Abstract
Burkholderia species are free-living bacteria with a versatile metabolic lifestyle. The genome of B. fungorum LB400 is predicted to encode three different pathways for formaldehyde oxidation: an NAD-linked, glutathione (GSH)-independent formaldehyde dehydrogenase; an NAD-linked, GSH-dependent formaldehyde oxidation system; and a tetrahydromethanopterin-methanofuran-dependent formaldehyde oxidation system. The other Burkholderia species for which genome sequences are available, B. mallei, B. pseudomallei, and B. cepacia, are predicted to contain only the first two of these pathways. The roles of the three putative formaldehyde oxidation pathways in B. fungorum LB400 have been assessed via knockout mutations in each of these pathways, as well as in all combinations of knockouts. The resulting mutants have the expected loss of enzyme activities and exhibit defects of varying degrees of severity during growth on choline, a formaldehyde-producing substrate. Our data suggest that all three pathways are involved in formaldehyde detoxification and are functionally redundant under the tested conditions.
The lifestyle of free-living organisms involves many challenges related to both seasonal and sudden changes in nutrient supply, temperature, salinity, etc., and in some cases it appears that a correlation exists between the versatility of the lifestyle and the genome size (8, 30). Larger genome sizes are correlated not only with the variety of the functions encoded but also with redundancy (2, 29), which has been implicated as playing a significant role in the genetic robustness of organisms (11, 30). Two major types of functional redundancy are known: the presence of (multiple) gene paralogs with overlapping functions and the presence of nonhomologous biochemical pathways that fulfill similar functions. These two kinds of genetic redundancy have been extensively studied in eukaryotes, and currently, the second type of redundancy appears to play a major role in genetic robustness (11, 16, 35). Little is known about the role of functional redundancy in prokaryotes. The recent emergence of the complete genomic sequences for a number of free-living microbes permits approaches for addressing this question.
One of the important biochemical necessities in life is the ability to detoxify highly toxic aldehydes, intermediates of many biochemical pathways, of which formaldehyde is the most toxic (4, 9). Four different pathways for formaldehyde detoxification are known in bacteria, encoded by unrelated or distantly related genes. The best characterized is the pathway involving glutathione (GSH)-dependent NAD-linked formaldehyde dehydrogenase (GSH-FDH) and formyl-GSH hydrolase (FGH), which is found in both prokaryotes and eukaryotes (10, 14, 15, 17, 25, 27). An FDH that does not require GSH for its activity has been characterized from Pseudomonas putida (31), and homologs of this enzyme are also widespread (30). However, the physiological function of this enzyme has not been tested by mutation. A third pathway for formaldehyde detoxification involves the enzymes that are characteristic of the ribulose-monophosphate cycle methylotrophs: hexulose phosphate synthase (HPS) and hexulose phosphate isomerase (HPI) (1, 7). More recently it has been shown that these are also present in heterotrophs, and it has been suggested that they play a role in formaldehyde handling (37). The fourth pathway is carried out by “archaeal” enzymes and is linked to the archaeal cofactors tetrahydromethanopterin (H4MPT) and methanofuran (MFR) (6). This pathway has so far been identified only in methylotrophic bacteria and has been characterized in detail in one methylotroph, Methylobacterium extorquens AM1 (12, 20, 21, 23, 24, 32-34). Based on mutant analysis in this organism, 18 genes have been implicated in this pathway, 16 that have homologs in archaea and 2 that do not (5, 6, 12, 21). Some methylotrophs have multiple formaldehyde oxidation pathways, and at least in some cases these do not appear to be redundant (7).
In this work it is shown that Burkholderia fungorum LB400 contains three of the four known routes for formaldehyde oxidation, including the H4MPT-MFR-dependent pathway. These pathways have been shown to be nonhomologous and redundant under the conditions tested, and each is differentially involved in growth on choline, a formaldehyde-generating substrate.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. B. fungorum LB400 (3) was grown at 30°C in a minimal medium (13) supplemented with one of the following carbon sources: succinate (0.4% [wt/vol]), choline (0.2% [wt/vol]), and citrate (0.2% [wt/vol]). To test for growth on methylotrophic substrates, methanol (0.1 to 0.5% [vol/vol]) or methylamine (0.5% [wt/vol]) was added to the same minimal medium in agar plates. Growth on methane or methanethiol was tested under a substrate-air atmosphere (50:50 and 20:80, respectively). In addition, a number of other potential methylotrophic substrates were tested in agar plates at 10 to 20 mM concentrations, including dimethyl sulfide, dimethyl sulfoxide, betaine, sarcosine, and dimethylglycine. Escherichia coli strains were cultured at 37°C in Luria-Bertani medium (18). Antibiotics were supplied at the indicated concentrations (micrograms per milliliter): ampicillin (50), chloramphenicol (10), kanamycin (E. coli, 50; B. fungorum LB400, 20 on solid medium and 10 in liquid), and tetracycline (10).
TABLE 1.
B. fungorum LB400 strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| B. fungorum LB400 | Wild type | 3 |
| CM359.1 | ΔmtdB | This study |
| CM359K.1 | ΔmtdB::kan | This study |
| CM359-363.1 | ΔmtdB ΔflhA | This study |
| CM359-363K.1 | ΔmtdB ΔflhA::kan | This study |
| CM359-363-409K.1 | ΔmtdB ΔflhA ΔfdhA::kan | This study |
| CM359-409K.1 | ΔmtdB ΔfdhA::kan | This study |
| CM363.1 | ΔflhA | 19 |
| CM363K.1 | ΔflhA::kan | 19 |
| CM363-409K.1 | ΔflhA ΔfdhA::kan | This study |
| CM409K.1 | ΔfdhA::kan | This study |
| Plasmids | ||
| pCM157 | Cre expression plasmid | 19 |
| pCM184 | Allelic-exchange vector | 19 |
| pCM356 | pCR2.1 with mtdB upstream | This study |
| pCM357 | pCR2.1 with mtdB downstream | This study |
| pCM358 | pCM184 with mtdB downstream | This study |
| pCM359 | pCM358 with mtdB upstream | This study |
| pCM363 | Construct for ΔflhA::kan mutations | 19 |
| pCM406 | pCR2.1 with fdhA upstream | This study |
| pCM407 | pCR2.1 with fdhA downstream | This study |
| pCM408 | pCM184 with fdhA upstream | This study |
| pCM409 | pCM408 with fdhA downstream | This study |
| pCR2.1 | PCR cloning vector | Invitrogen |
Sequence analysis.
The draft genomic sequences of B. fungorum LB400 and Burkholderia mallei have been assessed at www.jgi.doe.gov/JGI_microbial/html/index.html. The genomic sequences of Burkholderia cepacia and Burkholderia pseudomallei have been assessed at http://www.sanger.ac.uk/Projects/B_cepacia. The sequence of Methylococcus capsulatus Bath has been assessed at www.tigr.org/tdb/mdb/mdbinprogress.html. The sequences of the archaeal genes for formaldehyde oxidation in M. extorquens AM1 are available at http://www.ncbi.nlm.nih.gov under accession numbers AF032114, AY117134, and AY093431. To search the genomes of the four available Burkholderia species for the presence of formaldehyde handling functions, BLAST engines available at the genome sites listed were used. The following amino acid sequences were used as queries: the GSH-linked FDH and FGH from Paracoccus denitrificans (GenBank accession numbers L36327 and U34346, respectively), the GSH-independent FDH from P. putida (GenBank accession number D21201), the HPS and HPI from Methylomonas aminofaciens (GenBank accession number AB026428), and all 18 polypeptides involved in H4MPT-MFR-linked formaldehyde oxidation from M. extorquens AM1 (GenBank accession numbers are listed in reference 5).
Mutant generation.
B. fungorum LB400 mutants bearing deletions in mtdB (H4MPT-dependent formaldehyde oxidation) and fdhA (GSH-independent formaldehyde oxidation) were generated using the recently designed broad-host-range cre-lox system for antibiotic marker recycling (19). The constructs for allelic exchange were created by PCR amplification of regions of approximately 0.5 kb immediately upstream and downstream of mtdB and fdhA. These PCR products were first sequenced to ensure that no mutations were introduced and then cloned into pCR2.1 (Invitrogen) to yield pCM356 and pCM357 (mtdB) and pCM406 and pCM407 (fdhA). The construct for obtaining ΔmtdB::kan mutants was generated by introducing the 0.6-kb SacII-SacI fragment from pCM357 into the same sites of pCM184 to produce pCM358, into the same site of which the 0.5-kb EcoRI fragment from pCM356 was introduced, resulting in pCM359. The construct for obtaining ΔfdhA::kan mutants was generated by introducing the 0.5-kb BamHI-NdeI fragment from pCM406 between the BglII and NdeI sites of pCM184 to produce pCM408, into the same sites of which the 0.6-kb SacII-SacI fragment from pCM407 was cloned, yielding pCM409. These donor plasmids were conjugated into B. fungorum LB400 by use of the E. coli helper strain S17-1 (28), and null mutants were identified as previously described (19). Double-crossover recombination events were confirmed by diagnostic PCR. To generate mutants deficient in more than one formaldehyde oxidation pathway, additional mutations were introduced as described above in unmarked deletion backgrounds generated by Cre-mediated recombination as described previously (19).
Enzyme assays.
The activities of GSH-dependent and GSH-independent FDHs were measured in the following reaction mixture: 50 mM Tris-HCl (pH 8.0), 1 mM NAD, 10 mM formaldehyde, and 2 mM GSH for the former. To discriminate between the two enzymes, they were visualized in isoelectrofocusing gels (PhastSystem; Amersham) by activity staining (data not shown). The activity staining mixture was the same as that described above with phenazine methosulfate (0.5 mM) and nitroblue tetrazolium (0.1 mM) added for color development. The activities of methylene-H4MPT dehydrogenase and methenyl-H4MPT cyclohydrolase were determined as described in references 32 and 23, respectively. Protein concentrations were determined by spectrophotometric assay (36).
RT-PCR analysis.
RNA was isolated from 3-ml cultures of B. fungorum LB400 grown on choline or citrate to an optical density at 600 nm of 0.4, with the Qiagen RNeasy kit, including the DNase I (Qiagen) treatment. The preparations were repeatedly treated with DNase I (ZymoResearch, Orange, Calif.) and further purified and concentrated using the ZymoResearch RNA purification kit. First-strand cDNA synthesis was performed using the reverse transcription-PCR (RT-PCR) kit (avian myeloblastosis virus) from Roche, from a primer (Fae1) complementary to the 3′ part of the fae genes, followed by 30 cycles of PCR at the annealing temperature of 55°C from Fae1 and Fae2 (complementary to the 5′ part of fae).
Phenotypic analyses of B. fungorum LB400 strains.
Growth of wild-type B. fungorum LB400 and the mutant strains was monitored on both solid and liquid media, with all phenotypic analyses performed in at least two replicates. Growth on solid medium was determined by comparing the sizes and rates of colony formation. Growth curves in minimal media were obtained using 30 ml of culture in 250-ml flasks shaken at 250 rpm. Inoculum was obtained by pelleting exponentially growing cultures by centrifugation and resuspending cells in fresh growth medium to an optical density at 600 nm of 0.1.
RESULTS
Multiple formaldehyde oxidation pathways are encoded in the genomes of Burkholderia species.
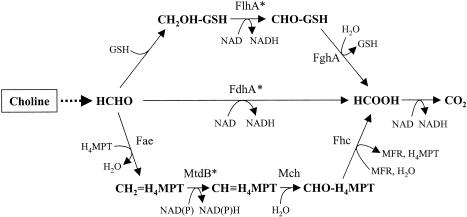
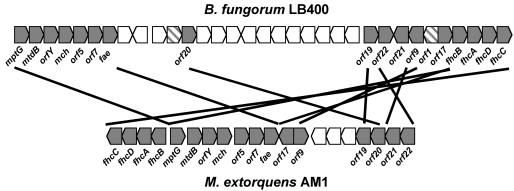
To assess the presence of genes encoding putative formaldehyde oxidation pathways in the four Burkholderia genomes, similarity searches with polypeptide query sequences indicative of each of the four known pathways were carried out as described in Materials and Methods. Putative genes for the GSH-linked pathway were identified in all four Burkholderia genomes by the high similarity of their encoded polypeptides with the queries. The two genes were found clustered together in all four genomes, in the order flhA-fghA. The gene for the GSH-independent FDH (fdhA) was similarly identified in all four genomes. None of the genomes contained the genes for HPS and HPI, and only one genome, of B. fungorum LB400, contained the genes for the archaeal pathway for formaldehyde oxidation (Fig. 1). Homologs of 17 of the 18 genes implicated in the archaeal formaldehyde oxidation pathway in M. extorquens AM1 (5) were detected in B. fungorum LB400, and as in M. extorquens AM1, they were found arranged in one cluster, with the gene order partially conserved between the two organisms (Fig. 2). One gene involved in the H4MPT-linked formaldehyde oxidation in M. extorquens AM1, dmrA, which is suggested to encode the final reaction in the biosynthesis of H4MPT (21), has no homolog in the B. fungorum LB400 genome. It is possible that this function could be encoded by a nonhomologous gene. The presence of the archaeon-like pathway in a bacterium not known to be a methylotroph prompted further study of its role in this bacterium.
FIG. 1.
Reactions of the three alternative pathways for formaldehyde oxidation in B. fungorum LB400. FlhA, GSH, NAD-linked FDH; FghA, FGH; FdhA, NAD-linked (GSH-independent) FDH; Fae, formaldehyde-activating enzyme; MtdB, NAD(P)-linked methylene-H4MPT dehydrogenase; Mch, methenyl-H4MPT cyclohydrolase; Fhc, formyltransferase-hydrolase complex. Genes subjected to mutation in this study are denoted by asterisks.
FIG. 2.
Alignment of the archaeal gene clusters from B. fungorum LB400 and M. extorquens AM1. Gray symbols connected by lines, gene homologs of archaeal nature; striped symbols, genes of B. fungorum LB400 having homologs in Archaea but not in M. extorquens AM1; open symbols, genes not considered in this study.
The three pathways for formaldehyde oxidation are functional in B. fungorum LB400.
The functionality of the three pathways potentially involved in formaldehyde oxidation was tested in B. fungorum LB400 by measuring specific enzyme activities characteristic of each pathway. As is known from previous work, formaldehyde detoxification pathways are often not expressed constitutively in bacteria but are induced in the presence of formaldehyde or formaldehyde-producing substrates (25, 37). Therefore, we searched the genome of B. fungorum LB400 for genes that potentially encode formaldehyde-producing enzymes and/or pathways. No genes predicted to encode homologs of typical methylotrophic formaldehyde-generating systems, such as methanol dehydrogenase, methylamine dehydrogenase, halomethane degradation enzymes, or methane oxidation systems, are present in the genome of B. fungorum LB400, and accordingly, growth tests on methane, methanol, or methylamine were negative. A number of other methylated substrates were also tested, with negative results. The only obvious formaldehyde-producing metabolic pathway deduced from the B. fungorum LB400 genome was the pathway for oxidation of choline, via betaine aldehyde, betaine, dimethylglycine, and sarcosine, which produces two formaldehydes per betaine or choline (http://www.expasy.ch/cgi-bin/searchbiochem-index). Therefore, we tested B. fungorum LB400 for growth on choline and betaine and observed strong growth on both substrates. Thus, we used a formaldehyde-producing substrate, choline, and a non-formaldehyde-producing substrate, citrate, to test for the enzyme activities in question. The GSH-independent FDH was present at high levels in choline-grown cultures, while no activity was detected in citrate-grown cultures (Table 2). In contrast, the GSH-dependent FDH was present at moderate levels in citrate-grown cultures (Table 2). In wild-type cells grown on choline, the GSH-linked activity was masked by the higher activity of GSH-independent FDH, but tests in the FDH-negative mutant (see below) indicated that GSH-FDH is present in choline-grown cells at slightly elevated levels. To test for the functionality of the archaeal pathway, we measured activities of two key enzymes in the pathway, methylene-H4MPT dehydrogenase (MtdB) and methenyl-H4MPT cyclohydrolase (Mch). Neither was detectable in citrate-grown cells, while both were present at low but significant levels in choline-grown cells (Table 2). In addition, we tested for the expression of a gene (fae) encoding another key enzyme of the archaeal pathway, formaldehyde-activating enzyme, which directly binds formaldehyde, via RT-PCR. fae-specific mRNA was detected in both choline-grown and citrate-grown cells but was more abundant in choline-grown cells (data not shown).
TABLE 2.
Enzyme activities measured in wild-type and mutant strains of B. fungorum LB400 grown on citrate and choline.
| Strain | Sp act (mU)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Citrate
|
Cholineb
|
|||||||
| FDHc | FDH(G)d | MtdB | Mch | FDHc | FDH(G)d | MtdB | Mch | |
| Wild type | <1 | 100 | <1 | <1 | 800 | 750 | 20 | 20 |
| flhA | <1 | <1 | NDe | <1 | 700 | 750 | ND | 22 |
| fdhA | <1 | 30 | ND | <1 | <1 | 200 | ND | 23 |
| mtdB | <1 | 45 | <1 | <1 | 650 | 700 | <1 | 25 |
| flhA fdhA | <1 | <1 | <1 | <1 | 25 | 25 | ND | 22 |
| flhA mtdB | <1 | <1 | ND | <1 | 660 | 590 | ND | 24 |
| mtdB fdhA | <1 | 45 | ND | <1 | <1 | 145 | ND | 23 |
| flhA fdhA mtdB | <1 | <1 | ND | <1 | <1 | <1 | ND | 25 |
Specific activities are in milliunits (nanomoles of substrate converted per minute per milligram of protein). Assays were carried out in triplicate, and activity values agreed between the replicates within 15%.
The triple mutant is severely defective for growth on choline. To test for enzyme activities normally induced during growth on choline, cells were grown in citrate-supplemented medium, pelleted, and incubated in the presence of choline overnight.
FDH assay with no GSH added (see Materials and Methods).
FDH assay with GSH added.
ND, not determined.
Mutant analysis suggests that all three pathways contribute to formaldehyde tolerance.
To test the potential physiological functions of the three formaldehyde oxidation pathways, mutants were generated that were defective in each of the pathways, defective in combinations of two different pathways, and defective in all three pathways. Deletions in the following genes were generated as described in Materials and Methods: fdhA to block the GSH-independent pathway, flhA to block the GSH-dependent pathway, and mtdB to block the archaeal pathway. All lesions were confirmed by diagnostic PCR (data not shown). As expected, the fdhA mutant lacked detectable GSH-independent FDH activity, and the mtdB mutant lacked detectable methylene-H4MPT dehydrogenase activity (Table 2). The flhA single mutant lacked the GSH-dependent activity in citrate-grown cells, but no change in activities in choline-grown cells could be detected due to masking by FdhA. However, in the flhA fdhA double mutant, the GSH-dependent activity was reduced to low levels compared to those in the fdhA single mutant, demonstrating loss of FlhA activity in that case (Table 2). A low-level GSH-independent activity was also detected in choline-grown cells of the double flhA fdhA mutant, suggesting the presence of a fourth, unknown aldehyde oxidation system.
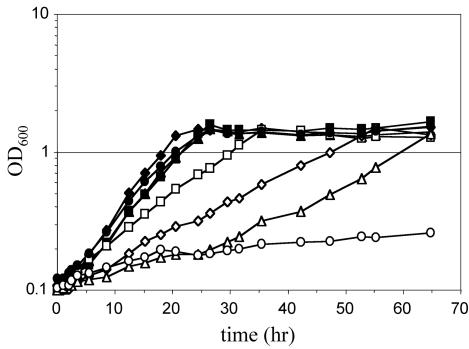
Growth characteristics of the mutants were tested on both citrate and choline and compared to those of the wild-type strain. All mutant strains grew on citrate at the same rate as did the wild-type strain (data not shown). Additionally, mutants defective in the GSH-linked pathway only or defective in the archaeal pathway only and the double mutant negative for both of these pathways displayed growth characteristics on choline similar to those of the wild-type strain (Fig. 3), suggesting that the GSH-independent FDH alone is sufficient to metabolize the formaldehyde produced during oxidation of choline. Growth of the mutant defective in the GSH-independent FDH was diminished compared to that of the wild-type strain, however, suggesting that this enzyme must play a significant role in formaldehyde oxidation. An even greater choline growth defect was observed for the double mutants lacking the GSH-independent FDH and either the archaeal pathway or the GSH-dependent FDH. These results indicate that these other two pathways can contribute to formaldehyde oxidation, but the contribution can be observed only in the absence of FDH. Finally, the mutant defective in all three formaldehyde oxidation systems exhibited the most dramatic growth defect on choline, resulting in almost complete arrest of growth.
FIG. 3.
Growth of B. fungorum LB400 strains on choline. The strains with GSH-independent FDH activity are shown by filled symbols: squares (wild type), diamonds (mtdB::Km), triangles (flhA::Km), and circles (mtdB flhA::Km). Strains lacking GSH-independent FDH activity are shown by open symbols: squares (fdhA), diamonds (mtdB fdhA::Km), triangles (flhA fdhA::Km), and circles (mtdB flhA fdhA::Km). OD600, optical density at 600 nm.
DISCUSSION
Functional annotation of bacterial genomes and metabolic reconstruction based on such annotations is difficult when redundant pathways and/or enzymes are present that are predicted to perform a similar function. In some cases, multiple pathways are differentially regulated and are not physiologically redundant (22), while in other cases true functional redundancy exists, at least under laboratory growth conditions (11, 26). One physiologically critical function that often involves redundancy is formaldehyde detoxification, and in this work, formaldehyde oxidation pathways were assessed in metabolically versatile free-living Burkholderia strains. Genomic analysis of four species of Burkholderia revealed a potential for multiple formaldehyde oxidation pathways in all the species. Further studies of one of these strains, B. fungorum LB400, demonstrated the functional presence of three of the four known formaldehyde oxidation systems: the GSH-linked pathway carried out via two specific enzymes, GSH- and NAD-linked FDH and FGH, encoded by the two genes tightly linked on the chromosomes of Burkholderia (flhA-fghA), the GSH-independent FDH pathway encoded by a single gene (fdhA), and the archaeal H4MPT-MFR-linked pathway involving several linked genes (Fig. 2).
The function of the GSH-linked pathway for formaldehyde detoxification has been addressed previously for both prokaryotes and eukaryotes (14, 17, 25), and in the case of P. denitrificans, a facultative methylotrophic autotroph, the pathway was demonstrated to play a role in growth not only on C1 compounds but also on choline (25). Therefore, a functional role of this pathway in choline utilization in B. fungorum LB400 was not surprising. However, the function of the GSH-independent FDH has not been tested by mutation in any strain, and we have confirmed a key role for this enzyme in formaldehyde detoxification. The predicted presence of this enzyme in all four Burkholderia species tested suggests that it may play a central role in formaldehyde metabolism in these bacteria.
In addition to the two pathways involving dehydrogenases of formaldehyde, an alternative, elaborate pathway is present in one of the Burkholderia species tested, B. fungorum LB400. It involves multiple C1 transfer reactions similar to those of methanogenesis, and it utilizes the archaeal cofactors H4MPT and MFR. Previously, the presence of this pathway has been reported only for methylotrophic proteobacteria. In methylotrophs, two distinct functions have been recognized for the pathway: a central role in energy generation during methylotrophic growth (6, 20, 21, 23, 32, 34) and an accessory role in formaldehyde detoxification (7, 20). However, in B. fungorum LB400 this pathway appears to play a minor detoxification role, at least during growth on choline. Since the attempts to discover a methylotrophic substrate for this strain were unsuccessful, it appears at this time to be a nonmethylotroph. The presence of the archaeal pathway in B. fungorum LB400 further expands the distribution of this pathway in the Proteobacteria. In methylotrophs, the enzyme activities of the H4MPT-MFR-linked pathway are at much higher levels than are those in B. fungorum LB400. Low expression of the archaeal pathway in B. fungorum LB400 is consistent with the proposed minor role in formaldehyde detoxification. However, it is possible that this pathway might play a more significant role in growth on a yet unknown formaldehyde-generating substrate.
One known gene required for the H4MPT-MFR-linked pathway in M. extorquens AM1 (dmrA) was not recognized in the genome. However, this gene is also missing from the genome of a gamma-proteobacterial methanotroph, Methylococcus capsulatus, suggesting that a functional alternative to dmrA must be present in this bacterium. One candidate for such a gene that shows homology to archaeal genes of unknown function is designated orf1 in Fig. 2. This gene is found in the archaeal-like gene cluster in both M. capsulatus and B. fungorum LB400 but not in M. extorquens AM1.
The results presented here suggest that the GSH-independent FDH alone is capable of fulfilling the necessary formaldehyde detoxification function for growth on choline independently of the presence of the two other systems. This correlates with the high activity of the FdhA during growth on choline, compared to the activities of FlhA and MtdB, enzymes indicative of the GSH-dependent and the archaeal pathways, respectively. In the absence of the GSH-independent pathway, however, the two other pathways take over the role, with the GSH-linked pathway being more effective than the H4MPT-MFR pathway in terms of supporting growth on choline. In the absence of all three pathways, B. fungorum LB400 is dramatically affected in its ability to use choline as a growth substrate. It is worth noting that even more redundancy must exist towards formaldehyde handling, as low FDH activity was detected in the triple mutant. This must be due to expression in this mutant of one or more other systems capable of formaldehyde oxidation at low levels.
Acknowledgments
We are grateful to R. K. Thauer for a generous gift of H4MPT and methenyl-H4MPT and to Rebecca Parales and David Gibson for providing B. fungorum LB400.
This work was supported by grants from the NIH (GM58933) to M.E.L. and from the NSF (MCB-0131957) to M.E.L. and L.C.
REFERENCES
- 1.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom.
- 2.Bansal, A. K. 1999. An automated comparative analysis of 17 complete microbial genomes. Bioinformatics 15:900-908. [DOI] [PubMed] [Google Scholar]
- 3.Bopp, L. H. 1986. Degradation of highly chlorinated PCBs by Pseudomonas strain LB400. J. Ind. Microbiol. 1:23-29. [Google Scholar]
- 4.Chang, C. C., and M. E. Gershwin. 1992. Perspectives on formaldehyde toxicity: separating fact from fantasy. Regul. Toxicol. Pharmacol. 16:150-160. [DOI] [PubMed] [Google Scholar]
- 5.Chistoserdova, L., S.-W. Chen, A. Lapidus, and M. E. Lidstrom. 2003. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chistoserdova, L., J. A. Vorholt, R. K. Thauer, and M E. Lidstrom. 1998. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic archaea. Science 281:99-102. [DOI] [PubMed] [Google Scholar]
- 7.Chistoserdova, L., L. Gomelsky, J. A. Vorholt, M. Gomelsky, Y. D. Tsygankov, and M. E. Lidstrom. 2000. Analysis of two formaldehyde oxidation pathways in Methylobacillus flagellatum KT, a ribulose monophosphate cycle methylotroph. Microbiology 146:3762-3769. [DOI] [PubMed] [Google Scholar]
- 8.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Baumer, C. Jacobi, H. Bruggemann, T. Lienard, A. Christmann, M. Bomeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453-461. [PubMed] [Google Scholar]
- 9.Feron, V. J., H. P. Til, F. de Vrijer, R. A. Woutersen, F. R. Cassee, and P. J. van Bladeren. 1991. Aldehydes: occurrence, carcinogenic potential, mechanism of action and risk assessment. Mutat. Res. 259:363-385. [DOI] [PubMed] [Google Scholar]
- 10.Giese, M., U. Bauer-Doranth, C. Langebartels, and H. Sandermann, Jr. 1994. Detoxification of formaldehyde by the spider plant (Chlorophytum comosum L.) and by soybean (Glycine max L.) cell-suspension cultures. Plant. Physiol. 4:1301-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu, Z., L. M. Steinmetz, X. Gu, C. Scharfe, R. W. Davis, and W. H. Li. 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421:63-66. [DOI] [PubMed] [Google Scholar]
- 12.Hagemeier, C. H., L. Chistoserdova, M. E. Lidstrom, R. K. Thauer, and J. A. Vorholt. 2000. Characterization of a second methylene tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1. Eur. J. Biochem. 267:3762-3769. [DOI] [PubMed] [Google Scholar]
- 13.Harder, W., M. Attwood, and J. R. Quayle. 1973. Methanol assimilation by Hyphomicrobium spp. J. Gen. Microbiol. 78:155-163. [Google Scholar]
- 14.Harms, N., J. Ras, W. N. Reijnders, R. J. van Spanning, and A. H. Stouthamer. 1996. S-Formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway for formaldehyde detoxification? J. Bacteriol. 178:6296-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoog, J. O., J. J. Hedberg, P. Stromberg, and S. Svensson. 2001. Mammalian alcohol dehydrogenase—functional and structural implications. J. Biomed. Sci. 8:71-76. [DOI] [PubMed] [Google Scholar]
- 16.Kitami, T., and J. H. Nadeau. 2002. Biochemical networking contributes more to genetic buffering in human and mouse metabolic pathways than does gene duplication. Nat. Genet. 32:191-194. [DOI] [PubMed] [Google Scholar]
- 17.Lee, B., H. Yurimoto, Y. Sakai, and N. Kato. 2002. Physiological role of the glutathione-dependent formaldehyde dehydrogenase in the methylotrophic yeast Candida boidinii. Microbiology 148:2697-2704. [DOI] [PubMed] [Google Scholar]
- 18.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Marx, C. J., and M. E. Lidstrom. 2002. A broad-host-range cre-lox system for antibiotic marker recycling in Gram-negative bacteria. BioTechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 20.Marx, C. J., L. Chistoserdova, and M. E. Lidstrom. 2003. Formaldehyde-detoxifying role of the tetrahydromethanopterin-linked pathway in Methylobacterium extorquens AM1. J. Bacteriol. 185:7160-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marx, C. J., B. N. O'Brien, J. Breezee, and M. E. Lidstrom. 2003. Novel methylotrophy genes of Methylobacterium extorquens AM1 identified by using transposon mutagenesis including a putative dihydromethanopterin reductase. J. Bacteriol. 185:669-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palacios, S., V. J. Starai, and J. C. Escalante-Semerena. 2003. Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol. J. Bacteriol. 185:2802-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pomper, B. K., J. A. Vorholt, L. Chistoserdova, M. E. Lidstrom, and R. K. Thauer. 1999. A methenyl tetrahydromethanopterin cyclohydrolase and a methenyl tetrahydrofolate cyclohydrolase in Methylobacterium extorquens AM1. Eur. J. Biochem. 261:475-480. [DOI] [PubMed] [Google Scholar]
- 24.Pomper, B. K., O. Saurel, A. Milon, and J. A. Vorholt. 2002. Generation of formate by the formyltransferase/hydrolase complex (Fhc) from Methylobacterium extorquens AM1. FEBS Lett. 17:133-137. [DOI] [PubMed] [Google Scholar]
- 25.Ras, J., P. W. Van Ophem, W. N. Reijnders, R. J. Van Spanning, J. A Duine, A. H. Stouthamer, and N. Harms. 1995. Isolation, sequencing, and mutagenesis of the gene encoding NAD- and glutathione-dependent formaldehyde dehydrogenase (GD-FALDH) from Paracoccus denitrificans, in which GD-FALDH is essential for methylotrophic growth. J. Bacteriol. 177:247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rizzitello, A. E., J. R. Harper, and T. J. Silhavy. 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J. Bacteriol. 183:6794-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanghani, P. C., H. Robinson, W. F. Bosron, and T. D. Hurley. 2002. Human glutathione-dependent formaldehyde dehydrogenase. Structures of apo, binary, and inhibitory ternary complexes. Biochemistry 41:10778-10786. [DOI] [PubMed] [Google Scholar]
- 28.Simon, R., U. Priefer, and A. Puhler. 1984. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 29.Snel, B., P. Bork, and M. A. Huynen. 1999. Genome phylogeny based on gene content. Nat. Genet. 21:108-110. [DOI] [PubMed] [Google Scholar]
- 30.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka, N., Y. Kusakabe, K. Ito, T. Yoshimoto, and K. T. Nakamura. 2002. Crystal structure of formaldehyde dehydrogenase from Pseudomonas putida: the structural origin of the tightly bound cofactor in nicotinoprotein dehydrogenases. J. Mol. Biol. 324:519-533. [DOI] [PubMed] [Google Scholar]
- 32.Vorholt, J. A., L. Chistoserdova, M. E. Lidstrom, and R. K. Thauer. 1998. The NADP-dependent methylene tetrahydromethanopterin dehydrogenase in Methylobacterium extorquens AM1. J. Bacteriol. 180:5351-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vorholt, J. A., L. Chistoserdova, S. M. Stolyar, R. K. Thauer, and M. E. Lidstrom. 1999. Distribution of tetrahydromethanopterin-dependent enzymes in methylotrophic bacteria and phylogeny of methenyl tetrahydromethanopterin cyclohydrolases. J. Bacteriol. 181:5750-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vorholt, J. A., C. J. Marx, M. E. Lidstrom, and R. K. Thauer. 2000. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J. Bacteriol. 182:6645-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner, A. 2000. Robustness against mutations in genetic networks of yeast. Nat. Genet. 24:355-361. [DOI] [PubMed] [Google Scholar]
- 36.Whitaker, J. R., and P. E. Granum. 1980. An absolute method for protein determination based on the difference in absorbance at 235 and 280 nm. Anal. Biochem. 109:156-159. [DOI] [PubMed] [Google Scholar]
- 37.Yasueda, H., Y. Kawahara, and S. Sugimoto. 1999. Bacillus subtilis yckG and yckF encode two key enzymes of the ribulose monophosphate pathway used by methylotrophs, and yckH is required for their expression. J. Bacteriol. 181:7154-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]