Abstract
BACKGROUND
Recent advances demonstrate a relationship between chronic/recurrent inflammation and prostate cancer (PCA). Among inflammatory regulators, toll-like receptors (TLRs) play a critical role in innate immune responses. However, it remains unclear whether variant TLR genes influence PCA risk among men of African descent. Therefore, we evaluated the impact of 32 TLR-associated single nucleotide polymorphisms (SNPs) on PCA risk among African-Americans and Jamaicans.
METHODS
SNP profiles of 814 subjects were evaluated using Illumina’s Veracode genotyping platform. Single and combined effects of SNPs in relation to PCA risk were assessed using age-adjusted logistic regression and entropy-based multifactor dimensionality reduction (MDR) models.
RESULTS
Seven sequence variants detected in TLR6, TOLLIP, IRAK4, IRF3 were marginally related to PCA. However, none of these effects remained significant after adjusting for multiple hypothesis testing. Nevertheless, MDR modeling revealed a complex interaction between IRAK4 rs4251545 and TLR2 rs1898830 as a significant predictor of PCA risk among U.S. men (permutation testing p-value = 0.001).
CONCLUSIONS
MDR identified an interaction between IRAK4 and TLR2 as the best two factor model for predicting PCA risk among men of African descent. However, these findings require further assessment and validation.
Keywords: prostate cancer, Toll-like receptor (TLR), single nucleotide polymorphisms (SNPs), gene-gene interactions, multifactor dimensionality reduction (MDR)
INTRODUCTION
Prostate cancer (PCA) is one of the most frequently diagnosed cancers among western men of African origin and is the second leading cause of their cancer-related deaths [1, 2]. In the United States (U.S.), African-American men are more likely to receive a PCA diagnosis and die from disease than any other racial or ethnic group. Between 2003 and 2007, the average annual PCA incidence and mortality rates among African-American men were 1.6 and 2.4 higher than Caucasian-American men, respectively. Moreover, several studies indicate similar PCA disparities between American men and Caribbean men [1, 3, 4]. Based on 2008 worldwide statistics, age-standardized death rates among Caribbean men were 2.65-fold higher than American men [4]. Ethnicity, age, and family history are a few risk factors that have been implicated in determining PCA risk. More recently, chronic inflammation has also been considered as an important contributor of PCA, yet the precise etiology of PCA still remains unknown [5–7].
Prostate inflammation may occur as the result of direct infection by microbial pathogens, chemical irritation caused by urine reflux, hormone imbalances and/or autoimmunity. Thus, any or all of the aforementioned factors may also play a role in inflammation-induced PCA. Evolutionarily conserved toll-like receptors (TLRs) play an essential role in regulating innate immune responses to harmful pathogens, as depicted in Figure 1 [8–10]. In humans, some TLRs (e.g. TLR1, TLR2, TLR4, TLR5, and TLR6) are located on the cell surface; whereas, others remain within the intracellular compartments (e.g., TLR3, TLR7, and TLR9) [11]. Cell surface TLRs recognize pathogens either directly or with the aid of extracellular accessory proteins (e.g. CD-14, MD-2). Upon pathogen recognition, the cytoplasmic domain of TLRs is activated, which enables their interaction with adapter molecules (e.g., MyD88, TRIM, TRIF). TLR-adapter molecule complexes then recruit downstream targets (e.g., IRAK4 and TRAF6), resulting in the activation of transcription factors, such as nuclear factor-κB (NF-κB) and interferon regulatory factors (IRFs) via mitogen-activated protein kinase (MAPKs) signaling integrators [8, 12–22]. Overall, TLR signaling cascades induce the expression of inflammatory chemokines, cytokines [tumor necrosis factor-α (TNF- α) and interleukins], and interferons, which prompt local inflammation.
Fig. 1.
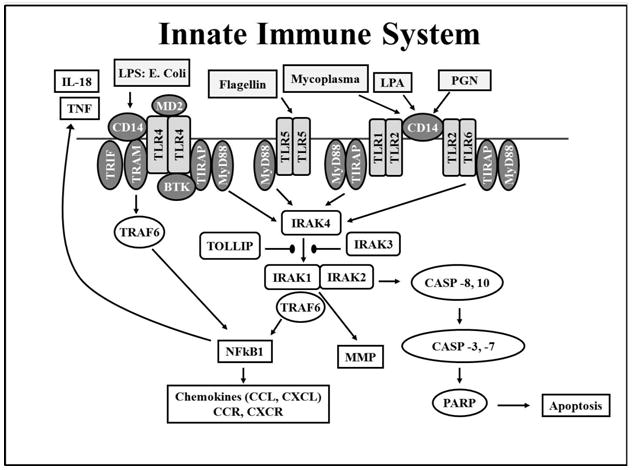
The Toll-like Receptor (TLR) signaling pathway is initiated via activation of TLRs, followed by adaptor complex formation, IRAK and/or TRAF6 activation to induce subsequent MAPK, NFκB, and interferon regulatory factor (IRF) activation, nuclear translocation and regulation of pro- or anti-inflammatory gene expression. Abbreviations: LPS, lipopolysaccharides; PGN, proteoglycans; LTA, lipoteichoic acid; TNF, tumor necrosis factor; IRAK, interleukin-1 receptor-associated kinase; MAPKs, mitogen-activated protein kinases.
The TLR signaling pathway also impacts apoptosis, a form of “programmed cell death” which influences cell differentiation, proliferation, and tumorigenesis. Specifically, TLR2, TLR4, IRAK1, IRAK2, IRAK4, and MyD88 are suggested to regulate both cell survival and cell death signaling pathways [23–27]. Genetic alterations in these TLR genes may dysregulate apoptosis, resulting in tumor escape from cell death, uncontrolled cell proliferation, an increase in cellular damage, accumulation of genetic alterations, and ultimately increased susceptibility to PCA [28–30].
TLR-related sequence variants have been evaluated in relation to a variety of human inflammatory and immune response-related diseases [9, 10, 31–37], including prostate cancer [38–46]. Among the individual TLR SNPs analyzed, TLR1 rs57430604, TLR10 (rs4274855, rs11096957 and rs11096955) and TLR4 rs1927911 have been linked to an increase in the risk of developing PCA [38–42]. At the same time, studies have also identified several TLR SNPs (TLR1 rs4833095, TLR1 rs5743595, TLR4 rs1927911, TLR4 rs2149356, TLR10 rs4274855, and TLR10 rs11096957) associated with a decrease in PCA risk [43, 44] or with no effect (TLR4 rs1927906, TLR4 rs1927911, TLR6 rs5743810, TLR6 rs3821985, TLR6 rs1039559, and TLR10 rs11466640) [39, 43–46]. Although the relationship between genetic alterations and PCA is controversial, available published reports typically focus on men of Asian or European descent. Yet, little or no data addresses the impact of TLR-related sequence variants on PCA risk among men of African descent, even though this population suffers disproportionately from this disease.
This study examined the role of TLR signaling pathway sequence variants on PCA among men of African descent. We evaluated the effects of 32 TLR SNPs individually or jointly on PCA susceptibility among 814 African-American and Jamaican men. Main effects and complex interactions were assessed using conventional and bioinformatic techniques, including logistic regression and entropy-based multi-factor dimensionality reduction (MDR). Investigation of genetic susceptibilities detected within the TLR signaling pathway will provide a better understanding of their influence on PCA risk among men of African descent.
RESULTS
Population Description
The demographic and other pertinent characteristics of cases and controls for the entire study population and each study center are summarized in Table 1 and Supplemental Tables AB. Overall, men diagnosed with prostate cancer were 14 years older and had higher PSA levels than controls (P < 0.0001). Among controls, Jamaican men were about 9 years older and had higher PSA levels (P < 0.0001) and as well as higher median Gleason scores (P = 0.018) than U.S. men. As summarized in Table 1, there were significant differences in family history of PCA with respect to the following: (1) cases to controls from the total population (P = 0.316) (Table 1), U.S. alone (P = 0.592) (Supplemental Table A), or Jamaica alone (P = 0.272) (Supplemental Table B), and (2) controls (P = 0.757) or cases (P = 0.830) comparing the two study centers (data not shown). Among African-Americans, the degree of West African ancestry did not vary by disease-status, as shown in Supplemental Table A [47–49]; however, no such data was collected for the Jamaican men.
Table 1.
Study population characteristics among men of African descent from the U.S. and Jamaica.
| Characteristics | Cases | Controls | p value |
|---|---|---|---|
|
| |||
| Number of Participants, n | 279 | 535 | --- |
|
| |||
| Age at enrollment (yrs), Median (range) | 67 (45–91) | 53 (27–89) | <0.0001a |
|
| |||
| Family History of Prostate Cancer, n (%) | |||
| Yes | 35 (16.1) | 21 (12.5) | |
| No | 182 (83.9) | 147 (87.5) | 0.316b |
| Missing | 62 (22.2) | 367 (68.6) | |
|
| |||
| PSA (ng/ml), median (range) | 11.7 (0.01–10,000) | 0.9 (0.0–4.0) | <0.0001a |
|
| |||
| PSA (ng/ml)b, n (%) | |||
| < 4 | 37 (13.8) | 517 (99.8) | <0.0001b |
| ≥ 4 | 231 (86.2) | 1 (0.2) | |
| Missing | 11 (0.04) | 17 (0.03) | |
|
| |||
| Gleason Score, n (%) | |||
| 4 | 12 (5.6) | ||
| 5 | 14 (6.5) | ||
| 6 | 74 (34.2) | ||
| 7 | 70 (32.4) | ||
| 8 | 18 (8.3) | ||
| 9 | 22(10.2) | ||
| 10 | 6 (2.8) | ||
| Missing | 63 (22.6) | ||
|
| |||
| Global WAA, median (range) | 0.79 (0.25–0.94) | 0.767 (0.25–0.95) | 0.107a |
Abbreviations: PSA, prostate specific antigen; WAA, West African Ancestry;
Wilcoxon Sum Rank Test was used to examine whether differences exists within median age (yrs),
Chi-square test of heterogeneity was used to determine whether the prevalence of family history or high PSA levels (PSA 4 ng/mL) vary between cases and controls, PSA (ng/ml) and Global WAA between cases and controls.
Minor allele and genotype frequency among men of African descent from the U.S. and Jamaica
Overall, the average minor allele frequency (MAF) for TLR-associated SNPs among disease-free U.S. and Jamaican men combined was 23.3% [standard deviation (SD) = 15.8], as shown in Supplemental Table C. When stratified by study site, the MAFs for U.S. and Jamaican men were 23.3% (SD = 16.1) and 21.9% (SD = 14.9), respectively. When the TLR-associated SNPs among the total population (i.e., U.S. and Jamaican men combined) were analyzed, 87.5% had minor allele frequencies ≥5%.
Association between TLR-associated sequence variants and prostate cancer risk
Three TLR-associated markers (TLR6 rs2381289, TOLLIP rs3168046, and TOLLIP rs5743899) were modestly linked with PCA susceptibility for the total population under the age-adjusted LR models, as shown in Table 2. In particular, possession of the TLR6 rs2381289 GA or TOLLIP rs5743899 AG+GG genotypes was marginally associated with a 1.46–1.49 fold increase in the risk of developing PCA. In contrast, TOLLIP rs3168046 AA carriers had a marginally significant 42% reduction in PCA susceptibility (ORage-adjusted = 0.58; 95%CI = 0.35, 0.98). However, none of these markers remained significant after adjusting for multiple hypothesis testing.
Table 2.
Relationship between TLR- associated SNPS and prostate cancer risk among men of African descent (total population).
| Gene (Alleles and position) | Genotype | Cases N (%) | Controls N (%) | Unadj OR (95% CI) | Age Adj OR (95% CI) | Chi- square P-value | Age Adj Chi- square P-value | p- trend | FDR p-value |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| TLR6 | GG | 140 (50.4) | 302 (56.7) | 1.00 (referent) | 1.00 (referent) | 0.164 | 0.198 | 1.000 | |
| rs2381289 | GA | 124 (44.6) | 200 (37.7) | 1.33 (0.98, 1.80) | 1.46 (1.02, 2.09) | 0.062 | 0.041 | ||
| 3′UTR | AA | 14 (5.04) | 30 (5.63) | 1.00 (0.52, 1.96) | 1.20 (0.52, 2.76) | 0.984 | 0.655 | ||
| GA+AA | 138 (49.64) | 230 (43.33) | 1.28 (0.96, 1.72) | 1.43 (1.00, 2.00) | 0.088 | 0.046 | 1.000 | ||
| AA vs (GG+AG) | 0.88 (0.46, 1.70) | 1.02 (0.46, 2.29) | 0.724 | 0.956 | 0.853 | ||||
|
| |||||||||
| TOLLIP | GG | 104 (37.3) | 189 (35.4) | 1.00 (referent) | 1.00 (referent) | 0.363 | 0.261 | 0.798 | |
| rs3168046 | GA | 131 (46.9) | 239 (44.8) | 1.00 (0.72, 1.40) | 0.85 (0.58, 1.25) | 0.981 | 0.415 | ||
| 3′UTR | AA | 44 (15.8) | 106 (19.9) | 0.75 (0.49, 1.15) | 0.58 (0.35, 0.98) | 0.194 | 0.042 | ||
| miRNA | GA+AA | 175 (62.7) | 345 (64.7) | 0.92 (0.68, 1.24) | 0.76 (0.54,1.10) | 0.596 | 0.155 | 1.000 | |
| AA vs (GG+AG) | 0.76 (0.51, 1.11) | 0.64 (0.40, 1.02) | 0.155 | 0.628 | 1.000 | ||||
|
| |||||||||
| TOLLIP | AA | 85 (30.8) | 187 (35.2) | 1.00 (referent) | 1.00 (referent) | 0.208 | 0.686 | 1.000 | |
| rs5743899 | AG | 140 (50.7) | 235 (44.2) | 1.31 (0.94, 1.82) | 1.59 (1.06, 2.38) | 0.109 | 0.023 | ||
| Intron 1 | GG | 51 (18.5) | 110 (20.7) | 1.02 (0.67, 1.55) | 1.29 (0.78, 2.14) | 0.926 | 0.031 | ||
| AG+GG | 191 (69.2) | 1.22 (0.89, 1.66) | 1.49 (1.02, 2.18) | 0.215 | 0.035 | 0.884 | |||
| GG vs (AA+AG) | 0.87 (0.60, 1.26) | 0.98 (0.64, 1.53) | 0.458 | 0.949 | 0.756 | ||||
Fisher’s P-value was calculated when expected genotype counts were < 5 for both cases and controls. Significant associations are indicated in boldface.
Abbreviations: UTR, untranslated region, miRNA, microRNA binding site.
Upon stratification by study center, six of the 32 candidate TLR-associated SNPs were identified as modestly significant determinants of PCA risk after adjusting for age, as shown in Table 3. Within the U.S. population, a nominal 40–66% reduction in PCA susceptibility was observed among men who possessed one or more IRAK4 (rs4251545A and rs4251473A) or TLR6 rs5743818C minor alleles; however, the relationship was strongest for carriers of the IRAK4 rs4251545 AA genotype (ORage-adjusted = 0.34; 95%CI = 0.13, 0.91). Moreover, the TOLLIP rs5743899 locus, under the recessive genetic model (GG versus AG+AA), was modestly associated with a 1.14 fold increase in the risk of developing PCA (ORage-adjusted = 1.14; 95%CI = 1.12, 1.18). Among Jamaican men, IRF3 rs2304206, TLR6 rs2381289, and TLR6 rs5743818 were marginally associated with PCA risk. For instance, inheritance of the IRF3 rs2304206 GG genotype (ORage-adjusted = 0.32; 95%CI = 0.10, 0.98) was linked to a modestly significant 68% reduction in PCA susceptibility. On the other hand, there was a 1.10–2.0-fold increase in PCA risk associated with inheriting the TLR6 rs2381289 GA+AA (ORage-adjusted = 2.05; 95%CI = 1.10, 3.78) or TLR6 rs5743818 AC+CC GA (ORage-adjusted = 1.10; 95%CI = 1.06, 1.14) genotypes. Notably, additive genetic models for IRF3 rs2304206 and TLR6 rs2381289 were significantly related to prostate cancer risk, which is modestly suggestive of a significant dose-response effect in relation to the number of inherited minor alleles (P-trend ≥ 0.0193). These modest associations; however, did not persist after adjustments for multiple hypothesis testing.
Table 3.
Relationship between TLR-associated SNPs and prostate cancer risk stratified by location.
| Gene (Allele and position) | Genotype | Unadjusted OR (95%CI) U.S. Men | Unadjusted OR (95%CI) Jam. Men | Age Adjusted OR (95%CI) U.S. Men | Age Adjusted OR (95%CI) Jam. Men | p-value U.S. Men | p-trend U.S. Men | p-value Jam. Men | p-trend Jam. Men |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| IRAK4 | CC | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 0.270* | 0.1009 | 1.000* | 0.9546 |
| rs4251473 | CA | 0.76 (0.51, 1.14) | 1.02 (0.55, 1.88) | 0.61 (0.38, 0.99) | 0.92 (0.47, 1.79) | 0.1809 | 0.9504 | ||
| Intron 5 | AA | 0.57 (0.21, 1.56) | 0.92 (0.28, 2.98) | 0.34 (0.09, 1.21) | 1.42 (0.32, 6.31) | 0.2738 | 0.8865 | ||
| CA+AA | 0.74 (0.50, 1.08) | 1.00 (0.56, 1.78) | 0.58 (0.36, 0.92) | 0.97 (0.52, 1.84) | 0.1189 | 0.9966 | |||
| AA vs (CC+CA) | 0.62 (0.22, 1.68) | 0.91 (0.28, 2.92) | 0.40 (0.11, 1.40) | 1.46 (0.34, 6.38) | 0.3472 | 0.8777 | |||
|
| |||||||||
| IRAK4 | GG | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 0.0723 | 0.0227 | 0.6845 | 0.4725 |
| rs4251545 | GA | 0.71 (0.48, 1.04) | 1.04 (0.58, 1.86) | 0.60 (0.38, 0.94) | 1.07 (0.56, 2.04) | 0.0759 | 0.9087 | ||
| Exon 8 or Exon 1 Splicing (ESE or ESS) | AA | 0.50 (0.23, 1.08) | 1.47 (0.60, 3.56) | 0.34 (0.13, 0.91) | 1.97 (0.70, 5.52) | 0.0785 | 0.3907 | ||
| GA+AA | 0.68 (0.47, 0.96) | 1.13 (0.66, 1.94) | 0.56 (0.36, 0.86) | 1.22 (0.67, 2.24) | 0.0331 | 0.6632 | |||
| AA vs (GG+GA) | 0.58 (0.28, 1.23) | 1.45 (0.62, 3.40) | 0.1575 | 0.39 | |||||
|
| |||||||||
| IRF3 | AA | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 0.767 | 0.5576 | 0.0791 | 0.0365 |
| rs2304206 | AG | 1.15 (0.78, 1.70) | 0.74 (0.42, 1.32) | 1.22 (0.76, 1.97) | 0.65 (0.34, 1.22) | 0.4754 | 0.314 | ||
| Intron 1 | GG | 1.12 (0.65, 1.94) | 0.32 (0.11, 0.90) | 1.30 (0.68, 2.49) | 0.32 (0.10, 0.98) | 0.666 | 0.0313 | ||
| TFBS | AG+GG | 1.14 (0.80, 1.66) | 0.65 (0.38, 1.12) | 1.24 (0.80, 1.95) | 0.57 (0.31, 1.06) | 0.4696 | 0.1195 | ||
| GG vs (AA+AG) | 1.04 (0.63, 1.70) | 0.37 (0.14, 1.00) | 1.15 (0.64, 2.07) | 0.40 (0.13, 1.16) | 0.8813 | 0.0508 | |||
| TLR6 | GG | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 0.770* | 0.8009 | 0.043* | 0.0193 |
| rs2381289 | GA | 1.13 (0.78, 1.64) | 1.98 (1.12, 3.50) | 1.28 (0.82, 2.00) | 2.00 (1.06, 3.76) | 0.5102 | 0.0174 | ||
| 3′ UTR | AA | 0.88 (0.40, 1.96) | 2.12 (0.48, 9.30) | 0.92 (0.34, 2.50) | 2.57 (0.50, 13.2) | 0.7683 | 0.3176 | ||
| GA+AA | 1.10 (0.76, 1.56) | 1.99 (1.51, 3.46) | 1.23 (0.80, 1.90) | 2.05 (1.10, 3.78) | 0.607 | 0.014 | |||
| AA vs (GG+GA) | 0.84 (0.38, 1.82) | 1.58 (0.36, 6.82) | 0.82 (0.31, 2.20) | 1.92 (0.38, 9.64) | 0.6634 | 0.5348 | |||
|
| |||||||||
| TLR6 | AA | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 0.170* | 0.1756 | 0.056* | 0.0767 |
| rs5743818 | AC+CC | 0.64 (0.38, 1.08) | 2.62 (0.90, 7.65) | 0.54 (0.28, 1.01) | 1.10 (1.06, 1.14) | 0.0986 | 0.0767 | ||
| Exon 1 | |||||||||
|
| |||||||||
| TOLLIP | GG | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 0.3264 | 0.1655 | 0.22 | 0.8106 |
| rs3168046 | GA | 0.77 (0.52, 1.14) | 1.56 (0.85, 2.83) | 0.73 (0.45, 1.18) | 1.15 (0.58, 2.26) | 0.2006 | 0.1492 | ||
| UTR’3 miRNA | AA | 0.73 (0.44, 1.20) | 0.89 (0.38, 2.06) | 0.63 (0.34, 1.16) | 0.56 (0.21, 1.46) | 0.2204 | 0.7891 | ||
| GA+AA | 0.76 (0.52, 1.10) | 1.37 (0.80, 2.41) | 0.70 (0.45, 1.08) | 0.99 (0.52, 1.88) | 0.1393 | 0.2831 | |||
| AA vs (GG+AG) | 0.84 (0.53, 1.32) | 0.69 (0.32, 1.46) | 0.74 (0.42, 1.30) | 0.52 (0.22, 1.22) | 0.449 | 0.3328 | |||
|
| |||||||||
| TOLLIP | AA | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 0.239 | 0.4904 | 0.5591 | 0.5343 |
| rs5743899 | AG | 1.40 (0.93, 2.10) | 1.11 (0.59, 2.08) | 1.67 (1.02, 2.73) | 1.35 (0.66, 2.74) | 0.103 | 0.7397 | ||
| Intron 1 | GG | 1.10 (0.66, 1.86) | 0.75 (0.35, 1.61) | 1.30 (0.69, 2.41) | 1.12 (0.47, 2.66) | 0.6974 | 0.4647 | ||
| AG+GG | 1.31 (0.89, 1.92) | 0.98 (0.54, 1.76) | 1.55 (0.98, 2.46) | 1.28 (0.66, 2.48) | 0.166 | 0.9517 | |||
| GG vs (AA+AG) | 0.90 (0.58, 1.43) | 0.70 (0.36, 1.37) | 1.14 (1.12, 1.18) | 0.93 (0.44, 1.96) | 0.6776 | 0.3061 | |||
Fisher’s P-value was calculated when expected genotype counts were < 5 for either cases and controls. Significant associations are indicated in boldface.
Abbreviations: UTR, untranslated region; TFBS, transcription factor binding site; ESE, exonic splicing enhancer; ESS, exonic splicing silencers; nsSNP, non-synonymous coding SNP; Jam., Jamaican
Analysis of gene-gene interactions using Multi-factor Dimensionality Reduction (MDR)
MDR modeling was used to efficiently assess and validate age-adjusted gene-gene interactions for the total population, U.S. men alone, and Jamaican men alone in relation to PCA risk. The top one-, two-, and three-way interaction models for the total population, involving U.S. and Jamaican men combined, displayed 100% cross validation consistency (CVC) values, 57–65% average testing accuracy (ATA) scores, and permutation p-values = 0.001. However, the three-way interaction among TLR6 rs2381289, TLR10 rs11096957, and IRF3 rs2304206 was selected as the best PCA predictor for men of African descent, since this model had the highest average testing accuracy (ATA = 0.6505). This three-way interaction was primarily driven by a synergistic relationship between TLR6 rs2381289 and IRF3 rs2304206 (data not shown).
CVC and ATA scores for all 1-, 2-, and 3-factor models among U.S. men were significant and characterized as 90–100% and 57–62%, respectively, as described in Table 4A-B. However, interaction between TLR2 rs1898830 and IRAK4 rs4251545 was chosen as the best PCA predictor, based on a higher average testing accuracy (ATA = 61.94%) and lower permutation testing value (P = 0.001) relative to the best three-factor model (P = 0.015). The two-way interaction, as shown in Figure 2, was highly synergistic since the joint information gain score (2.33%) exceeded the mutual information gain scores for TLR2 rs1898830 alone (0.04%) and IRAK4 rs4251545 alone (1.30%).
Table 4A. Interactions and main effects of TLR-associated SNPs as predictors of prostate cancer using age-adjusted MDR.
MDR models for U.S. and Jamaican men of African descent combined.
| Best Total Population Model (dbSNPID #) | Cross Validation Consistency (CVC) | Average Testing Accuracy (ATA) | Permutation Testing p-value |
|---|---|---|---|
|
| |||
| One Factor | 10/10 | 0.5722 | 0.001 |
| TLR6 rs2381289 | |||
|
| |||
| Two Factor | 10/10 | 0.6113 | 0.001 |
| TLR10 rs11096957 | |||
| TLR6 rs2381289 | |||
|
| |||
| Three Factor | 10/10 | 0.6505 | 0.001 |
| TLR10 rs11096957 | |||
| TLR6 rs2381289 | |||
| IRF3 rs2304206 | |||
Table 4B. Interactions and main effects of TLR-associated SNPs as predictors of prostate cancer using age-adjusted MDR.
MDR models for U.S. men.
| Best U.S. Men Model (dbSNPID #) | Cross Validation Consistency (CVC) | Average Testing Accuracy (ATA) | Permutation Testing p-value |
|---|---|---|---|
|
| |||
| One Factor | 10/10 | 0.5744 | 0.179 |
| IRAK4 rs4251473 | |||
|
| |||
| Two Factor | 10/10 | 0.6194 | 0.001 |
| TLR2 rs1898830 | |||
| IRAK4 rs4251545 | |||
|
| |||
| Three Factor | 9/10 | 0.6184 | 0.015 |
| TLR6 rs3821985 | |||
| TLR4 rs1927906 | |||
| TLR2 rs3804099 | |||
Fig. 2. Interaction entropy model of model of TLR-associated SNPs and prostate cancer risk for U.S. men of African descent.
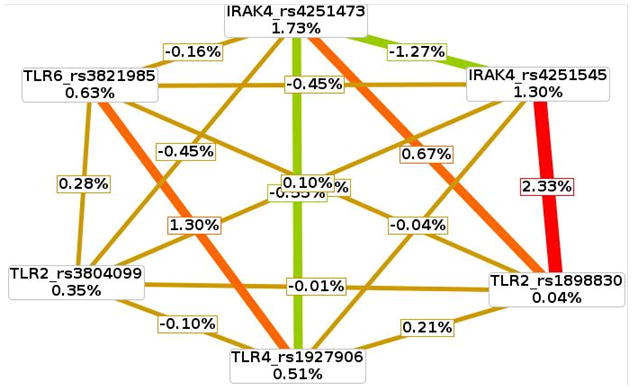
This graphical model describes the percent entropy as explained by each single TLR-related SNP or a combination of two loci within the U.S. population. Information gain or synergy is expressed as positive percent entropy. Redundant or missing information gain is expressed as negative percent entropy. A range from synergy (i.e. non-additive) to redundancy is represented using a schematic coloration in the visualization tool. The colors range from red representing a high degree of synergy (positive information gain), orange a lesser degree, and gold representing independence and a midway point between synergy and redundancy. Green represents redundancy.
For the Jamaican population, the one-way model containing the TLR6 rs2381289 loci was the best PCA-related MDR model based on a 62.7% prediction accuracy score and significant permutation p-value (P = 0.0018). Although the two- and three-way models both had high CVC scores (≥ 80%), these models failed to reach statistical significance.
DISCUSSION
Dysregulation and genetic alterations in immune system function are linked to many cancers. In particular, it is estimated that approximately 20% of all human cancers, including prostate cancer (PCA), are associated with chronic inflammation [28, 50]. TLR activation, a key initiator of inflammation and dysregulation of TLR-responsive pathways, has been associated with cancer susceptibility. The current study evaluated 32 TLR-associated sequence variants to determine their individual and joint modifying effects on PCA risk among 279 cases and 535 disease free men of African descent. Out of the 32 minor variant alleles, 7 were modestly associated with a 1.14–2.05-fold increase (TLR6 rs2381289, TOLLIP rs5743899) or 39–68% decrease (TOLLIP rs3168046, IRAK4 rs4251473, IRAK4 rs4251545, IRF3 rs2304206, TLR6 rs5743818) in the risk of developing prostate cancer either in the total population and/or the stratified analysis, after adjusting for age. Among U.S. men, there was a nominal 40–66% reduction in PCA susceptibility among those who possessed one or more IRAK4 (rs4251545 and rs4251473) or TLR6 rs5743818 minor alleles. We also found that the TOLLIP rs5743899 SNP under the recessive genetic model (GG versus AG+AA) was modestly associated with a 1.14 fold increase in the risk of developing PCA (ORage-adjusted = 1.14; 95%CI = 1.12, 1.18) among men of African descent from the U.S. The TOLLIP rs3168046 SNP was unique to the total population; whereas, 3 markers (IRAK4 rs4251473, IRAK4 rs4251545, TLR6 rs2381289) were unique to the U.S. men. Jamaican and U.S. sub-groups each had 1 SNP in common with the total population, namely TLR6 rs2381289 and TOLLIP rs5743899, respectively. Among Jamaicans, both the IRF3 rs2304206 and TLR6 rs2381289 loci were significant under the additive genetic model. However, only the TLR6 rs2381289 SNP for the Jamaican population remained statistically significant after adjusting for age and multiple hypothesis testing (Permutation p-value = 0.018).
Finally, we examined main effects and interactions of TLR-associated SNPs as predictors of prostate cancer using age-adjusted MDR and found that the best model varied depending on the composition of the study population. Among U.S. men of African descent, the best predictor of PCA risk was the two-factor interaction between IRAK4 rs4251545 and TLR2 rs1898830. Several investigators have evaluated the link between prostate cancer outcomes and toll-like receptor-associated (TLR1, TLR4, TLR6, TLR10, IRAK4) sequence variants. Collectively, 9 studies evaluated 14 out of the 32 SNPs considered in the current study with mixed findings [38–46, 51]. In the Cancer of the Prostate Study (CAPS), inheritance of one or more TLR1 rs4833095 and TLR10 (rs11096955, rs11096957) minor alleles were associated with an increase in prostate cancer risk among Caucasians. However, the American Cancer Society Cancer Prevention II Nutrition Cohort (CPS-II) study reported protective effects [44] and null findings were observed for Caucasians of the Health Professionals Follow-up Study [45, 46], PLCO Study [45] as well as a meta-analysis of 3142 cases and 2567 controls [45]. Consistent with the current study, five independent studies reported null findings in relation to prostate cancer risk and possession of TLR1 rs46224663 and TLR6 (rs5743814, rs5743810, rs1039559, rs3821985) variant alleles [44–46, 51]. Inheritance of one or more minor alleles were associated with either an increase (TLR1 rs5743604, TLR10 rs4274855) [45, 51] or decrease (TLR1 rs5743595, TLR1 rs2149356, TLR4 rs1927911) [43–45] in the risk of developing prostate cancer among Caucasians in four separate observational studies. However, these same markers were not significantly related to prostate cancer among men of African and European descent in the current and other independent studies [40, 42, 45, 46]. The TLR4 rs1927911CC genotype was linked to an increase in prostate cancer risk in one small Korean case-control study [38]; whereas, another Korean study did not reveal a statistically significant relationship [41]. Discrepant findings for these two Korean studies may be attributed to differences in methods used for allelic discrimination. Utilization of RFLP-PCR method used in the former study [38] may not have the same capacity to discern between homozygous and heterozygous genotypes, relative to a more advance method used in the study performed by and co-workers [41]. Other explanations for differences in the directionality of prostate cancer risk estimates in the aforementioned TLR-related SNPs may include: inadequate statistical power to detect significant differences between prostate cancer cases and controls among Koreans [38, 41]; failure to adjust for multiple hypothesis testing [38, 39, 41–46, 51]; and unknown environmental exposures/lifestyle factors (e.g., pathogens, environmental toxins, antioxidants) [38–46, 51]. Variations in population composition may account for the mildly suggestive yet distinct risk alleles noted in the men of African descent from the U.S. (IRAK rs4251473, IRAK rs425154, TOLLIP rs5743899) and Jamaica (IRF3 rs2304206, TLR6 rs2381289) in the current study.
This study is the first to address a relationship between select polymorphic TLR-related genes and PCA in general (i.e., TLR2, IRF3) and more specifically among men of African descent [i.e., TLR1,TLR 2,TLR 4,TLR 6,TLR10, IRAK4, IRF3, and TOLLIP]. TLR6 rs2381289 and TLR2 rs1898830 PCA-related SNPs identified in this study are likely to affect transcriptional regulation of TLR genes. TLRs 2 and 6 are controlled by the master regulatory transcription factor p53 [76]. TLR-associated SNPs may alter p53 interactions with these TLR genes. Furthermore, the IRAK4 rs4251545 SNP, in silco, codes for alterations in mRNA splicing, which in turn may influence mRNA stability, IRAK4 kinase activity, as well as TLR signaling protein-protein interactions. Such alterations may lead to biochemical conditions that favor cell death, decreases in matrix metalloproteinases linked to cell migration, decreases in pro-inflammatory cytokines/chemokines and ultimately abrogation of tumor growth. However, further molecular biological studies are needed to determine the exact impact of the aforementioned SNPs on prostate tumor biology.
We are aware of both strengths and limitations of our approach. Although we observed nominal relationships between selected TLR-associated SNPs and PCA risk, we cannot rule out the possibility that anomalies within the innate immune pathway will influence disease prognosis. Consequently, future studies will enable us to evaluate the relationship between TLR SNPs and Gleason score, tumor stage, biochemical/disease recurrence, and overall/disease specific mortality. Future multi-center pooled genetic studies with thousands of cases and controls may enable us to confirm and refine our small effect sizes. It is plausible that other targets immediately downstream of the TLRs in innate immune signaling pathways may play a role in PCA risk among men of African descent, as shown in Figure 1. Moreover, PCA susceptibility may also be influenced by polymorphisms of some genes even further down the TLR signaling pathway, including caspases (CASP 3, 7, 8, and 10), mitogen-activated protein kinases (MAPKs), interferon-regulatory factors (IRFs), interferons, inflammatory cytokines, and chemokines. This represents an area of active research within future collaborative studies in our lab.
Analysis of mRNA and protein levels of TLR-related SNPs and investigation of the relative expression and activity of downstream targets are needed to define the biological mechanisms that give rise to prostate cancer disparities among men of African and European descent. The genetic admixture among African-American men, as documented by other published reports, may modify the relationship between TLR SNPs and prostate cancer risk. For the current study, analyses were restricted to men with ≥25% West African Ancestry. Adjustment of our risk models for West African Ancestry ultimately had no significant bearing on calculated risk estimates among African-American men. Due to chance alone, it is estimated that 5% of the 744 SNP interactions among 32 TLR sequence variants will result in 37 significant relationships. However, we controlled for multiple hypothesis testing bias by adjusting our MDR findings with permutation testing. Given the low prediction accuracy (i.e., 61.9%) between TLR2 and IRAK4, our study findings require replication within independent study sets. However, recent simulation studies demonstrate that even modest disparities in genotype frequencies among study participants of independent study sets may interfere with the capacity to replicate complex interactions [52]. Consequently, to replicate our findings, it is critical that future replicate studies should have the same genetic architecture (i.e., ancestry identification markers and TLR SNPs) as the African-Americans in the current study. Caution is recommended in the interpretation of our study findings due to a modest marginal effect between TLR signaling sequence variants and PCA risk. However, enthusiasm for the relationship between PCA and the innate immune signaling pathway was slightly elevated in our exhaustive 2- and 3-way interactions. In particular, our exploratory analysis revealed a synergistic relationship between IRAK4 and TLR2 as significant PCA markers among men of African descent in the U.S. We speculate that genetic variations in TLR-related genes may influence the PCA risk by modulating cell survival, proliferation and/or inflammation. Mechanistic studies are needed in order to corroborate these findings and explore the functional consequences of TLR-related SNPs on PCA development. By combining our genetic variation analysis of TLR-related polymorphisms with biological studies, we hope to develop a level of understanding that will allow us to accurately predict and eventually offset the increased genetic risk factors for PCA that threaten men of African descent.
MATERIALS AND METHODS
Study Population
Two independent case-control study sets with participants from the Prostate Cancer Clinical Outcome (PC2O) Study and the Prostate Cancer Case-Control Study were used in the current study (Table 1 and Supplemental Tables A-B). Among all 814 men of African descent, germ-line DNA samples were collected for 279 PCA cases and 535 disease-free men, as shown in Table 1. In the PC2O Study, 603 unrelated men of African descent were recruited between 2001 and 2005 from Columbia, South Carolina and Howard University Hospital (HUH) Division of Urology in Washington, DC. Self-identified African-American, East African-American, West African-American, or Afro-Caribbean American men from the U.S. were participants of the PC2O Study, consisting of 170 incident PCA cases and 433 controls, as shown in Supplemental Table A. For the Prostate Cancer Case-Control Study, 211 unrelated Jamaican men (109 incident PCA cases, 102 controls) were consecutively enrolled between 2005 and 2007 during a first time urological clinic visit, as depicted in Supplemental Table B. The examination and inclusion criteria of all subjects have been described in detail previously [53, 54].
Genetic Analysis of Variant TLR-Associated SNPs
De-identified germ-line DNA was obtained from incident PCA male cases (n = 279) and controls (n = 535). SNPs detected in TLRs (1, 2, 4, 6, and 10), IRAK4, TOLLIP, and IRF3 were genotyped using Illumina’s GoldenGate genotyping assay system combined with Veracode Technology (Illumina, Inc., San Diego, CA). Allelic discrimination was performed using a BeadXpress Reader (Illumina, Inc., San Diego, CA) according to the manufacturer’s instructions [51]. Quality control analyses and data management were performed using Golden Helix’s SNP Variation Software 7.0 (Bozeman, MT). To ensure high quality data, SNPs were excluded if: genotype call rates were <95% (n = 1); genotypic distribution between controls deviated substantially from the HWE with a significance cut-off value of P ≤ 0.005 (n = 4); or the minor allele frequency was <1% (n = 5). Based on the above criteria, 32 TLR-related SNPs were examined among men of African descent, as listed in Supplemental Table A. Based on the above criteria, 32 TLR-related SNPs were examined among men of African descent, as listed in Supplemental Table A.
Ancestry Markers
Among the U.S. men, cases and controls were also genotyped with a set of 100 genome-wide ancestry informative markers to correct for potential population stratification among our admixed population, as previously described [47, 48]. Individual genetic ancestry (IA) was determined for each person using 100 AIMs for West African and European genetic ancestry. IA was estimated from the genotype data using the Bayesian Markov Chain-Monte Carlo (MCMC) method implemented in the program STRUCTURE 2.1, as detailed elsewhere [47, 49]. Study participants were grouped from lowest to highest genetic West African ancestry, with scores ranging from 0–100%. These 100 markers were evaluated using DNA from self-identified African-Americans (Coriell Institute for Medical Research, n = 96), Yoruban West Africans (HapMap, n = 60), West Africans (Bantu and Nilo Saharan speakers, n = 72), Europeans (New York City, n = 24), and CEPH Europeans (HapMap Panel, n = 60), as previously reported [48]. Individuals with a West African ancestry (WAA) score ≥25% and available TLR genotype data were included in the final analysis.
Statistical Analysis for Single Gene Effects
Univariate and multivariate statistical analyses were used to examine the relationship between TLR SNPs among men of African descent and PCA risk. For each TLR SNP, frequency differences in TLR genotypes between cases and controls were tested using the Chi-square (χ2) test of homogeneity. Odds ratios (OR) and corresponding 95% confidence intervals (CI) for PCA risk in association with TLR SNPs were estimated using unconditional multivariate logistic regression (LR) models after adjusting for age. LR analyses for genetic variants and PCA risk were conducted using the major or common genotype as the referent category. Chi-square test and LR analyses were performed using SAS 9.3 (SAS Institute Inc., Cary, NC) and SVS software (Golden Helix, Inc., Bozeman, MT). Adjustment for multiple hypothesis testing was achieved using false discovery rate (FDR). Statistically significant data was based on a p-value cut-off of 0.05.
Statistical Power for Single Gene Effects
We calculated the odds of developing PCA among carriers of at least one or more minor allele based on the average MAF 21.9–23.3% for the three study sets (i.e., U.S. men, Jamaican men, and U.S. and Jamaican men combined), a PCA disease prevalence of 0.740%, a significance level (α) of 5%, and 100% linkage disequilibrium (LD) between the casual and the predisposing variant. According to our sample size for the combined population (279 cases, 535 controls), U.S. (170 cases, 433 controls) and Jamaican men (109 cases, 102 controls), we had >80% power to detect odds ratios (ORs) of ≥1.4 ≥ 1.6, and ≥1.9 for PCA risk, respectively. Statistical power calculations were performed using Power for Genetic Association Version 2 Software, as described previously [55].
Analysis of Gene Interactions Using Multi-factor Dimensionality Reduction (MDR)
To evaluate the single- and joint- modifying effects of 32 candidate TLR-associated SNPs within a large dataset is computationally challenging. In order to overcome this problem, open source and freely available MDR 2.0 was used to detect and characterize all possible one-, two-, and three-way interaction models in relation to PCA (http://www.epistasis.org/) [56]. To reduce computation time needed to process thousands of SNP combinations in relation to PCA risk, we distributed MDR on a workstation with 12 hyper-threaded cores across two central processing units (total of 24 simultaneous threads of execution) and 24GB of RAM. Although MDR has been described elsewhere, for convenience we provide a brief summary. With MDR, reduced genetic information is reduced to a one-dimensional multi-locus genotype variable [57, 58]. Information from various disease loci were grouped and labeled as “high risk” or “low risk” based on whether or not the control ratio met or exceeded a particular threshold. Subsequently, the resulting one-dimensional multi-locus genotype variable was examined for its capability to categorize and predict disease outcome through cross-validation and permutation testing procedures. A 10-fold cross-validation was achieved by dividing the entire dataset into a training set and an independent testing set. The training set involved 9/10ths of the data; the remaining 1/10th, known as the independent testing set, was evaluated against the training set. Evaluation of each independent testing set predicted average testing accuracy values for each MDR model. The greatest cross validation consistency (i.e., CVC ≥ 8/10) and highest prediction accuracy [i.e., Average Testing Accuracy (ATA)] were selected as the best predictors of disease outcome. Sensitivity and specificity were determined as functions of true negatives (TN), false positives (FP), and false negatives (FN). Sensitivity, specificity and balanced accuracy values were calculated as follows: sensitivity = (TP)/(TP+FN); specificity = (TN)/(FP + TN); and balanced accuracy = (sensitivity + specificity)/2. ATAs were averaged across all 10 pieces of the data, in order to provide an estimate of the predictive ability of the loci in relation to the outcome of interest. We used cross-validation consistency (CVC) to determine the degree to which the same best MDR model is selected across the 10 divisions of the data. Models with a CVC of ≥8/10 using a 10-fold cross-validation were considered more carefully. CVCs and ATAs were calculated across 1,000 random seeds to ensure reproducibility in model selection. If the MDR model met the CVC criteria, we selected models that had the highest ATAs. Multiple hypothesis testing was controlled by CVC in combination with permutation testing. Permutation testing results ≤0.05, generated using random seed 500, were considered statistically significant. Age-group covariate effects were removed by integrating over- and under-sampling methods.
Visualization of Interaction Models Using Hierarchical Interaction Entropy Graphs
Hierarchical interaction entropy graphs, based on information theory, were used to visualize and interpret complex interactions among selected TLR SNPs and PCA risk [59–62]. With this approach, individual and all possible pairwise loci are assigned a joint or mutual information percentage score based on disease risk, respectively. Joint mutual information and mutual information gain scores are based on a number system, ranging from 0–100%. However, these scores rarely exceed 5–6%. When the pairwise or joint mutual information exceeds the mutual information gain scores, then the pairwise interaction is considered more informative in relation to prostate cancer risk when compared to each locus considered separately. Potential interactions are assessed using interaction entropy graphs, which uses a color-coding system to depict redundant or synergistic interactions. The distinction between the synergistic or redundant epistasis models is based on a color coding system. Within the entropy graph, lines depicted between SNP pairs that are color-coded red, orange, green, blue, and gold represent highly synergistic, moderately synergistic, moderately redundant, highly redundant, and neither synergistic/redundant pairwise interaction models, respectively. All entropy-based analyses were conducted using Orange software [63].
Supplementary Material
Acknowledgments
The authors thank Tiva T. VanCleave and Dr. Nicole A. Lavender for preparing DNA samples used in the current study. We also would like to thank Dr. Rick A. Kittles for providing U.S. African American DNA samples.
The authors appreciate the contract services of Expression Analysis, Inc. (http://expressionanalysis.com/) for the generation of genotype data.
This work was supported by the following grants: Clinical Translational Science Pilot Grant to LRK; the JGBCC Bucks for Brains “Our Highest Potential” in Cancer Research Endowment to LRK; and the P20-MD000175 NIH NCMHD to KSK.
Abbreviations
- PCA
prostate cancer
- TLR
toll-like receptor
- SNP
single nucleotide polymorphism
- MDR
multifactor dimensionality reduction
Footnotes
Conflict of Interest
The authors declare that they have no competing interests.
Reference List
- 1.American Cancer S. Cancer Facts and Figures 2012. Atlanta, Georgia: American Cancer Society; 2012. [Google Scholar]
- 2.Hsing AW, Tsao L, Devesa SS. International trends and patterns of prostate cancer incidence and mortality. Int J Cancer. 2000;85(1):60–67. doi: 10.1002/(sici)1097-0215(20000101)85:1<60::aid-ijc11>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 3.American Cancer S. Cancer Facts & Figures for African Americans 2011–2012. Atlanta: American Cancer Society; 2011. [Google Scholar]
- 4.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer Journal international du cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 5.Nickel CJ. Prostatitis syndromes: an update for urologic practice. Can J Urol. 2000;7(5):1091–1098. [PubMed] [Google Scholar]
- 6.Palapattu GS, Sutcliffe S, Bastian PJ, Platz EA, De Marzo AM, Isaacs WB, Nelson WG. Prostate carcinogenesis and inflammation: emerging insights. Carcinogenesis. 2005;26(7):1170–1181. doi: 10.1093/carcin/bgh317. [DOI] [PubMed] [Google Scholar]
- 7.Sfanos KS, De Marzo AM. Prostate cancer and inflammation: the evidence. Histopathology. 2012;60(1):199–215. doi: 10.1111/j.1365-2559.2011.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Latz E, Visintin A, Lien E, Fitzgerald KA, Monks BG, Kurt-Jones EA, Golenbock DT, Espevik T. Lipopolysaccharide rapidly traffics to and from the Golgi apparatus with the toll-like receptor 4-MD-2-CD14 complex in a process that is distinct from the initiation of signal transduction. J Biol Chem. 2002;277(49):47834–47843. doi: 10.1074/jbc.M207873200. [DOI] [PubMed] [Google Scholar]
- 9.Frantz S, Ertl G, Bauersachs J. Mechanisms of disease: Toll-like receptors in cardiovascular disease. Nat Clin Pract Cardiovasc Med. 2007;4(8):444–454. doi: 10.1038/ncpcardio0938. [DOI] [PubMed] [Google Scholar]
- 10.Tsujimoto H, Ono S, Efron PA, Scumpia PO, Moldawer LL, Mochizuki H. Role of Toll-like receptors in the development of sepsis. Shock. 2008;29(3):315–321. doi: 10.1097/SHK.0b013e318157ee55. [DOI] [PubMed] [Google Scholar]
- 11.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32(3):305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Akira S, Yamamoto M, Takeda K. Role of adapters in Toll-like receptor signalling. Biochem Soc Trans. 2003;31(Pt 3):637–642. doi: 10.1042/bst0310637. [DOI] [PubMed] [Google Scholar]
- 13.Beutler B. The Toll-like receptors: analysis by forward genetic methods. Immunogenetics. 2005;57(6):385–392. doi: 10.1007/s00251-005-0011-3. [DOI] [PubMed] [Google Scholar]
- 14.Beutler B. Innate immune responses to microbial poisons: discovery and function of the Toll-like receptors. Annual review of pharmacology and toxicology. 2003;43:609–628. doi: 10.1146/annurev.pharmtox.43.100901.135729. [DOI] [PubMed] [Google Scholar]
- 15.Gay NJ, Keith FJ. Drosophila Toll and IL-1 receptor. Nature. 1991;351(6325):355–356. doi: 10.1038/351355b0. [DOI] [PubMed] [Google Scholar]
- 16.Gay NJ, Packman LC, Weldon MA, Barna JC. A leucine-rich repeat peptide derived from the Drosophila Toll receptor forms extended filaments with a beta-sheet structure. FEBS Lett. 1991;291(1):87–91. doi: 10.1016/0014-5793(91)81110-t. [DOI] [PubMed] [Google Scholar]
- 17.Nakata T, Yasuda M, Fujita M, Kataoka H, Kiura K, Sano H, Shibata K. CD14 directly binds to triacylated lipopeptides and facilitates recognition of the lipopeptides by the receptor complex of Toll-like receptors 2 and 1 without binding to the complex. Cell Microbiol. 2006;8(12):1899–1909. doi: 10.1111/j.1462-5822.2006.00756.x. [DOI] [PubMed] [Google Scholar]
- 18.Muzio M, Polentarutti N, Bosisio D, Prahladan MK, Mantovani A. Toll-like receptors: a growing family of immune receptors that are differentially expressed and regulated by different leukocytes. J Leukoc Biol. 2000;67(4):450–456. doi: 10.1002/jlb.67.4.450. [DOI] [PubMed] [Google Scholar]
- 19.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130(6):1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Jin X, Qin Q, Tu L, Zhou X, Lin Y, Qu J. Toll-like receptors (TLRs) expression and function in response to inactivate hyphae of Fusarium solani in immortalized human corneal epithelial cells. Mol Vis. 2007;13:1953–1961. [PMC free article] [PubMed] [Google Scholar]
- 21.Farhat K, Riekenberg S, Heine H, Debarry J, Lang R, Mages J, Buwitt-Beckmann U, Roschmann K, Jung G, Wiesmuller KH, et al. Heterodimerization of TLR2 with TLR1 or TLR6 expands the ligand spectrum but does not lead to differential signaling. J Leukoc Biol. 2008;83(3):692–701. doi: 10.1189/jlb.0807586. [DOI] [PubMed] [Google Scholar]
- 22.Beutler B, Hoebe K, Georgel P, Tabeta K, Du X. Genetic analysis of innate immunity: identification and function of the TIR adapter proteins. Adv Exp Med Biol. 2005;560:29–39. doi: 10.1007/0-387-24180-9_4. [DOI] [PubMed] [Google Scholar]
- 23.Salaun B, Romero P, Lebecque S. Toll-like receptors’ two-edged sword: when immunity meets apoptosis. Eur J Immunol. 2007;37(12):3311–3318. doi: 10.1002/eji.200737744. [DOI] [PubMed] [Google Scholar]
- 24.Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nature reviews Cancer. 2009;9(1):57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- 25.Creighton CJ, Benham AL, Zhu H, Khan MF, Reid JG, Nagaraja AK, Fountain MD, Dziadek O, Han D, Ma L, et al. Discovery of novel microRNAs in female reproductive tract using next generation sequencing. PLoS One. 2010;5(3):e9637. doi: 10.1371/journal.pone.0009637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinz LX, Rebsamen M, Rossi DC, Staehli F, Schroder K, Quadroni M, Gross O, Schneider P, Tschopp J. The death domain-containing protein Unc5CL is a novel MyD88-independent activator of the pro-inflammatory IRAK signaling cascade. Cell death and differentiation. 2012;19(4):722–731. doi: 10.1038/cdd.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romoser AA, Chen PL, Berg JM, Seabury C, Ivanov I, Criscitiello MF, Sayes CM. Quantum dots trigger immunomodulation of the NFkappaB pathway in human skin cells. Molecular immunology. 2011;48(12–13):1349–1359. doi: 10.1016/j.molimm.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Marzo AM, Platz EA, Sutcliffe S, Xu J, Gronberg H, Drake CG, Nakai Y, Isaacs WB, Nelson WG. Inflammation in prostate carcinogenesis. Nature reviews Cancer. 2007;7 (4):256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang B, Zhao J, Unkeless JC, Feng ZH, Xiong H. TLR signaling by tumor and immune cells: a double-edged sword. Oncogene. 2008;27(2):218–224. doi: 10.1038/sj.onc.1210904. [DOI] [PubMed] [Google Scholar]
- 30.Uno K, Kato K, Atsumi T, Suzuki T, Yoshitake J, Morita H, Ohara S, Kotake Y, Shimosegawa T, Yoshimura T. Toll-like receptor (TLR) 2 induced through TLR4 signaling initiated by Helicobacter pylori cooperatively amplifies iNOS induction in gastric epithelial cells. American journal of physiology Gastrointestinal and liver physiology. 2007;293(5):G1004–1012. doi: 10.1152/ajpgi.00096.2007. [DOI] [PubMed] [Google Scholar]
- 31.Song Z, Yin J, Yao C, Sun Z, Shao M, Zhang Y, Tao Z, Huang P, Tong C. Variants in the Toll-interacting protein gene are associated with susceptibility to sepsis in the Chinese Han population. Crit Care. 2011;15(1):R12. doi: 10.1186/cc9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian FH, Zhang Q, Zhou LF, Jin GF, Bai JL, Yin KS. Polymorphisms in the toll-like receptor 2 subfamily and risk of asthma: a case-control analysis in a Chinese population. J Investig Allergol Clin Immunol. 2010;20(4):340–346. [PubMed] [Google Scholar]
- 33.Slattery ML, Herrick JS, Bondurant KL, Wolff RK. Toll-like receptor genes and their association with colon and rectal cancer development and prognosis. Int J Cancer. 2011 doi: 10.1002/ijc.26314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandey S, Mittal RD, Srivastava M, Srivastava K, Singh S, Srivastava S, Mittal B. Impact of Toll-like receptors [TLR] 2 (−196 to −174 del) and TLR 4 (Asp299Gly, Thr399Ile) in cervical cancer susceptibility in North Indian women. Gynecologic oncology. 2009;114(3):501–505. doi: 10.1016/j.ygyno.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 35.Hold GL, Rabkin CS, Chow WH, Smith MG, Gammon MD, Risch HA, Vaughan TL, McColl KE, Lissowska J, Zatonski W, et al. A functional polymorphism of toll-like receptor 4 gene increases risk of gastric carcinoma and its precursors. Gastroenterology. 2007;132(3):905–912. doi: 10.1053/j.gastro.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 36.Rajaraman P, Brenner AV, Neta G, Pfeiffer R, Wang SS, Yeager M, Thomas G, Fine HA, Linet MS, Rothman N, et al. Risk of meningioma and common variation in genes related to innate immunity. Cancer Epidemiol Biomarkers Prev. 2010;19(5):1356–1361. doi: 10.1158/1055-9965.EPI-09-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srivastava K, Srivastava A, Kumar A, Mittal B. Gallbladder cancer predisposition: a multigenic approach to DNA-repair, apoptotic and inflammatory pathway genes. PLoS One. 2011;6(1):e16449. doi: 10.1371/journal.pone.0016449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song J, Kim DY, Kim CS, Kim HJ, Lee DH, Lee HM, Ko W, Lee G. The association between Toll-like receptor 4 (TLR4) polymorphisms and the risk of prostate cancer in Korean men. Cancer Genet Cytogenet. 2009;190(2):88–92. doi: 10.1016/j.cancergencyto.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 39.Sun J, Wiklund F, Hsu FC, Balter K, Zheng SL, Johansson JE, Chang B, Liu W, Li T, Turner AR, et al. Interactions of sequence variants in interleukin-1 receptor-associated kinase4 and the toll-like receptor 6-1-10 gene cluster increase prostate cancer risk2. Cancer Epidemiol Biomarkers Prev. 2006;15(3):480–485. doi: 10.1158/1055-9965.EPI-05-0645. [DOI] [PubMed] [Google Scholar]
- 40.Cheng I, Plummer SJ, Casey G, Witte JS. Toll-like receptor 4 genetic variation and advanced prostate cancer risk1. Cancer Epidemiol Biomarkers Prev. 2007;16(2):352–355. doi: 10.1158/1055-9965.EPI-06-0429. [DOI] [PubMed] [Google Scholar]
- 41.Kim HJ, Bae JS, Chang IH, Kim KD, Lee J, Shin HD, Lee JY, Kim WJ, Kim W, Myung SC. Sequence variants of toll-like receptor 4 (TLR4) and the risk of prostate cancer in Korean men. World journal of urology. 2011 doi: 10.1007/s00345-011-0690-3. [DOI] [PubMed] [Google Scholar]
- 42.Zheng SL, ugustsson-Balter K, Chang B, Hedelin M, Li L, Adami HO, Bensen J, Li G, Johnasson JE, Turner AR, et al. Sequence variants of toll-like receptor 4 are associated with prostate cancer risk: results from the CAncer Prostate in Sweden Study3. Cancer Res. 2004;64(8):2918–2922. doi: 10.1158/0008-5472.can-03-3280. [DOI] [PubMed] [Google Scholar]
- 43.Chen YC, Giovannucci E, Lazarus R, Kraft P, Ketkar S, Hunter DJ. Sequence variants of Toll-like receptor 4 and susceptibility to prostate cancer. Cancer Res. 2005;65 (24):11771–11778. doi: 10.1158/0008-5472.CAN-05-2078. [DOI] [PubMed] [Google Scholar]
- 44.Stevens VL, Hsing AW, Talbot JT, Zheng SL, Sun J, Chen J, Thun MJ, Xu J, Calle EE, Rodriguez C. Genetic variation in the toll-like receptor gene cluster (TLR10-TLR1-TLR6) and prostate cancer risk. Int J Cancer. 2008;123(11):2644–2650. doi: 10.1002/ijc.23826. [DOI] [PubMed] [Google Scholar]
- 45.Lindstrom S, Hunter DJ, Gronberg H, Stattin P, Wiklund F, Xu J, Chanock SJ, Hayes R, Kraft P. Sequence variants in the TLR4 and TLR6-1-10 genes and prostate cancer risk. Results based on pooled analysis from three independent studies. Cancer Epidemiol Biomarkers Prev. 2010;19(3):873–876. doi: 10.1158/1055-9965.EPI-09-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen YC, Giovannucci E, Kraft P, Lazarus R, Hunter DJ. Association between Toll-like receptor gene cluster (TLR6, TLR1, and TLR10) and prostate cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(10):1982–1989. doi: 10.1158/1055-9965.EPI-07-0325. [DOI] [PubMed] [Google Scholar]
- 47.Giri VN, Egleston B, Ruth K, Uzzo RG, Chen DY, Buyyounouski M, Raysor S, Hooker S, Torres JB, Ramike T, et al. Race, genetic West African ancestry, and prostate cancer prediction by prostate-specific antigen in prospectively screened high-risk men. Cancer Prev Res(Phila Pa) 2009;2(3):244–250. doi: 10.1158/1940-6207.CAPR-08-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian C, Hinds DA, Shigeta R, Kittles R, Ballinger DG, Seldin MF. A genomewide single-nucleotide-polymorphism panel with high ancestry information for African American admixture mapping. Am J Hum Genet. 2006;79(4):640–649. doi: 10.1086/507954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang QX, Cheng XY, Mao ZC, Wang YS, Zhao LL, Yan X, Ferris VR, Xu RM, Xie BY. MicroRNA discovery and analysis of pinewood nematode Bursaphelenchus xylophilus by deep sequencing. PLoS One. 2010;5(10):e13271. doi: 10.1371/journal.pone.0013271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun J, Wiklund F, Zheng SL, Chang B, Balter K, Li L, Johansson JE, Li G, Adami HO, Liu W, et al. Sequence variants in Toll-like receptor gene cluster (TLR6-TLR1-TLR10) and prostate cancer risk4. J Natl Cancer Inst. 2005;97(7):525–532. doi: 10.1093/jnci/dji070. [DOI] [PubMed] [Google Scholar]
- 52.Greene CS, Penrod NM, Williams SM, Moore JH. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS One. 2009;4 (6):e5639. doi: 10.1371/journal.pone.0005639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kidd LC, Vancleave TT, Doll MA, Srivastava DS, Thacker B, Komolafe O, Pihur V, Brock GN, Hein DW. No association between variant N-acetyltransferase genes, cigarette smoking and Prostate Cancer susceptibility among men of African descent. Biomark Cancer. 2011;2011(3):1–13. doi: 10.4137/BIC.S6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jackson MD, Walker SP, Simpson-Smith CM, Lindsay CM, Smith G, McFarlane-Anderson N, Bennett FI, Coard KC, Aiken WD, Tulloch T, et al. Associations of whole-blood fatty acids and dietary intakes with prostate cancer in Jamaica. Cancer causes & control : CCC. 2012;23(1):23–33. doi: 10.1007/s10552-011-9850-4. [DOI] [PubMed] [Google Scholar]
- 55.Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case-control genetic association analyses. BMC Genet. 2008;9:36. doi: 10.1186/1471-2156-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gui J, Andrew AS, Andrews P, Nelson HM, Kelsey KT, Karagas MR, Moore JH. A simple and computationally efficient sampling approach to covariate adjustment for multifactor dimensionality reduction analysis of epistasis. Hum Hered. 2010;70 (3):219–225. doi: 10.1159/000319175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greene CS, Penrod NM, Kiralis J, Moore JH. Spatially uniform relieff (SURF) for computationally-efficient filtering of gene-gene interactions. BioData mining. 2009;2 (1):5. doi: 10.1186/1756-0381-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moore JH, Asselbergs FW, Williams SM. Bioinformatics challenges for genome-wide association studies. Bioinformatics. 2010;26(4):445–455. doi: 10.1093/bioinformatics/btp713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden T, Barney N, White BC. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol. 2006;241(2):252–261. doi: 10.1016/j.jtbi.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 60.Jakulin A, Bratko I, Smrike D, Demsar J, Zupan B. Protarus: Cyprus. 2003. Attribute interactions in medical data analysis; pp. 229–238-238. [Google Scholar]
- 61.Jakulin A, Bratko I. Analyzing attribute interations. Lecture Notes in Artificial Intelligence. 2003;2838:229. [Google Scholar]
- 62.McGill WL. Multivariate information transmission. Psychometrika. 1954;19:97–116. [Google Scholar]
- 63.Mramor M, Leban G, Demsar J, Zupan B. Visualization-based cancer microarray data classification analysis. Bioinformatics. 2007;23(16):2147–2154. doi: 10.1093/bioinformatics/btm312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


