Abstract
The HOX genes encode a family of transcription factors that are dysregulated in several malignancies and have been implicated in oncogenesis and cancer cell survival. Disruption of HOX protein function using the peptide HXR9 has shown anti-tumor effects against melanoma, lung cancer, and renal cancer. In this report we evaluated the expression of all 39 HOX genes in a panel of six malignant B-cell lines, including multiple myeloma cells, and found different levels of expression of HOX family members suggesting that they also play a role in malignant B-cell survival. We show that disrupting HOX function using the peptide HXR9 induces significant cytotoxicity in the entire panel of cell lines. Importantly, we found that the cytotoxic effects of HXR9 can be enhanced by combining it with ch128.1Av, an antibody-avidin fusion protein specific for the human transferrin receptor 1 (CD71). Iron starvation induced by the fusion protein contributes to the enhanced effect and involves, at least in part, the induction of a caspase-independent pathway. These results demonstrate the relevance of the HOX proteins in malignant B-cell survival and suggest that our therapeutic strategy may be effective in the treatment of incurable B-cell malignancies such as multiple myeloma.
Keywords: antibody fusion protein, HOX genes, cytotoxicity, transferrin receptor, hematopoietic malignancies, HXR9
Introduction
Multiple myeloma (MM) is a malignant plasma cell disorder that accounts for approximately 10% of all hematological malignancies and is the second most frequent blood cell cancer in the United States (US), after non-Hodgkin’s lymphoma (1). According to the American Cancer Society, in the US there were 20,580 estimated new cases for 2009 and about 10,580 myeloma deaths (2). Since 2001, the overall survival rate of myeloma has increased dramatically due to the advent of new therapies including thalidomide and its derivative lenalidomide as well as the proteosome inhibitor bortezomib (3). With or without autologous stem cell transplantation, combination therapies of these new drugs with corticosteroids (such as dexamethasone and prednisone), alkylating agents (such as melphalan), or anthracyclines have dramatically improved survival of myeloma patients (reviewed in (4)). However, some of these combination therapies are associated with severe toxicities and no complete cures have been reported (4). Therefore, the continued development of novel agents for the treatment of MM is a necessity.
Early genetic studies in Drosophilia melanogaster led to the discovery of the HOX genes and their role in determining embryonic identity along the anterior-posterior axis (5). Various HOX genes have also been suggested to act as oncogenes and may play a central role in both carcinogenesis and leukemic transformation (6–8). The HOX genes have been found to be aberrantly expressed in both myeloid and lymphoblastic leukemic cells (7–9). HOXC5 and HOXC6 were previously found to be expressed in biopsies of Non-Hodgkin’s lymphoma patients (10). In addition, abnormal HOXA expression has been observed in the malignant plasma cells isolated from a subset of MM patients (11). HOX genes are a family of homeobox containing genes that encode DNA binding transcription factors that have key regulatory roles (7). There are 39 mammalian HOX genes that are organized into four paralogue clusters A–D (11 genes for HOXA, 10 genes for HOXB, 9 genes for HOXC, and 9 genes for HOXD) located on four different chromosomes (7). Due to the large number of genes and considerable functional overlap, defined roles for each gene remain unknown. The altered expression of HOX genes in tumorigenesis has been implicated in the dysregulation of cell cycle progression, terminal differentiation, metastasis, and angiogenesis making them potential targets for cancer therapy (6–8).
In order to better understand the role of HOX genes in neoplastic progression a small, membrane permeable peptide (HXR9) was developed to inhibit the interaction between the HOX proteins and a second transcription factor PBX (12). The interaction of PBX with the HOX proteins increases their DNA-binding affinity and thus enhances the transcriptional activity of the HOX proteins (13). More importantly, the disruption of the HOX/PBX dimer has been shown to be cytotoxic in solid tumors such as melanoma, renal cell carcinoma, and non-small-cell lung cancers in vitro and in vivo (12, 14, 15).
We have previously reported the cytotoxic effects of a mouse/human chimeric antibody fusion protein that consists of an IgG3 antibody specific for the human transferrin receptor 1 (TfR1, CD71) that is genetically fused to chicken avidin (ch128.1Av, previously known as anti-hTfR1 IgG3-Av) in malignant B cells including myeloma (16–19). The TfR1, through its interaction with iron-loaded transferrin (Tf), is responsible for iron uptake by cells (20). Iron is a required co-factor for many key cellular enzymes including the ribonucleotide reductase that is needed for DNA synthesis (20). For this reason, the TfR1 is highly expressed on the surface of many cancer cells including MM and in some cases its expression level can be associated with tumor stage and poor prognosis (20). Thus, the TfR1 has been extensively used as a therapeutic target (20, 21). ch128.1Av was initially developed to serve as a general delivery vector for biotinylated anti-cancer agents to target malignancies that overexpress the TfR1. However, ch128.1Av alone was found to be cytotoxic through the induction of lethal iron starvation due to a significant decrease in cell surface TfR1 expression (17, 19). This intrinsic activity can be enhanced through its conjugation with biotinylated toxins (22) or by its combination with non-biotinylated agents, including gambogic acid (a xanthone used in traditional Chinese medicine) (19) and the chemotherapeutic agent cisplatin (23). Importantly, ch128.1Av was recently shown to inhibit the NF-κB pathway (23), which is constitutively activated in MM and as been identified as one of the most important pathways driving myeloma development and progression (24).
Despite the suggested central role of the HOX genes in the development of certain hematopoietic malignancies, a complete evaluation of HOX gene expression in malignant B cells, especially myeloma, has not been conducted. Previous studies are limited to the study of HOXA gene expression in MM (11) and HOXC (specifically HOXC4, HOXC5, and HOXC6) in non-Hodgkin’s lymphoma (10). Therefore, the goals of this study were to evaluate the expression of all 39 HOX genes in a panel of malignant B cells, including myeloma, and explore the functional consequences of blocking their activity using the HXR9 peptide in these cells. Here we show that HXR9 is highly cytotoxic to all cells tested, including U266 myeloma cells that are resistant to apoptosis due to the high expression level of the survival protein Bcl-XL (25). We also show that ch128.1Av enhances the effects of HXR9 alone and that the mechanism of synergy is due, at least in part, to the induction of a caspase-independent pathway mediated by iron deprivation. Our studies indicate that the HOX genes can be targeted for therapy of B-cell malignancies including myeloma due to the abnormal expression of some of these genes.
Materials and Methods
Human cell lines
The following human cell lines were purchased from ATCC (American Type Culture Collection, Manassas, VA): IM-9 (an EBV-transformed lymphoblastoid cell line isolated from the peripheral blood of a patient with multiple myeloma), ARH-77 (an EBV-transformed lymphoblastoid cell line isolated from the peripheral blood blood of a patient with plasma cell leukemia), U266 (myeloma cell line), and RPMI-8226 (myeloma cell line). MM.1S (myeloma cell line) was a kind gift from Drs. Kenneth Anderson and Darminder Chauhan (Harvard University). The above cell lines were maintained in RPMI 1640 (Invitrogen Corporation, Carlsbad, CA) supplemented with 100 U/ml penicillin, 10 µg/ml streptomycin, and 10% (v/v) heat inactivated fetal bovine serum (FBS) (Atlanta Biologicals, Atlanta, GA). The human myeloma cell line KMS-11 was a kind gift from Lawrence Boise (University of Miami) and was cultured in Iscove’s Modified Dulbecco’s Medium (IMDM; Invitrogen) supplemented with 100 U/ml penicillin, 10 µg/ml streptomycin, and 10% FBS. All cell lines were grown in 5% CO2 at 37○C.
Peptides and antibody fusion protein
Both the HOX-PBX-targeted HXR9 peptide and the control peptide CXR9 have been described previously (12, 14). HXR9 is a peptide consisting of the previously identified hexapeptide sequence that binds to PBX and nine C-terminal arginine residues (R9) that facilitate cell entry. CXR9 is a control peptide that lacks a functional hexapeptide sequence but includes the R9 sequence. Both peptides were synthesized using conventional column-based chemistry and purified to at least 80% (Biosynthesis Inc., Lewisville, TX).
The ch128.1Av fusion protein contains chicken avidin genetically fused to the CH3 domains of a chimeric mouse/human IgG3 and has been described previously (16, 17). ch128.1Av, containing the variable regions of the murine monoclonal anti-human TfR IgG1 antibody 128.1 (16, 26), was purified from cell culture supernatants using affinity chromatography (16, 27). Fusion protein integrity and purity was assessed by non-reducing SDS-PAGE analysis using 5% phosphate gels. Purified ch128.1Av was dialyzed into buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.8) and protein concentration was determined by bicinchoninic acid based protein assay (BCA Protein Assay, Pierce Biotechnology, Inc., Rockford, IL). The antibody fusion protein was stored at −80○C in snap frozen aliquots.
Semi-quantitative PCR
HOX or iron-related gene expression relative to the housekeeping control gene glyceraldehyde 3-phosphate dehyrdrogenase (GAPDH) was determined by semi-quantative real time PCR. Total RNA from each cell line was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA was then prepared using the SuperScript III First-Strand synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). Semi-quantitative reverse transcriptase-PCR was performed using the Stratagene MX4000 real-time PCR Machine (Stratagene, La Jolla, CA), which measures product accumulation during the exponential phase of the reaction. The Brilliant SYBR Green QPCR Master Mix (Sigma Aldrich Co., St. Louis, MO) and oligonucleotide primers (sequences provide in Supplementary Table S1) designed to facilitate the unique amplification of each HOX and iron-related gene were used. For the analysis of changes in iron-related gene expression, changes above 2-fold are considered to be relevant changes.
To obtain accurate counts of HOX gene expression relative to GAPDH, the Ct value for GAPDH in a given cell line was subtracted from each of the Ct values of the HOX genes (dCt). This quantity was used in the formula V= (2dCt)*1000 to provide actual counts of gene expression (V) relative to GAPDH. This data was then Log base 2 (Log2) transformed using the Excel software and subject to clustering based on a Eucledian distance algorithm with centroid linkage for both genes and cell lines provided by the Gene Cluster program version 3.0 (28) and visualized using Java Tree View software (29).
Proliferation assay and morphology study
The effect of HXR9 on cell proliferation was determined by the [3H]-thymidine incorporation assay as previously described (22). Briefly, cells were treated in triplicate with various concentrations of HXR9 or the control peptide (CXR9) for a total of 48 hours and radioactivity determined. Data is expressed as a percent of the [3H]-thymidine incorporation of control cells. For morphological analysis, cells were treated with 50 µM HXR9 or CXR9 for 48 hours. Cell images were captured using a Zeiss Axiovert 40 CFL PlasDIC Inverted Microscope using a 20X objective (Mikron Instruments Inc., San Marcos, CA) and a Canon PowerShot A620 digital camera (Mikron Instruments Inc.).
Cytotoxicity of the combination treatments
In order to determine the cytotoxic effect of the combination of the ch128.1Av and HXR9, cell proliferation and the level of apoptosis were measured simultaneously in the same cell population. IM-9 cells were seeded in 24-well plates at a density of 1.6 × 105 cells per well in a total volume of 1.6 mL and treated with 20 µM HXR9, 2.5 nM ch128.1Av, or the combination of both agents for a total of 48 hours. For the proliferation assay, cells were gently mixed and 100 µL was transferred to one well of a 96-well plate (in triplicates) and the amount of [3H]-thymidine incorporation was determined as described above. The cells remaining in the 24-well plate were used to determine the level of apoptosis using the Vybrant® Apoptosis Assay Kit #2 (Invitrogen Corporation, Carlsbad, CA) following procedures suggested by the manufacturer. This kit consists of the Annexin V Alexa Fluor 488 conjugate and propidium iodide (PI) stain. Samples were analyzed on a BD-LSR Analytic Flow Cytometer (BD Biosciences, San Jose, CA) in the UCLA Jonsson Comprehensive Cancer Center and Center for AIDS Research Flow Cytometry Core Facility.
In order to determine the dependence of caspases or iron in HXR9-mediated cell death (alone and combined with ch128.1Av), IM-9 cells were pretreated with 50 µmol/L Z-VAD-FMK methyl ester (Enzo Life Sciences Inc., Plymouth Meeting, PA) for 1 hour before the addition of the other treatments or treated simultaneously with 25 µM ferric ammonium citrate (FAC; Sigma Aldrich Co.) and assayed as described above for proliferation and induction of apoptosis.
Synergy study of the combination treatment
IM-9 cells were treated simultaneously with various concentrations of ch128.1Av (2.5, 5, 10, 20, or 30 nM) and HXR9 (10 or 20 µM) for a total of 48 hours. Cytoxicity of each agent alone and in the combination treatment was monitored using the [3H]-thymidine incorporation assay as described above. The degree of synergism between the two compounds was determined using the CalcuSyn Software (BioSoft, Ferguson, MO) that was developed by Chou and Hayball (30), The combination index (CI) for each combination was calculated at a non-constant ratio. By this method, CI equal to 1 indicate an additive effect, less than 1 indicate synergism, and greater than 1 indicate antagonism (30).
Caspase activity assays
IM-9 cells were seeded at a density of 104 cells per well of black, clear-bottomed 96-well tissue culture plates (ThermoFisher Scientific, Fremont, CA). Cells were treated with 2.5 nM ch128.1Av, 20 or 40 µM HXR9, or the combination of 2.5 nM ch128.1Av and 20 µM HXR9 for various times. After treatment cells were assayed for caspase activity using fluorogenic substrates in a one-step assay as described previously (22, 31). Fluorogenic substrates (all from Axxora Life Sciences, Inc., San Diego, CA) specific for caspase-2 (Ac-VDVAD-AMC), caspase-9 (Ac-LEHD-AMC), caspase-8 (Ac-IETD-AMC), and caspase-3 (Ac-DMQD-AMC) were used. The plate was then read at excitation and emission wavelengths of 380 and 460 nm, respectively, using a DTX880 Multimode Detector (Beckman Coulter). Background fluorescence, measured in wells containing only medium and one-step assay buffer with substrate, was subtracted from each sample. To calculate the fold increase in activation of each caspase, the average relative fluorescence intensity of the treated wells was divided by the average relative fluorescence intensity of the control wells.
Human colony forming (progenitor) assay
The colony forming assay was performed as described previously (19, 22). Bone marrow mononuclear cells (BMMC) were purchased from StemCell Technologies, Inc. (Vancouver, British Columbia, Canada) and plated in quadruplicate according to the manufacturer’s instructions. Mononuclear cells were seeded in 35 mm dishes in MethoCult GF H4434 (‘Complete’ Methylcellulose Medium with Recombinant Cytokines and Erythropoietin; StemCell Technologies) in the presence of various concentrations of HXR9, 2.5 nM ch128.1Av, or the combination of 20 µM HXR9 with 2.5 nM ch128.1Av for 14 days at 37 °C in 5% CO2. Control treatments consisted of non-treated cells as well as those treated with diluent alone (buffer as the diluent for ch128.1Av or water as the diluent for HXR9 or CXR9). Colonies were identified and counted using an Olympus CK2 inverted microscope (Olympus America Inc., Center Valley, PA) and the criteria defined by StemCell Technologies for each colony type. Colony types identified were: CFU-E (colony forming unit-erythroid, mature erythroid progenitors), BFU-E (burst forming unit-erythroid, more primitive progenitor than CFU-E), CFU-GM (colony forming unit-granulocyte/macrophage, more mature than CFU-GEMM) and CFU-GEMM (colony forming unit granulocyte/erythroid/macrophage/ megakaryocyte).
Statistical analysis
All statistical analyses were done using Microsoft Excel 2000 SR-1 Standard. p values < 0.05 were considered to be significant and were calculated using the Student’s t test (unpaired samples, two-tailed, unequal variance).
Results
HOX gene expression in a panel of human malignant B-cell lines
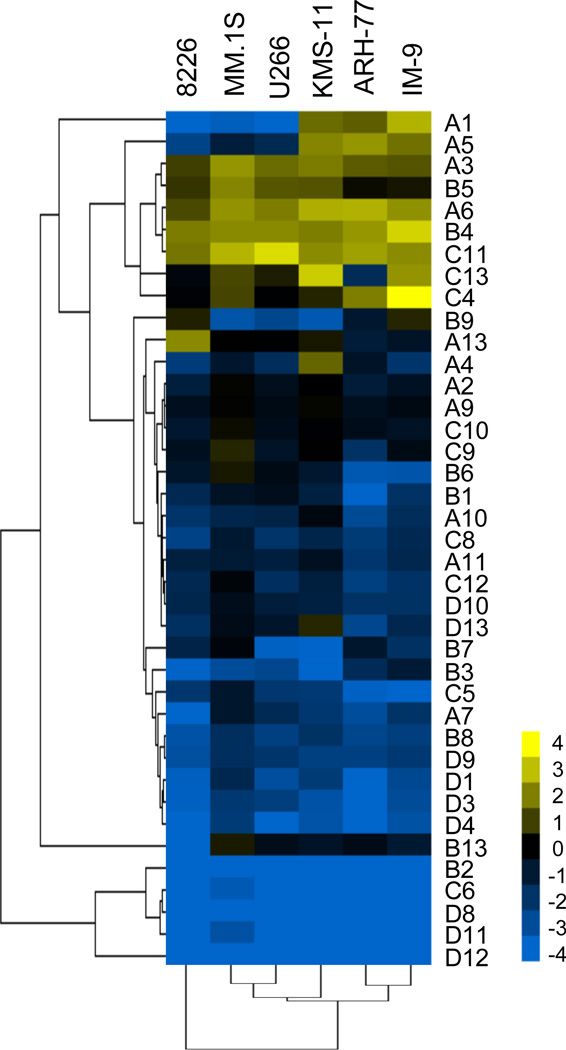
In order to fully evaluate the expression of all HOX genes in malignant B cells, including myeloma cells, we examined the mRNA levels of HOXA-D (39 genes) in a panel of six cell lines by semi-quantitative PCR. Analysis of the HOXA genes (Supplementary Data, Figure S1) shows high levels of expression in all cell lines, although the expression of each gene varies among the cell lines. The HOXB genes (Supplementary Data, Figure S1) are expressed in general at low levels except for HOXB4 and to a lesser extent HOXB5. HOXC11 shows high expression in all cell lines and HOXC4 is expressed at exceptionally high levels in IM-9 cells (Supplementary Data, Figure S2). Expression of the other HOXC genes is low. Overall the expression of the HOXD genes is very low in all cell lines tested (Supplementary Data, Figure S2). Clustering analysis (Figure 1) of the HOX gene expression data for all six cell lines shows that the B-cell lymphoblastoid cell lines, ARH-77 and IM-9, demonstrate the most similar HOX gene expression pattern. KMS-11, U266, and MM.1S are similar to each other, with RPMI 8226 showing the most different pattern of HOX gene expression. These data show that many of the HOX genes are highly expressed in malignant B cells and therefore, suggest that the HOX genes represent meaningful therapeutic targets.
Figure 1. Hierarchical clustering of differential expression of HOX genes in a panel of hematopoietic malignant B cells.
The clustergram shows expression of an array of HOX genes for each cell line in the panel. Linkage trees represent similarities among data points for either groups of genes (left) or cell lines (below). The colormap represents expression counts relative to GAPDH in a Log base 2 scale. Values range from -4 (blue), indicating gene expression lower than that of GAPDH, to 4 (yellow), indicating gene expression higher than that of GAPDH.
Cytotoxicity of HXR9 against malignant B-cell lines
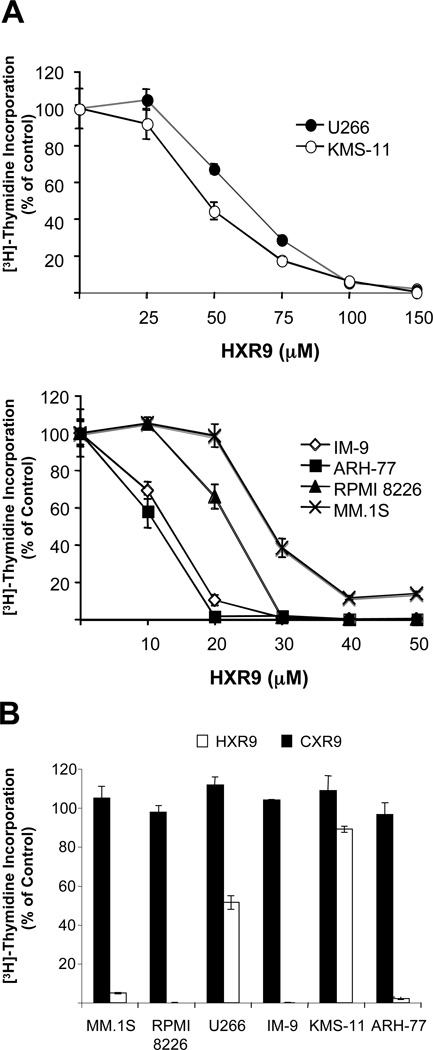
In order to block HOX protein transcriptional activity, we used the membrane-permeable peptide HXR9 that blocks the interaction of the HOX proteins with PBX, a second transcription factor that binds to the HOX proteins and enhances their DNA binding activity (12). HXR9 demonstrated dose-dependent anti-proliferative effects in all 6 of the cells lines tested (Figure 2A). The sensitivities varied slightly among the cell lines with ARH-77 and IM-9 being the most sensitive, RPMI 8226 and MM.1S demonstrating modest sensitivity, and U266 and KMS-11 being the least sensitive. As expected, the control peptide showed no anti-proliferative effects (Figure 2B). Cells treated with 50 µM HXR9 also showed changes in morphology including cell shrinkage and fragmentation consistent with the induction of cell death (Supplementary Data, Figure S3). These data suggest that HOX protein activity is important for malignant B-cell survival and interference with this function results in malignant cell death.
Figure 2. Anti-proliferative effects of HXR9 in malignant B cells.
A) Cells were treated in triplicate with various concentrations of HXR9 for 48 hours. The [3H]-thymidine incorporation assay was used to monitor cell proliferation. Least sensitive cell-lines, U266 and KMS-11 (top panel); sensitive cell lines, IM-9, ARH-77, RPMI 8226, and MM.1S (bottom panel). B) Specificity of HXR9 was confirmed by testing all cell lines with the control peptide CXR9. Cells were treated with 50 µM CXR9 or HXR9 for 48 hours. Data is expressed as a percent of the [3H]-thymidine incorporation of control cells. Standard deviation is indicated for each treatment. Data is representative of two independent experiments.
Synergistic effects of HXR9 and ch128.1Av
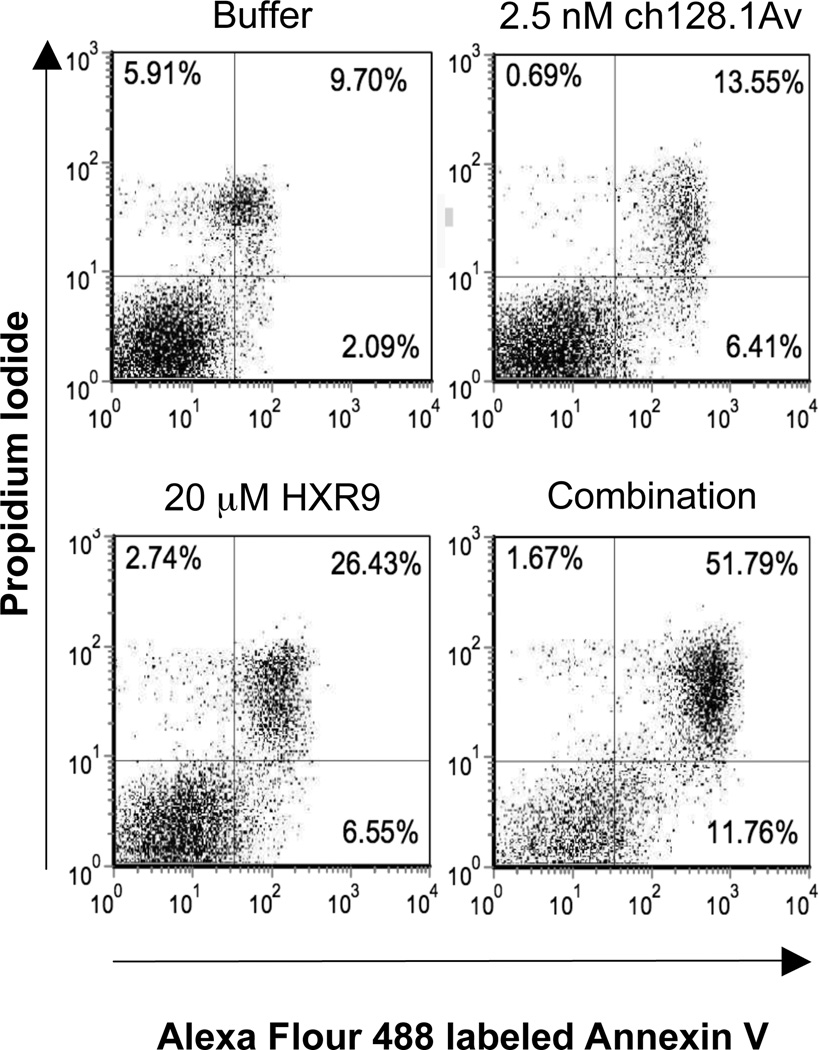
In order to explore the potential of enhancing the anti-tumor effect of HXR9, we evaluated the cytotoxic effects of the HXR9 peptide combined with ch128.1Av. We have previously shown that ch128.1Av at concentrations of 10 nM and above has a dramatic intrinsic in vitro cytotoxic activity against malignant B cells (17, 19) that can be enhanced through the combination treatment with other anti-cancer agents (19, 23). The combination of 20 µM HXR9 and a suboptimal dose of 2.5 nM ch128.1Av in IM-9 cells showed a greater anti-cancer effect compared to either agent alone (Figure 3). HXR9 alone showed a small increase in the induction of apoptosis compared to control cells, but as expected at the low concentration used, the effect of ch128.1Av was minimal. The simultaneous combination of the two agents demonstrated a dramatic increase in apoptosis as evidenced by the increase in Annexin V positive cells in both the upper and lower right quadrants (Figure 3). It is important to note that there is a shift to the right of the “healthy” population (lower left quadrant) in cells treated with the combination treatment compared to control cells, indicating that numbers shown in the right quadrants do not adequately reflect the effect of the combination treatment and that apoptosis is being induced in most if not all of the treated cells. In order to evaluate whether the enhanced effects of the combination treatment were synergistic or additive, combination index (CI) analysis was performed and showed that the combination of the two compounds resulted in synergistic anti-proliferative effects at all concentrations tested (Table 1) as evidenced by CI values that are less than 1. The synergy between the two compounds was stronger with the use of 20 µM HXR9 compared to 10 µM. In addition, longer incubation times (greater than 24 hours) were required for the synergistic effects to be apparent in IM-9 cells (data not shown). This is most likely due to the fact that the effects of ch128.1Av take time to occur and are visualized mostly after 24 hours. The effects of HXR9 in IM-9 cells alone can be observed within 30 minutes (data not shown).
Figure 3. Combination treatment of HXR9 and ch128.1Av increases the level of apoptosis in IM-9 cells.
Apoptosis was measured by flow cytometry analysis of IM-9 cells stained with Alexa flour 488 labeled Annexin V and counterstained with PI after 48 hours with each indicated treatment. Percentage of cells is shown in the corner of each quadrant. Results are representative of four independent experiments.
Table 1.
Combination Index analysis of ch128.1Av combined with HXR9 at a non-constant ratio in IM-9 cells.
| [ch128.1Av] nM | [HXR9] µM | Fa | CI | Description |
|---|---|---|---|---|
| 2.5 | 20 | 0.987 | 0.503 | Synergism |
| 5 | 20 | 0.997 | 0.382 | Synergism |
| 10 | 20 | 0.980 | 0.551 | Synergism |
| 20 | 20 | 0.986 | 0.515 | Synergism |
| 30 | 20 | 0.983 | 0.541 | Synergism |
| 2.5 | 10 | 0.781 | 0.862 | Moderate Synergism |
| 5 | 10 | 0.852 | 0.705 | Moderate Synergism |
| 10 | 10 | 0.899 | 0.609 | Synergism |
| 20 | 10 | 0.906 | 0.764 | Moderate Synergism |
| 30 | 10 | 0.928 | 0.667 | Synergism |
Analysis was performed using the Calcusyn software (BioSoft). Fa = fraction affected as tested by the [3H]-thymidine incorporation assay, CI = combination index. Descriptions are based on CI values and the recommendations of BioSoft: < 0.1 = very strong synergism, 0.1–0.3 = strong synergism, 0.3–0.7 = synergism, 0.7–0.85 – moderate synergism, 0.85–0.9 = slight synergism.
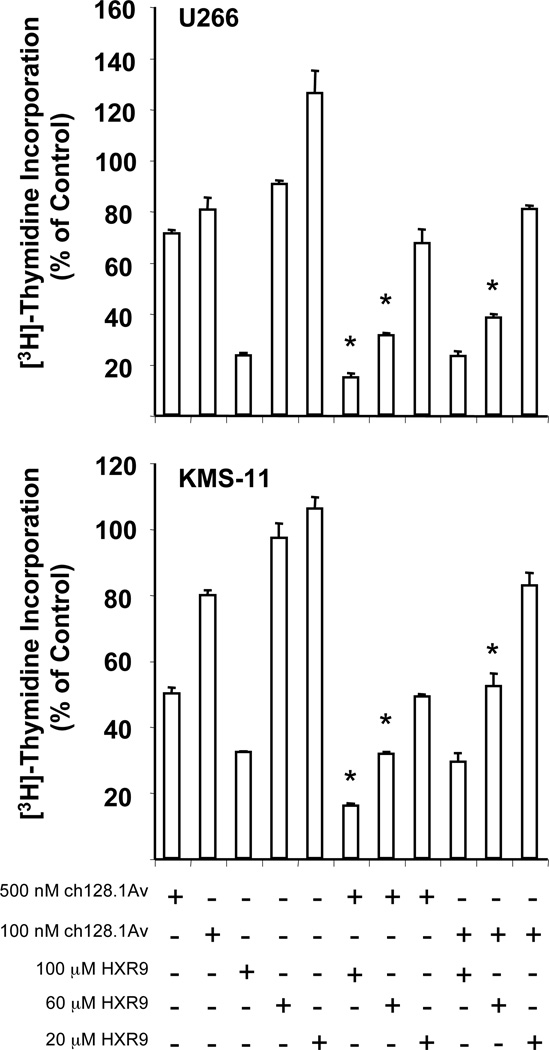
In order to determine if the enhanced effects of the combination treatment of HXR9 and ch128.1Av also occurred in other cell lines, we examined the effects in the myeloma cell lines U266 and KMS-11. U266 has demonstrated low sensitivity to the cytotoxic effects of ch128.1Av alone (17, 22) as well as to several other therapeutics; resistance to the latter has been associated with a high level of Bcl-XL expression (25). KMS-11 also shows low sensitivity to the fusion protein (Daniels et al., in preparation), but is more sensitive than U266 cells. Therefore, high doses (500 and 100 nM) of ch128.1Av and longer incubation times were required for the enhanced effects to occur. In both cell lines, the combination of 500 nM ch128.1Av with either 100 or 60 µM HXR9 for 96 hours showed a significant enhancement of anti-proliferative effects when compared to either agent alone (Figure 4). Enhanced effects were also observed with the combination of 100 nM ch128.1Av and 60 µM HXR9 in both cell lines (Figure 4). These findings indicate that the enhanced effects of the combination treatment are observed in multiple cell lines, including those that are highly resistant to other therapeutics.
Figure 4. Enhanced anti-proliferative effects of HXR9 and ch128.1Av in U266 and KMS-11 cells.
Cells were treated in triplicates with the indicated concentrations of HXR9, ch128.1Av, or the various combinations of the two compounds for 96 hours. Cytotoxicity was determined by the [3H]-thymidine incorporation assay. Results are representative of two independent experiments. * p-value < 0.05 when compared to both agents alone used at the same concentrations.
Mechanism of synergy
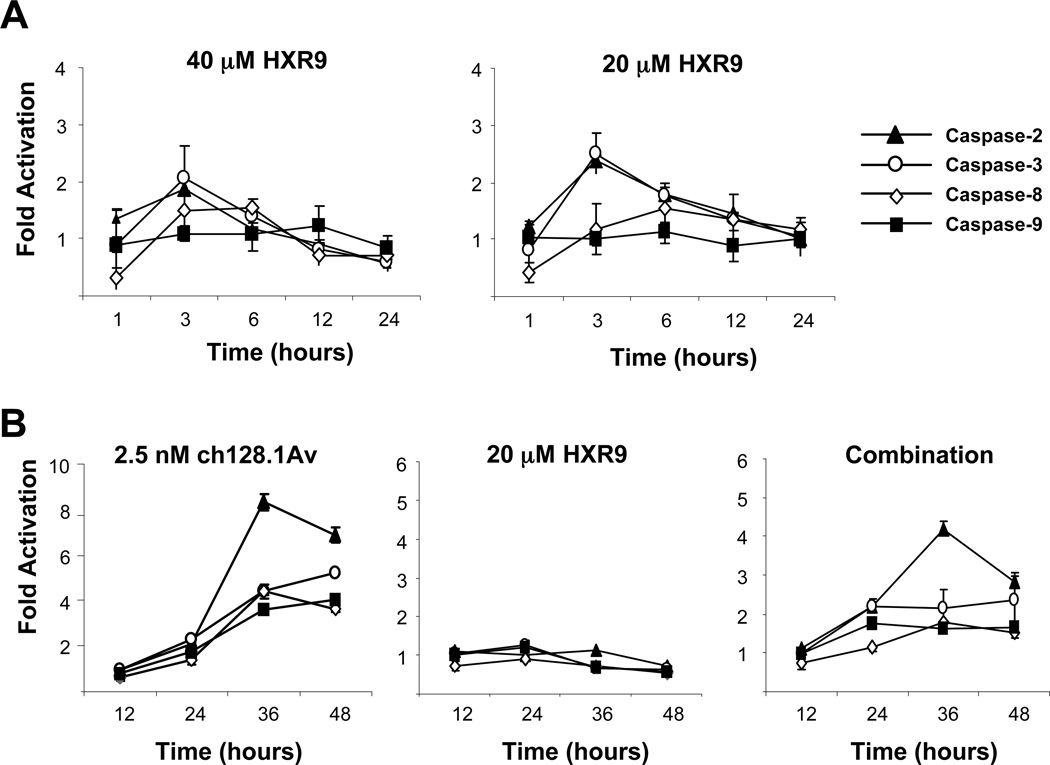
Since the combination of HXR9 and ch128.1Av results in synergistic cytotoxic effects compared to either agent alone, we wanted to explore the mechanism of this synergy. Due to the high sensitivity of IM-9 cells to ch128.1Av, we chose to use this cell line for the mechanistic studies. We first examined the activation of various caspases to determine their role in the cell death induced by HXR9 alone and combined with ch128.1Av. Treatment of IM-9 cells with HXR9 alone resulted in only low levels of caspase activation within the first three hours of treatment (Figure 5A). Caspases 2 and 3 showed the highest levels of activation by HXR9, while caspase 9 was not activated at all. As expected and shown previously (22), ch128.1Av alone at longer time points induced the activation of all caspases tested, especially caspase 2 (Figure 5B). Interestingly, the combination treatment resulted in the activation of caspases at about half the level they were with ch128.1Av alone, suggesting that a caspase-independent pathway is involved in HXR9-mediated cell death as well as the cell death induced by the combination treatment. In order to explore this further, we determined the effect of caspase blockade on the cytotoxicity induced by the combination treatment. Z-VAD-FMK (Z-VAD) was used to block the activation of all caspases. As previously shown, Z-VAD only partially blocked the anti-proliferative effects of ch128.1Av alone (Figure 6A) (17). Z-VAD also partially blocked the anti-proliferative effects of HXR9 alone (Figure 6A), but had no effect on the level of apoptosis (Figure 6B). For the combination treatment, Z-VAD partially protected the cells from the induction of apoptosis (Figure 6B). These results are consistent with the morphology of treated cells (Supplementary Data, Figure S4). Therefore, a caspase-dependent pathway involving caspase-2 is activated upon treatment with both ch128.1Av and HXR9. However, caspase-independent pathways or pathways involving other proteases may also be involved since cell death still occurs in the absence of caspase activation.
Figure 5. Caspase activation in IM-9 cells treated with HXR9 alone and in combination with ch128.1Av.
A) Caspase activity in IM-9 cells treated with 40 µM HXR9 or 20 µM HXR9 alone at various time points. At each time point cells were lysed and caspase activation was determined using flourogenic caspase substrates: Ac-IETD-AMC (caspase-8), Ac-LEHD-AMC (capase-9), Ac-DMQD-AMC (caspase-3), or Ac-VDVAD-AMC (caspase-2). B) Cells treated with 2.5 nM ch128.1Av, 20 µM HXR9, or the two in combination at the times indicated. Fold activation is expressed as the ratio of treated cells over buffer alone treated cells. Data are representative of three independent experiments.
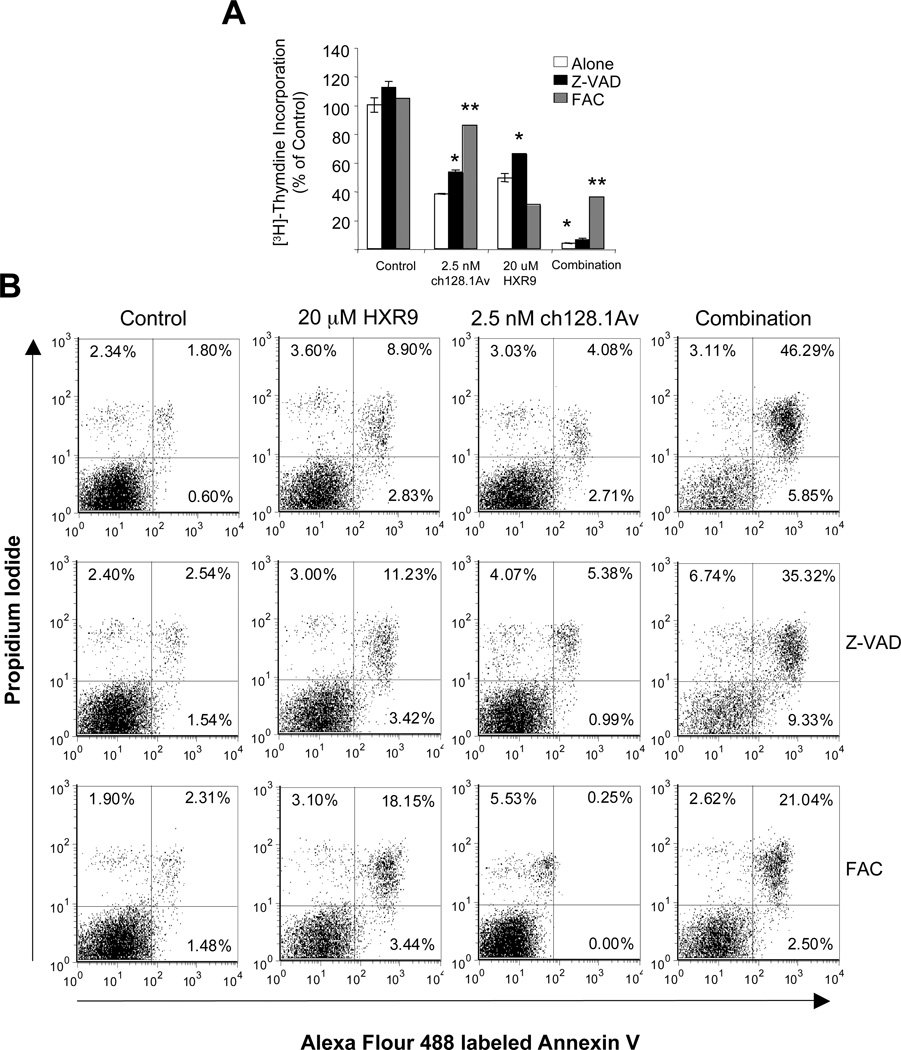
Figure 6. Effects of caspase inhibition and iron supplementation on the cytotoxicity of HXR9 and its combination with ch128.1Av.
IM-9 cells were treated with 2.5 nM ch128.1Av, 20 µM HXR9, or their combination for 48 hours; pre-treated with 50 µM ZVAD for 1 hour followed by the above treatments; or treated simultaneously with 25 µM FAC and the above treatments. A) [3H]-thymidine incorporation was used to monitor proliferation. Data are shown as the percent of control cells and represent the mean of triplicate samples. Error bars represent the standard deviation (* p < 0.05, ** p < 0.001). B) Apoptosis in cells treated as in A) was measured by flow cytometry analysis of cells stained with Annexin V and counterstained with PI after 48 hours. Percentage of cells is shown in the corner of each quadrant. Results are representative of two independent experiments.
Since ch128.1Av targets the TfR1 and has been previously shown to lead to iron starvation due to decreased cell surface TfR1 levels (17, 18), we explored the role of iron in the mechanism of synergy of the combination treatment. Iron supplementation using ferric ammonium citrate (FAC) was able to block the anti-proliferative effects of ch128.1Av as expected, however, it did not block the effects of HXR9 alone (Figure 6A). In fact, it sightly enhanced the cell death of HXR9 alone (Figure 6B). Interestingly, FAC significantly blocked the anti-proliferative and apoptotic effects of the combination treatment (Figure 6A and 6B). These data were corroborated by the cell morphology of treated cells (Supplementary data, Figure S4). Taken together these data suggest that iron starvation induced by ch128.1Av plays a role in the synergistic effects of the combination treatment.
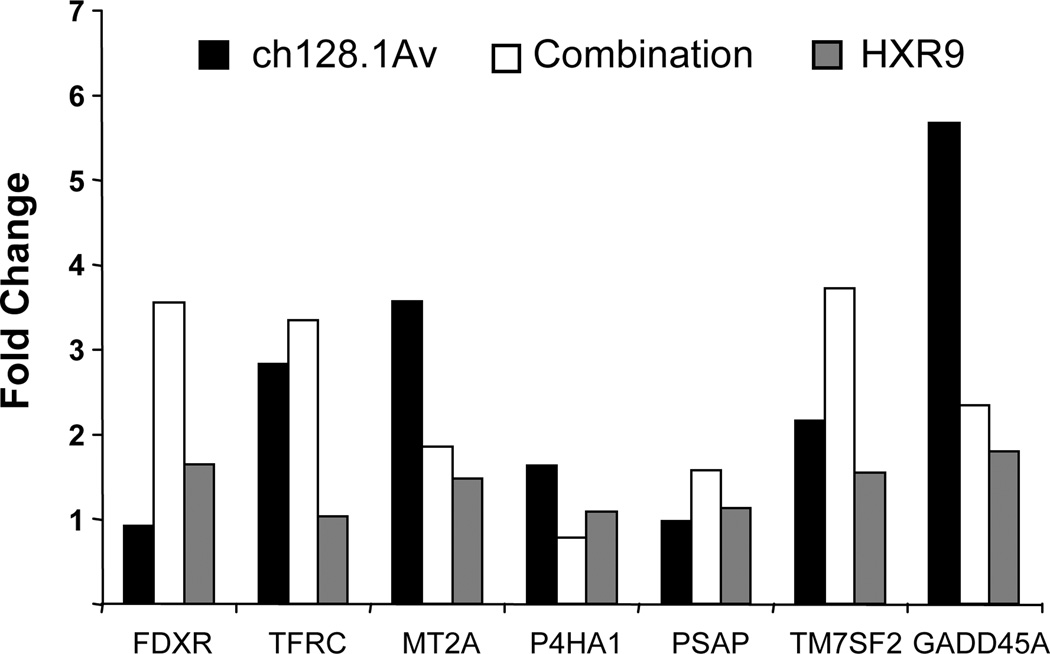
The role of iron and other stress-related pathways was further explored by gene expression analysis using semi-quantitative PCR. The genes selected for this analysis have been shown to be significantly modulated in IM-9 cells treated with 10 nM ch1281.Av for 24 hours as determined previously by microarray analysis (Rodriguez et al., manuscript in preparation), and are also listed in the IronChip V5.0 genes (European Molecular Biology Laboratory), a gene expression array composed of genes involved in iron uptake, storage and recycling, as well as genes involved in a number of interlinked pathways (32–34). Of the seven genes that were tested, no changes in gene expression were detected in cells treated with 20 µM HXR9 alone (Figure 7). As expected TfRC (CD71), MT2A (metallothionein 2A), and GADD45A (growth arrest and DNA-damage-inducible protein alpha) were shown to be upregulated in cells treated with 2.5 nM ch12.1Av alone (Figure 7). As shown in Figure 7, cells treated with both agents showed upregulation of FDXR (ferredoxin reductase), TFRC, and TM7SF2 (transmembrane 7 superfamily member 2, 3β-hydroxysterol Δ14-reductase). No changes in expression were observed with any of the treatments for P4HA1 (prolyl 4-hydroxylase α subunit), which has been shown to be increased in hypoxic conditions (35) and in the small intestines of iron-deficient rats (36) or PSAP (prosaposin) a precursor for the lysosomal saposins that are required for the degradation of sphingolipids and inhibition of metastasis (37).
Figure 7. Expression analysis of a subset of iron and stress-related genes in IM-9 cells treated with HXR9, ch128.1Av, or the combination treatment.
IM-9 cells were treated with 2.5 nM ch128.1Av, 20 µM HXR9, or the two agents in combination for 48 hours in triplicate at which time mRNA was isolated, cDNA synthesized, and the expression of each gene analyzed by semi-quantitative PCR. The average fold change of the triplicate wells relative to control cells is shown for each gene.
Toxicity to normal hematopoietic progenitor cells
Since both HXR9 and ch128.1Av were evaluated for their therapeutic potential against B-cell malignancies, their effect on normal hematopoiesis was also studied. The human colony-forming assay was used to determine the toxic effects of either treatment alone or in combination as evidenced by the inhibition of the development of colonies of different blood cell progenitor types in a semi-solid medium. Data between diluent alone and untreated cells was not significantly different (data not shown). The HOX proteins are known to play a role in hematopoiesis (38), thus it is expected that treatment with HXR9 alone would demonstrate toxicity to progenitor cells. As shown in Table 2, the toxicity of HXR9 is dose-dependent and also donor dependent. A previous study showed that 1 nM ch128.1Av decreases colony formation in this assay (19), therefore, it is not surprising that 2.5 nM also showed toxic effects as reported here (Table 2). The combination treatment showed toxicity as well, which is greater than either agent alone as evidenced by the further decrease of the formation of each colony type.
Table 2.
Toxicity to hematopoietic progenitors cultured in MethoCult for 14 days in the presence of HXR9 alone and in combination with ch128.1Av.
| Colony Type | Donor 1 | Donor 2 | Donor 3 | |
|---|---|---|---|---|
| Control (Buffer alone) | ||||
| CFU-E | 31 ± 3.3 | 12 ± 4.9 | 19 ± 3.6 | |
| BFU-E | 40 ± 5.7 | 16 ± 6.4 | 28 ± 5.0 | |
| CFU-GM | 54 ± 5.2 | 29 ± 0.6 | 38 ± 3.3 | |
| CFU-GEMM | 5 ± 0.8 | 3 ± 0.6 | 6 ± 3.4 | |
| 50 µM HXR9 | ||||
| CFU-E | 4 ± 3.0** | 4 ± 2.2* | 2 ± 1.9** | |
| BFU-E | 5 ± 2.6** | 5 ± 2.8* | 3 ± 1.6** | |
| CFU-GM | 6 ± 1.7** | 8 ± 1.6** | 6 ± 2.6** | |
| CFU-GEMM | 0 ± 0.0* | 1 ± 1.0* | 1 ± 0.8* | |
| 20 µM HXR9 | ||||
| CFU-E | 14 ± 2.2** | 10 ± 2.2 | 5 ± 1.9** | |
| BFU-E | 24 ± 5.4* | 8 ± 1.9 | 8 ± 1.5** | |
| CFU-GM | 52 ± 5.5 | 21 ± 2.2* | 21 ± 5.3** | |
| CFU-GEMM | 3 ± 1.4 | 1 ± 0.6* | 1 ± 0.8* | |
| 10 µM HXR9 | ||||
| CFU-E | 17 ± 2.2** | 9 ± 2.2 | 9 ± 3.8* | |
| BFU-E | 30 ± 6.7 | 12 ± 4.7 | 16 ± 3.4* | |
| CFU-GM | 60 ± 9.0 | 27 ± 2.1 | 21 ± 5.8* | |
| CFU-GEMM | 4 ± 3.4 | 3 ± 1.3 | 4 ± 3.5* | |
| 2.5 nM ch128.1Av | ||||
| CFU-E | 13 ± 9.8* | 4 ± 0.8* | 8 ± 3.7* | |
| BFU-E | 26 ± 7.4* | 3 ± 1.4* | 7 ± 1.8** | |
| CFU-GM | 26 ± 4.5** | 14 ± 3.9* | 18 ± 2.8** | |
| CFU-GEMM | 1 ± 1.4* | 0 ± 0.0* | 0 ± 0.0* | |
| Combination (20 µM HXR9 + 2.5 nM ch128.1Av) |
||||
| CFU-E | 3 ± 1.0** | 4 ± 2.2* | 4 ± 3.6* | |
| BFU-E | 3 ± 1.6** | 2 ± 2.7* | 4 ± 2.7** | |
| CFU-GM | 5 ± 2.3** | 8 ± 2.2** | 11 ± 0.6** | |
| CFU-GEMM | 0 ± 0.0* | 0 ± 0.5* | 1 ± 0.5* | |
CFU-E: colony forming units-erythroid; BFU-E: burst forming units-erythroid; CFU-GM: colony forming units-granulocyte/machrophage; CFU-GEMM: colony forming units-granulocyte/erythrocyte/macrophage/megakaryocyte. Data represents the mean of quadruplicates ± the standard deviation
p < 0.001,
p < 0.05 compared with control cells treated with diluent alone.
Discussion
The HOX genes are important in development as well as normal hematopoiesis. Disruption of normal HOX expression during blood cell development due to either translocations or irregular gene expression has been shown to lead to leukemogenesis (38). However, dysregulation of these genes has not been fully evaluated in malignant B cells. Therefore, we evaluated the expression levels of all 39 HOX genes in a panel of malignant B cells, including myeloma cells. We found that all of the cell lines tested showed high levels of HOXA6, HOXA3, HOXB4, and HOXC11. A previous study focusing on the HOXA genes showed that the malignant plasma cells in 9.4% (3 out of 32) of multiple myeloma patients tested showed abnormal HOXA gene expression (11). This study showed aberrant expression of HOXA4, HOXA7, and HOXA9 each in one patient. Our study confirms that the HOXA genes are abnormally expressed, however, the individual genes differ in each cell line tested. Our study shows aberrant expression of HOXA6 and HOXA3 in all cell lines tested. Other HOXA genes are highly expressed in certain cell lines but not all six. In addition, HOXB4 and HOXC11 showed high expression in all cell lines. HOXB4 is a positive regulator of hematopoietic stem cell renewal (39). Its expression is induced by the NF-Y transcription factor that regulates the transcription of numerous cell cycle control and proliferation-related genes (40). ZHX-2 is a negative regulator of NF-Y and its reduced expression has been shown to be associated with poor survival in MM patients (41). Therefore, it has been previously suggested that the loss of ZHX-2 leads to the upregulation of HOXB4 that allows myeloma cells to develop stem-cell like attributes and resistance to chemotherapy (41). Our results are consistent with this hypothesis and suggest that the targeting of the HOX genes (specifically HOXB4) may be a meaningful therapeutic option for aggressive, chemoresistant MM. The heterogeneity of expression between the cell lines (as shown in this report), as well as the heterogeneity of expression between our cell lines and cells previously isolated from patients (11), cannot be explained at this time. Although different HOX genes may be critical for malignant B-cell survival and their expression may vary from patient to patient, the fact that HXR9 targets multiple HOX genes suggests that HXR9 will be effective without regards to which HOX gene is aberrantly expressed.
Our data show that the panel of cell lines tested demonstrates variable sensitivities to HXR9. The most sensitive cell lines are IM-9 and ARH-77, which are both Epstein Barr Virus transformed. KMS-11 and U266 were the least sensitive, but not resistant to the cytotoxic effects of HXR9. HOXB5 was not expressed in the most sensitive cell lines, however, specific HOX genes associated with HXR9 sensitivity could not be identified. IM-9 cells treated with HXR9 showed very low caspase activation even when treated with 50 µM, a dose that induces a high level of cell death (Supplementary Data Figure S3). In addition, the caspase inhibitor Z-VAD only slightly rescued the cells from the effects of HXR9 alone. This suggests the possible involvement of other caspase-independent cell death pathways in HXR9-mediated cytotoxicity in malignant B cells. Caspase activation was higher in cells treated with the combination treatment compared to HXR9 alone, most likely due to the activation of caspases by ch128.1Av. This is corroborated by the fact that Z-VAD partially protected cells from the effects of the combination treatment indicating that the role of ch128.1Av in the mechanism of synergy is significant.
Interestingly, we have observed a slight increase in cytotoxicity when FAC was added to IM-9 cells in the presence of HXR9. Although further studies are needed to understand this phenomenon, iron-induced oxidative stress (42, 43) might reduce HOX protein expression and/or activity as described for ethanol-induced oxidative stress that decreases the expression of several HOX genes in certain neuronal cells (44). As shown previously the exogenous addition of iron through the addition of FAC was able to rescue cells from the cytotoxic effects induced by ch128.1Av alone (17, 22). Iron supplementation also partially protected IM-9 cells from the effects of the combination treatment suggesting that the iron-related stress induced by ch128.1Av plays a significant role in the synergy between the two compounds. The role of iron-induced stress by ch128.1Av was further confirmed by the semi-quantitative PCR analysis. Upregulation of TFRC, which encodes the TfR1, most likely occurs as a cellular response to low iron levels caused by TfR1 degradation that occurs with ch128.1Av treatment (17). Metallothionein 2A (MT2A) is involved in metal homeostasis and protection from oxidative stress (45) and its upregulation may also be in response to the iron deprivation induced by ch128.1Av (17, 22). The growth arrest and DNA-damage-inducible, alpha (GADD45α) protein is upregulated in response to many stress stimuli and drug therapies (46). Since iron is a key element in most of the cytochrome enzymes involved in the oxidative phosphorylation of the citric acid cycle (47), GADD45α, a p53 target gene (46), may be upregulated in IM-9 cells (p53 wildtype (48)) due to the metabolic stress that results from iron deprivation. Interestingly, the increase in GADD45α and MT2A levels observed with ch128.1A alone were not detected in cells treated with the combination treatment. HXR9 and ch128.1Av together in IM-9 cells induced the expression of TfRC (as did ch128.1Av alone) as well as FDXR and TM7SF2. FDXR, another p53 target gene, encodes for ferredoxin reductase and is a mitochondrial respiratory chain protein that has been shown to contribute to apoptosis induced by 5-fluorouracil through the induction of oxidative stress in the mitochondria (49). FDXR has also been shown to be upregulated in response to irradiation or UV treatment (50). The transmembrane 7 superfamily member 2 (TM7SF2, 3β-hydroxysterol Δ14-reductase) is localized to the endoplasmic reticulum and is involved in cholesterol biosynthesis (51). Its relation to iron or the induction of apoptosis is unknown, but its upregulation by the combination treatment may be a result of endoplasmic reticulum-induced stress that has not yet been elucidated. Although the above studies focus on IM-9 cells, we also found an enhancement of cytotoxicity with the combination of HXR9 and ch128.1Av in two MM cells lines (U266 and KMS-11), which both show lower sensitivities to either agent alone. Further studies are needed to evaluate the mechanism of this enhancement in these two cell lines. However, our data suggest the utility of using our approach to for the treatment of resistant B-cell malignancies, including MM.
With the evaluation of any potential therapeutic, the toxicity to normal cells is always a concern. Since the two agents in this study are being evaluated as potential treatments for hematopoietic malignancies, especially B-cell malignancies including MM, the effects on normal hematopoietic cells were evaluated. The colony forming assays show that HXR9 alone significantly inhibited hematopoietic progenitor colony formation. These effects were dose dependent and are consistent with previous data that showed decreased proliferation of CD133+ cells upon treatment with 60 µM HXR9 (12). Although this suggests that a transient myelosuppression may result in vivo in response to HXR9 treatement, no toxicities were observed in previous studies in mice that were treated daily with 15 mg/kg doses of HXR9 for 10 days, conditions where anti-tumor effects were also observed (12). Blood counts in these mice were within the expected ranges and no toxicity to the liver was observed. Since HXR9 is a small peptide it is expected that its half-life in vivo would be very short due to the rapid clearance through the kidneys. This may be an advantage since any myelosupression that may occur would be very short in duration or possibly non-existent.
Significant toxicity to human progenitor cells was also observed with the treatment of 2.5 nM ch128.1Av. These results are consistent with a previous report that showed toxicity of the antibody-fusion protein using this assay (19). As discussed in a previous report (22), early hematopoietic progenitor cells express low to no TfR1 (52–55). As these early progenitors differentiate into late progenitor cells they acquire TfR1 expression (52, 54). Thus, if the BMMC are treated continuously with ch128.1Av for 14 days, it is expected that the toxicity would be greater due to the increase in TfR1 expression as the cells differentiate into late progenitor cells. If the BMMC are treated with ch128.1Av for a short period prior to differentiation (1 hour in liquid culture), and then allowed to grow for 14 days in the semi-solid MethoCult medium, the effect on the early progenitor population could be evaluated and it was previously shown that under these conditions, ch128.1Av did not inhibit colony formation (22). In addition, no toxic effect on early progenitor cells was observed in the Long-Term Culture-Initiating Cell (LTC-IC) assay (Daniels et al., manuscript in preparation). Thus, the effect of ch128.1Av is on the late progenitor population, which could be repopulated by the early progenitors upon treatment cessation.
The present study shows dysregulation of the HOX genes through gene expression analysis of all 39 HOX genes in a panel of six malignant B-cell lines. This analysis validates the use of HOX proteins as potential targets for myeloma therapy. To this end, we disrupted HOX protein function through the use of a synthetic peptide (HXR9) that blocks the interaction of the HOX proteins with one of its co-transcription factors. Anti-cancer activity was observed in all cell lines tested, confirming that the HOX proteins can be successfully targeted for malignant B-cell therapy. This study also shows that the activity of HXR9 can be enhanced by its combination with ch128.1Av. Taken together, this study shows that the HOX proteins are important for malignant B-cell survival and inhibition of their function through the use of HXR9 alone or combined with ch128.1Av shows great utility as potential treatments for B-cell malignancies, including myeloma, in humans.
Supplementary Material
Acknowledgements
We would like to thank Dr. Lawrence Boise (University of Miami) for providing the KMS-11 cell line and Drs Kenneth Anderson and Darminder Chauhan (Harvard University) for providing the MM.1S cell line. This work was supported in part by NIH/NCI grants R01CA107023, K01CA138559, R01 supplement CA57152-13S1, and the training grant T32-CA009120. Our work was also supported by the Howard Hughes Medical Institute Gilliam Fellowship, the Whitcome Fellowship of the Molecular Biology Interdepartmental Ph.D. Program at UCLA, The Prostate Project (UK), and Cancer Research UK (C7822/A3832). The UCLA Jonsson Comprehensive Cancer Center and Center for AIDS Research Flow Cytometry Core Facility is supported by the NIH Awards CA-16042 and AI-28697, the Jonsson Cancer Center, the UCLA AIDS Institute, and the UCLA School of Medicine.
Footnotes
Supplementary information is available at the Leukemia website (www.nature.com/leu/index.html).
Conflicts of Interest
None of the authors have any conflicts of interest to disclose at this time.
References
- 1.Kyle RA, Rajkumar SV. Treatment of multiple myeloma: a comprehensive review. Clin Lymphoma Myeloma. 2009;9:278–288. doi: 10.3816/CLM.2009.n.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward Y, Hao JX, Thun MJ. Cancer Statistics. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Kumar SK, Rajkumar SV, Dispenzieri A, Lacy MQ, Hayman SR, Buadi FK, Zeldenrust SR, Dingli D, Russell SJ, Lust JA, Greipp PR, Kyle RA, Gertz MA. Improved survival in multiple myeloma and the impact of novel therapies. Blood. 2008;111:2516–2520. doi: 10.1182/blood-2007-10-116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palumbo A, Rajkumar SV. Treatment of newly diagnosed myeloma. Leukemia. 2009;23:449–456. doi: 10.1038/leu.2008.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veraksa A, Del Campo M, McGinnis W. Developmental patterning genes and their conserved functions: from model organisms to humans. Mol Genet Metab. 2000;69:85–100. doi: 10.1006/mgme.2000.2963. [DOI] [PubMed] [Google Scholar]
- 6.Grier DG, Thompson A, Kwasniewska A, McGonigle GJ, Halliday HL, Lappin TR. The pathophysiology of HOX genes and their role in cancer. J Pathol. 2005;205:154–171. doi: 10.1002/path.1710. [DOI] [PubMed] [Google Scholar]
- 7.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 8.Sitwala KV, Dandekar MN, Hess JL. HOX Proteins and Leukemia. Int J Clin Exp Pathol. 2008;1:461–474. [PMC free article] [PubMed] [Google Scholar]
- 9.Abramovich C, Pineault N, Ohta H, Humphries RK. Hox genes: from leukemia to hematopoietic stem cell expansion. Ann N Y Acad Sci. 2005;1044:109–116. doi: 10.1196/annals.1349.014. [DOI] [PubMed] [Google Scholar]
- 10.Bijl JJ, van Oostveen JW, Walboomers JM, Horstman A, van den Brule AJ, Willemze R, Meijer CJ. HOXC4, HOXC5, and HOXC6 expression in non-Hodgkin's lymphoma: preferential expression of the HOXC5 gene in primary cutaneous anaplastic T-cell and oro-gastrointestinal tract mucosa-associated B-cell lymphomas. Blood. 1997;90:4116–4125. [PubMed] [Google Scholar]
- 11.Hudlebusch HR, Lodahl M, Johnsen HE, Rasmussen T. Expression of HOXA genes in patients with multiple myeloma. Leuk Lymphoma. 2004;45:1215–1217. doi: 10.1080/10428190310001625836. [DOI] [PubMed] [Google Scholar]
- 12.Morgan R, Pirard PM, Shears L, Sohal J, Pettengell R, Pandha HS. Antagonism of HOX/PBX dimer formation blocks the in vivo proliferation of melanoma. Cancer Res. 2007;67:5806–5813. doi: 10.1158/0008-5472.CAN-06-4231. [DOI] [PubMed] [Google Scholar]
- 13.Chang CP, Shen WF, Rozenfeld S, Lawrence HJ, Largman C, Cleary ML. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 14.Plowright L, Harrington KJ, Pandha HS, Morgan R. HOX transcription factors are potential therapeutic targets in non-small-cell lung cancer (targeting HOX genes in lung cancer) Br J Cancer. 2009;100:470–475. doi: 10.1038/sj.bjc.6604857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shears L, Plowright L, Harrington K, Pandha HS, Morgan R. Disrupting the interaction between HOX and PBX causes necrotic and apoptotic cell death in the renal cancer lines CaKi-2 and 769-P. J Urol. 2008;180:2196–2201. doi: 10.1016/j.juro.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Ng PP, Dela Cruz JS, Sorour DN, Stinebaugh JM, Shin SU, Shin DS, Morrison SL, Penichet ML. An anti-transferrin receptor-avidin fusion protein exhibits both strong proapoptotic activity and the ability to deliver various molecules into cancer cells. Proc Natl Acad Sci U S A. 2002;99:10706–10711. doi: 10.1073/pnas.162362999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng PP, Helguera G, Daniels TR, Lomas SZ, Rodriguez JA, Schiller G, Bonavida B, Morrison SL, Penichet ML. Molecular events contributing to cell death in malignant human hematopoietic cells elicited by an IgG3-avidin fusion protein targeting the transferrin receptor. Blood. 2006;108:2745–2754. doi: 10.1182/blood-2006-04-020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez JA, Helguera G, Daniels TR, Neacato II, Lopez-Valdes HE, Charles AC, Penichet ML. Binding specificity and internalization properties of an antibody-avidin fusion protein targeting the human transferrin receptor. J Control Release. 2007;124:35–42. doi: 10.1016/j.jconrel.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 19.Ortiz-Sanchez E, Daniels TR, Helguera G, Martinez-Maza O, Bonavida B, Penichet ML. Enhanced cytotoxicity of an anti-transferrin receptor IgG3-avidin fusion protein in combination with gambogic acid against human malignant hematopoietic cells: functional relevance of iron, the receptor, and reactive oxygen species. Leukemia. 2009;23:59–70. doi: 10.1038/leu.2008.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniels TR, Delgado T, Rodriguez JA, Helguera G, Penichet ML. The transferrin receptor part I: Biology and targeting with cytotoxic antibodies for the treatment of cancer. Clin Immunol. 2006;121:144–158. doi: 10.1016/j.clim.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Daniels TR, Delgado T, Helguera G, Penichet ML. The transferrin receptor part II: targeted delivery of therapeutic agents into cancer cells. Clin Immunol. 2006;121:159–176. doi: 10.1016/j.clim.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Daniels TR, Ng PP, Delgado T, Lynch MR, Schiller G, Helguera G, Penichet ML. Conjugation of an anti transferrin receptor IgG3-avidin fusion protein with biotinylated saporin results in significant enhancement of its cytotoxicity against malignant hematopoietic cells. Mol Cancer Ther. 2007;6:2995–3008. doi: 10.1158/1535-7163.MCT-07-0330. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki E, Daniels TR, Helguera G, Penichet ML, Kazuo U, Bonavida B. Inhibition of NF-κB and Akt pathways by an antibody-avidin fusion protein sensitizes malignant B cells to cisplatin-induced apoptosis. Int J Onc. 2010;36:1299–1307. doi: 10.3892/ijo_00000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernandez-Luna JL, Nunez G, Dalton WS, Jove R. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 26.White S, Taetle R, Seligman PA, Rutherford M, Trowbridge IS. Combinations of anti-transferrin receptor monoclonal antibodies inhibit human tumor cell growth in vitro and in vivo: evidence for synergistic antiproliferative effects. Cancer Res. 1990;50:6295–6301. [PubMed] [Google Scholar]
- 27.Helguera G, Penichet ML. Antibody-cytokine fusion proteins for the therapy of cancer. Methods Mol Med. 2005;109:347–374. doi: 10.1385/1-59259-862-5:347. [DOI] [PubMed] [Google Scholar]
- 28.de Hoon MJ, Makita Y, Imoto S, Kobayashi K, Ogasawara N, Nakai K, Miyano S. Predicting gene regulation by sigma factors in Bacillus subtilis from genome-wide data. Bioinformatics. 2004;20(Suppl 1):101–108. doi: 10.1093/bioinformatics/bth927. [DOI] [PubMed] [Google Scholar]
- 29.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 30.Chou TC, Hayball MP. CalcuSyn: windows software for dose effect analysis. Cambridge, Unitied Kingdom: Biosoft; 1996. [Google Scholar]
- 31.Carrasco RA, Stamm NB, Patel BK. One-step cellular caspase-3/7 assay. Biotechniques. 2003;34:1064–1067. doi: 10.2144/03345dd02. [DOI] [PubMed] [Google Scholar]
- 32.Muckenthaler M, Richter A, Gunkel N, Riedel D, Polycarpou-Schwarz M, Hentze S, Falkenhahn M, Stremmel W, Ansorge W, Hentze MW. Relationships and distinctions in iron-regulatory networks responding to interrelated signals. Blood. 2003;101:3690–3698. doi: 10.1182/blood-2002-07-2140. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez M, Galy B, Dandekar T, Bengert P, Vainshtein Y, Stolte J, Muckenthaler MU, Hentze MW. Iron regulation and the cell cycle: identification of an iron-responsive element in the 3'-untranslated region of human cell division cycle 14A mRNA by a refined microarray-based screening strategy. J Biol Chem. 2006;281:22865–22874. doi: 10.1074/jbc.M603876200. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez M, Galy B, Muckenthaler MU, Hentze MW. Iron-regulatory proteins limit hypoxia-inducible factor-2alpha expression in iron deficiency. Nat Struct Mol Biol. 2007;14:420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- 35.Brooks JT, Elvidge GP, Glenny L, Gleadle JM, Liu C, Ragoussis J, Smith TG, Talbot NP, Winchester L, Maxwell PH, Robbins PA. Variations within oxygen-regulated gene expression in humans. J Appl Physiol. 2009;106:212–220. doi: 10.1152/japplphysiol.90578.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collins JF. Gene chip analyses reveal differential genetic responses to iron deficiency in rat duodenum and jejunum. Biol Res. 2006;39:25–37. [PMC free article] [PubMed] [Google Scholar]
- 37.Kang SY, Halvorsen OJ, Gravdal K, Bhattacharya N, Lee JM, Liu NW, Johnston BT, Johnston AB, Haukaas SA, Aamodt K, Yoo S, Akslen LA, Watnick RS. Prosaposin inhibits tumor metastasis via paracrine and endocrine stimulation of stromal p53 and Tsp-1. Proc Natl Acad Sci U S A. 2009;106:12115–12120. doi: 10.1073/pnas.0903120106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Oostveen J, Bijl J, Raaphorst F, Walboomers J, Meijer C. The role of homeobox genes in normal hematopoiesis and hematological malignancies. Leukemia. 1999;13:1675–1690. doi: 10.1038/sj.leu.2401562. [DOI] [PubMed] [Google Scholar]
- 39.Morgan R. Hox genes: a continuation of embryonic patterning? Trends Genet. 2006;22:67–69. doi: 10.1016/j.tig.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Zhu J, Giannola DM, Zhang Y, Rivera AJ, Emerson SG. NF-Y cooperates with USF1/2 to induce the hematopoietic expression of HOXB4. Blood. 2003;102:2420–2427. doi: 10.1182/blood-2003-01-0251. [DOI] [PubMed] [Google Scholar]
- 41.Harousseau JL, Shaughnessy J, Jr, Richardson P. Multiple myeloma. Hematology Am Soc Hematol Educ Program. 2004:237–256. doi: 10.1182/asheducation-2004.1.237. [DOI] [PubMed] [Google Scholar]
- 42.Fernaeus S, Land T. Increased iron-induced oxidative stress and toxicity in scrapie-infected neuroblastoma cells. Neurosci Lett. 2005;382:217–220. doi: 10.1016/j.neulet.2005.03.069. [DOI] [PubMed] [Google Scholar]
- 43.Ryter SW, Kim HP, Hoetzel A, Park JW, Nakahira K, Wang X, Choi AM. Mechanisms of cell death in oxidative stress. Antioxid Redox Signal. 2007;9:49–89. doi: 10.1089/ars.2007.9.49. [DOI] [PubMed] [Google Scholar]
- 44.Wentzel P, Eriksson UJ. Altered gene expression in neural crest cells exposed to ethanol in vitro. Brain Res. 2009;1305(Suppl):S50–S60. doi: 10.1016/j.brainres.2009.08.057. [DOI] [PubMed] [Google Scholar]
- 45.Andrews GK. Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem Pharmacol. 2000;59:95–104. doi: 10.1016/s0006-2952(99)00301-9. [DOI] [PubMed] [Google Scholar]
- 46.Siafakas AR, Richardson DR. Growth arrest and DNA damage-45 alpha (GADD45a) Int J Biochem Cell Biol. 2009;41:986–989. doi: 10.1016/j.biocel.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 47.Oexle H, Gnaiger E, Weiss G. Iron-dependent changes in cellular energy metabolism: influence on citric acid cycle and oxidative phosphorylation. Biochim Biophys Acta. 1999;1413:99–107. doi: 10.1016/s0005-2728(99)00088-2. [DOI] [PubMed] [Google Scholar]
- 48.Liu Q, Hilsenbeck S, Gazitt Y. Arsenic trioxide-induced apoptosis in myeloma cells: p53-dependent G1 or G2/M cell cycle arrest, activation of caspase-8 or caspase-9, and synergy with APO2/TRAIL. Blood. 2003;101:4078–4087. doi: 10.1182/blood-2002-10-3231. [DOI] [PubMed] [Google Scholar]
- 49.Hwang PM, Bunz F, Yu J, Rago C, Chan TA, Murphy MP, Kelso GF, Smith RA, Kinzler KW, Vogelstein B. Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nat Med. 2001;7:1111–1117. doi: 10.1038/nm1001-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okumura H, Ishii H, Pichiorri F, Croce CM, Mori M, Huebner K. Fragile gene product, Fhit, in oxidative and replicative stress responses. Cancer Sci. 2009;100:1145–1150. doi: 10.1111/j.1349-7006.2009.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennati AM, Castelli M, Della Fazia MA, Beccari T, Caruso D, Servillo G, Roberti R. Sterol dependent regulation of human TM7SF2 gene expression: role of the encoded 3beta-hydroxysterol Delta14-reductase in human cholesterol biosynthesis. Biochim Biophys Acta. 2006;1761:677–685. doi: 10.1016/j.bbalip.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 52.Lansdorp PM, Dragowska W. Long-term erythropoiesis from constant numbers of CD34+ cells in serum-free cultures initiated with highly purified progenitor cells from human bone marrow. J Exp Med. 1992;175:1501–1509. doi: 10.1084/jem.175.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gross S, Helm K, Gruntmeir JJ, Stillman WS, Pyatt DW, Irons RD. Characterization and phenotypic analysis of differentiating CD34+ human bone marrow cells in liquid culture. Eur J Haematol. 1997;59:318–326. doi: 10.1111/j.1600-0609.1997.tb01693.x. [DOI] [PubMed] [Google Scholar]
- 54.Andrews RG, Singer JW, Bernstein ID. Precursors of colony-forming cells in humans can be distinguished from colony-forming cells by expression of the CD33 and CD34 antigens and light scatter properties. J Exp Med. 1989;169:1721–1731. doi: 10.1084/jem.169.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bender JG, Unverzagt K, Walker DE, Lee W, Smith S, Williams S, Van Epps DE. Phenotypic analysis and characterization of CD34+ cells from normal human bone marrow, cord blood, peripheral blood, and mobilized peripheral blood from patients undergoing autologous stem cell transplantation. Clin Immunol Immunopathol. 1994;70:10–18. doi: 10.1006/clin.1994.1003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.