries JP, Kranendonk SE, Zeebregts CJ, van Sterkenburg SM, Geelkerken RH, van Rij AM, Williams MJ, Boll AP, Kostic JP, Jonasdottir A, Jonasdottir A, Walters GB, Masson G, Sulem P, Saemundsdottir J, Mouy M, Magnusson KP, Tromp G, Elmore JR, Sakalihasan N, Limet R, Defraigne JO, Ferrell RE, Ronkainen A, Ruigrok YM, Wijmenga C, Grobbee DE, Shah SH, Granger CB, Quyyumi AA, Vaccarino V, Patel RS, Zafari AM, Levey AI, Austin H, Girelli D, Pignatti PF, Olivieri O, Martinelli N, Malerba G, Trabetti E, Becker LC, Becker DM, Reilly MP, Rader DJ, Mueller T, Dieplinger B, Haltmayer M, Urbonavicius S, Lindblad B, Gottsater A, Gaetani E, Pola R, Wells P, Rodger M, Forgie M, Langlois N, Corral J, Vicente V, Fontcuberta J, Espana F, Grarup N, Jorgensen T, Witte DR, Hansen T, Pedersen O, Aben KK, de GJ, Holewijn S, Folkersen L, Franco-Cereceda A, Eriksson P, Collier DA, Stefansson H, Steinthorsdottir V, Rafnar T, Valdimarsson EM, Magnadottir HB, Sveinbjornsdottir S, Olafsson I, Magnusson MK, Palmason R, Haraldsdottir V, Andersen K, Onundarson PT, Thorgeirsson G, Kiemeney LA, Powell JT, Carey DJ, Kuivaniemi H, Lindholt JS, Jones GT, Kong A, Blankensteijn JD, Matthiasson SE, Thorsteinsdottir U, Stefansson K. Genome-wide association study identifies a sequence variant within the DAB2IP gene conferring susceptibility to abdominal aortic aneurysm. Nat Genet. 2010;42:692–697. doi: 10.1038/ng.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, Gudjonsson SA, Jonasdottir A, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Lochen ML, Kong A, Thorsteinsdottir U, Stefansson K. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- Jeff JM, Brown-Gentry K, Buxbaum SG, Sarpong DF, Taylor HA, George AL, Jr, Roden DM, Crawford DC. SCN5A Variation is Associated with Electrocardiographic Traits in the Jackson Heart Study. Circ Cardiovasc Genet. 2011;4:139–144. doi: 10.1161/CIRCGENETICS.110.958124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LP, Deng CL, Moss AJ, Kass RS, Liang CS. Polymorphism of the gene encoding a human minimal potassium ion channel (minK) Gene. 1994;151:339–340. doi: 10.1016/0378-1119(94)90685-8. [DOI] [PubMed] [Google Scholar]
- Marroni F, Pfeufer A, Aulchenko YS, Franklin CS, Isaacs A, Pichler I, Wild SH, Oostra BA, Wright AF, Campbell H, Witteman JC, Kaab S, Hicks AA, Gyllensten U, Rudan I, Meitinger T, Pattaro C, van Duijn CM, Wilson JF, Pramstaller PP. A genome-wide association scan of RR and QT interval duration in 3 European genetically isolated populations: the EUROSPAN project. Circ Cardiovasc Genet. 2009;2:322–328. doi: 10.1161/CIRCGENETICS.108.833806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, Li R, Masys DR, Ritchie MD, Roden DM, Struewing JP, Wolf WA. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, Bis JC, Marciante K, Rivadeneira F, Noseworthy PA, Sotoodehnia N, Smith NL, Rotter JI, Kors JA, Witteman JC, Hofman A, Heckbert SR, O’Donnell CJ, Uitterlinden AG, Psaty BM, Lumley T, Larson MG, Stricker BH. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Cheh C, Guo CY, Wang TJ, O’Donnell CJ, Levy D, Larson MG. Genome-wide association study of electrocardiographic and heart rate variability traits: the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S7. doi: 10.1186/1471-2350-8-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergrass SA, Dudek S, Crawford DCC, Ritchie M. Synthesis-View: visualization and interpretation of SNP association results for multi-cohort, multi-phenotype data and meta-analysis. BioData Mining. 2010;3:10. doi: 10.1186/1756-0381-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeufer A, van NC, Marciante KD, Arking DE, Larson MG, Smith AV, Tarasov KV, Muller M, Sotoodehnia N, Sinner MF, Verwoert GC, Li M, Kao WH, Kottgen A, Coresh J, Bis JC, Psaty BM, Rice K, Rotter JI, Rivadeneira F, Hofman A, Kors JA, Stricker BH, Uitterlinden AG, van Duijn CM, Beckmann BM, Sauter W, Gieger C, Lubitz SA, Newton-Cheh C, Wang TJ, Magnani JW, Schnabel RB, Chung MK, Barnard J, Smith JD, Van Wagoner DR, Vasan RS, Aspelund T, Eiriksdottir G, Harris TB, Launer LJ, Najjar SS, Lakatta E, Schlessinger D, Uda M, Abecasis GR, Muller-Myhsok B, Ehret GB, Boerwinkle E, Chakravarti A, Soliman EZ, Lunetta KL, Perz S, Wichmann HE, Meitinger T, Levy D, Gudnason V, Ellinor PT, Sanna S, Kaab S, Witteman JC, Alonso A, Benjamin EJ, Heckbert SR. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post W, Shen H, Damcott C, Arking DE, Kao WH, Sack PA, Ryan KA, Chakravarti A, Mitchell BD, Shuldiner AR. Associations between genetic variants in the NOS1AP (CAPON) gene and cardiac repolarization in the old order Amish. Hum Hered. 2007;64:214–219. doi: 10.1159/000103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez AH, Schildcrout JS, Blakemore DL, Masys DR, Pulley JM, Basford MA, Roden DM, Denny JC. Modulators of normal electrocardiographic intervals identified in a large electronic medical record. Heart Rhythm. 2011;8:271–277. doi: 10.1016/j.hrthm.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, Masys DR. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11(5):356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JG, Lowe JK, Kovvali S, Maller JB, Salit J, Daly MJ, Stoffel M, Altshuler DM, Friedman JM, Breslow JL, Newton-Cheh C. Genome-wide association study of electrocardiographic conduction measures in an isolated founder population: Kosrae. Heart Rhythm. 2009;6(5):634–641. doi: 10.1016/j.hrthm.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JG, Magnani JW, Palmer C, Meng YA, Soliman EZ, Musani SK, Kerr KF, Schnabel RB, Lubitz SA, Sotoodehnia N, Redline S, Pfeufer A, Muller M, Evans DS, Nalls MA, Liu Y, Newman AB, Zonderman AB, Evans MK, Deo R, Ellinor PT, Paltoo DN, Newton-Cheh C, Benjamin EJ, Mehra R, Alonso A, Heckbert SR, Fox ER. Genome-wide association studies of the PR interval in African Americans. PLoS Genet. 2011;7(2):e1001304. doi: 10.1371/journal.pgen.1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotoodehnia N, Isaacs A, de Bakker PI, Dorr M, Newton-Cheh C, Nolte IM, van der HP, Muller M, Eijgelsheim M, Alonso A, Hicks AA, Padmanabhan S, Hayward C, Smith AV, Polasek O, Giovannone S, Fu J, Magnani JW, Marciante KD, Pfeufer A, Gharib SA, Teumer A, Li M, Bis JC, Rivadeneira F, Aspelund T, Kottgen A, Johnson T, Rice K, Sie MP, Wang YA, Klopp N, Fuchsberger C, Wild SH, Mateo LI, Estrada K, Volker U, Wright AF, Asselbergs FW, Qu J, Chakravarti A, Sinner MF, Kors JA, Petersmann A, Harris TB, Soliman EZ, Munroe PB, Psaty BM, Oostra BA, Cupples LA, Perz S, de Boer RA, Uitterlinden AG, Volzke H, Spector TD, Liu FY, Boerwinkle E, Dominiczak AF, Rotter JI, van HG, Levy D, Wichmann HE, van Gilst WH, Witteman JC, Kroemer HK, Kao WH, Heckbert SR, Meitinger T, Hofman A, Campbell H, Folsom AR, van Veldhuisen DJ, Schwienbacher C, O’Donnell CJ, Volpato CB, Caulfield MJ, Connell JM, Launer L, Lu X, Franke L, Fehrmann RS, te MG, Groen HJ, Weersma RK, van den Berg LH, Wijmenga C, Ophoff RA, Navis G, Rudan I, Snieder H, Wilson JF, Pramstaller PP, Siscovick DS, Wang TJ, Gudnason V, van Duijn CM, Felix SB, Fishman GI, Jamshidi Y, Stricker BH, Samani NJ, Kaab S, Arking DE. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010;42(12):1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CC, Su Z, Donnelly P, Marchini J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009;5(5):e1000477. doi: 10.1371/journal.pgen.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splawski I, Timothy KW, Tateyama M, Clancy CE, Malhotra A, Beggs AH, Cappuccio FP, Sagnella GA, Kass RS, Keating MT. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science. 2002;297(5585):1333–1336. doi: 10.1126/science.1073569. [DOI] [PubMed] [Google Scholar]
- Turner SD, Berg RL, Linneman JG, Peissig PL, Crawford DC, Denny JC, Roden DM, McCarty CA, Ritchie MD, Wilke RA. Knowledge-driven multi-locus analysis reveals gene-gene interactions influencing HDL cholesterol level in two independent EMR-linked biobanks. PLoS One. 2011;6(5):e19586. doi: 10.1371/journal.pone.0019586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, Shen J, Timothy KW, Vincent GM, de JT, Schwartz PJ, Toubin JA, Moss AJ, Atkinson DL, Landes GM, Connors TD, Keating MT. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet. 1996;12(1):17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- Wang Q, Shen J, Li Z, Timothy K, Vincent GM, Priori SG, Schwartz PJ, Keating MT. Cardiac sodium channel mutations in patients with long QT syndrome, an inherited cardiac arrhythmia. Hum Mol Genet. 1995a;4(9):1603–1607. doi: 10.1093/hmg/4.9.1603. [DOI] [PubMed] [Google Scholar]
- Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995b;80(5):805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F- Statistics for the Analysis of Population Structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Xiao R, Boehnke M. Quantifying and correcting for the winner’s curse in genetic association studies. Genet Epidemiol. 2009;33(5):453–462. doi: 10.1002/gepi.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Prentice RL. Correcting “winner’s curse” in odds ratios from genomewide association findings for major complex human diseases. Genet Epidemiol. 2010;34(1):78–91. doi: 10.1002/gepi.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann K, Leffler A, Babes A, Cendan CM, Carr RW, Kobayashi J, Nau C, Wood JN, Reeh PW. Sensory neuron sodium channel Nav1.8 is essential for pain at low temperatures. Nature. 2007;447(7146):855–858. doi: 10.1038/nature05880. [DOI] [PubMed] [Google Scholar]
- Zuvich RL, Armstrong LL, Bielinski SJ, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, de AM, Doheny KF, Haines JL, Hayes MG, Jarvik GP, Jiang L, Kullo IJ, Li R, Ling H, Manolio TA, Matsumoto ME, McCarty CA, McDavid AN, Mirel DB, Olson LM, Paschall JE, Pugh EW, Rasmussen LV, Rasmussen-Torvik LJ, Turner SD, Wilke RA, Ritchie MD. Pitfalls of merging GWAS data: lessons learned in the eMERGE network and quality control procedures to maintain high data quality. Genet Epidemiol. 2011;35(8):887–898. doi: 10.1002/gepi.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. SNP-trait associations significant in African Americans (p<0.05) but trend in the opposite direction compared with previously identified association. We compared association results in African Americans to the association results from the original study accounting for the coded allele. This table only represents associations that are statistically significant in African Americans (p<0.05) but have discordant effect sizes (expressed as a beta coefficient) compared to the original study. Abbreviations: CAF= coded allele frequency.
Supplementary Figure 1. Genome-wide association results for heart rate in African Americans (n= 455). (A) Manhattan plot of GWAS results for heart rate. Association results are plotted for each maker on the x-axis across the genome, and the −log of the p-value is plotted on the y axis. (B)Locus Zoom Plot for the most significant association. We plotted the local association results around 100kb of the most significant SNP rs1015003 from our GWAS of heart rate. The −log of the p-value is plotted on the y-axis, and each SNP is plotted by location on the x-axis. Pairwise linkage disequilibrium with the index SNP, measured by r2, is indicated by color. (C) Locus Zoom Plot for the SERPINI1 region. We plotted the local association results around 100kb of the most significant SNP (rs13090836) region in the SERPINI1 region.
Supplementary Figure 2. Comparison of SCN5A/SCN10A Locus Zoom plots between African Americans (top panel) and European Americans from Denny et al (Denny et al. 2010) (bottom panel). Using locus specific plots, we compared the index SNP (indicated at the top of each plot) identified in African Americans to previously identified association results in European Americans from VGER.
Supplementary Table 2. Effect size and p-values for GWAS-identified associations with a consistent direction of effect in African Americans compared to European descent populations. We compared the association results from GWAS-identified variants to African American association results for SNPs that have a consistent direction of effect but did not meet our significance threshold (p<0.05) for all ECG traits.
Supplementary Table 3. Coded Allele Frequencies for GWAS-identified SNPs in African Americans compared to the original study population. We calculated allele frequencies for all GWAS-identified SNPs in African Americans after accounting for the coded allele.
Supplementary Table 4. Replication of previous associations for PR interval in VGER/NUgene African Americans. We tested previously reported associations with PR interval in African Americans from Smith et al. for replication in our study population (Smith et al. 2011). Using the same coded allele as the original study, we performed tests of association for all reported SNPs genotyped in our study population using an additive genetic model. Abbreviations: CAF= coded allele frequency
Supplementary Table 5. Replication of previous associations for QT interval in VGER/NUgene African Americans. We tested previously reported associations with QT interval in African Americans from Avery et al. for replication in our study population (Avery et al. 2012). Using the same coded allele as the original study, we performed tests of association for all reported SNPs that were also genotyped in our study population using an additive genetic model. Abbreviations: CAF= coded allele frequency, *= Novel association in African Americans for PR interval reported by Avery et al
Supplementary Figure 3. Linkage Disequilibrium plots around the SCN10A region in YRI HapMap samples compared to CEU samples. Pairwise linkage disequilibrium was calculated for all SNPs with a MAF of >0.05 within 100kb of the SCN10A gene using Seattle SNPs Genome Server (http://gvs.gs.washington.edu/GVS/index.jsp).
Supplementary Figure 4. Linkage Disequilibrium plots in the NOS1AP gene in YRI HapMap samples compared to CEU samples. Pairwise linkage disequilibrium was calculated for all SNPs with a MAF of >0.05 within the NOS1AP gene using Seattle SNPs Genome Server (http://gvs.gs.washington.edu/GVS/index.jsp).