Abstract
The complex genome of Rhodobacter sphaeroides 2.4.1, composed of chromosomes I (CI) and II (CII), has been sequenced and assembled. We present data demonstrating that the R. sphaeroides genome possesses an extensive amount of exact DNA sequence duplication, 111 kb or ∼2.7% of the total chromosomal DNA. The chromosomal DNA sequence duplications were aligned to each other by using MUMmer. Frequency and size distribution analyses of the exact DNA duplications revealed that the interchromosomal duplications occurred prior to the intrachromosomal duplications. Most of the DNA sequence duplications in the R. sphaeroides genome occurred early in species history, whereas more recent sequence duplications are rarely found. To uncover the history of gene duplications in the R. sphaeroides genome, 44 gene duplications were sampled and then analyzed for DNA sequence similarity against orthologous DNA sequences. Phylogenetic analysis revealed that ∼80% of the total gene duplications examined displayed type A phylogenetic relationships; i.e., one copy of each member of a duplicate pair was more similar to its orthologue, found in a species closely related to R. sphaeroides, than to its duplicate, counterpart allele. The data reported here demonstrate that a massive level of gene duplications occurred prior to the origin of the R. sphaeroides 2.4.1 lineage. These findings lead to the conclusion that there is an ancient partnership between CI and CII of R. sphaeroides 2.4.1.
Rhodobacter sphaeroides 2.4.1, a purple nonsulfur photosynthetic eubacterium, belongs to the α-3 subgroup of the Proteobacteria (40, 41). This species, along with other members of the class Proteobacteria, represents one of the largest divisions within the prokaryotes (41) and comprises a large number of gram-negative bacteria. R. sphaeroides is also one of the most metabolically versatile and diverse subgroups of the α-3 subgroup of the Proteobacteria, which includes Rhizobium, Agrobacterium, Caulobacter, Brucella, and Rickettsia (40, 41). A few examples of the metabolic diversity are the diversity in assembly and regulation of the light-harvesting apparatus, in nitrogen fixation, in carbon dioxide fixation, in hydrogen metabolism, in electron transport, in oxyanion reduction, and in tetrapyrrole biosynthesis (9, 19).
R. sphaeroides 2.4.1 contains one of the most complex genomes found in members of the Proteobacteria (4, 18, 21). This species was the first bacterial species shown to possess a complex genome consisting of two circular chromosomes, one ∼3.0 Mbp long (chromosome I [CI]) and one ∼0.9 Mbp long (chromosome II [CII]), and five additional endogenous replicons (36, 37). The existence of multiple chromosomes in bacteria is now no longer an exception and has been instrumental in setting aside the long-held dogma that all bacterial species have a single circular chromosome.
Currently there is an extensive list of prokaryotic genomes that have been sequenced, including that of R. sphaeroides 2.4.1. As a result, R. sphaeroides is an ideal system for the study of genome complexity because the genome has been assembled and annotated (www.rhodobacter.org). CI and CII are 3,188,631 and 943,022 bp long, respectively, and contain approximately 3,106 and 874 open reading frames, respectively. Preliminary genome analyses (5, 6, 25, 26) have revealed that a wide variety of essential and housekeeping genes are present on both CI and CII.
Gene duplication followed by DNA sequence divergence plays a major role in genome evolution (33). Besides generating the genetic diversity that allows a species a wider spectrum of metabolic capabilities, gene duplication also contributes to the production of biodiversity by promoting genome divergence and further speciation events (24). The R. sphaeroides genome possesses a high degree of gene duplication (25, 26). Studies involving the genetic and biochemical characterization of a number of gene duplications in R. sphaeroides have been conducted previously (8, 13, 14, 28, 29, 30).
Duplications can arise from single-gene duplications, duplication of short chromosomal fragments, duplication of an entire chromosome, or duplication of the whole genome; these events are thought to be major sources of evolutionary novelties (33). In order to uncover both the nature and the amount of exact DNA sequence duplication within and between the two chromosomes, we aligned the CI and CII DNA sequences to each other and also against their own sequences. To examine the evolutionary history of gene duplications and to further understand the relationship between gene duplication events and the separation of the R. sphaeroides lineage from its ancestor, we compared the DNA sequences for each duplicate gene pair with orthologues from species and genera closely related to R. sphaeroides. On the basis of the inferred phylogeny of each set of duplicate genes and their orthologues, a relative age for CI and CII was derived.
MATERIALS AND METHODS
DNA duplication analysis.
The MUMmer 2.13 program (7) was used to identify the exact DNA duplications, tandem arrays, and small repeats present on the two chromosomes of the R. sphaeroides 2.4.1 genome. Plasmid sequences were excluded from this analysis. In order to analyze DNA sequence duplications, the assembled CI and CII sequences were run through the repeat match program to find all repeats within and between the two chromosomes. The MUMer output results show the coordinates of each pair of exact matches and the length of the match. The letter r attached to a number indicates that the sequence is on the reverse strand. All internal matches longer than 20 bp for CI-CII, 22 bp for CII, and 23 bp for CI were selected as cut off. We used 23 bp as the cutoff for CI because CI is larger than CII and a higher cutoff value removes some of the noise due to small repeats.
NUCmer was used to cluster all the matches found by MUMer and to construct larger regions of alignment. Clusters of these matches were subsequently grouped into larger sequence blocks. The NUCmer output file contains multiple columns that show several features of the sequence match, including the coordinates of the matching segments, the lengths of the matching segments, and the level of identity of a match between the two sequences.
Identification of gene duplications and orthologous sequences.
Gene duplications were identified as described previously (26), and similarity searches were carried out by using BLASTP (1). The amino acid sequences of the duplicated gene pairs were run through the databases to identify orthologues. The DNA sequences corresponding to the corresponding gene duplications and their orthologues were obtained from closely related species and genera, such as Rhodobacter capsulatus, Paracoccus denitrificans, Sinorhizobium meliloti, Bradyrhizobum japonicum, Agrobacterium tumefaciens, and Caulobacter crescentus, as these sequences are available in the National Center for Biotechnology Information database. The DNA and protein sequences of all duplicate gene pairs and their orthologues were saved in a local sequence file, and then these sequences were used for multiple alignment with CLUSTALW (38).
Phylogenetic tree construction.
Phylogenetic relationships were analyzed by using each pair of duplicated alleles and the orthologous DNA sequences from several species and genera closely related to R. sphaeroides. Phylogenetic and molecular analyses were conducted by using MEGA, version 2.1 (22). Phylogenetic tree construction was performed by using the neighbor-joining (NJ) method (35) because of its known accuracy. The distances between DNA sequences used for building the NJ tree were computed by using Jukes-Cantor corrections (17). The NJ method produced a unique final tree based on the assumption of minimum evolution with the correct tree topology. Bootstrap values for the consensus tree were calculated by using 1,000 replications.
RESULTS
Exact DNA duplication.
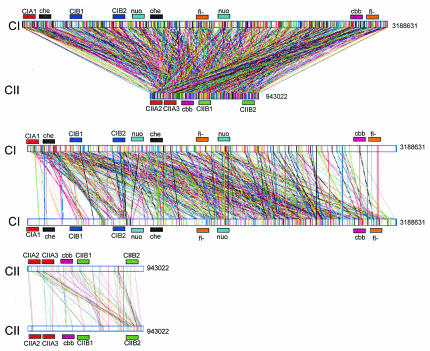
The DNA sequence analysis revealed an extensive amount of exact DNA duplication within the R. sphaeroides chromosomes, as shown in Fig. 1. Aside from four large duplicated segments, intrachromosomal duplications were less abundant than interchromosomal duplications.
FIG. 1.
Schematic representation of chromosomal duplications within and between CI and CII. CI and CII are depicted as horizontal bars from left (5′ DNA end) to right (3′ DNA end). Connecting vertical lines represent the locations on the chromosome(s) where the sequence matches perfectly. Genes involved in flagellum biosynthesis (fl-), electron transport (nuo), chemotaxis (che), and carbon assimilation (cbb) are duplicated as gene clusters, and they are indicated by different colors.
The chromosomal content duplicated within and between the two chromosomes is described in Table 1. The total amount of exactly duplicated sequences was 111.7 kb, representing ∼2.7% of the total chromosomal content. The amounts of CI-CI and CII-CII sequence duplications were ∼39 and ∼18 kb, respectively, which does not reflect the relative sizes of the chromosomes as CI is approximately three times larger than CII. The amount of interchromosomal sequence duplication was ∼55 kb. The degree of sequence duplication was identified by using a high stringency criterion for exact matches, a perfect 20-nucleotide match. The criterion used in this analysis was more stringent than the criterion used in analyses in which DNA sequences that are >100 nucleotides long with 50% mismatches are used. Thus, the criterion applied in this study provided only a conservative estimate of the amount of DNA sequence duplication.
TABLE 1.
DNA exact duplication in R. sphaeroides 2.4.1
| Chromosome | No. of nucleotides | No. of duplicated nucleotides (%)
|
|
|---|---|---|---|
| CI | CII | ||
| CI | 3,188,631 | 39,056 (1.22) | 54,967 (1.33) |
| CII | 943,022 | 54,967 (1.33) | 17,726 (1.88) |
| Total | 4,131,653 | 111,749 (2.70) | |
The frequency distribution of the sizes of the duplicated sequences is shown in Table 2. Of the 2,880 duplications, 1,509 (>50%) were interchromosomal. Of the intrachromosomal duplications, 1,034 and 337 were the CI-CI and CII-CII types, respectively, which is consistent with the relative chromosome sizes. Furthermore, the ratios of CI-CI DNA sequence duplications to CII-CII DNA sequence duplications for all small sizes (<200 nucleotides) were approximately 3:1. Large duplications (>0.5 kb) were rarely present, and small duplications ranging from 20 to100 nucleotides long were found to be the most prevalent duplications in the R. sphaeroides genome.
TABLE 2.
Frequency distribution of sequence duplications for CI and CII of R. sphaeroides 2.4.1
| Length of duplication (nucleotides) | No. of duplications
|
|||
|---|---|---|---|---|
| CI-CI | CII-CII | CI-CII | Total | |
| 20-25 | 346 | 112 | 1,082 | 1,540 |
| 26-50 | 530 | 170 | 317 | 1,017 |
| 51-100 | 130 | 44 | 83 | 257 |
| 101-200 | 24 | 9 | 13 | 46 |
| 201-500 | 3 | 0 | 8 | 11 |
| 501-1,000 | 1 | 1 | 2 | 4 |
| >1,000 | 0 | 1 | 4 | 5 |
| Total | 1,034 | 337 | 1,509 | 2,880 |
Assuming that mutations are random, older duplications should have accumulated more changes over time, and therefore there should be a higher frequency of identical duplications of shorter DNA sequences. The distribution of the number of duplications as a function of the length of the duplication is shown in Fig. 2 and in Table 2. Of the 1,509 interchromosomal duplications, 1,082 (∼72%) were 20 to 25 nucleotides long. In contrast, most of the intrachromosomal duplications were also small, but the most common duplications were 26 to 50 nucleotides long, suggesting that these duplications are more recent than the interchromosomal duplications.
FIG. 2.
Frequency distribution of the number of intra- and interchromosomal DNA sequence duplications. The two panels have different scales for the x and y axes.
Large duplicated regions.
The cluster analysis identified seven large duplicated regions on CI and CII, which were described as types A and B, as shown in Table 3 and Fig. 1. Three of the seven large duplicated regions are type A regions (CIA1, CIIA2, and CIIA3) that are ∼5.5 kb long and encode rRNA operons (rrnA, rrnB, and rrnC, respectively). This finding has been published previously (8).
TABLE 3.
Large duplicated regions of CI and CII
| Duplicated region | Coordinates on chromosome | Length (kb) |
|---|---|---|
| CIA1 (rrnA) | 0-5350 | 5.35 |
| CIB1 | 222609-253700 | 31.1 |
| CIB2 | 737289-762734 | 25.45 |
| CIIA2 (rrnC) | 0-5350 | 5.35 |
| CIIA3 (rrnB) | 33674-39022 | 5.35 |
| CIIB1 | 645366-673966 | 28.6 |
| CIIB2 | 922523-891191 reverse | 34.8 |
Four large duplicated regions, CIB1 and CIB2 in CIB and CIIB1 and CIIB2 in CIIB, are located on CI and CII, respectively. These four duplications are approximately 30 kb long and are located 391 and 128 kb apart on CI and CII, respectively. In relation to each other, the duplicated blocks present on CI, CIB1 and CIB2, are in the same orientation, whereas the duplicated blocks located on CII, CIIB1 and CIIB2, are in the reverse orientation. All four duplicated regions encode numerous phage-related proteins, such as integrase/recombinase, portal protein, phage tail and capsid protein, head-tail preconnector protein, DNA methyltransferase, and other phage proteins. In addition to the phage-related functions, each of these duplicated regions present on each chromosome also encodes non-phage-related proteins, such as dGTP triphosphohydrolase, transcriptional regulators, and transposases. Derived protein sequences for most of the corresponding genes in these duplicated regions show <50% amino acid identity to the corresponding homologues in the database (data not shown).
Phylogenetic relationship between duplicate gene copies and their orthologues.
In addition to a computational analysis performed by using the sizes and frequencies of exact DNA sequence duplications within and between CI and CII, an independent, phylogenetic analysis was used to infer the evolutionary origins of CI and CII. The DNA sequences of a number of duplicated gene pairs and their orthologous DNA sequences from closely related organisms were used for phylogenetic tree construction to determine which of the alternative gene sequences they most closely matched, the other sequence of the duplicate pair (type B tree) or an orthologous sequence (type A tree). Two types of phylogenetic relationships, type A and type B, were expected based on the assumptions adopted from a previous study (23). The derived amino acid similarity for each duplicate gene pair (homologue) and the similarity to the orthologous sequence are shown in Table 4. The species with which a copy of the duplicate gene showed the best match is also listed. The tree topology and the bootstrap value for each consensus tree are indicated in Table 4. It was found that the bootstrap values varied for different gene trees. In general, the bootstrap value is a function of the sequence length and the divergence time for the two DNA sequences, which in this case was strongly affected by the timing of gene duplication.
TABLE 4.
Similarity analysis of gene paralogues and orthologues
| Duplicated gene(s) | No. of copies in R. sphaeroides | Function | % Identitya | Close match
|
Treec
|
||
|---|---|---|---|---|---|---|---|
| Organism | % Identityb | Type | Bootstrap value | ||||
| CI-CI | |||||||
| pucB1/pucB2 | 2 | Light entrapment | 100 | Rhodovulum sulfidophilum | 80 | B | 94 |
| pucA1/pucA2 | 2 | Light entrapment | 58 | R. sulfidophilum | 54 | A | 55 |
| dxsI/dxsII | 2 | Isoprenoid synthesis | 66 | Rhodobacter capsulatus | 75 | A | 100 |
| flgG | 2 | Flagellum biosynthesis | 42 | Caulobacter crescentus | 47 | A | 82 |
| flgI | 2 | Flagellum biosynthesis | 42 | C. crescentus | 50 | A | 43 |
| flhA | 2 | Flagellum biosynthesis | 32 | C. crescentus | 32 | A | 100 |
| fliI | 2 | Flagellum biosynthesis | 36 | C. crescentus | 44 | A | 76 |
| fliP | 2 | Flagellum biosynthesis | 39 | Sinorhizobium meliloti | 40 | A | 57 |
| fliQ | 2 | Flagellum biosynthesis | 47 | S. meliloti | 40 | B | 62 |
| fliR | 2 | Flagellum biosynthesis | 33 | S. meliloti | 27 | A | 42 |
| fliN | 2 | Flagellum biosynthesis | 35 | C. crescentus | 37 | A | 79 |
| fliF | 2 | Flagellum biosynthesis | 30 | C. crescentus | 30 | B | 73 |
| flhB | 2 | Flagellum biosynthesis | 33 | C. crescentus | 32 | B | 98 |
| flgH | 2 | Flagellum biosynthesis | 30 | C. crescentus | 42 | A | 100 |
| flgF | 2 | Flagellum biosynthesis | 33 | C. crescentus | 42 | B | 57 |
| flgE | 2 | Flagellum biosynthesis | 25 | S. meliloti | 29 | A | 90 |
| flgC | 2 | Flagellum biosynthesis | 28 | Bradyrhizobium japonicum | 39 | A | 100 |
| flgB | 2 | Flagellum biosynthesis | 31 | Pseudomonas aeruginosa | 46 | B | 57 |
| nuoA | 2 | Electron transport protein | 36 | R. capsulatus | 85 | A | 85 |
| nuoB | 2 | Electron transport protein | 53 | R. capsulatus | 88 | A | 80 |
| nuoD | 2 | Electron transport protein | 42 | R. capsulatus | 83 | A | 98 |
| nuoF | 2 | Electron transport protein | 38 | R. capsulatus | 88 | A | 99 |
| nuoH | 2 | Electron transport protein | 42 | R. capsulatus | 82 | A | 100 |
| nuoI | 2 | Electron transport protein | 43 | R. capsulatus | 88 | A | 96 |
| nuoL | 2 | Electron transport protein | 30 | R. capsulatus | 33 | A | 98 |
| fnrL | 2 | Anaerobic regulator | 33 | R. capsulatus | 77 | A | 41 |
| rpoN | 2 | Sigma factor | 41 | R. capsulatus | 50 | A | 100 |
| hemN/Z | 4 | Coproporphyrinogen III oxidase | 24 | S. meliloti | 43 | B | 54 |
| cheA | 4 | Chemotaxis histidine kinase | 35 | C. crescentus | 47 | A | 99 |
| cheB | 2 | MCPd-glutamate methylesterase | 41 | C. crescentus | 45 | A | 70 |
| cheR | 3 | MCP-glutamate methyltransferase | 33 | C. crescentus | 48 | A | 100 |
| cheW | 4 | Chemotaxis protein | 33 | C. crescentus | 48 | A | 100 |
| CI-CII | |||||||
| rdxA/rdxB | 2 | Electron transport protein | 67 | R. capsulatus | 67 | A | 97 |
| qoxA | 2 | Electron transport protein | 45 | S. meliloti | 48 | A | 62 |
| qor | 2 | Electron transport protein | 31 | R. capsulatus | 54 | A | 100 |
| cbbGI/cbbGII | 2 | Carbon assimilation | 84 | Paraccocus denitrificans | 84 | B | 77 |
| hemA/hemT | 2 | ALAe synthase (tetrapyrrole biosynthesis) | 54 | R. capsulatus | 76 | A | 100 |
| cbbTI/cbbTII | 2 | Carbon assimilation | 58 | R. capsulatus | 66 | A | 100 |
| cbbAI/cbbAII | 2 | Carbon assimilation | 78 | R. capsulatus | 89 | A | 78 |
| cbbFI/cbbFII | 2 | Carbon assimilation | 67 | R. capsulatus | 69 | A | 99 |
| cbbPI/cbbPII | 2 | Carbon assimilation | 86 | R. capsulatus | 86 | B | 71 |
| cbbMI/cbbMII | 2 | Carbon assimilation | 31 | R. capsulatus | 94 | A | 100 |
| groEL | 3 | HSP60 | 40 | R. capsulatus | 70 | A | 100 |
| groES | 2 | HSP10 | 35 | R. capsulatus | 84 | A | 99 |
Amino acid identity for duplicate pair.
Amino acid identity with orthologue.
Phylogenetic relationship.
MCP, membrane-spanning chemoreceptor proteins.
ALA, 5-aminolevulinic acid.
Four phylogenetic trees, two type A trees and two type B trees, are presented as examples in Fig. 3. These gene phylogenies represent the data for rdxA/B, hemA/T, pucB1/pucB2, and flhB1/flhB2. Thirty-five (∼80%) of the 44 gene duplications shown in Table 4 represent type A relationships with the orthologous sequences. The inferred phylogeny from a type A tree shows that the duplicate alleles are less similar to each other than to an orthologous sequence from a species closely related to R. sphaeroides. In contrast, in type B phylogenetic relationships there is greater DNA sequence similarity between the duplicated alleles than between the alleles and their orthologues. Nine of 44 duplicated genes showed a type B phylogenetic relationship. These nine gene pairs were cbbGI/cbbGII, cbbPI/cbbPII, flgB1/flgB2, flgF1/flgF2, flhB1/flhB2, fliF1/fliF2, fliQ1/fliQ2, hemN/hemZ, and pucB1/pucB2. Three of these duplications, cbbGI/cbbGII, cbbPI/cbbPII, and pucB1/pucB2, also showed a high level of genetic identity (>80%) both between the duplicated alleles and with the orthologous sequences, as shown in Table 4.
FIG. 3.
Phylogenetic relationships of duplicated gene paralogues of R. sphaeroides and the orthologous sequences from closely related species or genera. As examples, consensus phylogenetic trees representing four gene pairs, rdxA/rdxB, hemA/hemT, pucB1/pucB2, and flhB1/flhB2, and their orthologous sequences are shown. The relationships reflect the two types of topology (type A and type B), and the strength of branching support is indicated by the bootstrap values at the nodes. Scale bars represent genetic distances.
DISCUSSION
In the last 5 years, many bacterial genomes have been completely or partially sequenced (www.jgi.doe.gov; www.tigr.org), and the sequences have been subjected to extensive analyses. Prokaryotic genome analyses have also revealed a >20-fold variation in genome size, with sizes ranging from approximately 0.6 to 13 Mb (10, 15). At present, the most commonly held view is that bacterial genome size increases through the transfer of genetic material (31, 32), gene duplication (2, 16), and duplication of transposable elements; however, these different genetic events are not mutually exclusive. It is very likely that bacteria spread and encounter different ecological niches; thus, their genome sizes can increase through the acquisition of habitat-relevant genes from other species and/or by duplication of genes in the preexisting genome that subsequently evolve and provide a new gene function. G+C composition can be a predictor of lateral gene transfer (32). Preliminary genome analysis of the R. sphaeroides genome revealed that the two chromosomes have nearly identical G+C contents (26). Also, both di- and trinucleotide repeats and codon preferences are shared by the two chromosomes (26). This remarkable similarity between the two chromosomes suggests that R. sphaeroides and other GC-rich organisms which have matching duplicated genes occupy similar ecological niches, which may reflect the selection of such genes by codon preferences. The unequal usage of synonymous codons is thought to have evolved due to natural selection to match the most abundant class of isoaccepting tRNA, resulting in increased translation efficiency. The ecological factors that help to maintain genome size are the selective pressures imposed by the need to develop greater physiologic specialization and/or diversity.
CI-CII duplications are older than CI-CI duplications.
Genomes of most prokaryotic (20) and eukaryotic (34) species examined to date show a high degree of gene duplication, which is an ongoing process. Recently, analysis of the Arabidopsis genome revealed that this genome contains extensive duplications (3) and has gone through several successive rounds of duplication (44) that resulted in different types of duplications. The recent duplications are the least altered in the present genome. The oldest duplications have undergone repeated modifications by a number of DNA-modifying events during evolution of the genome, leading to shortened remnants of the original duplicated sequence blocks. Therefore, an older duplication event results in a higher frequency of small stretches of perfect DNA sequence matches. In contrast, more recent duplication events result in perfect DNA sequence identity over longer lengths of the DNA since the duplications have had less opportunity to be modified.
The rarity of exact large duplications (>1 kb) in the R. sphaeroides 2.4.1 genome validates the assumption that most sequence duplications found in the genome are older duplications. Therefore, most large duplications within or between CI and CII occurred a long time ago, during the evolution and derivation of R. sphaeroides 2.4.1 as a species. The high frequency of the smallest duplications (20 to 25 nucleotides) between CI and CII suggests that a major interchromosomal duplication appeared as a single event, which occurred earlier than most of the intrachromosomal duplications. Hence, CI and CII have existed together over a very long period of evolutionary time.
Both chromosomes are integral to species formation.
Gene duplication is common in plants, animals, and microorganisms (27, 39, 42). Based on the inferred phylogeny of each set of gene duplications in several closely related species, the relative timing of these gene duplications can be estimated. It has been shown that the yeast genome duplication occurred as a single event before separation of the Saccharomyces cerevisiae lineage from its ancestor (23). Two possible phylogenetic trees, type A and type B, predict two different outcomes in time, describing gene duplication events prior to or after speciation. The relationship expected in the type A trees predicts that the gene duplication occurred before the formation of the R. sphaeroides 2.4.1 lineage. In contrast, the relationship shown in the type B trees indicates that the gene duplication occurred after separation of the R. sphaeroides 2.4.1 lineage from its ancestor.
Approximately 80% of all the gene duplications sampled showed a type A phylogenetic relationship. The other nine gene duplications displayed a type B relationship, as indicated in Table 4 (also see Fig. S1 in the supplemental material). If gene duplication occurred after species formation, the duplicated gene pair should exhibit a high level of genetic identity, unless the duplicate copies have diverged rapidly. cbbGI/cbbGII, cbbPI/cbbPII, and pucBA1/pucBA2 are reflective of a type B phylogenetic tree, and the duplicated protein sequences have >80% amino acid sequence identity. Six gene duplications, fliQ1/fliQ2, flgB1/flgB2, flgE1/flgE2, fliF1/fliF2, flhB1/flhB2, and hemN/hemZ, also displayed a type B phylogenetic relationship, but the levels of genetic identity between the amino acid sequences encoded by the corresponding duplicated alleles were lower (<50%). This result might have been possible if the gene duplications occurred after the formation of the new lineage, followed by rapid DNA sequence divergence.
To gain some quantitative insights into gene divergence, we can determine the bootstrap value. The bootstrap value signifies the phylogenetic topology (type A or type B), as indicated in Table 4; however, it might be affected by the timing of the gene duplication event. If gene duplication occurred long before speciation, the observed type A topology would have a relatively high bootstrap value (70 or 80). Similarly, there would be a high bootstrap value for the type B topology if gene duplication occurred long after speciation.
Thirty-four (77%) of the 44 gene duplications exhibited either a type A or type B phylogenetic relationship with a bootstrap value of >70. Ten of the gene duplications reflected either a type A or type B relationship with a bootstrap value of <70. If we exclude the 10 gene duplications with low bootstrap values, there are 34 definitive phylogenetic trees, and 29 (∼85%) of these trees exhibited a type A topology with a high bootstrap value (>70). Furthermore, 27 (60%) of the gene duplications exhibited a more definitive tree topology with a bootstrap value of >80, and 92% of these trees exhibited a type A topology. Therefore, the majority (80 to 92%) of the definitive and more robust phylogenetic trees had a type A topology, which suggests that a copy of the duplicate pair is more related to its orthologue than to its homologue. This indicates that these duplications are very old and likely occurred prior to the development of the R. sphaeroides lineage.
In summary, two different methods were used to decipher the evolutionary relationship of CI and CII. In the first method we analyzed the length and the frequency distribution of exact DNA sequence duplications in the R. sphaeroides 2.4.1 genome. In the other independent approach we performed a phylogenetic analysis of the duplicated gene pairs and orthologues from closely related species. Tree topology was used to predict the relative timing of intra- and interchromosomal gene duplications. The data from both analyses yielded similar interpretations, that interchromosomal DNA sequence duplications are older than intrachromosomal duplications. Therefore, CI and CII have existed together for a very long time, even before the appearance of R. sphaeroides or a distantly related species. Some of the duplicated genes present in the R. sphaeroides genome are also duplicated in the chromosome and the megaplasmid in other closely related genera, such as Sinorhizobium (data not shown). In contrast, many duplicate genes in R. sphaeroides exist as single copies in the genome of Brucella melitensis, which also possesses two chromosomes. In R. capsulatus, a species reported to be closely related to R. sphaeroides, gene duplications for a number of the gene loci described for R. sphaeroides are not observed. Therefore, the distributions of gene duplications in other related organisms appear to be independent from each other and from the distribution in R. sphaeroides, suggesting that the origins of the complex genomes were independent. However, a more detailed analysis is required.
On the basis of a phylogenetic analysis of several photosynthesis genes, the Proteobacteria emerged as the earliest lineage among the photosynthetic prokaryotes (43). However, phylogenies based on several genes from widely different metabolic pathways provide evidence that the cyanobacteria constitute one of the earliest prokaryotic lineages (11), having evolved about 2.5 billion years ago (12). If we subscribe to the hypothesis that the anaerobic photosynthetic bacteria existed prior to the oxygen-evolving cyanobacteria, then the heterotrophic purple bacteria may have arisen before the cyanobacteria. If this is true, CI and CII have been together for an extended period of evolutionary time. Therefore, we concluded that CI and CII have been partners in the R. sphaeroides genome since it separated from its ancestral lineage.
Supplementary Material
Acknowledgments
This work was supported by Department of Energy grant DOE-ER 63232-1018220-0007203 to S.K. and by National Institutes of Health grant GM50428 to Y.F.
We thank Steven L. Salzberg, Institute of Genomic Research, for the MUMer analysis. We also thank Haipeng Li and Xiaoming Liu, Human Genetics Center, University of Texas School of Public Health, for providing help with the phylogenetic analysis.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Blanc, G., A. Barakat, R. Guyot, and M. Delseny. 2000. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12:1093-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casjens, S. 1998. The diverse and dynamic structures of bacterial genomes. Annu. Rev. Genet. 32:339-377. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary, M., C. Mackenzie, K. S. Nereng, E. Sodergren, G. M. Weinstock, and S. Kaplan. 1994. Multiple chromosomes in bacteria: structure and function of chromosome II of Rhodobacter sphaeroides 2.4.1T. J. Bacteriol. 176:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choudhary, M., C. Mackenzie, K. S. Nereng, E. Sodergren, G. M. Weinstock, and S. Kaplan. 1997. Low resolution sequencing of Rhodobacter sphaeroides 2.4.1T: chromosome II is a true chromosome. Microbiology 143:3085-3099. [DOI] [PubMed] [Google Scholar]
- 7.Delcher, A. L., S. Kasif, R. D. Fleischmann, J. Peterson, O. White, and S. L. Salzberg. 1999. Alignment of whole genomes. Nucleic Acids Res. 27:2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dryden, S., and S. Kaplan. 1990. Localization and structural analysis of the ribosomal RNA operons of Rhodobacter sphaeroides. Nucleic Acids Res. 18:7267-7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gest, H. 1972. Energy conservation and generation of reducing power in bacterial photosynthesis. Adv. Microb. Physiol. 7:243-282. [Google Scholar]
- 10.Graur, D., and W.-H. Li. 1999. Fundamentals of molecular evolution. Sinauer, Sunderland, Mass.
- 11.Gupta, R. S. 1998. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62:1435-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta, R. S., T. Mukhtar, and B. Singh. 1999. Evolutionary relationships among photosynthetic prokaryotes (Heliobacterium chlorum, Chloroflexus aurantiacus, cyanobacteria, Chlorobium tepidum and proteobacteria): implication regarding the origin of photosynthesis. Mol. Microbiol. 32:893-906. [DOI] [PubMed] [Google Scholar]
- 13.Hallenbeck, P. L., R. Lerchen, P. Hessler, and S. Kaplan. 1990. Phosphoribulose kinase activity and the regulation of CO2 fixation critical for photosynthetic growth of Rhodobacter sphaeroides. J. Bacteriol. 172:1749-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallenbeck, P. L., R. Lerchen, P. Hessler, and S. Kaplan. 1990. The role of CFXA, CFXB, and the external electron acceptors in the regulation of ribulose 1,5-bis phosphate carboxylase/oxygenase expression in Rhodobacter sphaeroides. J. Bacteriol. 172:1736-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herdman, M. 1985. The evolution of bacterial genomes, p. 37-68. In T. Cavalier-Smith (ed.), The evolution of genome size. John Wiley and Sons, Chichester, United Kingdom.
- 16.Jordan, I. K., K. S. Makarova, J. L. Spouge, Y. I. Wolf, and E. V. Koonin. 2001. Lineage-specific gene expansions in bacterial and archaeal genomes. Genome Res. 11:555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, New York, N.Y. [Google Scholar]
- 18.Jumas-Bilak, E., S. Michaux-Charachon, G. Bourg, M. Ramuz, and A. Allardet-Servent. 1998. Unconventional genomic organization in the alpha subgroup of the Proteobacteria. J. Bacteriol. 180:2749-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiley, P., and S. Kaplan. 1988. Molecular genetics of photosynthetic membrane biosynthesis in Rhodobacter sphaeroides. Microbiol. Rev. 52:50-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin, E. V., and M. Y. Galperin. 1997. Prokaryotic genomes: the emerging paradigm of genome-based microbiology. Curr. Opin. Genet. Dev. 7:757-763. [DOI] [PubMed] [Google Scholar]
- 21.Krawiec, S., and M. Riley. 1990. Organization of the bacterial chromosome. Microbiol. Rev. 54:502-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. Molecular Evolutionary Genetics Analysis software. Arizona State University, Tempe. [DOI] [PubMed]
- 23.Langkjaer, R., P. F. Cliften, M. Johnston, and J. Piskur. 2003. Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature 421:848-852. [DOI] [PubMed] [Google Scholar]
- 24.Lynch, M., and A. G. Force. 2000. Gene duplication and the origin of interspecific genomic incompatibility. Am. Nat. 156:590-605. [DOI] [PubMed] [Google Scholar]
- 25.Mackenzie, C., A. E. Simmon, and S. Kaplan. 1999. Multiple chromosomes in bacteria. The yin and yang of trp gene localization in Rhodobacter sphaeroides 2.4.1. Genetics 153:525-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackenzie, C., M. Choudhary, F. W. Larimer, P. F. Predki, S. Stilwagen, J. P. Armitage, R. D. Barber, T. J. Donohue, J. P. Hosler, J. E. Newman, J. P. Shapleigh, R. E. Sockett, J. Zeilstra-Ryalls, and S. Kaplan. 2001. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth. Res. 70:19-41. [DOI] [PubMed] [Google Scholar]
- 27.McLysaght, A., K. Hokamp, and K. H. Wolfe. 2002. Extensive genomic duplications during early chordate evolution. Nat. Genet. 31:200-204. [DOI] [PubMed] [Google Scholar]
- 28.Neidle, E., and S. Kaplan. 1992. Rhodobacter sphaeroides rdxA, a homolog of Rhizobium meliloti fixG, encodes a membrane protein which may bind cytoplasmic (4Fe-4S) clusters. J. Bacteriol. 74:6444-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neidle, E., and S. Kaplan. 1993. Expression of Rhodobacter sphaeroides hemA and hemT genes encoding two aminolevulininc acid synthase isozymes. J. Bacteriol. 175:2292-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neidle, E., and S. Kaplan. 1993. 5-Aminolevulinic acid availability and control of spectral complex formation in HemA and HemT mutants of Rhodobacter sphaeroides. J. Bacteriol. 175:2304-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 32.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 33.Ohno, S. 1970. Evolution by gene duplication. Springer-Verlag, Heidelberg, Germany
- 34.Rubin, G. M., M. D. Yandell, J. R. Wortman, G. L. Gabor Miklos, C. R. Nelson, I. K. Hariharan, M. E. Fortini, P. W. Li, R. Apweiler, W. Fleischmann, J. M. Cherry, S. Henikoff, M. P. Skupski, S. Misra, M. Ashburner, E. Birney, M. S. Boguski, T. Brody, P. Brokstein, S. E. Celniker, S. A. Chervitz, D. Coates, A. Cravchik, A. Gabrielian, R. F. Galle, W. M. Gelbart, R. A. George, L. S. Goldstein, F. Gong, P. Guan, N. L. Harris, B. A. Hay, R. A. Hoskins, J. Li, Z. Li, R. O. Hynes, S. J. Jones, P. M. Kuehl, B. Lemaitre, J. T. Littleton, D. K. Morrison, C. Mungall, P. H. O'Farrell, O. K. Pickeral, C. Shue, L. B. Vosshall, J. Zhang, Q. Zhao, X. H. Zheng, and S. Lewis. 2000. Comparative genomics of the eukaryotes. Science 287:2204-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. E vol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 36.Suwanto, A., and S. Kaplan. 1989. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: genome size, fragment identification, and gene localization. J. Bacteriol. 171:5840-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suwanto, A., and S. Kaplan. 1989. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: presence of two unique circular chromosomes. J. Bacteriol. 171:5850-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vision, T. J., D. G. Brown, and S. D. Tanksley. 2000. The origin of genomic duplication in Arabidopsis. Science 290:2114-2117. [DOI] [PubMed] [Google Scholar]
- 40.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woese, C. R., E. Stackebrandt, W. G. Weisburg, B. J. Paster, M. T. Madigan, C. R. M. Fowler, C. M. Hahn, P. Blanz, R. Gupta, K. H. Nealson, and G. E. Fox. 1984. The phylogeny of the purple bacteria: the alpha subdivision. Syst. Appl. Microbiol. 5:315-326. [DOI] [PubMed] [Google Scholar]
- 42.Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708-713. [DOI] [PubMed] [Google Scholar]
- 43.Xiong, Jin, W. M. Fischer, K. Inoue, M. Nakahara, and C. E. Bauer. 2000. Molecular evidence for the early evolution of photosynthesis. Science 289:1724-1730. [DOI] [PubMed] [Google Scholar]
- 44.Ziolkowaski, P. A., G. Blanc, and J. Sadowski. 2003. Structural divergence of chromosomal segments that arose from successive duplication events in the Arabidopsis genome. Nucleic Acids Res. 31:1339-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.