Abstract
Leptospira species colonize a significant proportion of rodent populations worldwide and produce life-threatening infections in accidental hosts, including humans. Complete genome sequencing of Leptospira interrogans serovar Copenhageni and comparative analysis with the available Leptospira interrogans serovar Lai genome reveal that despite overall genetic similarity there are significant structural differences, including a large chromosomal inversion and extensive variation in the number and distribution of insertion sequence elements. Genome sequence analysis elucidates many of the novel aspects of leptospiral physiology relating to energy metabolism, oxygen tolerance, two-component signal transduction systems, and mechanisms of pathogenesis. A broad array of transcriptional regulation proteins and two new families of afimbrial adhesins which contribute to host tissue colonization in the early steps of infection were identified. Differences in genes involved in the biosynthesis of lipopolysaccharide O side chains between the Copenhageni and Lai serovars were identified, offering an important starting point for the elucidation of the organism's complex polysaccharide surface antigens. Differences in adhesins and in lipopolysaccharide might be associated with the adaptation of serovars Copenhageni and Lai to different animal hosts. Hundreds of genes encoding surface-exposed lipoproteins and transmembrane outer membrane proteins were identified as candidates for development of vaccines for the prevention of leptospirosis.
The genus Leptospira comprises a heterogeneous group of pathogenic and saprophytic species belonging to the order Spirochaetales (32). Leptospires are thin, helically coiled, motile bacteria, serologically classified into hundreds of pathogenic serovars (14). Leptospiral serovar diversity results from structural heterogeneity in the carbohydrate component of lipopolysaccharides (7). Many serovars are adapted for specific mammalian reservoir hosts, which harbor the organisms in their renal tubules and shed them in their urine. Because of the large spectrum of animal species that serve as reservoirs, leptospirosis is considered to be the most widespread zoonotic disease (32). Transmission to humans occurs through contact with wild or domestic animals or exposure to contaminated soil or water. In the urban setting, L. interrogans serovars that colonize the brown rat (Rattus norvegicus) population are the predominant cause of disease in humans. Fever, chills, headache, and severe myalgias characterize the early phase of the disease. Progression to multi-organ system complications occurs in 5 to 15% of cases, with mortality rates of 5 to 40% (14, 15). Leptospirosis is also a major economic burden as a cause of disease in livestock and domestic animals (14). Environmental control measures are difficult to implement, and currently available inactivated whole-cell vaccines have a number of disadvantages (14, 32).
An initial description of the L. interrogans serovar Lai genome has recently been released (42). Here we report the related serovar Copenhageni genome sequence, contrast it with the serovar Lai sequence, and present a more complete analysis of how the sequence elucidates the unique physiology of this versatile spirochetal pathogen. It is anticipated that the availability of leptospiral genomic sequences will illuminate our understanding of the molecular mechanisms of leptospiral pathogenesis and facilitate the identification of novel vaccine candidates.
MATERIALS AND METHODS
The sequenced strain, Fiocruz L1-130, was isolated from a patient with severe leptospirosis during an epidemic burst in 1996 (1). Biochemical and serotyping analyses identified the isolate as L. interrogans serovar Copenhageni (1, 30). Frozen aliquots of the clinical isolate were thawed, cultured in albumin-Tween 80 media (14), and used to inoculate Golden Syrian hamsters. Virulent leptospires were reisolated from the kidney of a diseased hamster and passaged three times in culture media to provide sufficient material to prepare genomic DNA. The 50% lethal dose of the strain used for DNA preparations was 104 in the hamster infection model (34).
Library construction, sequencing, assembly, and finishing were carried out by Agronomical and Environmental Genomes (AEG), a research initiative funded by FAPESP (http://aeg.lbi.ic.unicamp.br), Instituto Butantan, and Fundação Instituto Oswaldo Cruz. Genome annotation and comparative genomics were done as previously described (46). Whole-genome sequence comparison was performed at the nucleotide level using the program MUMmer (8) with default values. Detection of potential surface-exposed integral membrane proteins was carried out by software developed especially to detect lipoproteins in spirochetes, and the results were combined with those from TMHMM (http://www.cbs.dtu.dk/services/TMHMM), Psort (http://psort.nibb.ac.jp), and signalP (http://www.cbs.dtu.dk/services/SignalP).
Description of developed software method for finding surface-exposed lipoproteins.
The specially developed software is a program designed to detect lipoproteins in spirochetes. Briefly, the program is based on a set of 28 experimentally confirmed lipoproteins in spirochetes and provides a set of somewhat loose rules for lipobox recognition in these organisms, as described by Haake (24). Based on Haake's description and rules, we created a program that uses the technique of weight matrices (12) to evaluate potential lipoboxes in the first 70 amino acids of every predicted coding sequence. Those that have a putative lipobox (positive score) are then evaluated in terms of their putative hydrophobic region composition and cleavage site position, resulting in a rescoring. Putative lipoproteins are taken as those that have a final score greater than or equal to three. Putative lipoproteins are classified as probable and possible, reflecting what is known about the occurrence of certain amino acids in the detected lipobox. Certain coding sequences get high scores but are nevertheless classified as possible because they contain amino acids not yet seen in experimentally confirmed spirochetal lipoproteins but, at the same time, not forbidden according to Haake's rules. The weight matrix that is at the heart of the program was derived from the described set of 28 lipoproteins (the training set). There is a manuscript in preparation describing this program and, once it is accepted for publication, the program will be available.
Nucleotide similarity between serovars Copenhageni and Lai.
Based on the MUMmer alignment of the genomes, we calculated that the average percent identity of the nucleotides is at least 95%. This number was obtained as follows. All MUMmer matches on the two main diagonals of the chromosome I alignment were added and then divided by its length. This number actually represents a lower bound of percent identity because MUMmer finds only exact matches of 20 bp or higher and therefore may miss additional alignments that would increase the total. A similar computation was done for chromosome II, and in that case only matches on the lower-left to upper-right main diagonal were used, since there are no inversions. In both computations, the percent identity came out as 95%.
The percent identity of pairs of ortholog genes calculated for the two serovars is 99%. The methodology was as follows. From a BLASTP of all against all (Lai times Copenhageni) and applying the bidirectional best-hit rule, we obtained pairs of predicted genes that are putative orthologs. BLASTN was applied to each of these pairs; the percent identity was retrieved from each alignment that included at least 80% coverage of each predicted coding sequence. The average was then computed from these alignments. The total number of pairs was 3,340.
Nucleotide sequence accession number.
The sequences have been deposited in GenBank under accession numbers AE016823 (chromosome I) and AE016824 (chromosome II).
RESULTS AND DISCUSSION
Genome features.
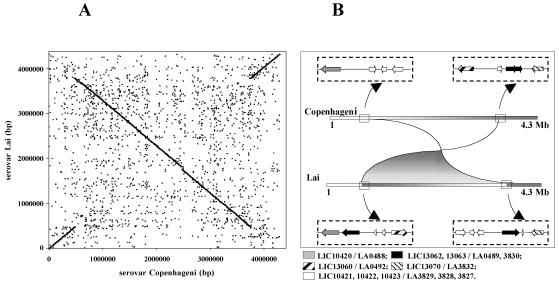
We have sequenced the genome of L. interrogans serovar Copenhageni, strain Fiocruz L1-130 (1). Table 1 summarizes the genome features of serovars Copenhageni and Lai. The average percent identity of the nucleotides between the two genomes is 95%. The average percent identity of the nucleotides between pairs of predicted protein coding genes that are orthologs is 99%. The numbers of ortholog pairs are 3,079 and 261 for CI and CII, respectively. The CII chromosomes in both serovars are colinear, but a large inversion exists in the CI chromosome (Fig. 1A). In the Lai genome, there are two identical copies of an insertion sequence (IS) element (LA0489 and LA3830) in opposite orientation flanking the inversion breakpoints (Fig. 1B). This finding suggests that the rearrangement took place in serovar Lai. We believe that the inversion is not an assembly artifact. In serovar Copenhageni, the assembly is unambiguously confirmed by 56 shotgun clones that span the inversion breakpoints and are anchored in nonrepetitive portions of the sequence. In addition, inversions around the replication origin have been observed in several other bacterial species (13). As previously described (19), rRNA genes in L. interrogans are not organized in operons, as in most other bacteria, but are scattered over the CI chromosome. Serovar Copenhageni has one rrf gene, two rrl genes, and two rrs genes encoding 5S, 23S, and 16S rRNA, respectively. One of the rrl copies is missing in serovar Lai, but a remnant is found in this serovar in the region corresponding to that where the extra copy appears in Copenhageni. From the description of the serovar Lai genome, it would appear that the serovar Copenhageni sequence has fewer putative genes than that of serovar Lai (Table 1). However, the difference in the number of structural genes occurred mainly because we did not consider predicted coding sequences less than or equal to 150 bp in length that lacked significant homologs. The serovar Lai genome has 718 predicted genes of this kind. Table 2 summarizes genes greater than or equal to 180 bp in length which are unique to serovars Copenhageni or Lai (see Table S1 in the supplemental material). Many of the genes unique to serovar Lai are located in a 54-kb insertion not found in serovar Copenhageni, containing 81 mostly hypothetical structural genes.
TABLE 1.
General features of the L. interrogans serovar Copenhageni and serovar Lai genomes
| Feature | Copenhageni
|
Lai
|
||
|---|---|---|---|---|
| CI | CII | CI | CII | |
| Size (bp) | 4,277,185 | 350,181 | 4,332,241 | 358,943 |
| G+C content (%) | 35.1 | 35.0 | 36.0 | 36.1 |
| Protein-coding genes | ||||
| With assigned function | 1,811 | 161 | 1,901 | 159 |
| Conserved and hypothetical | 1,643 | 113 | 2,459 | 208 |
| Total | 3,454 | 274 | 4,360 | 367 |
| Transfer RNA | 37 | 0 | 37 | 0 |
| Ribosomal RNA genes | ||||
| 23S | 2 | 0 | 1 | 0 |
| 16S | 2 | 0 | 2 | 0 |
| 5S | 1 | 0 | 1 | 0 |
| Insertion sequences | 26 | 0 | 48 | 9 |
| tmRNAa | 0 | 0 | 0 | 0 |
tmRNA, transfer-messenger RNA.
FIG. 1.
Inversion of L. interrogans serovars Copenhageni and Lai CI chromosomes. (A) Nucleotide alignment obtained by using MUMmer, which relies on exact matches of at least 20 bp. Each dot in the figure is one such match. The dark lines on the two main diagonals result from the high density of points with sequence identity along chromosome I of the two serovars. The scattered points outside the main diagonals represent other short regions of sequence identity. (B) Scheme showing predicted genes flanking the inversion breakpoints. Pairs of ortholog genes have the same pattern code. The black arrows represent IS elements.
TABLE 2.
Specific and extra-copy genesa
| Serovar | No. of genes |
|---|---|
| Copenhageni | |
| Hypothetical | 30 |
| Conserved hypothetical | 14 |
| Lipoproteins | 7 |
| Tautomerase | 1 |
| DegT proteins (extra copies) | 2 |
| Large cytoplasmic membrane-associated (extra copy) | 1 |
| Cytoplasmic membrane-associated | 1 |
| Transcriptional regulator, LexA family (extra copy) | 1 |
| Diguanilate phosphodiesterase | 1 |
| N-acetylglutamate synthase | 1 |
| Acryltransferase, GNAT family | 1 |
| RNA polymerase ECF-type sigma factor | 1 |
| Ribosomal RNA 23S (extra copy) | 1 |
| RNA binding protein | 1 |
| BigL/LigA (extra copy) | 1 |
| Lai | |
| Hypothetical | 108 |
| Conserved hypothetical | 3 |
| Lipoprotein | 1 |
| Aminoglycoside N6′-acetyltransferase | 1 |
| Methyl-accepting chemotaxis | 1 |
| Outer membrane | 1 |
| Transcriptional regulator, AraC family | 1 |
| Serine O-acetyltransferase | 1 |
| Acetyltransferase | 1 |
Hypothetical genes shorter than 60 amino acids as well as IS have been excluded. Some genes are specific, and some are extra copies.
Energy metabolism and transport.
Leptospira species utilize beta-oxidation of long-chain fatty acids as the major energy and carbon source (26) instead of the more common sugar oxidative pathways. As expected, a complete beta-oxidation route was found. Genes for glycerol metabolism are also present, including those encoding a glycerol-3-phosphate transporter, a glycerol uptake facilitator protein, glycerokinases, and a glycerol-3-phosphate dehydrogenase, suggesting that glycerol and fatty acids are obtained through phospholipid degradation.
Although under normal laboratory conditions L. interrogans cannot utilize glucose as a carbon and energy source, genome sequence and comparative analysis revealed that the glucose utilization pathway is complete, with one noticeable change: the presence of a pyrophosphate-fructose-6-phosphate 1-phosphotransferase instead of phosphofructokinase. We identified a glucose kinase (EC 2.7.1.2) that can replace hexokinase in the first step of the glycolytic pathway in both serovars Copenhageni (LIC12312) and Lai (LA1437). This finding is in contrast to what has recently been proposed by Ren et al. (42), who suggested that Leptospira species would not use the glycolytic pathway as a source of energy because they lack hexokinase. We note that Leptospira species have only one glucose uptake system, a glucose-sodium symporter (LIC12908) that is dependent on a sodium gradient across the bacterial membrane. In addition, no sugar ABC transporter was identified and an incomplete phosphoenolpyruvate-protein phosphotransferase system is present with no B or C sugar permease components. Since glucose uptake by the glucose-sodium symporter does not involve sugar phosphorylation, the role of the identified glucose kinase is critical for production of glucose-6-phosphate in Leptospira species, as opposed to other bacteria that import glucose via the phosphotransferase system. We suggest that the difficulties in utilization of glucose as an energy source under certain growth conditions may be the result of a limited uptake system rather than an incapacity to generate glucose-6-phosphate, as previously proposed (42).
Respiratory electron transport chain components are present, in agreement with the bacterium's oxygen requirements as an obligate aerobe. The cytochrome c oxidase subunit III is replaced by an electron transport protein that is most similar to cytochrome O ubiquinol oxidase subunit III of Escherichia coli, an enzyme that has lower O2 affinity than cytochrome c. Also, L. interrogans has four copies of genes encoding cytochrome c peroxidase (LIC10972, LIC10682, LIC12927, and LIC11088), which reduces hydrogen peroxide to water by using its c-type heme as an oxidizable substrate. However, since they possess two, instead of one, heme prosthetic groups, bacterial cytochrome c peroxidases reduce hydrogen peroxide without the need to generate semistable free radicals (20). Serovar Lai has the same four copies of cytochrome c peroxidase, although they had previously been identified either as putative lipoproteins or as methylamine utilization proteins (42). Thus, it is apparent that L. interrogans is able to use hydrogen peroxide as an alternative final acceptor of reducing power for respiration; in addition, this pathway may contribute to peroxide detoxification.
The generation of ATP is achieved via an F0F1-type ATPase that is encoded in a single operon, atpBEFHAGDC, which has the same organization as that of most of the eubacteria, in contrast to the that of the ATP synthases found in Borrelia burgdorferi and Treponema pallidum, which are of the V1V0-type (17, 18). Virtually all leptospiral ATP synthase subunits have the highest similarity to the sodium ion-specific ATP synthase of Propionigenium modestum that uses sodium ions instead of protons as the physiologic coupling ion (9).
Of the 3,731 predicted Leptospira species proteins, a total of 1,496 proteins (41%) were found to have at least one transmembrane segment, comparable to the relative number of transmembrane proteins predicted for T. pallidum (40%) and B. burgdorferi (42%). A total of 346 protein-coding genes (9.3%) were found to have four or more transmembrane segments, which is in the range found for most bacteria (6 to 13%) (39). Interestingly, there are 111 proteins having four or more transmembrane segments for which no function could be assigned (see Table S2 in the supplemental material), including one new Leptospira protein with 18 predicted transmembrane segments. It is known that between 40 and 60% of proteins with four or more transmembrane segments in the fully sequenced prokaryotes are related to transport (35).
DNA repair and oxygen tolerance.
Leptospires are exposed to UV light during the free-living stage of their life cycles. The ability to tolerate this radiation is probably facilitated by RecA and the SOS-inducible system of DNA repair, transcriptionally regulated by LexA. In addition, two identified photolyases, Phr and SplB, may mediate photoreactivation of DNA. Leptospires have a full complement of nucleotide excision repair proteins (UvrA, UvrB, UvrC, and UvrD, as well as Mfd) and enzymes linked to the repair of alkylated bases. In contrast, enzymes normally responsible for the repair of oxidative base damage, notably Fpg, Nfo, and Nei orthologs, are not found in the genome of L. interrogans. Exonuclease III orthologs, ExoA, Nth, and other enzymes with nuclease function may help the cells repair oxidative base damage. While a catalase gene is present, superoxide dismutase (SOD) orthologs are absent, and two important regulons normally responsible for the defense against oxidative stress, SoxRS and OxyR, are also missing. These findings are difficult to interpret considering that the organism grows in both aerobic and microaerophilic environments. Although SOD is also absent in T. pallidum, a superoxide reductase has recently been described in that microaerophilic spirochete (29, 33), but no superoxide reductase ortholog was found in the Leptospira genome of both serovars Copenhageni and Lai. The lack of SOD would increase the leptospires' susceptibility to the oxidative burst generated by macrophages during the infection process. However, this susceptibility may be overcome by the presence of metalloporphyrins with SOD-like properties (47). Alternatively, leptospires might have an as-yet-unknown mechanism to cope with oxidative stress.
Four of the five genes of the Bacteroides fragilis BatI operon were found. The absence of these genes from a B. fragilis mutant resulted in reduced aerotolerance and impaired growth (45). In L. interrogans, they may provide an additional oxygen stress defense. Because two of these genes, batA and batB (LIC20040 and LIC20041), encode proteins having a von Willebrand factor type A domain, it was recently inferred that these proteins were involved in loss of hemostasis caused by L. interrogans during infection (42). While the von Willebrand factor type A domain is known to be distributed in more than 20 different human proteins and is implicated in the immune and hemostatic systems, cell adhesion, or matrix assembly and mediates ligand binding in other proteins (6, 22), its function in prokaryotes remains unclear.
Regulatory functions.
The leptospiral life cycle requires the ability to respond to a complex array of environmental conditions. Accordingly, signal transduction mechanisms are mediated by at least 79 genes encoding two-component sensor histidine kinase-response regulator proteins. These include fourteen sensor-regulator pairs, six probable operons encoding hybrid sensor-regulators, and one or more of the sensors or regulators. Sigma-70, sigma-28, and sigma-54 factors are present, along with 11 extra cytoplasmic function (ECF) sigma factors (36). Accordingly, there are 9 anti-sigma factors and 19 anti-sigma factor antagonists, though none are cotranscribed with a specific sigma factor of the ECF family. There are only three predicted sigma-54 activators, suggesting that nitrogen starvation is responsible for regulating a relatively small set of functions. Although there is no sigma-32 factor, there is an HrcA heat-inducible repressor, indicating that the heat shock response is mediated by a derepression mechanism. Cyclic nucleotides appear to have a major regulatory role in Leptospira species, given the finding of orthologs of adenylate-guanylate cyclases, phosphodiesterases, cyclic nucleotide-binding proteins, and genes encoding proteins of the GGDEF family of signal transduction proteins with diguanylate cyclase activity (21). GGDEF regulatory proteins are more commonly found in nonobligate parasitic bacteria than in obligate parasites, indicating their importance in responding to environmental signals. Two copies of genes encoding BolA-like proteins were identified in the L. interrogans genomes but not in the genomes of two other spirochetes, T. pallidum and B. burgdorferi. BolA has recently been linked to maintenance of cell shape in extreme conditions, such as starvation (43). BolA appears to function by regulating transcription levels of genes encoding beta-lactamases and penicillin-binding proteins, both of which are also present in L. interrogans genomes.
We identified 12 L. interrogans genes that encode proteins with 27- to 33-amino-acid ankyrin repeat domains. In eukaryotes, ankyrin repeat domains are believed to be involved in several functions, including intracellular signaling. Genes encoding ankyrin-like proteins have been found in bacterial genomes located in close proximity to genes involved in either nutrient acquisition or tolerance or resistance to antibiotics (10, 27).
Motility and chemotaxis.
Virulence mechanisms of pathogenic leptospires, such as motility and chemotaxis responses, enable them to penetrate host tissue barriers during infection (4, 42). The L. interrogans genomes of both Copenhageni and Lai appear to contain at least 79 motility-associated genes, including orthologs of gldA (designated bcrA in Lai), gldG, and gldF of Flavobacterium johnsoniae, which form ABC transporters required for gliding motility (28). Motility and chemotaxis genes are well conserved among the spirochetes L. interrogans, T. pallidum, and B. burgdorferi, and 42 genes were found to be common to all three. Here we report that the chemotaxis apparatus of L. interrogans is likely much more complex, as its genome contains approximately twice as many methyl-accepting chemotaxis proteins (MCP) as either T. pallidum or B. burgdorferi. We identified 11 MCPs in serovar Copenhageni, compared to 12 in serovar Lai. The reason for the higher number of motility-associated genes, notably the MCPs, is not clear, but it may be that it reflects the survival and adaptation of pathogenic Leptospira to a variety of environments and hosts either by differential expression or not.
Pathogenicity.
The multiorgan dissemination of leptospires is probably a result of rapid translocation across host cell monolayers (2). In addition to motility and chemotaxis, leptospiral invasion may be mediated by secretion of enzymes capable of degrading host cell membranes (42). Both serovars Copenhageni and Lai have five genes encoding secreted sphingomyelinase C-type hemolysins and one ortholog of phospholipase D. Orthologs of the Serpulina hyodysenteriae tlyABC hemolysins were also identified. Proteolytic degradation of extracellular matrix proteins may also facilitate invasion of host tissues. Genes encoding proteases, including a collagenase, a metalloprotease, and several thermolysin orthologs, were identified. Table 3 summarizes predicted genes potentially involved in pathogenesis. A great number of them appear to be exported to the leptospiral surface as lipoproteins, and the transport systems discussed below may constitute an important target to control pathogenicity.
TABLE 3.
Genes with a potential role in pathogenesis and virulence
| Putative gene | No. of genes |
|---|---|
| LPS | 23 |
| Motility and chemotaxis | 79 |
| Adhesion and/or surface protein | |
| Afimbrial adhesin | |
| Bacterial immunoglobulin-like domain | 3 |
| FG-GAP repeat domain | 3 |
| Hemagglutinin-like protein | 9 |
| Lipoprotein | 184 |
| Capsular polysaccharide and/or secreted | |
| exopolysaccharide | 33 |
| Host cell membrane degradation | |
| Sphingomyelinase C-type hemolysin | 5 |
| Phospholipase D ortholog | 1 |
| Hemolysin (tlyABC ortholog) | 3 |
| Protease | |
| Collagenase | 1 |
| Metalloprotease | 1 |
| Thermolysin | 1 |
| Ankyrin-like protein | 12 |
| Resistant to oxidative stress | 8 |
Secretion of extracellular hemolysins and enzymes presumably occurs by the type I and/or type II secretion systems. In addition to numerous ABC transporters and membrane fusion proteins, there are two orthologs of the type I secretion protein TolC. The main terminal branch of the general secretory pathway is represented in the leptospiral genome by the gspCDEFG operon, encoding the key components of the type II secretion system. As expected, the full complement of flagellar assembly components is present; however, there is no clear evidence for the related type III secretion system. The L. interrogans genome contains at least 263 predicted genes encoding potential surface-exposed integral membrane proteins, 250 of which were previously unknown. Export of membrane proteins to the leptospiral surface is mediated by proteins of the Sec pathway, combined with signal peptidase I (LepB), the lipoprotein biosynthesis pathway (Lgt, LspA, and Lnt), and the proteins involved in transport and incorporation of lipoproteins into the outer membrane (LolA, LolC, and LolD). The specificity of leptospiral lipoprotein signal peptidase (LspA) differs from that of most of its orthologs in gram-negative bacteria (24), a difference that is reflected in the lipoboxes of the 184 identified surface lipoproteins (see Table S3 in the supplemental material). Eighty-four outer membrane proteins with transmembrane domains were found, including the two TolC orthologs, two orthologs of outer membrane factor CzcC, and a number of TonB-dependent outer membrane proteins and porins (25).
Colonization factors.
Host tissue colonization is essential for disease establishment. Leptospira has two families of afimbrial adhesins not previously described for serovar Lai, which may contribute to the early steps of infection. The first family consists of three paralogous genes (ligA, ligB, and ligC) recently identified in L. interrogans and L. kirschneri (34), encoding proteins with bacterial immunoglobulin-like (Big) repeat domains. LigB and LigC contain 90-amino-acid Big repeats followed by unique carboxy terminal domains that may be involved in host-pathogen interactions. In addition, it was recently shown that LigA and LigB are expressed by low-passage, virulent strains but not by highly-passaged, culture-attenuated strains during infection in mammalian hosts (34, 37). Genome analysis of serovar Copenhageni reveals that ligA (LIC10465) and ligB (LIC10464) are organized in tandem and that ligC (LIC11022; LIC11021) contains a stop codon in frame that interrupts the gene, indicating that ligC is a pseudogene in serovar Copenhageni. In contrast, L. interrogans serovar Lai harbors integral copies of ligB and ligC genes but lacks a copy of the ligA gene. The differences in the lig genes between serovars Copenhageni and Lai may reflect adaptation to their distinct reservoir hosts, namely Rattus norvegicus in the case of serovar Copenhageni and Apodemus agrarius in the case of serovar Lai (14). Mechanisms of host adaptation are also likely to include modifications in surface polysaccharides.
We identified a second family of candidate leptospiral adhesins consisting of three integrin alpha-like proteins (LIC12259, LIC10021, and LIC13101), each containing seven FG-GAP repeats (44). These integrin alpha-like proteins are predicted to be integral membrane proteins, which would support their potential role in ligand-binding interactions on the leptospiral surface. These protein-coding genes are also present in serovar Lai (LA1499, LA0022, and LA3881), previously assigned as conserved hypothetical proteins (42). Among the fully sequenced spirochetes, integrin alpha-like proteins are unique to L. interrogans.
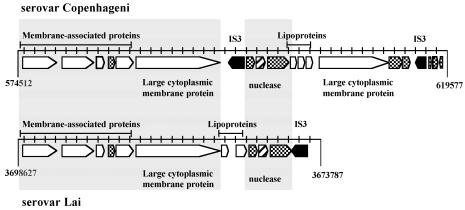
There is an interesting family of genes clustered in a region of ∼45 kb (position 574512 to 619577) encoding a high percentage (10 of 20 predicted genes) of membrane-associated proteins (Fig. 2). This region has several other notable features, including the presence of two distinct transposases related to the IS3 family, a nuclease gene similar to that for staphylococcal thermonuclease, lipoproteins with internal repeat sequences, and, finally, two unusually large genes (6.7 and 8.4 kb) encoding cytoplasmic-membrane proteins (LIC10502 and LIC10510; LIC10510 may be considered a nearly identical copy of LIC10502, even though the two genes differ in size). There is a similar region in serovar Lai that lacks one of the two genes encoding the large cytoplasmic-membrane proteins (Fig. 2). Both serovars contain yet another gene outside this region (LIC12896 and LA709) encoding a paralog of the large cytoplasmic-membrane protein containing an additional peptidase domain.
FIG. 2.
Schematic representation of a region with clusters of several predicted genes encoding membrane-associated proteins in the genome of serovar Copenhageni (top) and the equivalent region in the genome of serovar Lai (bottom). The open arrows represent genes encoding membrane-associated proteins, and the cross-hatched arrows represent hypothetical genes; the hatched arrow represents a predicted nuclease gene, and the filled arrows represent transposases unique to these regions. The large gray box encompasses identical equivalent regions in both serovars.
Surface polysaccharides.
Lipopolysaccharides (LPSs) distinguish the leptospiral surface from those of the other invasive spirochetes. The 40-kb rfb or O antigen biosynthesis gene locus of L. interrogans is likely to have been acquired through lateral transfer (1, 7). Changes in genes involved in the LPS polysaccharide biosynthesis apparatus are thought to account for serovar diversity among leptospires. Flexibility in leptospiral LPS biosynthesis is thought to be a mechanism of adaptation to new animal host species and probably accounts for the remarkable diversity of pathogenic leptospiral serovars (7, 14). Comparison of the rfb loci of serovars Copenhageni and Lai reveals only minor nucleotide discrepancies not reflected in the protein sequence. This finding is consistent with the fact that serovars Copenhageni and Lai belong to the same serogroup, icterohaemorrhagiae (32), but implies that antigenic differences between the serovars are due to genes outside the rfb locus. In this regard, it is of interest that the genome of serovar Copenhageni contains nine genes in the degT family compared to seven for serovar Lai. DegT proteins are involved in the biosynthesis of the LPS O side chain and may account for serological characteristics (23, 38). The two unique degT genes in serovar Copenhageni may determine the serovar difference between the Copenhageni and Lai LPS, reflecting evolutionary adaptation to different animal hosts. The genome sequence should facilitate elucidation of the complete leptospiral LPS structure, which will be essential for understanding why leptospiral LPS is several orders of magnitude less potent in the activation of macrophages than gram-negative endotoxin (48).
Although there is no experimental evidence that L. interrogans produces a capsule or forms a biofilm (14), a number of genes related to the biosynthesis of covalently-linked cell wall capsular polysaccharides (49) and secreted exopolysaccharides (31) were found in its genome. For example, several orthologs of algI, a gene which encodes the enzyme responsible for O acetylation of the mucoid exopolysaccharide alginate, were identified (16). The finding of genes related to biofilm formation may shed light on the mechanism of leptospiral colonization of the renal tubular epithelium surface in the reservoir host. Cell surface capsular polysaccharides and exopolysaccharides may also be important in the survival of pathogenic leptospires in the environment outside the host, protecting them from hydric and osmotic stresses (11).
ISs.
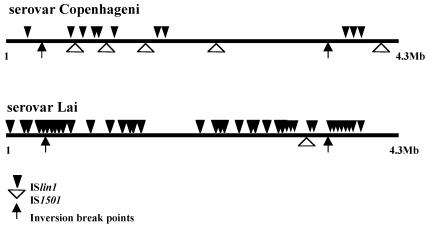
Bacterial ISs generate diversity in prokaryotic genomes through proliferation and movement from one insertion site to another. Our analysis of the Copenhageni and Lai genomes revealed the presence of the previously described IS elements IS1500, IS1501, IS1502, and IS1533 (3, 5, 51) and identified a new IS we designated ISlin1. The number and distribution of IS1501 and ISlin1 differed dramatically, highlighting the impact these elements have in Leptospira species strain genome differentiation. For the purpose of this analysis, we considered an integral copy of an IS element only when it had more than 95% nucleotide identity, with more than 90% coverage. The serovar Lai genome is subjected to more IS proliferation, since it has 57 highly conserved elements, while serovar Copenhageni has only 26 IS elements. Chromosome II of serovar Lai contains nine insertions of transposable elements (one copy of IS1500, two copies of IS1501, and six copies of ISlin1), while the Copenhageni CII replicon has none. Insertions on the Lai CII have no mutagenic effect at gene level, as all the insertions are found in intergenic regions. On the other hand, the genes corresponding to LIC10590, LIC10952, and LIC12901 in Copenhageni CI have been mutagenized by IS elements in Lai CI. Although some domains can be recognized, the precise function of the proteins encoded by these genes and the consequence of the disruption have not been determined. Of the previously described IS elements in Leptospira species, only IS1501 (gi2961088) is found as a completely conserved unit. This element has been reported to be able to transpose, as judged by the heterogeneity in copy number detected among different strains of L. interrogans (3). Differences in IS1501 genomic position and copy number in the two serovars reinforce its transposition activity, as serovar Copenhageni contains five copies found exclusively in CI, while serovar Lai presents three, one copy located in CI and two copies located in CII (Fig. 3). One of the copies in serovar Copenhageni is inserted in a highly repetitive 115-bp region within both the Copenhageni genome and other Leptospira species strains. IS1500 (3) is equally distributed in both genomes (eight copies), the only difference being that in serovar Lai one copy invaded the CII replicon, which has not previously been described. Reminiscent insertions of IS1533 (51) and IS1502 (52) are found at the same relative locations in both genomes.
FIG. 3.
Comparative distribution of IS1501 and ISlin1 on CI chromosome of L. interrogans serovars Copenhageni and Lai.
The newly identified IS element, named ISlin1, is the most polymorphic between the two genomes. Serovar Lai harbors 40 copies, and Copenhageni harbors only 11 copies of this element (Fig. 3). Transposable element activity is usually associated with a stress response (5). We propose that the differences observed in the IS distribution between these two genomes indicate that, although harboring similar genomes, these two serovars have been subjected to different environmental pressures that resulted in a particular pattern of IS amplification in each genome. From this analysis, it is possible to map old insertions common to both serovars which occurred prior to serovar specialization, such as the IS1533 and IS1502 insertion sites, and very recent IS activation, such as that of ISlin1, which resulted in a large genomic rearrangement in the Lai genome. Analyses of phage-related regions indicate that these genomes probably contain two ancient prophage insertions, with no major differences between the serovars.
Conclusion.
The comparative genomics of L. interrogans presented here reveals new insights into the serovar biosynthesis pathway, adaptation, colonization, and pathogenesis. A large number of exported lipoproteins and transmembrane outer membrane proteins which may be involved in leptospiral pathogenesis and protective immunity were identified. Currently available vaccines have low efficacy, are serovar-specific, and do not induce long-term protection against infection (14, 32). The large number of pathogenic serovars (>200) and the cost of producing a multiserovar vaccine have been the major limitations. Outer membrane proteins that are conserved among pathogenic serovars might be used in a recombinant vaccine without the limitations of currently available whole-cell preparations. It is anticipated that examination of these candidate protective immunogens will provide new approaches for vaccine development (40, 41, 50).
Additional information is contained in the project website: http://aeg.lbi.ic.unicamp.br/world/lic/.
Supplementary Material
Acknowledgments
We are deeply indebted to I. Raw (Fundação Butantan, São Paulo, Brazil) and M. G. Reis (Fiocruz, Bahia, Brazil) for use of laboratory facilities and valuable support. We thank all the technicians in the sequencing laboratories of the ONSA network and C. A. de Pian for his administrative coordination.
Project funding was from Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Barocchi, M. A., A. I. Ko, S. R. Ferrer, M. T. Faria, M. G. Reis, and L. W. Riley. 2001. Identification of new repetitive element in Leptospira interrogans serovar Copenhageni and its application to PCR-based differentiation of Leptospira serogroups. J. Clin. Microbiol. 39:191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barocchi, M. A., A. I. Ko, M. G. Reis, K. L. McDonald, and L. W. Riley. 2002. Rapid translocation of polarized MDCK cell monolayers by Leptospira interrogans, an invasive but nonintracellular pathogen. Infect. Immun. 70:6926-6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boursaux-Eude, C., I. Saint Girons, and R. Zuerner. 1995. IS1500, an IS3-like element from Leptospira interrogans. Microbiology 141:2165-2173. [DOI] [PubMed] [Google Scholar]
- 4.Charon, N. W., and S. F. Goldstein. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu. Rev. Genet. 36:47-73. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C. W., C. H. Huang, H. H. Lee, H. H. Tsai, and R. Kirby. 2002. Once the circle has been broken: dynamics and evolution of Streptomyces chromosomes. Trends Genet. 18:522-529. [DOI] [PubMed] [Google Scholar]
- 6.Colombatti, A., P. Bonaldo, and R. Doliana. 1993. Type A modules: interacting domains found in several non-fibrillar collagens and in other extracellular matrix proteins. Matrix 13:297-306. [DOI] [PubMed] [Google Scholar]
- 7.de la Pena-Moctezuma, A., D. M. Bulach, T. Kalambaheti, and B. Adler. 1999. Comparative analysis of the LPS biosynthetic loci of the genetic subtypes of serovar Hardjo: Leptospira interrogans subtype Hardjoprajitno and Leptospira borgpetersenii subtype Hardjobovis. FEMS Microbiol. Lett. 177:319-326. [DOI] [PubMed] [Google Scholar]
- 8.Delcher, A. L., S. Kasif, R. D. Fleischmann, J. Peterson, O. White, and S. L. Salzberg. 1999. Alignment of whole genomes. Nucleic Acids Res. 27:2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimroth, P., H. Wang, M. Grabe, and G. Oster. 1999. Energy transduction in the sodium F-ATPase of Propionigenium modestum. Proc. Natl. Acad. Sci. USA 96:4924-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolata, M. M., J. J. Van Beeumen, R. P. Ambler, T. E. Meyer, and M. A. Cusanovich. 1993. Nucleotide sequence of the heme subunit of flavocytochrome c from the purple phototrophic bacterium, Chromatium vinosum. A 2.6-kilobase pair DNA fragment contains two multiheme cytochromes, a flavoprotein, and a homolog of human ankyrin. J. Biol. Chem. 268:14426-14431. [PubMed] [Google Scholar]
- 11.Dunne, W. M., Jr. 2002. Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 15:155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durbin, R., S. R. Eddy, A. Krogh, and G. Mitchison. 1998. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge University Press, Cambridge, United Kingdom.
- 13.Eisen, J. A., J. F. Heidelberg, O. White, and S. L. Salzberg. 4 December 2000, posting date. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 1 [Online.] http://genomebiology.com/2000/1/6/RESEARCH/0011. [DOI] [PMC free article] [PubMed]
- 14.Faine, S., B. Adler, C. Bolin, and P. Perolat. 1999. Leptospira and leptospirosis. MediSci, Melbourne, Australia.
- 15.Farr, R. W. 1995. Leptospirosis. Clin. Infect. Dis. 21:1-6; 7-8. [DOI] [PubMed] [Google Scholar]
- 16.Franklin, M. J., and D. E. Ohman. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184:3000-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. C. Venter, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 18.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, J. C. Venter, et al. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 19.Fukunaga, M., and I. Mifuchi. 1989. Unique organization of Leptospira interrogans rRNA genes. J. Bacteriol. 171:5763-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulop, V., C. J. Ridout, C. Greenwood, and J. Hajdu. 1995. Crystal structure of the di-haem cytochrome c peroxidase from Pseudomonas aeruginosa. Structure 3:1225-1233. [DOI] [PubMed] [Google Scholar]
- 21.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 22.Gilges, D., M. A. Vinit, I. Callebaut, L. Coulombel, V. Cacheux, P. H. Romeo, and I. Vigon. 2000. Polydom: a secreted protein with pentraxin, complement control protein, epidermal growth factor and von Willebrand factor A domains. Biochem. J. 352:49-59. [PMC free article] [PubMed] [Google Scholar]
- 23.Godfroid, F., B. Taminiau, I. Danese, P. Denoel, A. Tibor, V. Weynants, A. Cloeckaert, J. Godfroid, and J. J. Letesson. 1998. Identification of the perosamine synthetase gene of Brucella melitensis 16M and involvement of lipopolysaccharide O side chain in Brucella survival in mice and in macrophages. Infect. Immun. 66:5485-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haake, D. A. 2000. Spirochaetal lipoproteins and pathogenesis. Microbiology 146:1491-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haake, D. A., C. I. Champion, C. Martinich, E. S. Shang, D. R. Blanco, J. N. Miller, and M. A. Lovett. 1993. Molecular cloning and sequence analysis of the gene encoding OmpL1, a transmembrane outer membrane protein of pathogenic Leptospira spp. J. Bacteriol. 175:4225-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henneberry, R. C., and C. D. Cox. 1970. Beta-oxidation of fatty acids by Leptospira. Can. J. Microbiol. 16:41-45. [DOI] [PubMed] [Google Scholar]
- 27.Howell, M. L., E. Alsabbagh, J. F. Ma, U. A. Ochsner, M. G. Klotz, T. J. Beveridge, K. M. Blumenthal, E. C. Niederhoffer, R. E. Morris, D. Needham, G. E. Dean, M. A. Wani, and D. J. Hassett. 2000. AnkB, a periplasmic ankyrin-like protein in Pseudomonas aeruginosa, is required for optimal catalase B (KatB) activity and resistance to hydrogen peroxide. J. Bacteriol. 182:4545-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunnicutt, D. W., M. J. Kempf, and M. J. McBride. 2002. Mutations in Flavobacterium johnsoniae gldF and gldG disrupt gliding motility and interfere with membrane localization of GldA. J. Bacteriol. 184:2370-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jovanovic, T., C. Ascenso, K. R. Hazlett, R. Sikkink, C. Krebs, R. Litwiller, L. M. Benson, I. Moura, J. J. Moura, J. D. Radolf, B. H. Huynh, S. Naylor, and F. Rusnak. 2000. Neelaredoxin, an iron-binding protein from the syphilis spirochete, Treponema pallidum, is a superoxide reductase. J. Biol. Chem. 275:28439-28448. [DOI] [PubMed] [Google Scholar]
- 30.Ko, A. I., M. Galvao Reis, C. M. Ribeiro Dourado, W. D. Johnson, Jr., and L. W. Riley. 1999. Urban epidemic of severe leptospirosis in Brazil. Salvador Leptospirosis Study Group. Lancet 354:820-825. [DOI] [PubMed] [Google Scholar]
- 31.Lamothe, G. T., L. Jolly, B. Mollet, and F. Stingele. 2002. Genetic and biochemical characterization of exopolysaccharide biosynthesis by Lactobacillus delbrueckii subsp. bulgaricus. Arch. Microbiol. 178:218-228. [DOI] [PubMed] [Google Scholar]
- 32.Levett, P. N. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lombard, M., D. Touati, M. Fontecave, and V. Niviere. 2000. Superoxide reductase as a unique defense system against superoxide stress in the microaerophile Treponema pallidum. J. Biol. Chem. 275:27021-27026. [DOI] [PubMed] [Google Scholar]
- 34.Matsunaga, J., M. A. Barocchi, J. Croda, T. A. Young, Y. Sanchez, I. Siqueira, C. A. Bolin, M. G. Reis, L. W. Riley, D. A. Haake, and A. I. Ko. 2003. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol. Microbiol. 49:929-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meidanis, J., M. D. Braga, and S. Verjovski-Almeida. 2002. Whole-genome analysis of transporters in the plant pathogen Xylella fastidiosa. Microbiol. Mol. Biol. Rev. 66:272-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28:1059-1066. [DOI] [PubMed] [Google Scholar]
- 37.Palaniappan, R. U., Y. F. Chang, S. S. Jusuf, S. Artiushin, J. F. Timoney, S. P. McDonough, S. C. Barr, T. J. Divers, K. W. Simpson, P. L. McDonough, and H. O. Mohammed. 2002. Cloning and molecular characterization of an immunogenic LigA protein of Leptospira interrogans. Infect. Immun. 70:5924-5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paton, A. W., and J. C. Paton. 1999. Molecular characterization of the locus encoding biosynthesis of the lipopolysaccharide O antigen of Escherichia coli serotype O113. Infect. Immun. 67:5930-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paulsen, I. T., L. Nguyen, M. K. Sliwinski, R. Rabus, and M. H. Saier, Jr. 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301:75-100. [DOI] [PubMed] [Google Scholar]
- 40.Pizza, M., V. Scarlato, V. Masignani, M. M. Giuliani, B. Arico, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Storni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandi, and R. Rappuoli. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816-1820. [DOI] [PubMed] [Google Scholar]
- 41.Rappuoli, R. 2001. Reverse vaccinology, a genome-based approach to vaccine development. Vaccine 19:2688-2691. [DOI] [PubMed] [Google Scholar]
- 42.Ren, S. X., G. Fu, X. G. Jiang, R. Zeng, Y. G. Miao, H. Xu, Y. X. Zhang, H. Xiong, G. Lu, L. F. Lu, H. Q. Jiang, J. Jia, Y. F. Tu, J. X. Jiang, W. Y. Gu, Y. Q. Zhang, Z. Cai, H. H. Sheng, H. F. Yin, Y. Zhang, G. F. Zhu, M. Wan, H. L. Huang, Z. Qian, S. Y. Wang, W. Ma, Z. J. Yao, Y. Shen, B. Q. Qiang, Q. C. Xia, X. K. Guo, A. Danchin, I. Saint Girons, R. L. Somerville, Y. M. Wen, M. H. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888-893. [DOI] [PubMed] [Google Scholar]
- 43.Santos, J. M., M. Lobo, A. P. Matos, M. A. De Pedro, and C. M. Arraiano. 2002. The gene bolA regulates dacA (PBP5), dacC (PBP6) and ampC (AmpC), promoting normal morphology in Escherichia coli. Mol. Microbiol. 45:1729-1740. [DOI] [PubMed] [Google Scholar]
- 44.Springer, T. A. 1997. Folding of the N-terminal, ligand-binding region of integrin alpha-subunits into a beta-propeller domain. Proc. Natl. Acad. Sci. USA 94:65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang, Y. P., M. M. Dallas, and M. H. Malamy. 1999. Characterization of the Batl (Bacteroides aerotolerance) operon in Bacteroides fragilis: isolation of a B. fragilis mutant with reduced aerotolerance and impaired growth in in vivo model systems. Mol. Microbiol. 32:139149. [DOI] [PubMed] [Google Scholar]
- 46.Van Sluys, M. A., M. C. de Oliveira, C. B. Monteiro-Vitorello, C. Y. Miyaki, L. R. Furlan, L. E. Camargo, A. C. da Silva, D. H. Moon, M. A. Takita, E. G. Lemos, M. A. Machado, M. I. Ferro, F. R. da Silva, M. H. Goldman, G. H. Goldman, M. V. Lemos, H. El-Dorry, S. M. Tsai, H. Carrer, D. M. Carraro, R. C. de Oliveira, L. R. Nunes, W. J. Siqueira, L. L. Coutinho, E. T. Kimura, E. S. Ferro, R. Harakava, E. E. Kuramae, C. L. Marino, E. Giglioti, I. L. Abreu, L. M. Alves, A. M. do Amaral, G. S. Baia, S. R. Blanco, M. S. Brito, F. S. Cannavan, A. V. Celestino, A. F. da Cunha, R. C. Fenille, J. A. Ferro, E. F. Formighieri, L. T. Kishi, S. G. Leoni, A. R. Oliveira, V. E. Rosa, Jr., F. T. Sassaki, J. A. Sena, A. A. de Souza, D. Truffi, F. Tsukumo, G. M. Yanai, L. G. Zaros, E. L. Civerolo, A. J. Simpson, N. F. Almeida, Jr., J. C. Setubal, and J. P. Kitajima. 2003. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 185:1018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vujaskovic, Z., I. Batinic-Haberle, Z. N. Rabbani, Q. F. Feng, S. K. Kang, I. Spasojevic, T. V. Samulski, I. Fridovich, M. W. Dewhirst, and M. S. Anscher. 2002. A small molecular weight catalytic metalloporphyrin antioxidant with superoxide dismutase (SOD) mimetic properties protects lungs from radiation-induced injury. Free Radic. Biol. Med. 33:857-863. [DOI] [PubMed] [Google Scholar]
- 48.Werts, C., R. I. Tapping, J. C. Mathison, T. H. Chuang, V. Kravchenko, I. Saint Girons, D. A. Haake, P. J. Godowski, F. Hayashi, A. Ozinsky, D. M. Underhill, C. J. Kirschning, H. Wagner, A. Aderem, P. S. Tobias, and R. J. Ulevitch. 2001. Leptospiral lipopolysaccharide activates cells through a TLR2-dependent mechanism. Nat. Immunol. 2:346-352. [DOI] [PubMed] [Google Scholar]
- 49.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 50.Wizemann, T. M., J. H. Heinrichs, J. E. Adamou, A. L. Erwin, C. Kunsch, G. H. Choi, S. C. Barash, C. A. Rosen, H. R. Masure, E. Tuomanen, A. Gayle, Y. A. Brewah, W. Walsh, P. Barren, R. Lathigra, M. Hanson, S. Langermann, S. Johnson, and S. Koenig. 2001. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 69:1593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuerner, R. L. 1994. Nucleotide sequence analysis of IS1533 from Leptospira borgpetersenii: identification and expression of two IS-encoded proteins. Plasmid 31:1-11. [DOI] [PubMed] [Google Scholar]
- 52.Zuerner, R. L., and W. M. Huang. 2002. Analysis of a Leptospira interrogans locus containing DNA replication genes and a new IS, IS1502. FEMS Microbiol. Lett. 215:175-182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.