Abstract
The moaABCDE operon of Escherichia coli encodes enzymes essential for the biosynthesis of the molybdenum cofactor (Moco). However, the role of the moaB gene within this operon has remained enigmatic. Here, we have investigated the effect of moaB defects on two phenotypes diagnostic for Moco-deficiency: chlorate-resistance and sensitivity to the base analog 6-N-hydroxylaminopurine (HAP). We found that transposon insertions in moaB caused partial Moco-deficiency associated with chlorate-resistance, but not for HAP-sensitivity. On the other hand, in-frame deletions of moaB, or moaB overexpression, had no effect on either phenotype. Our combined data are consistent with the lack of any role for MoaB in Moco biosynthesis in E. coli.
Keywords: Molybdenum cofactor, moaABCDE operon, mogA gene, 6-N-hydroxylaminopurine, Chlorate resistance
1. Introduction
Molybdenum cofactor (Moco) is an essential prosthetic group for the activity of a range of pro- and eukaryotic oxidoreductases (Schwarz et al., 2009). In its simplest form, Moco is a pterine derivative, named molybdopterin or metal-binding pterin (MPT), that contains a molybdenum atom coordinated by a dithiolene group (Mo-MPT). Biosynthesis of the Moco is an evolutionary conserved, multistep process, which has been studied in detail (Schwarz et al., 2009) (Fig. 1A). The first step is conversion of GTP to a cyclic pyranopterin monophosphate (cPMP), performed in E. coli by the moaA and moaC gene products. In the second step, the dithiolene group is introduced into cPMP by the moaD, moaE, and moeB gene products, yielding MPT. Finally, the molybdenum atom is inserted onto the dithiolene sulphurs via an MPT-AMP intermediate synthesized by the mogA gene product. This intermediate is then converted to Mo- MPT by the moeA gene product. Mo-MPT can be further modified by the mobA gene product to yield a molybdopterin guanine dinucleotide form (MGD). The MGD form is utilized by at least ten E. coli molybdoenzymes, including three nitrate reductases, three formate dehydrogenases, two trimethylamine-N-oxide reductases, dimethyl sulfoxide reductase, and biotin sulfoxide oxidoreductase (Iobbi-Nivol and Leimkühler, 2012). Thus far, five mobA-independent molybdoactivities have also been described in E. coli, including PaoABC aldehyde oxidoreductase and XdhABC xanthine oxidase (which both use molybdopterin cytosine dinucleotide), YedY S(N)-oxide reductase (which utilizes Mo-MPT), and two 6-N- hydroxylaminopurine (HAP)-reductases, YcbX and YiiM (for which the precise form of Moco unknown) (Iobbi-Nivol and Leimkühler, 2012; Kozmin and Schaaper, 2007; Kozmin et al., 2008).
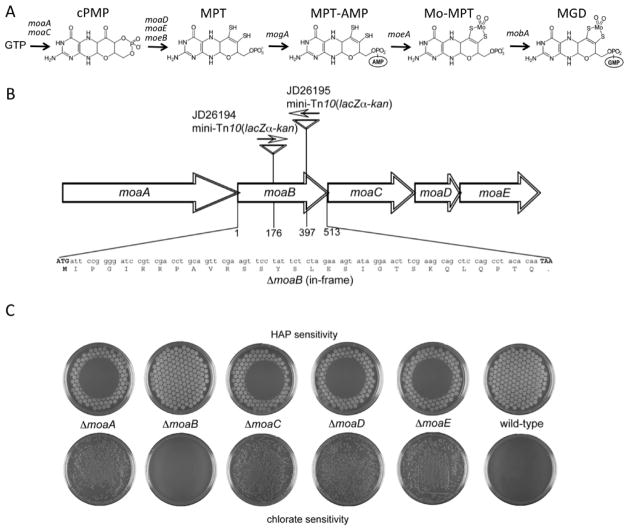
Fig. 1.
Effect of in-frame deletions of the individual genes of the E. coli moaABCDE operon on Moco-dependent activities. A. Steps in Moco biosynthesis and the responsible genes (Schwarz et al., 2009). B. Organization of the moaABCDE operon. Indicated are the locations of the moaB::mini-Tn10(lacZα-kan) insertions as present in strains JD26194 and JD26195, as well as the endpoints of the moaB deletion showing the ATG start and TAA stop codon as well as the remaining in-frame “scar” of 28 residues (see Materials and methods). All other moa genes were deleted in identical manner. C. HAP- or chlorate-sensitivity of the in-frame deletion mutants (NR17378 - NR17382) and the wild-type strain (NR10836). For the HAP-sensitivity test, cells were transferred using a multi-prong replicator to a minimal-medium plate, and 50 μg of HAP was applied onto the center of the plate. Plates were incubated overnight at 37 °C. For the chlorate-sensitivity test, cells were plated on LB plates containing 5 mM potassium chlorate and incubated 48 h at 37 °C under anaerobic conditions.
The moaABCDE operon (Fig. 1B) is the largest operon involved in Moco biosynthesis E. coli. As noted above, it contains the genes essential for the first (moaA, moaC) and the second (moaD, moaE) steps of Moco biosynthesis. However, the role of the moaB gene in this operon has remained enigmatic (Magalon and Mendel, 2008). All known Moco-deficient E. coli mutants display a common phenotype - resistance to chlorate under anaerobic conditions, which is due to lack of nitrate reductase activity responsible for conversion of chlorate to toxic chlorite (Stewart and MacGregor, 1982). However, no chlorate-resistant (chl) mutants in the moaB gene have ever been isolated (Magalon and Mendel, 2008; Shanmugam et al., 1992). In the present work, we have investigated the effect of several moaB defects using two established markers of Moco deficiency in E. coli: chlorate resistance and sensitivity to the base analog 6-N-hydroxylaminopurine (HAP) (Kozmin et al., 2000; Kozmin and Schaaper, 2007). HAP sensitivity is due primarily to the lack of two Moco-dependent activities, YcbX and YiiM, which mediate the conversion of toxic HAP to non-toxic adenine (Kozmin et al., 2008). A minor role in HAP detoxification is also played by a third molybdoenzyme, biotin sulfoxide oxidoreductase (BisC) (Kozmin et al., 2008). Our results, using precise in-frame deletions of the individual genes of the moaABCDE operon, indicate that moaB is fully dispensable for both Moco-dependent phenotypes and that, hence, MoaB is not required for Moco biosynthesis.
2. Materials and Methods
2.1. Media and chemicals
Bacteria were cultivated in LB broth (Miller, 1972) or minimal Vogel-Bonner medium (VB) (Vogel and Bonner, 1956) containing 0.2 % glucose as carbon source and supplemented with 1 μg/ml of thiamine. Solid media contained 1.5% agar. For selection of antibiotic-resistant clones, media were supplemented with 35 μg/ml of kanamycin, 15 μg/ml of tetracycline, or 100 μg/ml of ampicillin. 6-N-hydroxylaminopurine (HAP) was purchased from Midwest Research Institute (Kansas City, USA). All other chemicals were from Sigma-Aldrich.
2.2. Bacterial strains and plasmids
The E. coli strains used in this study are listed in Table 1, along with their source or derivation. In-frame deletion alleles of moaA-E genes were generated using a PCR-mediated one-step gene-replacement procedure (Datsenko and Wanner, 2000). In each case, the PCR primers contained 50-nt extensions complementary to the left or right end of the gene as well as 20-nt 3′-ends complementary to the kanamycin-resistance (Kanr) module of plasmid pKD13 (Datsenko and Wanner, 2000), generating kanamycin-resistant deletion/insertions. Initial deletions were made in strain NR17368 (Table 1). After elimination of the Kanr markers using plasmid pCP20 (Datsenko and Wanner, 2000), the markerless in-frame deletions were verified by DNA sequencing. In this manner, each gene of the moa operon was deleted precisely from start to stop codon, as exemplified in Fig. 1B for the moaB deletion. Finally, the deletions were transferred into strains NR10836 and KP7600 by P1 transduction using the closely-linked zbi-29::Tn10 insertion [17.7 min (Nichols et al., 1998)] as a selective (tetracycline-resistance) marker (Table 1). Transfer of each moa deletion was verified by PCR. To obtain MoaB- overexpressing constructs, moaB gene was amplified from strain NR10836 (Table 1) using primers moaB-SacI (5′-cta gag ctc aat gag tca ggt aag cac tga a-3′) and moaB-BamHI (5′-ata ggg atc cat ggg tca gtt gcg aca tac-3′) or moaB-NdeI (5′-cta cca tat gag tca ggt aag cac tga a-3′), and cloned into the SacI/BamHI site of a multicopy plasmid pBluescript II SK(+) (Stratagene), or into the NdeI/BamHI site of the pET3a expression vector (Novagen).
Table 1.
E. coli strains used in this study.
| Strain | Genotype | Source or derivation |
|---|---|---|
| KP7600 | lacIQ lacZΔM15 galK2 galT22 λ− IN(rrnD-rrnE)1 | Miki et al., 2008 |
| JD26193 | KP7600, but moaA::mini-Tn10(lacZα-kan)(−)a | Miki et al., 2008 |
| JD26194 | KP7600, but moaB::mini-Tn10(lacZα-kan)(+)a | Miki et al., 2008 |
| JD26195 | KP7600, but moaB::mini-Tn10(lacZα-kan)(−) a | Miki et al., 2008 |
| JD26196 | KP7600, but moaC::mini-Tn10(lacZα-kan)(+) a | Miki et al., 2008 |
| JD26197 | KP7600, but moaD::mini-Tn10(lacZα-kan)(−) a | Miki et al., 2008 |
| JD26198 | KP7600, but moaE::mini-Tn10(lacZα-kan)(+) a | Miki et al., 2008 |
| BW25113 | lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | Datsenko and Wanner, 2000 |
| KP7600-ΔB | KP7600, but ΔmoaB zbi-29::Tn10 | This study |
| CAG18493 | zbi-29::Tn10 | Singer et al., 1989 |
| NR10836 | ara thi Δ (p ro-lac ) F′CC106 | Kozmin et al., 2000 |
| NR17368 | BW25113, but zbi-29::Tn10 | BW25113 × P1/CAG18493 |
| NR17378 | NR10836, but ΔmoaA zbi-29::Tn10 | This study |
| NR17379 | NR10836, but ΔmoaB zbi-29::Tn10 | This study |
| NR17380 | NR10836, but ΔmoaC zbi-29::Tn10 | This study |
| NR17381 | NR10836, but ΔmoaD zbi-29::Tn10 | This study |
| NR17382 | NR10836, but ΔmoaE zbi-29::Tn10 | This study |
| NR17383 | NR10836, but ΔmogA::kan | Kozmin and Schaaper, 2007 |
| NR17391 | NR10836, but λ(DE3) ΔguaB ΔmogA::kan | Laboratory collection |
| NR17427 | NR10836, but moaB::mini-Tn10(lacZα-kan)(+)a | NR10836 × P1/JD26194 |
| NR17428 | NR10836, but moaB::mini-Tn10(lacZα-kan)(−)a | NR10836 × P1/JD26195 |
(+) or (−) indicate the direction of the mini-Tn10(lacZα-kan) on the MG1655 genome.
2.3. Spot-test for HAP-sensitivity
Stationary E. coli cultures grown in LB were diluted 30-fold in 0.9% NaCl and transferred to VB plates using a multi-prong replicator device (approximately 0.1 ml total per plate) (Kozmin et al. 2000). After the spots had dried, 5 microliters of a 10-mg/ml solution of HAP in dimethyl sulfoxide were spotted onto the center of the plate. The plates were incubated overnight at 37 °C and inspected the next day for zones of inhibition.
2.4. Test for chlorate sensitivity
Approximately 103 cells were plated on LB plates containing various concentrations of KClO3. The plates were incubated for 48 h at 37° C under anaerobic conditions using a Becton Dickinson BBL gas pack anaerobic system.
3. Results and Discussion
3.1. A split phenotype for moaB transposon insertions
Our investigation into the role of moaB was triggered by an initial observation of a “split” phenotype for two moaB::mini-Tn10 mutants (JD26194 and JD26195) obtained from the National Institute of Genetics of Japan (see Table 1). These JD strains are part of a genome-wide collection of mini-Tn10(lacZα-kan)-mediated gene knockouts (Miki et al., 2008). Other strains from this collection carrying insertions in the moaA, moaC, moaD, and moaE genes were, as expected, both HAP-sensitive and chlorate-resistant (Table 2). In contrast, the two moaB::mini-Tn10(lacZα-kan) JD26194 and JD26195 (they differ in orientation of the mini-Tn10 insert, see Fig. 1) were fully HAP-resistant indicating normal status of the YcbX/YiiM enzymes, but were resistant to 15 mM potassium chlorate [the standard concentration used for selection of chl mutants (Miller 1972; Stewart and MacGregor, 1982)], indicating an impairment of nitrate reductase activity (Table 2). Thus, the strains exhibited a split phenotype.
Table 2.
HAP- and chlorate-sensitivity of wild-type, moa, and mogA strains.
| Strain | Relative genotype | HAP sensitivitya | Chlorate sensitivityb KClO3 (mM) | ||
|---|---|---|---|---|---|
| 15 | 5 | 0.5 | |||
| KP7600 derivatives: | |||||
| KP7600 | wild-type | R | S | R | R |
| JD26193 | moaA::mini-Tn10kan | S | R | R | R |
| JD26196 | moaC::mini-Tn10kan | S | R | R | R |
| JD26197 | moaD::mini-Tn10kan | S | R | R | R |
| JD26198 | moaE::mini-Tn10kan | S | R | R | R |
| JD26194 | moaB::mini-Tn10kan | R | R | R | R |
| JD26195 | moaB::mini-Tn10kan | R | R | R | R |
| KP7600-ΔB | ΔmoaB (in-frame) | R | S | R | R |
| NR10836 derivatives: | |||||
| NR10836 | wild-type | R | S | S | I |
| NR17378 | ΔmoaA (in-frame) | S | R | R | R |
| NR17380 | ΔmoaC (in-frame) | S | R | R | R |
| NR17381 | ΔmoaD (in-frame) | S | R | R | R |
| NR17382 | ΔmoaE (in-frame) | S | R | R | R |
| NR17427 | moaB::mini-Tn10kan | R | S | R | R |
| NR17428 | moaB::mini-Tn10kan | R | S | R | R |
| NR17379 | ΔmoaB (in-frame) | R | S | S | I |
| NR17383 | ΔmogA::kan | S | R | R | R |
| NR17383-p[moaB]c | ΔmogA:: kan, p[moaB]c | S | R | R | R |
| NR10836-p[moaB]c | wt, p[moaB]c | R | S | S | I |
The sensitivity to HAP-induced killing was tested in spot-tests as described in Materials and methods. “R” indicates HAP-resistant phenotype (as “wild-type” in Fig. 1C), “S” indicates HAP-sensitive phenotype (as ΔmoaA,C,D,E in Fig. 1C).
Cells were plated on LB-chlorate plates and incubated 48 h at 37° C under anaerobic conditions (see Materials and methods). Under these conditions, chlorate-sensitive strains do not form colonies (chlorate-sensitive phenotype, “S”) or form very small colonies (chlorate-inhibited phenotype, “I”), whereas chlorate-resistant strains (chlorate-resistant phenotype, “R”) form colonies of the same size as on regular LB plates.
p[moaB] indicates vector pBluescript II SK(+) containing the moaB gene (see Materials and methods). Identical results were obtained with strain NR17391 carrying plasmid pET3a-moaB (data not shown). All tests with plasmid-containing strains were performed in the presence of ampicillin and 0.5 mM IPTG.
To further investigate the phenotypes, we transferred the two moaB::mini-Tn10 alleles by P1 transduction into the NR10836 strain background, which has been used routinely in our laboratory to investigate the effects of Moco deficiencies (Kozmin et al., 2000). As shown in Table 2, the NR10836 moaB::mini-Tn10 derivatives were, again, HAP resistant, but a different result was obtained for chlorate resistance. The strains were sensitive to 15 mM potassium chlorate, while resistant to the 5 mM and 0.5 mM chlorate concentrations (Table 2). Note that 5 mM chlorate is sufficient to kill essentially 100% of the cells of the parental NR10836 strain (see Fig. 1C), while on 0.5 mM chlorate plates very small colonies are seen, which may be termed “chlorate-inhibited” (see Stewart and MacGregor, 1982) (Table 2). Thus, two conclusions are apparent: (i) the moaB::mini-Tn10 insertions provide an observable level of chlorate resistance, and (ii) the moaB::mini-Tn10 derivatives of the JD series are resistant to higher chlorate concentrations than those of the NR10836 background. This latter difference between the two strain sets is likely a background-specific issue. Indeed, the parent to the JD strains, KP7600, was chlorate resistant up to 5 mM chlorate (Table 2). This elevated basal level of resistance may be due to some uncharacterized mutation affecting nitrate reductase activity in KP7600. This is likely specific to KP7600, because its parental strain, W3110, displayed a pattern of chlorate-sensitivity similar to NR10836 (data not shown).
3.2. Lack of phenotype for in-frame moaB deletion
Regardless of the strain background difference, an explanation was sought that could account for the split phenotype of the moaB::mini-Tn10 insertions: an increased level of chlorate-resistance along with unaltered HAP-sensitivity. Two possibilities were considered. The Tn10 insertions in moaB could have a polar effect on expression of the downstream moaCDE genes, leading to a reduction of the total Moco level in the cell. Such reduction might have a more pronounced effect on chlorate-sensitivity than it has on HAP sensitivity. Alternatively, the moaB defects could specifically reduce the level of MGD, which is required for the nitrate reductase activities, but would not affect the level of Mo-MPT, required for the mobA-independent YcbX and YiiM activities (Kozmin and Schaaper, 2007; Kozmin et al., 2008). To test these proposals, we constructed an in-frame deletion allele of moaB (along with in-frame deletions of the other genes of the operon, see Materials and Methods). As expected, in-frame deletions of the moaA, moaC, moaD, and moaE genes led to clear HAP-sensitivity and chlorate-resistance (Fig. 1C and Table 2). The strains carrying the in-frame moaB deletion were resistant to HAP (Fig. 1C and Table 2), and yielded a pattern of chlorate-sensitivity that was indistinguishable from that of the respective parental strains, NR10836 or KP7600 (Fig. 1C and Table 2). Thus, while the NR10836 and KP7600 backgrounds have an intrinsic difference in chlorate sensitivity, the moaB deletion does not further affect this intrinsic sensitivity and, hence, moaB does not confer a split-phenotype. These results argue against any significant role of MoaB in Moco biosynthesis.
We also constructed a ΔmoaB ΔycbX ΔyiiM triple mutant and observed that it did not display any altered HAP-sensitivity compared to the ΔycbX ΔyiiM double mutant (data not shown). As the ΔycbX ΔyiiM strain still carries some level of HAP resistance due to BisC activity (Kozmin et al., 2008), we conclude that, in addition, the BisC biotin sulfoxide reductase activity is also independent of moaB.
3.3. Possible polar effects of the moaB::mini-Tn10 insertions
As the moaB gene resides in the middle of the moaABCDE operon, it seems plausible to assume that the increased chlorate-resistance of the moaB::mini-Tn10 insertion mutants, accounting for the split phenotype, is due to a polar effect of the transposon insertions on the downstream moaCDE genes, leading to at least a partial Moco deficiency. While we have not pursued this, gene expression measurements may be used to confirm this effect directly. It is an interesting question why a partial Moco deficiency would have a stronger effect on chlorate resistance than on HAP-sensitivity. It is possible that the presumed impairment of cofactor biosynthesis restricts the availability of MGD (for nitrate reductase) more severely than that of Mo-MPT (for YcbX and YiiM). On the other hand, the effect may simply reflect the relative sensitivity of the two phenotypic assays used: a given level of Moco deficiency may lead to a reduction in nitrate reductase activity sufficient for a demonstrable level of chlorate resistance, whereas a more drastic reduction in HAP-detoxification would be needed to confer HAP-sensitivity. Another interesting case of Moco limitation was reported for a molybdopterin synthase sulfurylase mutant (moeBA228T), which was able to produce an essentially normal level of active nitrate reductase (NarGHI) but little or no active DMSO reductase (DmsABC) (Sambasivarao et al., 2002). As both reductases use the same (Mo-bisMGD) cofactor, the differential sensitivity to Moco limitation was ascribed to a much greater affinity of NarGHI for the cofactor, allowing it to more effectively scavenge the available cofactor pool (Sambasivarao et al., 2002).
A recent study in E. coli described a novel tellurate reductase activity, which was reportedly molybdopterin-dependent (Theisen et al., 2013). The activity was dependent on the moaA and moaB genes, but, interestingly, not on the moaC or moaD genes (Theisen et al., 2013). The strains used were of the Keio collection (Baba et al., 2006), and carry a kanamycin resistance cassette in each inactivated gene. Hence, a polar effect of the moaB insertion is to be expected. However, the apparent lack of effect of the moaC and moaD deletions remains to be investigated as the MoaC and MoaD proteins are known to be essential for molybdopterin biosynthesis (Iobbi-Nivol and Leimkühler, 2012; Schwarz et al., 2009).
3.4. Physiological role of MoaB?
Having demonstrated that there is no obvious role for the moaB gene product in the investigated molybdoactivities, the question still remains what is the possible physiological role for MoaB in E. coli. Both the primary sequence and crystal structure of MoaB show strong similarities with the MogA protein (Bader et al., 2004; Bevers et al., 2008; Sanishvili et al., 2004), which catalyzes formation of the MPT-AMP intermediate during the Mo-insertion step into Moco. Indeed, in the archaeon Pyrococcus furiosus, which lacks a MogA protein, its MoaB enzyme is responsible for the MPT adenylylation (Bevers et al., 2008). However, the E. coli MoaB protein, in contrast to the P. furiosus MoaB, was shown incapable of forming the MPT-AMP intermediate (Bevers et al., 2008). The fact that E. coli mogA mutants display clear Moco-deficiency associated phenotypes (Kozmin and Schaaper, 2007; Shanmugam et al., 1992) also indicates that the chromosomal copy of moaB cannot substitute for the lack of mogA. In addition, the loss of nitrate reductase activity in mogA-deficient E. coli could not be restored by overexpression of MoaB protein (Bevers et al., 2008). Likewise, we did not detect any change in HAP- or chlorate-sensitivity in the ΔmogA strains carrying moaB-overexpressing vectors (Table 2), clearly indicating that MoaB cannot substitute for MogA. Based on the ability of MoaB protein to bind GTP and, presumably, other intermediates of Moco biosynthesis (Sanishvili et al., 2004), it has been proposed that MoaB may play a yet unknown regulatory role in the Moco biosynthesis pathway, for example, by sensing the Moco status in cell (Bevers et al., 2008) or functioning as a Moco transporting/storage protein (Sanishvili et al., 2004). It has also been proposed that MoaB and MogA might form functional hetero-oligomers (Sanishvili et al., 2004). However, if such oligomers occur, their lack (as in the moaB mutant) does not appear to have major consequences, at least for the molybdoactivities tested here. Also, MoaB overproduction in an otherwise wild-type strain did not show any negative consequences (see Table 2), which might have resulted if such hetero-oligomers would be involved in a mechanism for MoaB-mediated regulation of Moco biosynthesis.
As demonstrated by Bevers and coworkers, residue D56 in P. furiosus MoaB is essential for its catalytic activity (Bevers et al., 2008). As E. coli MoaB contains a Glu residue (E52) at the corresponding position, this could be a reason for its inactive state. Sequence comparisons revealed several other amino acid substitutions in E. coli MoaB (as well as in several other Enterobacteriaceae species that have both a MogA and MoaB protein), such as those corresponding to P. furiosus MoaB residues D57, R87 and T90, which could also be responsible for the inactivate state of E. coli MoaB (Bevers et al., 2008). On the other hand, we have noted the presence of potentially active MoaB proteins in several Serratia species (YP_004499735.1; ZP_06641453.1; YP_001477553.1; ZP_06189083.1), because they retain the four D, D, R, and T residues at the positions homologous to D56, D57, R87, and T90 of P. furiosus MoaB. These species also express potentially active MogA proteins (data not shown), thus weakening the perceived connection between the active status of MoaB and the absence of MogA. Interestingly, an alignment of the moa operons among Enterobacteriaceae genomes in the EcoCyc database (www.ecocyc.org) (Keseler et al., 2011) shows the specific absence of moaB from the moa operon (moaACDE) in several genera, including Edwardsiella, Photorhabdus, Proteus, Providencia, Xenorhabdus, and Yersinia (all of which also have a mogA gene) (data not shown). Based on these observations, one might speculate that moaB is the more ancient gene in Enterobacteriaceae; mogA may have arisen later by moaB duplication or horizontal gene transfer. Possibly, MogA had certain evolutionary advantages over MoaB leading to accumulation of deleterious mutations in moaB in certain species (as in E. coli) or to a complete loss of moaB in others (see above).
Acknowledgments
We thank Drs. K. Chan and M. Itsko of the NIEHS for their helpful comments on the manuscript for this paper. This work was supported by project ES050170 of the Intramural Research Program of the National Institute of Environmental Health Sciences (NIEHS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baba T, Ara T, Hasegawa M, Takao Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:20060008. doi: 10.1038/MSB4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G, Gomez-Ortiz M, Haussmann C, Bacher A, Huber R, Fischer M. Structure of the molybdenum-cofactor biosynthesis protein MoaB of Escherichia coli. Acta Crystallogr D Biol Crystallogr. 2004;60:1068–1075. doi: 10.1107/S0907444904007164. [DOI] [PubMed] [Google Scholar]
- Bevers LE, Hagedoorn PL, Santamaria-Araujo JA, Magalon A, Hagen WR, Schwarz G. Function of MoaB proteins in the biosynthesis of the molybdenum and tungsten cofactors. Biochemistry. 2008;47:949–956. doi: 10.1021/bi7020487. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iobbi-Nivol C, Leimkühler S. Molybdenum enzymes, their maturation and molybdenum cofactor biosynthesis in Escherichia coli. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbabio.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muñiz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, Kaipa P, Spaulding A, Pacheco J, Latendresse M, Fulcher C, Sarker M, Shearer AG, Mackie A, Paulsen I, Gunsalus RP, Karp PD. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39 (Database issue):D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmin SG, Pavlov YI, Dunn RL, Schaaper RM. Hypersensitivity of Escherichia coli Δ(u vrB-bio ) mutants to 6-hydroxylaminopurine and other base analogs is due to a defect in molybdenum cofactor biosynthesis. J Bacteriol. 2000;182:3361–3367. doi: 10.1128/jb.182.12.3361-3367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmin SG, Schaaper RM. Molybdenum cofactor-dependent resistance to N-hydroxylated base analogs in E. coli is independent of MobA function. Mutat Res. 2007;619:9–15. doi: 10.1016/j.mrfmmm.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmin SG, Leroy P, Pavlov YI, Schaaper RM. YcbX and yiiM, twonovel determinants for resistance of Escherichia coli to N-hydroxylated base analogues. Mol Microbiol. 2008;68:51–65. doi: 10.1111/j.1365-2958.2008.06128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalon A, Mendel RR. Chapter 3.6.3.13, Biosynthesis and insertion of the Molybdenum Cofactor. In: Böck A, Curtiss R III, Kaper JB, Karp PD, Neidhardt FC, Nyström T, Slauch JM, Squires CL, Ussery D, editors. EcoSal - Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, DC: Jan 18, 2008. posting date. http://www.ecosal.org. [Google Scholar]
- Miki T, Yamamoto Y, Matsuda H. A novel, simple, high-throughput method for isolation of genome-wide transposon insertion mutants of Escherichia coli K-12. Methods Mol Biol. 2008;416:195–204. doi: 10.1007/978-1-59745-321-9_13. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1972. [Google Scholar]
- Nichols BP, Shafiq O, Meiners V. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J Bacteriol. 1998;180:6408–6411. doi: 10.1128/jb.180.23.6408-6411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivarao D, Turner RJ, Bilous PT, Rothery RA, Shaw G, Weiner JH. Differential effects of a molybdopterin synthase sulfurylase (moeB) mutation on Escherichia coli molybdoenzyme maturation. Biochem Cell Biol. 2002;80:435–443. doi: 10.1139/o02-131. [DOI] [PubMed] [Google Scholar]
- Sanishvili R, Beasley S, Skarina T, Glesne D, Joachimiak A, Edwards A, Savchenko A. The crystal structure of Escherichia coli MoaB suggests a probable role in molybdenum cofactor synthesis. J Biol Chem. 2004;279:42139–42146. doi: 10.1074/jbc.M407694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G, Mendel RR, Ribbe MW. Molybdenum cofactors, enzymes and pathways. Nature. 2009;460:839–847. doi: 10.1038/nature08302. [DOI] [PubMed] [Google Scholar]
- Shanmugam KT, Stewart V, Gunsalus RP, Boxer DH, Cole JA, Chippaux M, DeMoss JA, Giordano G, Lin EC, Rajagopalan KV. Proposed nomenclature for the genes involved in molybdenum metabolism in Escherichia coli and Salmonella typhimurium. Mol Microbiol. 1992;6:3452–3454. doi: 10.1111/j.1365-2958.1992.tb02215.x. [DOI] [PubMed] [Google Scholar]
- Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V, MacGregor CH. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982;151:788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen J, Zylstra GJ, Yee N. Genetic evidence for a molybdopterin-containing tellurate reductase. Appl Environ Microbiol. 2013;79:3171–3175. doi: 10.1128/AEM.03996-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel HJ, Bonner DM. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]