Abstract
The conjugative element pRS01 from Lactococcus lactis encodes the putative relaxase protein LtrB. The ltrB gene is interrupted by the functional group II intron Ll.ltrB. Accurate splicing of the two ltrB exons is required for synthesis of the mRNA encoding the LtrB conjugative relaxase and subsequent plasmid transfer. A conjugation-based genetic assay was developed to identify Ll.ltrB mutations that affect splicing. In this assay a nonsplicing, transfer-defective pRS01 derivative (pM1014) and a shuttle vector carrying the ltrB region, including the Ll.ltrB intron (pCOM9), are used. pCOM9 provides splicing-dependent complementation of the transfer defect of pM1014. Site-directed mutations within Ll.ltrB, either in the catalytic RNA or in the intron-encoded protein gene ltrA, were generated in the context of pCOM9. When these mutants were tested in the conjugation-based assay, significantly reduced mating was observed. Quantitative molecular analysis of in vivo splicing activity confirmed that the observed mating defects resulted from reduced splicing. Once the system was validated for the engineered mutants, random mutagenesis of the intron followed by genetic and molecular screening for splicing defects resulted in identification of point mutations that affect splicing.
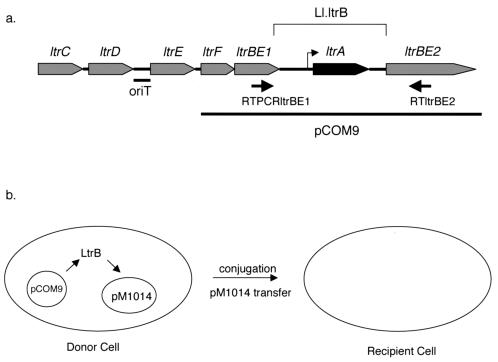
In Lactococcus lactis, high-frequency transfer of the lactose fermentation ability (Lac+) phenotype by conjugation can occur as a result of insertion element-mediated cointegration between a nonconjugative lactose plasmid (pSK08) and the conjugative element pRS01 (1, 25). Previously, a region of pRS01 encoding conjugative DNA processing functions (Tra I-II), which is physically separated from the Tra III-IV region (which appears to encode the proteins involved in formation of functional mating pairs), was identified (17). The genetic determinants in the Tra I-II segment (Fig. 1a) include ltrB, encoding a putative relaxase enzyme responsible for strand-specific nicking of the plasmid to initiate DNA transfer, several genes (ltrC, -D, -E, and -F) likely to encode accessory proteins for relaxosome formation, and a noncoding sequence which comprises the functional origin of transfer (oriT) for pRS01 (18). A striking feature of this system is that the ltrB determinant is interrupted by a group II intron, Ll.ltrB (Fig. 1a) (18). Splicing of Ll.ltrB from the full-length ltrB transcript fuses the two exons (ltrBE1 and ltrBE2) in frame to allow the synthesis of functional LtrB protein, which is required for pRS01 transfer.
FIG. 1.
Intron region and the splicing-dependent conjugation assay. (a) Map of the intron region of pRS01 (not to scale). As described by Mills et al. (18), the ltr genes are involved in transfer of pRS01. ltrF has not been described previously, but it is predicted to encode a protein translated from the UUG alternative start codon. The bar under the map indicates the location of the pRS01 origin of transfer (oriT). Intron Ll.ltrB disrupts the ltrBE1 and ltrBE2 exons and encodes the ltrA gene. A bent arrow indicates the ltrA promoter internal to the intron. The arrows under the map indicate the positions of primers RTPCRltrBE1 and RTltrBE2 used in the RT-PCR splicing assay. A portion of the transfer region is carried by the shuttle vector pCOM9, as indicated by the line below the map. (b) Schematic diagram of the conjugation-based genetic assay for splicing. Splicing of Ll.ltrB from pCOM9 allows production of functional LtrB protein. LtrB protein acts in trans to nick pM1014 oriT and promote pM1014 transfer to the recipient cell.
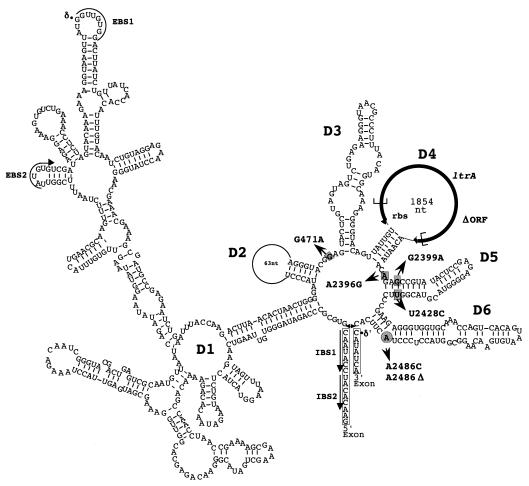
Group II introns are an abundant class of self-splicing, mobile RNAs found in bacteria and the organelles of plants and lower eukaryotes. Although group II introns show considerable sequence variation, they are characterized by a distinctive architecture. The intron RNA is comprised of six secondary-structure domains (D1 to D6) (Fig. 2; for a review see reference 26). D1, D5, and D6 are essential components of the catalytic core of the ribozyme, while D2 and D3 contribute to the overall structure and D4 is looped out of the functional ribozyme and often contains an open reading frame. In addition to the conserved secondary structure, a number of critical tertiary interactions have been found within group II introns (26). Furthermore, conserved base pairing interactions occur between complementary intron and exon sequences (EBS1 and IBS1, EBS2 and IBS2, δ and δ′) (Fig. 2).
FIG. 2.
Predicted folded structure of Ll.ltrB. Secondary-domain structures (D1 to D6) are indicated. Exon sequences are enclosed in boxes. Complementary intron and exon sequences (EBS1 and IBS1, EBS2 and IBS2, δ and δ′) are indicated. Most residues mutated in this study are indicated by gray rectangles; the only exception is the bulging A branch point residue, which is indicated by a gray circle. The extent of the ΔORF mutation is indicated by brackets in the ltrA coding sequence. nt, nucleotides; rbs, ribosome binding site.
In the primary pathway for group II intron splicing there are two sequential transesterification reactions that join the two exons and release a lariat intron product. Biochemically, this mechanism is identical to that of splicing of nuclear spliceosomal introns of eukaryotes, suggesting that group II intron invasion of nuclear genes might have initiated the evolution of present-day nuclear introns in eukaryotes (15). Group II introns can function as mobile genetic elements and are capable of insertion into an intronless allele of the native exon gene (homing) or insertion into ectopic sites (transposition) at lower frequencies. Mobility of Ll.ltrB proceeds via a reverse splicing reaction and relies on an RNA intermediate (6, 7). Recent studies demonstrating that the Ll.ltrB intron can be retargeted for insertion into novel sites by directed changes in the EBS sequences make this element a powerful tool for genetic manipulation of many different types of organisms (9, 10, 20).
The in vivo splicing and mobility functions of several group II introns require the presence of an intron-encoded protein (IEP) encoded in D4. In the case of Ll.ltrB, the IEP is the product of the ltrA gene (Fig. 1a). LtrA exhibits many of the structural motifs found in most group II IEPs, including separate domains which have been shown to encode reverse transcriptase, DNA endonuclease, and splicing maturase activities (14, 18). The splicing maturase activity results from the ability of the protein to bind to the intron RNA and induce conformational changes in the RNA that lead to important tertiary interactions required for ribozyme function (13, 27). Interestingly, it has been suggested that the major source of mRNA for LtrA synthesis in lactococci is not the unspliced full-length ltrB transcript, but a message produced from an internal ltrA promoter (PltrA) within the intron (Fig. 1a) (29).
Ll.ltrB is the first group II intron from prokaryotes that has been shown to be fully functional in vivo, although many additional group II introns have now been identified in various bacteria (11). Much of our understanding of the molecular events involved in group II intron splicing and mobility reactions has come from biochemical studies of introns from mitochondrial or chloroplast genes of eukaryotes (15). Because of the inherent difficulties involved in genetic analyses (especially random mutagenesis) of organellar group II introns, in most in vivo studies the workers have used directed mutagenesis of a few highly conserved positions to test predicted structure-function relationships in these catalytic RNAs (3, 21).
One of our long-term objectives is to identify and characterize important structural features of Ll.ltrB that are not readily predictable by comparative sequence analysis, including those that may be unique to this intron. To this end, we have developed a conjugation-based genetic assay and a molecular real-time reverse transcription (RT)-PCR splicing assay, both of which measure Ll.ltrB splicing in vivo. We tested the ability of these two assays to measure splicing proficiency by analyzing a set of engineered mutations that disrupt conserved sites of the intron and are predicted to have detrimental effects on intron splicing. Using both of these assays, we found that splicing of all of the mutations tested was significantly reduced, as predicted. We then employed the two assays in a screening analysis to identify splicing-deficient intron mutations following random mutagenesis of the intron. Using this approach, we identified residues affecting intron domain 5 that are important for Ll.ltrB function.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
Bacterial strains used in this study are listed in Table 1. Note that L. lactis strain MMS370 used in the conjugation and splicing assays does not contain a chromosomal copy of the intron. Escherichia coli strains were routinely grown in Luria-Bertani (LB) medium (16) at 37°C with shaking. L. lactis strains were grown in M17 medium (28) supplemented with 0.5% glucose (GM17) at 30°C. The concentrations of antibiotics used for growth of lactococci are as follows: erythromycin, 10 μg/ml; spectinomycin, 300 μg/ml; fuscidic acid, 25 μg/ml; and rifampin, 100 μg/ml. For growth of E. coli, spectinomycin was used at a concentration of 50 μg/ml. All antibiotics were obtained from Sigma.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant phenotype | Source or reference(s) |
|---|---|---|
| E. coli strains | ||
| DH5α | F−φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Gibco-BRL |
| XL1-Red | endA1 gyrA9 thi-1 hsdR17 supE44 relA1 lac mutD5 mutS mutT Tn10 (Tetr) | Stratagene |
| L. lactis strains | ||
| MMS370 | Rec−, Strr | 1 |
| LM2301RF | Recipient strain, Rec+ Rifr Fusr Strr | 18 |
| Plasmids | ||
| pCOM9 | ltrB region cloned into pDS281, Specr | 29 |
| pCOMT-Kan | pCOM9 with oriT from pRS01 and Kanr gene from pUC4K, Specr Kanr | This study |
| pM1014 | pRS01-based plasmid, ltrA disrupted by pTRK28 cointegration, Ermr | 17, 18 |
| pM2036 | pRS01-based plasmid, pTRK28 cointegrated downstream of ltrBE2, Ermr | 17, 18 |
| pDL281 | Shuttle vector, Specr | 29 |
| pCOM-ex | pCOM9 derivative containing intronless allele of ltrB | 29 |
| pCOM-G471A | pCOM9 derivative with a substitution at residue 471 of intron | This study |
| pCOM-G2399A | pCOM9 derivative with a substitution at residue 2399 of intron | This study |
| pCOM-A2486C | pCOM9 derivative with a substitution of the bulging A residue | This study |
| pCOM-A2486Δ | pCOM9 derivative with a deletion of the bulging A residue | This study |
| pCOM-ΔORF | pCOM9 derivative with a deletion of most of the LtrA coding region | This study |
| pCOM-A30-6 | pCOMT-Kan derivative isolated during random mutagenesis screening containing the mutations A2396G and U2428C | This study |
| pCOM-A2396G | pCOM9 derivative with a substitution at residue 2396 of intron | This study |
| pCOM-U2428C | pCOM9 derivative with a substitution at residue 2428 of intron | This study |
| pCOM-A2396G-U2428C | pCOM9 derivative with substitutions at residues 2396 and 2428 of intron | This study |
DNA analysis and manipulation.
To isolate plasmid DNA from E. coli for sequencing, a 10-ml overnight culture was grown, and the plasmid was isolated by using a Qiagen plasmid minikit. Automatic cycle sequencing was performed by the Advanced Genetic Analysis Center at the University of Minnesota. To isolate plasmid DNA from L. lactis, cells from a 5- to 10-ml overnight culture were pelleted and treated with lysozyme to digest the cell wall (30 mg of lysozyme per ml in 25% sucrose for 30 min at 37°C). The plasmid was isolated with the Qiagen plasmid minikit. Electroporation was carried out as previously described (29). Electrocompetent E. coli cells were made as described by Ausubel et al. (2), and electrocompetent L. lactis cells were made as previously described (29). DNA digestion and ligation were carried out by using standard procedures (2).
Plasmids used in this study were constructed as follows. Plasmid pCOMT-Kan was created by first cloning oriT from plasmid pLE12 (19) into the PacI-BamHI region of pCOM9 to create plasmid pCOMT. Then the aphIII kanamycin resistance gene was inserted into the intron of pCOMT, so that the gene was located immediately downstream of the LtrA coding region. Plasmids pCOM-G471A and pCOM-G2399A were constructed by oligonucleotide-directed mutation by using a Bio-Rad Mutagene kit, were confirmed by DNA sequencing, and were moved into pCOM9 on a HindIII fragment. Plasmids pCOM-A2486C and pCOM-2486Δ contained the intron mutation cloned from a pET11a-ltrA-based vector (M. Belfort, unpublished data) into the pCOM9 vector on a HindIII fragment. Plasmid pCOM-ΔORF was constructed by subcloning the SnaBI-BsrGI fragment from pL12-ΔORF (14) into pCOM9 and was essentially a deletion of LtrA amino acids 40 to 572. We found that subcloning mutations identified in the random mutagenesis screening analysis into the parent pCOMT-Kan vector was difficult. Therefore, all mutations were subcloned into pCOM9, and the conjugation and RT-PCR phenotypes of the subclone and the-wild type pCOM9 vector were subsequently compared. The A2396G mutation from mutant A30-6 was subcloned into pCOM9 on a BsrGI-BsiWI fragment to create pCOM-A2396G. The U2428C mutation from mutant A30-6 was subcloned into pCOM9 on a BsiWI-HpaI fragment to create pCOM-U2428C. Both A2396G and U2428C were subcloned together into pCOM9 on a BsrGI-HpaI fragment to create plasmid pCOM-A2396G-U2428C. The presence of the mutation in each subclone was determined by sequencing.
Quantitative conjugation assay.
Mating assays with engineered mutants and mutants identified in the random mutagenesis screening analysis were performed as previously described (29).
Random mutagenesis and conjugation assay screening.
The E. coli mutator strain XL1-Red (Stratagene) was transformed with 50 to 200 ng of plasmid pCOM9 (three separate transformations) or plasmid pCOMT-Kan (two separate transformations) by following the manufacturer's directions. Each transformation mixture was plated on LB medium plates containing spectinomycin and grown for 24 to 60 h. Transformants were scraped off the plates, resuspended in LB medium containing spectinomycin, and grown until saturation. Plasmid DNA was isolated at this point, and the remaining cells were passaged by subculturing them (1:100) in fresh medium and growing the cultures to saturation again. Plasmid DNA was isolated after each passage, and this DNA represented a separate pool of mutants. Plasmids were transformed into MMS370(pM1014) by electroporation and selected on GM17 plates containing spectinomycin. Transformants were screened for conjugation defects by using them as the donor strains in a cross-streak mating protocol. Transformants were picked into individual wells of a 48-well plate containing 500 μl of GM17 supplemented with spectinomycin and erythromycin and grown overnight. The overnight cultures were subcultured (1:10) in 900 μl of medium without antibiotics and grown for 4 h. Recipient strain LM2301RF and control donor strain MMS370(pM1014)(pCOMT-Kan) were grown similarly. Recipient strain LM2301 was swabbed down the center of GM17 plates, and donor strains were swabbed perpendicularly. These mating plates were incubated for 16 to 20 h, after which the area of intersection between the donor and the recipient on each plate was swabbed and the material was transferred to a selective plate. Transconjugants appeared after 2 to 3 days. Strains that displayed significantly fewer transconjugants than the wild type were retested with the same mating assay, and strains that exhibited the same mating-defective phenotype again were analyzed further by real-time RT-PCR and a quantitative mating assay. Plasmid DNA from interesting mutants was sequenced. To generate high-quality plasmid DNA for sequencing, plasmids were isolated from L. lactis and transformed into E. coli DH5α. Plasmid DNA isolated from E. coli was then used as the template in sequencing reactions.
RNA isolation and real-time RT-PCR.
Lactococcal strains from which RNA was isolated were grown overnight in medium containing the appropriate antibiotics. Cells were subcultured (1:10) and grown to an optical density at 600 nm of approximately 0.5 to 1.0, and 1.5 ml of each culture was pelleted and stored at −80°C or used immediately for RNA isolation. Prior to RNA isolation, the cells were treated for 30 min at 37°C with 100 μl of a 30-mg/ml lysozyme solution prepared in 25% sucrose. RNA was then isolated with RNeasy minicolumns (Qiagen) by following the manufacturer's directions. The amount of RNA was quantitated by measuring the absorbance at 260 nm.
Real-time RT-PCR was performed by using a one-tube format in which the RT reaction and the PCR were carried out in a single tube by using reagents from a QuantiTect SYBR Green RT-PCR kit (Qiagen). The sequence of the downstream primer (RTltrBE2) used for both RT and PCR is 5′-TTTGCGCCATAACGTGAAGA. The sequence of the upstream PCR primer (RTPCRltrBE1) used to measure the amount of spliced RNA is 5′-CAGGTGGCGAATATGAATTTGTG. The sequences of the PCR primers used to measure the amount of total spliced and unspliced mRNA are as follows: RTPCRltrBE1L4, 5′-GTGCTTGGTCATCACCTCATCCA; and RTPCRLtrBE1R3, 5′-CGACGTGGGTTGCAATCACAA. Prior to RT-PCR analysis, 1.5 μg of RNA was treated with 4 U of RQ1 DNase (Promega) for 30 min at 37°C. The enzyme was inactivated by using the supplied stop solution, followed by extraction with phenol-chloroform-isoamyl alcohol and ethanol precipitation. Immediately before RT-PCR analysis, the RNA was resuspended in RNase-free water, and one-half to one-fourth of the suspension was used as the template in the RT-PCR. RT-PCR mixtures (25 μl) containing 1× master mixture, 200 nM upstream primer, 200 nM downstream primer, and DNase-treated RNA template were prepared in 96-well optical plates (ABI). The reaction was performed with an ABI5700 thermal cycler (ABI) by using the following conditions: incubation at 50°C for 30 min and then at 95°C for 15 min, followed by 40 cycles of 95°C for 45 s, 54°C for 1 min, and 72°C for 1 min. A dissociation analysis ramping from 60 to 95°C over 20 min was performed to calculate dissociation curves.
For each strain tested, at least three RT-PCRs were performed, by using RNA isolated on at least two separate occasions. For each reaction, the cycle threshold (CT) (the cycle at which fluorescence reached 0.8 U) for pCOM9 and the CT for the mutant were calculated, and the difference between the CT values (ΔCT) was calculated. The ΔCT value was converted to a ratio of template amounts (pCOM9/mutant) by using the following formula: 2−ΔCT. This value represents the relative amount of spliced mRNA isolated from each mutant compared to the amount isolated from pCOM9. The values were averaged for each of the trials, and the standard deviation was determined. To confirm that the relative fluorescence measured was due to a single amplification product, dissociation curves were plotted for each reaction, and each of these curves was observed to have a single peak at the appropriate melting temperature for the spliced product. To control for amplification caused by DNA carryover, each template was used in a reaction lacking the RT enzyme. Amplification was not observed in these reactions.
RESULTS
Two-plasmid system to monitor splicing-dependent conjugation.
We developed a conjugation-based genetic assay for complementation of Ll.ltrB function using two plasmids (29). Plasmid pM1014 is a pRS01-based plasmid in which ltrA has been disrupted by insertion element-mediated cointegration of plasmid pTRK28 at the 28th codon of the gene. Due to the loss of LtrA expression, the Ll.ltrB intron is not efficiently spliced, and LtrB is not produced. While pM1014 carries all of the other genes required for conjugative plasmid transfer, loss of the LtrB relaxase results in a low plasmid transfer frequency (<2 × 10−9 transconjugant per donor [29]). The second plasmid, pCOM9, contains the cloned ltrB region of pRS01 on a shuttle vector (pDL281). pCOM9 was designed to facilitate site-directed or random mutagenesis of the intron sequences on a relatively small and easily manipulated plasmid without affecting the other genes required for conjugative plasmid transfer. Splicing of the intron in this context allows synthesis of the LtrB protein, which can act in trans to nick the oriT gene of pRS01 or its derivatives (Fig. 1b). The transfer defect of pM1014 is complemented by the pCOM9 construct, which increases the pM1014 transfer rate >2.5 × 106-fold (5 × 10−3 transconjugant per donor [29]). Furthermore, a pCOM9 derivative that lacks the intron so that the two ltrB exons are fused together (pCOM-ex) complements pM1014 transfer in a splicing-independent manner (1.3 × 10−2 transconjugant per donor [29]). However, a mutation in pCOM9 that disrupts the LtrA promoter, thereby reducing intron splicing and subsequent expression of LtrB, reduced the plasmid transfer rate approximately 1,000-fold (5.5 × 10−6 transconjugant per donor [29]). These initial experiments suggested that plasmid transfer efficiency in this two-plasmid genetic assay could be used as a measure of splicing efficiency.
Conjugation effects of mutating conserved Ll.ltrB residues.
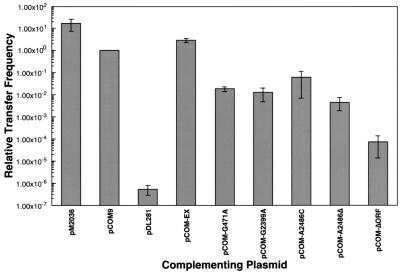
In order to validate the conjugation assay as a measure of intron splicing efficiency, we tested additional pCOM9 mutant derivatives. We engineered mutations in a number of residues of Ll.ltrB that correspond to critical positions in previously characterized fungal intron RNAs (Fig. 2). A few of the same changes have also been shown to inhibit Ll.ltrB splicing in vitro (Peebles, unpublished data), when the intron is overexpressed in E. coli (14), and in a coupled splicing-mobility assay with E. coli (8). The point mutations tested included deletion of the bulging A branch point residue (A2486Δ), which is involved in initiation of the splicing reaction, and substitution of a cytosine at this position (A2486C). The G2399A mutation substitutes a critical conserved G residue in domain 5 (22), while the G471A mutation alters another conserved critical base immediately adjacent to the γG residue in the joiner region between domains 2 and 3 (24). We also examined the effects of a large in-frame deletion of most of the ltrA coding sequence (ΔORF), which was predicted to eliminate any LtrA function in the cell while leaving the folded intron RNA structure relatively intact. In all of the pCOM9 mutant derivatives tested, the frequency of pM1014 transfer observed in the complementation assays was substantially reduced (Fig. 3 and Table 2). The G471A and G2399A mutations reduced transfer 60- and 80-fold, respectively. Substitution of cytosine for the bulging A residue in A2486C reduced transfer about 15-fold, while deletion of cytosine in A2486Δ resulted in a still greater reduction (about 200-fold). Not surprisingly, the ΔORF mutation had the most severe effect, reducing plasmid transfer >10,000-fold. However, the sharply reduced level of transfer was at least 100-fold higher than the limit of detection for the assay. This suggests that a very low but readily detectable level of splicing occurs in L. lactis in the absence of LtrA. The results of these complementation assays suggest that it is feasible to use the conjugation-based system to study intron mutations that affect the efficiency of in vivo splicing over a 106-fold range.
FIG. 3.
Complementation of pRS01-based plasmid transfer by pCOM9 and mutant derivatives. Conjugation assays were carried out to measure the transfer frequency of pM2036 (pM2036 bar) or pM1014 (other bars). pM1014 transfer frequency was complemented with the pCOM9-based plasmids indicated, as described in the text. The plasmid transfer frequencies represent the number of transconjugants per donor and are standardized to the level of pCOM9 transfer, which was defined as 1. The data are the averages for three or more trials.
TABLE 2.
Comparison of plasmid transfer frequencies and mRNA splicing for pCOM9 and intron mutants
| Plasmida | Relative plasmid transfer frequencyb | Relative amt of spliced mRNAc |
|---|---|---|
| pCOM9 | 100 (12) | 100 |
| pCOM-G471A | 1.83 ± 0.49 (4) | 2.73 ± 1.80 |
| pCOM-G2399A | 1.27 ± 0.79 (4) | 4.22 ± 0.75 |
| pCOM-A2486C | 6.15 ± 5.44 (3) | 1.43 ± 1.44 |
| pCOM-A2486Δ | 0.45 ± 0.27 (4) | 0.35 ± 0.15 |
L. lactis MMS370(pM1014) carried the plasmids indicated.
Frequency of pM1014 plasmid transfer from donor to recipient as determined by quantitative mating assays. The plasmid transfer frequency achieved by complementation with pCOM9 was defined as 100, and the transfer frequency for each of the mutants is expressed relative to the frequency observed for pCOM9. The values are means ± standard deviations for the actual numbers of replicate experiments indicated in parentheses.
Relative amount of spliced mRNA as determined by real-time RT-PCR. The values are means ± standard deviations for three replicate experiments.
Molecular analysis of splicing in pCOM9 and derivatives.
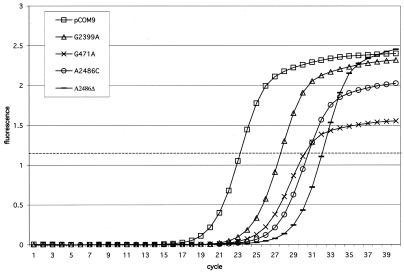
In order to confirm that the reduced plasmid transfer frequencies exhibited by the intron mutants resulted from reduced splicing in vivo, we analyzed RNA isolated from cells carrying pCOM9 mutant derivatives using real-time RT-PCR. In the RT-PCR assay we used a primer (RTltrBE2) complementary to a segment of ltrBE2 located 154 bp downstream from the splice junction to reverse transcribe RNA into cDNA. The cDNA was subsequently used as a template in a PCR with primer RTltrBE2 and a primer complementary to a segment of ltrBE1 (RTPCRltrBE1) (Fig. 1a). Under the specific cycling conditions used, a short (214-bp) RT-PCR product was obtained from spliced RNA, but a longer (∼3-kb) RT-PCR product from unspliced pre-mRNA was not detected (data not shown). The amount of RT-PCR product was quantitated by a real-time assay by employing the fluorescent DNA binding dye SYBR Green and measuring the fluorescence after each PCR cycle. We compared the amount of RT-PCR product amplified from RNA samples isolated from lactococcal strains carrying pCOM9 to the amounts of RT-PCR product amplified from strains containing each of the pCOM9 point mutation derivatives examined in the experiments described above. (The pCOM-ΔORF mutation was not analyzed since the product from unspliced RNA [1.1 kb] was routinely amplified along with the splicing-specific product and therefore confounded quantitation of the amount of spliced RNA.) Similar to the conjugation assay results, we found that each of the point mutations significantly delayed the appearance of the RT-PCR product compared to the results obtained for the wild-type intron (Fig. 4). A product was observed to accumulate from mutant samples four to eight cycles later than the product accumulated from pCOM9, indicating that the intron mutants had significantly less spliced RNA. The cycle at which the product reached a threshold of 0.8 U of fluorescence (a level arbitrarily set in the exponential phase of amplification) was designated the CT. The ΔCT values were used to calculate the relative amounts of spliced RNA for pCOM9 and the intron mutant derivatives (Table 2). Compared to pCOM9, the mutants had 20 to 300 times less spliced RNA. Nucleotide substitutions had less severe defects, while deletion of the bulging A residue had the most severe defect.
FIG. 4.
Real-time RT-PCR analysis of intron splicing from pCOM9 and intron mutant derivatives. The relative fluorescence at the end of each PCR cycle was plotted for each sample, and a threshold of 0.8 U of fluorescence was selected (dashed line) to calculate the CT values. The amplification plot is the plot for one RT-PCR trial and is representative of the results for the three trials used to calculate the relative amount of spliced mRNA.
To demonstrate that the reduced amounts of spliced products observed in the RT-PCR assays were due to reduced levels of splicing and not to defects in transcription that had indirect effects on the amounts of spliced templates, internal control RT-PCRs were performed. We measured the total amount of spliced and unspliced RNA by performing RT-PCR with each of the mutant samples using two primers within ltrBE1. We observed that the amplification products all crossed the CT within one or two cycles (data not shown), indicating that similar amounts of ltrB transcripts were present in the samples and suggesting that the reduced levels of spliced product were indeed due to a splicing defect.
Since quantitative real-time RT-PCR analysis could not be performed with the ΔORF derivative, RT-PCR products obtained with this template were analyzed by gel electrophoresis. A product whose size corresponded to the size of the spliced exons was consistently observed (data not shown). Together with the results from the conjugation assay, these results suggest that the intron can splice to some extent in the absence of LtrA.
The splicing-dependent conjugation assay and the RT-PCR splicing assay revealed comparable deficits for several intron variants (Table 2). We used these two assays together in a screening analysis to identify additional intron residues critical for splicing.
Isolation of splicing-defective intron mutants through random mutagenesis.
A random mutagenesis approach was used to identify splicing-defective intron mutants. A pCOM9-based plasmid was passed through an E. coli mutator strain to generate mutations within the intron that should have included transitions, transversions, or single-nucleotide insertions or deletions. Pools of mutated plasmids were then collected, and the mutated plasmids were introduced into a RecA− lactococcal strain containing pM1014. Transformants with reduced levels of plasmid transfer to recipient strain LM2301 were identified by using the conjugation assay.
Two different plasmids, pCOM9 and pCOMT-Kan, were used as targets for in vivo mutagenesis. The pCOMT-Kan plasmid differs from pCOM9 in that it contains a 446-bp segment of pRS01 that carries the functional origin of transfer (oriT). The presence of oriT provides additional flexibility in experimental design by allowing pCOMT-Kan plasmids to be mobilized from the donor cell to the recipient cell. This plasmid also carries a kanamycin resistance cassette cloned just downstream of the LtrA coding sequence in intron domain 4. The resistance gene provides a second selectable marker useful in screening and cloning, and previous studies have suggested that insertions can be made at this location without eliminating intron function (14).
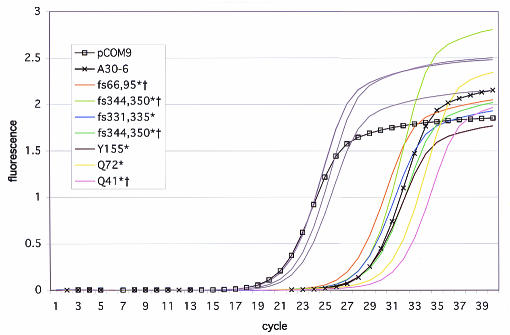
Using the conjugation assay, we screened between 45 and 90 transformants from each of 11 different mutant plasmid pools and identified between one and eight conjugation-defective mutants in each pool. Reduced levels of plasmid transfer could have resulted from plasmid backbone mutations, from defects in the LtrB relaxase encoded by the exons, or from splicing defects caused by mutations in the intron-encoded protein LtrA or the intron RNA ribozyme. Consequently, we tested these conjugation mutants for splicing deficiency using RT-PCR. Of 14 mutants screened by RT-PCR, 8 were defective for splicing (Fig. 5). Not surprisingly, among the mutants that were defective for plasmid transfer but not defective for splicing, we found deleterious sequence changes within the ltrB exons (data not shown). When the mutants defective in both plasmid transfer and splicing were examined, sequencing revealed that seven mutants had frameshift or nonsense mutations in LtrA (Fig. 5). Three of these mutants had additional nucleotide changes in the intron or exon sequences. We assumed that the splicing defect of these mutants was due primarily to loss of the LtrA protein, and any effect of these secondary mutations on splicing is unknown at this point. We found that one splicing-defective mutant, A30-6, had sequence changes located in the intron RNA ribozyme.
FIG. 5.
Real-time RT-PCR analysis of intron splicing by pCOM9 intron mutants identified in the random mutagenesis screening analysis. The amplification curves for 12 mutants defective in plasmid transfer are shown. Three mutants (gray lines) had wild-type levels of splicing. Eight mutants (other lines) had significantly delayed appearance of the RT-PCR product compared to the time of appearance of the wild-type intron in pCOM9, indicating that there was significantly less spliced RNA. One splicing-defective mutant, A30-6, had mutations in the intron RNA. Seven mutants had deleterious mutations in the LtrA coding region. For example, mutant Y155* had a nonsense mutation for LtrA amino acid 155, and mutant fs331,335* had a frameshift mutation for amino acid 331 that resulted in a stop codon at amino acid 335. Two independent isolates with the fs344,350* mutation were observed. Mutants indicated by a dagger also had additional mutations in the intron or exons.
The plasmid transfer efficiency of donors carrying A30-6 was more than 1,000-fold less than the transfer efficiency of the original unmutated plasmid (Table 3). The splicing efficiency of mutant A30-6 was about 1% of the splicing efficiency of the wild-type intron, as measured by RT-PCR analysis (Table 3). Sequencing of this mutant revealed two nucleotide substitutions in or near domain 5 of the intron RNA (A2396G and U2428C) (Fig. 2). The two mutations were subcloned into an unmutated vector (to separate them from any additional mutations in the plasmid that may have affected transcript synthesis, stability, or plasmid copy number) and were found to have splicing and conjugation defects similar to those of the original A30-6 plasmid (Table 3). The two mutated residues were subcloned individually into an unmutated vector; however, neither single mutation caused the splicing defect observed in the original double mutant (Table 3). From this analysis we concluded that the combined A2396G and T2428C mutations are both sufficient and necessary for the observed splicing defects.
TABLE 3.
Plasmid transfer frequencies and mRNA splicing of A30-6 and subclones
| Intron mutation(s) | Relative plasmid transfer frequencya | Relative amt of spliced mRNAb |
|---|---|---|
| A2396G, U2428Cc | 0.06 ± 0.02 | 1.4 ± 1.3 |
| A2396Gd | 72.4 ± 4.2 | 130.0 ± 31.7 |
| U2428Cd | 32.8 ± 9.9 | 174.9 ± 39.9 |
| A2396G, U2428Cd | 0.08 ± 0.04 | 0.22 ± 0.05 |
Frequency of pM1014 plasmid transfer from donor to recipient as determined by quantitative mating assays. The plasmid transfer frequency achieved by complementation with A30-6 is expressed relative to the level of parent vector pCOMT-Kan transfer. The transfer frequencies of the remaining plasmids are expressed relative to the pCOM9 transfer frequency. The values are means ± standard deviations for two replicate experiments.
Relative amount of spliced mRNA as determined by real-time RT-PCR. The values are means ± standard deviations for four replicate experiments.
L. lactis MMS370 (pM1014) carried the pCOM-A30-6 vector.
L. lactis MMS370 (pM1014) carried a pCOM9-based vector with the mutation(s) indicated in the intron.
DISCUSSION
In the present study we validated a conjugation-based assay for splicing of the Ll.ltrB intron, developed a molecular assay for in vivo intron splicing, and demonstrated that the two assays can be used for straightforward testing of designed intron mutants and reasonably convenient isolation of new intron variants with splicing defects.
Using the pCOM9 system described previously (29), we engineered mutations within the intron that were predicted to have defects in Ll.ltrB splicing based on sequence conservation and previous in vitro studies. We analyzed these constructs both by conjugation and by quantitative real-time RT-PCR. We found that the mutants were defective in both of these assays, and we concluded that the residues tested are critical for splicing of the Ll.ltrB intron in vivo. Two mutations of the branch point residue in D6, A2486Δ and A2486C, were tested. The bulging A residue provides a 2′-hydroxyl residue that is the nucleophile in the first transesterification reaction of splicing that leads to production of a branched intron lariat molecule. Proper branch point selection is critical for efficient splicing, and in the yeast ai5γ group II intron, deletion, base pairing, or substitution of the branch point adenosine usually eliminates branching (12, 23). Likewise, our results suggest that the branch point adenosine is critical for splicing of the Ll.ltrB intron. The G2399A mutation is located in D5, a site critical for the catalytic activity of group II introns (26). While D5 is not perfectly conserved in the primary sequence in group II introns, the lower helix does contain the highly conserved AGC sequence. Previous research has demonstrated that mutation of these residues, particularly the G residue, results in reduced splicing activity of the yeast a5γ intron in vitro and in vivo (3, 22). We found that changing this G residue to an A (G2399A) in the Ll.ltrB intron has a significant detrimental effect on splicing. Many tertiary interactions contribute to group II intron function; for example, tertiary interactions presumably position D5 close to the joiner between D2 and D3 of the yeast intron a5γ (24). We tested this interaction in the Ll.ltrB intron by creating the G471A mutation, and we found that this replacement impairs splicing in the lactococcal system. By using biochemical assays and an E. coli expression system for splicing, it has been established that LtrA, the intron-encoded protein of Ll.ltrB, is required for the splicing and mobility functions of the intron (14). We tested a large deletion in the LtrA open reading frame (pCOM-ΔORF) in the conjugation and RT-PCR assays and found a similar requirement for LtrA for in vivo splicing in lactococci.
The second conclusion which we made from the combined conjugation and RT-PCR data was that the conjugation assay is a feasible method for measuring splicing. While the conjugation assay provides an indirect measure of splicing since it measures the functionality of the LtrB product of splicing, it does accurately and sensitively reflect the splicing ability of the intron. Therefore, the pCOM9 conjugation system provides an excellent genetic tool for analyzing group II intron function that has not been available previously. To date, in studies of various group II introns workers have utilized biochemical methodologies to investigate intron structure and function. In genetic studies workers have relied on biochemical data or comparative sequence analyses to identify and mutate interesting residues. In particular, in studies of the Ll.ltrB intron researchers have utilized an E. coli expression system to perform biochemical studies (14). This system has been very useful for determining the mechanism of Ll.ltrB splicing and homing, but it is limited by the fact that it relies on unphysiologically high levels of intron expression. In addition, aspects of intron expression or function specific to L. lactis may be missed by the E. coli expression system.
To take a genetic approach towards understanding Ll.ltrB structure and function, we used a combination of the conjugation assay and quantitative RT-PCR to screen for residues critical for intron splicing. Using the conjugation assay, we screened hundreds of randomly mutagenized introns for transfer deficiency. We then used RT-PCR to determine whether the conjugation-defective mutants were able to undergo splicing. This step was designed to segregate mutations that caused transfer defects due to their locations within ltrB or the plasmid backbone (oriT) from mutations that were transfer deficient due to splicing defects. After sequencing splicing-defective mutants, we found two classes of mutations: mutations in the LtrA coding sequence and mutations in the intron RNA. Of interest was mutant A30-6, which had two mutations located in or near D5. The U2428C mutation is located in the lower helix of D5, opposite the G2399 residue described above. This change imposes Watson-Crick base pairing on residue 2399. The equivalent mutation in the yeast intron a5γ did not have a large effect on in vitro or in vivo splicing (3, 22), which is consistent with our observation that the U2428C mutation, when subcloned into the wild-type intron, was not substantially defective in either the conjugation or RT-PCR assay for splicing. The second change identified in mutant A30-6 (A2396G) is located 1 nucleotide upstream of D5 and could allow base pairing with the C2431 residue immediately after D5. Such a base-pairing interaction extends the lower helix of D5 by 1 bp and shortens the joiner region between D4 and D5, as well as the joiner region between D5 and D6. Work with the yeast a5γ intron has shown that intron mutations in which the D5-D6 joiner region is eliminated or in which the length of the D5-D6 joiner region is reduced to 1 nucleotide result in defective splicing (4). In another study of the a5γ intron the workers suggested that the length of the joiner region affects branch site selection (5). Since the A2396G mutation alone does not lead to a substantial splicing defect, the role of the joiner region in Ll.ltrB splicing is still unclear. Although neither of these mutations caused a splicing defect alone, we found that when both mutations were present, splicing was severely inhibited. One possible explanation for this observation is that the two mutations together significantly alter the structure and/or orientation of D5. Further experiments are needed to investigate the exact effect that these mutations have on Ll.ltrB function. However, the isolation of the double mutant demonstrates the potential of the genetic screening method to identify splicing-defective mutants that could not be predicted by sequence analysis. Our current efforts are directed towards using a more efficient approach to individually target the protein-encoding region or the ribozyme component of the intron for saturation mutagenesis.
Acknowledgments
We thank Joanne Bartkus at the Minnesota Department of Health for assistance with the real-time RT-PCR assays.
J.K. was supported by NIH training grants HD 07381-12 and T32 DEO 07288-07A1. This work was supported by grant GM58279 from the NIH.
REFERENCES
- 1.Anderson, G. G., and L. L. McKay. 1984. Genetic and physical characterization of recombinant plasmids associated with cell aggregation and high-frequency conjugal transfer in Streptococcus lactis ML3. J. Bacteriol. 158:954-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 3.Boulanger, S. C., S. M. Belcher, U. Schmidt, S. D. Dib-Hajj, T. Schmidt, and P. S. Perlman. 1995. Studies of point mutants define three essential paired nucleotides in the domain 5 substructure of a group II intron. Mol. Cell. Biol. 15:4479-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulanger, S. C., P. H. Faix, H. Yang, J. Zhuo, J. S. Franzen, C. L. Peebles, and P. S. Perlman. 1996. Length changes in the joining segment between domains 5 and 6 of a group II intron inhibit self-splicing and alter 3′ splice site selection. Mol. Cell. Biol. 16:5896-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, V. T., C. Adamidi, Q. Liu, P. S. Perlman, and A. M. Pyle. 2001. Control of branch-site choice by a group II intron. EMBO J. 20:6866-6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cousineau, B., S. Lawrence, D. Smith, and M. Belfort. 2000. Retrotransposition of a bacterial group II intron. Nature 404:1018-1021. [DOI] [PubMed] [Google Scholar]
- 7.Cousineau, B., D. Smith, C. S. Lawrence, J. E. Mueller, J. Yang, D. Mills, D. Manias, G. Dunny, A. M. Lambowitz, and M. Belfort. 1998. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell 94:451-462. [DOI] [PubMed] [Google Scholar]
- 8.D'Souza, L. M., and J. Zhong. 2002. Mutations in the Lactococcus lactis Ll.ltrB group II intron that retain mobility in vivo. BMC Mol. Biol. 3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, H., M. Karberg, M. Long, J. R. Jones, B. Sullenger, and A. M. Lambowitz. 2000. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science 289:452-457. [DOI] [PubMed] [Google Scholar]
- 10.Karberg, M., H. Guo, J. Zhong, R. Coon, J. Perutka, and A. M. Lambowitz. 2001. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat. Biotechnol. 19:1162-1167. [DOI] [PubMed] [Google Scholar]
- 11.Klein, J. R., and G. M. Dunny. 2002. Bacterial group II introns and their association with mobile genetic elements. Frontiers Biosci. 7:d1843-1856. [Online.] http://www.bioscience.org. [DOI] [PubMed] [Google Scholar]
- 12.Liu, Q., J. B. Green, A. Khodadadi, P. Haeberli, L. Beigelman, and A. M. Pyle. 1997. Branch-site selection in a group II intron mediated by active recognition of the adenine amino group and steric exclusion of non-adenine functionalities. J. Mol. Biol. 267:163-171. [DOI] [PubMed] [Google Scholar]
- 13.Matsuura, M., J. W. Noah, and A. M. Lambowitz. 2001. Mechanism of maturase-promoted group II intron splicing. EMBO J. 20:7259-7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuura, M., R. Saldanha, H. Ma, H. Wank, J. Yang, G. Mohr, S. Cavanagh, G. M. Dunny, M. Belfort, and A. M. Lambowitz. 1997. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 11:2910-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michel, F., and J. L. Ferat. 1995. Structure and activities of group II introns. Annu. Rev. Biochem. 64:435-461. [DOI] [PubMed] [Google Scholar]
- 16.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Mills, D. A., C. K. Choi, G. M. Dunny, and L. L. McKay. 1994. Genetic analysis of regions of the Lactococcus lactis subsp. lactis plasmid pRS01 involved in conjugative transfer. App. Environ. Microbiol. 60:4413-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills, D. A., L. L. McKay, and G. M. Dunny. 1996. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J. Bacteriol. 178:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mills, D. A., T. G. Phister, G. M. Dunny, and L. L. McKay. 1998. An origin of transfer (oriT) on the conjugative element pRS01 from Lactococcus lactis subsp. lactis ML3. Appl. Environ. Microbiol. 64:1541-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohr, G., D. Smith, M. Belfort, and A. M. Lambowitz. 2000. Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes Dev. 14:559-573. [PMC free article] [PubMed] [Google Scholar]
- 21.Peebles, C. L., S. M. Belcher, M. Zhang, R. C. Dietrich, and P. S. Perlman. 1993. Mutation of the conserved first nucleotide of a group II intron from yeast mitochondrial DNA reduces the rate but allows accurate splicing. J. Biol. Chem. 268:11929-11938. [PubMed] [Google Scholar]
- 22.Peebles, C. L., M. Zhang, P. S. Perlman, and J. S. Franzen. 1995. Catalytically critical nucleotide in domain 5 of a group II intron. Proc. Natl. Acad. Sci. 92:4422-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Podar, M., V. T. Chu, A. M. Pyle, and P. S. Perlman. 1998. Group II intron splicing in vivo by first-step hydrolysis. Nature 391:915-918. [DOI] [PubMed] [Google Scholar]
- 24.Podar, M., J. Zhuo, M. Zhang, J. S. Franzen, P. S. Perlman, and C. L. Peebles. 1998. Domain 5 binds near a highly conserved dinucleotide in the joiner linking domains 2 and 3 of a group II intron. RNA 4:151-166. [PMC free article] [PubMed] [Google Scholar]
- 25.Polzin, K. M., and M. Shimizu-Kadota. 1987. Identification of a new insertion element, similar to gram-negative IS26, on the lactose plasmid of Streptococcus lactis ML3. J. Bacteriol. 169:5481-5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin, P. Z., and A. M. Pyle. 1998. The architectural organization and mechanistic function of group II intron structural elements. Curr. Opin. Struct. Biol. 8:301-308. [DOI] [PubMed] [Google Scholar]
- 27.Saldanha, R., B. Chen, H. Wank, M. Matsuura, J. Edwards, and A. M. Lambowitz. 1999. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry 38:9069-9083. [DOI] [PubMed] [Google Scholar]
- 28.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou, L., D. A. Manias, and G. M. Dunny. 2000. Regulation of intron function: efficient splicing in vivo of a bacterial group II intron requires a functional promoter within the intron. Mol. Microbiol. 37:639-651. [DOI] [PubMed] [Google Scholar]