Abstract
Plasmid pTC-F14 contains a plasmid stability system called pas (plasmid addiction system), which consists of two proteins, a PasA antitoxin and a PasB toxin. This system is closely related to the pas of plasmid pTF-FC2 (81 and 72% amino acid identity for PasA and PasB, respectively) except that the pas of pTF-FC2 contains a third protein, PasC. As both pTC-F14 and pTF-FC2 are highly promiscuous broad-host-range plasmids isolated from bacteria that share a similar ecological niche, the plasmids are likely to encounter each other. We investigated the relative efficiencies of the two stability systems and whether they had evolved apart sufficiently for each pas to stabilize a plasmid in the presence of the other. The three-component pTF-FC2 pas was more efficient at stabilization of a heterologous tester plasmid than the two component pas of pTC-F14 in Escherichia coli host cells (±92% and ±60% after 100 generations, respectively). The PasA antidote of each pas was unable to neutralize the PasB toxin of the other plasmid. The pas proteins of each plasmid autoregulated their own expression as well as that of the pas of the other plasmid. The pas of pTF-FC2 was more effective at repressing the pas operon of pTC-F14 than the pas of pTC-F14 was able to repress itself or the pas of pTF-FC2. This increased efficiency was not due to the PasC of pTF-FC2. The effect of this stronger repression was that pTF-FC2 displaced pTC-F14 when the two plasmids were coresident in the same E. coli host cell. Plasmid curing resulted in the arrest of cell growth but did not cause cell death, and plasmid stability was not influenced by the E. coli mazEF genes.
Plasmid pTC-F14 is a 14-kb broad-host-range mobilizable plasmid isolated from the moderately thermophilic (optimum, 45°C), sulfur-oxidizing, acidophilic bacterium Acidithiobacillus caldus (previously Thiobacillus caldus) strain f (8). It bears a number of similarities to a 12.2-kb broad-host-range plasmid, pTF-FC2, which was isolated from a different, mesophilic (30 to 35°C), iron- and sulfur-oxidizing, chemolithotrophic, acidophilic bacterium, Acidithiobacillus ferrooxidans (21). Both plasmids have IncQ-like plasmid replicons consisting of an oriV with three or more 22-bp iterons as well as repB (primase-encoding), repA (helicase-encoding), and repC (DNA-binding protein-encoding) genes. A further striking similarity is that both plasmids have genes for a plasmid addiction system (pas) located between the repB and repA genes (8, 26).
The pas genes of pTF-FC2 have been reported to act as a post-segregational killing (PSK) plasmid stability system so that daughter cells that fail to inherit a plasmid are killed (26). Typical plasmid PSK systems consist of two genes, one encoding a highly expressed but short-lived antitoxin and a second gene encoding a poorly expressed but long-lived toxin (14). The pas of pTF-FC2 is unusual in having three genes: pasA which encodes an antitoxin, pasB which encodes a toxin, and pasC which was thought to enhance the ability of the antitoxin to neutralize the toxin (26). Plasmid pTC-F14 is more typical of other plasmid addiction systems in having only two genes (pasA and pasB). In all protein type toxin-antitoxin (TA) plasmid stability systems studied to date, the antitoxins have a second autoregulatory function (11). This autoregulatory role has been found to be essential for the proper functioning of the pas of pTF-FC2 (28). When placed under control of the tac promoter, abnormal expression of the pas genes inhibited host growth and they failed to stabilize the plasmid. Although there are clear examples of conservation between toxins and also antitoxins (e.g., RelB antitoxin, RelE toxin, and ChpAK toxin homologues) (11), in general, there is a large amount of amino acid sequence diversity between toxins and also between antitoxins which function as PSK stability systems (17, 22). Presumably, this sequence diversity allows individual PSK systems to enhance the stability of the plasmids on which they occur rather than that of coresident plasmids that may compete for limited host resources. Plasmids pTF-FC2 and pTC-F14 are unusual in that the toxins and antitoxins are highly conserved and share 81 and 72% amino acid sequence identity, respectively (8).
The discovery of closely related TA systems on two related but compatible plasmids presents an opportunity to ask interesting questions on the evolution of such systems. The bacteria from which plasmids pTC-F14 and pTF-FC2 were isolated are physiologically related and grow in a similar low-pH, inorganic mineral-rich environment. Since both plasmids are broad host range and highly mobilizable, there is a possibility that the plasmids could frequently encounter each other, as they share the same ecological replication space. If both plasmids were to occur in the same cell, then the possession of a TA system that cross-reacts with a second closely related system may not enhance the stability of each individual plasmid species in the presence of the other. We therefore wished to discover whether the two TA systems have diverged sufficiently for them to act without interference by the other. More specifically, we wished to investigate whether the antitoxin from one plasmid could neutralize the toxin of the other plasmid and whether each antitoxin could autoregulate the expression of the related system. Furthermore, we wished to determine whether a plasmid that possessed the three-component pas had a stability advantage over a plasmid with the related two-component pas, when each plasmid was on its own in a host cell or when coresident with the other.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Escherichia coli strains CSH50, CSH50-Iq (27), JM105 (29), and DH5α (Promega Corp.) were grown in Luria-Bertani broth (LB) or Luria agar (LA) (23), and ampicillin (100 μg/ml), kanamycin (30 μg/ml), tetracycline (15 μg/ml), and chloramphenicol (25 μg/ml) were added as required. E. coli MC4100 relA+ (wild type [WT]) and its derivatives, MC4100 relA+ mazEF::kan (ΔmazEF), MC4100 relA1 (Kms), and MC4100 relA1 mazEF::kan were a kind gift from Hanna Engelberg-Kulka.
DNA techniques, PCR, sequencing, and analysis.
Plasmids and primers are shown in Table 1. Standard methods were used for plasmid isolation, digestion, ligation, agarose gel electrophoresis, and preparation and transformation of competent E. coli cells (23). The PCR was carried out in a PCR Sprint temperature cycling system (Hybaid) by using the Expand high-fidelity PCR system DNA polymerase (Roche Molecular Biochemicals), and all constructs were sequenced by the dideoxy-chain termination method with an ABI PRISM 377 automated DNA sequencer to verify the integrity of PCR-amplified fragments. Sequence manipulations were performed by using DNAMAN (version 4.1; Lynnon BioSoft), and homology searches were performed by using the gapped-BLAST program of the National Center for Biotechnology Information (2).
TABLE 1.
Plasmid vectors, pas-containing constructs, and primers used in this study
| Plasmid or primer | Description or sequencea | Source or reference |
|---|---|---|
| Plasmid vectors | ||
| pBR322 | Apr, cloning vector | 3 |
| pGEM-T | Apr, T-tailed PCR product cloning vector | Promega Corp. |
| pACYC184 | Tcr, Cmr, p15A replicon, cloning vector | 6 |
| pKK223-3 | Apr, ColE1 replicon, tac promoter | 4 |
| pMC1403 | Apr, ColE1 replicon, lacZYA* | 5 |
| pOU82 | Apr, R1 replicon, lacZYA | 10 |
| Plasmid constructs | ||
| pTac-F14pasA | Apr, pKK223-3 vector, pTC-F14 pasA nt 1163-1405 | This study |
| pTac-F14pasB | Apr, pKK223-3 vector, pTC-F14 pasB nt 1364-1662 | This study |
| pTac-F14pasAB | Apr, pKK223-3 vector, pTC-F14 pasAB nt 1163-1662 | This study |
| pTac-F14Pr-pasAB | Apr, pKK223-3 vector, pTC-F14 pasAB own promoter nt 900-1662 | This study |
| pGEM-F14pasA | Apr, pGEM-T vector, pTC-F14 pas region nt 900-1405 | This study |
| pGEM-F14pasAB | Apr, pGEM-T vector, pTC-F14 pas region nt 900-1662 | This study |
| pMC-F14Pr(pas) | Apr, pMC1403 vector, pTC-F14 pas promoter region nt 900-1207 | This study |
| pMC-FC2Pr(pas) | Apr, pMC1403 vector, pTF-FC2 pas promoter region | This study |
| pOU-F14pasA | Apr, pOU82 replicon, pTC-F14 pas region nt 900-1405 | This study |
| pOU-F14pasAB | Apr, pOU82 replicon, pTC-F14 pas region nt 900-1662 | This study |
| pOU-FC2pasABC | Apr, pOU82 replicon, pTF-FC2 pas region, previously pOU-pasABC | 26 |
| pOU-FC2pasABC* | Apr, pOU82 replicon, pTF-FC2 pas region with frameshift mutation in pasC, previously pOU-pasABC | 26 |
| pTac-FC2pasA | Apr, pKK223-3, pTF-FC2 pasA with tac promoter, previously pTac-pasA | 26 |
| pTac-FC2pasB | Apr, pKK223-3, pTF-FC2 pasB with tac promoter, previously pTac-pasB | 26 |
| pTac-FC2pasA-ACYC | Apr, pACYC184 vector, pTF-FC2 pasA with tac promoter, previously pTac-pasA-ACYC | 28 |
| pFC2-pasABC | Kmr, pTF-FC2 minimal replicon, previously pKmM0 | 26 |
| pFC2-pasA*BC | pFC2-pasABC with frameshift mutation in pasA, previously pKmM1 | 26 |
| pFC2-pasAB*C | pFC2-pasABC with frameshift mutation in pasB, previously pKmM2 | 26 |
| pFC2-pasABC* | pFC2-pasABC with frameshift mutation in pasC, previously pKmM3 | 26 |
| pFC2-ΔpasABC | pFC2-pasA*BC with spontaneous deletion of pasABC, previously pKmM1del | 26 |
| pF14-pasAB (Km) | Kmr, pTC-F14 minimal replicon, previously pTC-F101 | 8 |
| pF14-pasA*B* | pF14-pasAB (Km) with frameshift mutations in pasA and pasB | This study |
| pF14-pasΔAB | pF14-pasAB (Km) with StuI-XbaI deletion in pasAB | This study |
| pF14-pasAB (Cm) | Cmr, pTC-F14 minimal replicon, previously pTC-F200 | 9 |
| pKGCm | Cmr, pACYC184 replicon | 27 |
| Primers | ||
| Primer 1 (EcoRI) | 5′-TATTGAATTCGAGCAGGAGCTAAACATGC | This study |
| Primer 2 (HindIII) | 5′-AATGAAGCTTAACTCAATCCGCCAAGCC | This study |
| Primer 3 (EcoRI) | 5′-ATACGAATTCCTAGAGGAAGTGGAGCGCG | This study |
| Primer 4 (HindIII) | 5′-TCGCAAGCTTGTTTACTTTCGGTATACCTCTCG | This study |
| Primer 5 (EcoRI) | 5′-TACTGAATTCTACCAGTGTGCCCCATCG | This study |
| Primer 6 (BamHI) | 5′-GTAGGGATCCACTTCGGTGGGTAATCGG | This study |
| Primer 7 (StuI) | 5′-TGAGGCCTTGGCTGCAGGCCACAGGACG | This study |
| TACPRI | 5′-GACAATTAATCATCGGCTCG | 25 |
| LACZPRI | 5′-CGCCAGCTGGCGAAAGGGGG | |
| FP2 (BamHI) | 5′-AGTAGGGATCCACTTCGGCGGGCAGTCGG | 28 |
| FC2PasAPr (EcoRI) | 5′-TCATGAATTCGAGGGCGCTATCCGC | This study |
Plasmid constructs.
Primer pairs 1 and 2, 3 and 4, and 1 and 4 were used to amplify the pasA, pasB, and pasAB genes, respectively, with pK13 (a subclone of pTC-F14) (8) as a template for the PCR. Amplified products were cloned in E. coli DH5α behind the tac promoter of pKK223-3 by using the EcoRI and HindIII restriction sites on the primers. Sequencing with primer TACPRI verified the integrity of the pTac-F14pasA and pTac-F14pasAB constructs. A number of attempts at the PCR, cloning, and sequencing of the tac-pasB construct revealed nucleotide changes in the pasB gene that presumably made it less lethal (and therefore cloneable). Two clones that resulted in translucent-looking E. coli colonies, capable of growth only at 30°C, were found to have single-base-pair deletions in the codon preceding the pasB stop codon. These deletions caused a frameshift, which extended PasB by 29 amino acids. To correct this, a 622-bp XbaI-SalI fragment containing the front portion of pasB (correctly fused to the tac promoter), was excised and ligated into the XbaI-SalI fragment of pTac-F14pasAB (containing the end of pasB plus the vector pKK223-3). This ligation mixture was transformed into E. coli CSH50-Iq but only gave colonies when the cells also contained pF14-pasAB (Km) (i.e., pasA) in trans. The tac-pasB plasmid DNA (pTac-F14pasB) was finally purified by electrophoresis of the mixed plasmid preparation (pTac-F14pasB plus pF14-pasAB [Km]) on a preparative gel, followed by excision and GFX column (Amersham) isolation of the pTac-F14pasB plasmid alone. Primer 5 (which binds 276 bp upstream of the ATG start of pasA) was paired with primer 2 or 4 to amplify pasA or pasAB, respectively, to include the putative promoter region known to be important for the autoregulation of the pas operon (28). These PCR products were cloned in pGEM-T (giving constructs pGEM-F14pasA and pGEM-F14pasAB), sequenced in both directions, and then excised with EcoRI and HindIII for cloning in pBR322. From here, the pas genes were excised with EcoRI and BamHI and cloned into the EcoRI-BamHI sites of pOU82 to give constructs pOU-F14pasA and pOU-F14pasAB.
Fusion of pasA promoter region to LacZ reporter gene and β-galactosidase assays.
An in-frame translational fusion of pasA to a lacZ reporter gene was made by cloning a PCR amplification product (primers 5 and 6) into the EcoRI-BamHI sites of pMC1403 to give construct pMC-F14Pr(pas). Similarly, a pasA-lacZ reporter gene construct [pMC-FC2Pr(pas)] was made for pTF-FC2, containing a pasA upstream region of approximately 298 bp (primers FP2 and FC2PasAPr). This pMC-FC2Pr(pas) construct was regulated in the same manner as its predecessor pP2H (28), which contained a smaller upstream region of pasA.
The PasA-LacZ fusion constructs were transformed into E. coli CSH50-Iq, which was then transformed by various other constructs to be tested in trans. β-Galactosidase assays were performed by the method of Miller (18) by using, where possible, the following fixed parameters. The culture volume sampled was 50 μl, which was added to 950 μl of Z buffer, and the cells were made permeable by vortexing with 10 μl of toluene. A fixed assay time (usually 5 to 10 min) was chosen for each comparative experiment, after which all sample reactions were terminated. Cultures were incubated in 50 ml of LB at 37°C with the appropriate antibiotic selection.
Plasmid stability assays.
Stability assays were performed by growing plasmid-containing E. coli cells without selection in 5 ml of LB at 37°C for 100 generations, with transfer of approximately 1,000 cells to fresh LB at 20-generation intervals. Samples taken at 20-generation intervals were diluted in saline, plated onto LA plates (to give 150 to 350 colonies per plate), and incubated at 37°C overnight in the absence of selection. Fifty colonies were patched onto LA plates with appropriate antibiotic selection, and the percentage survival was used to calculate plasmid retention. The stability assay for the pOU82 constructs was performed as described above, except that cultures were incubated at 30°C, samples taken at 20-generation time intervals were plated directly onto LA plates containing 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)/ml, and blue and white colonies were scored as plasmid-containing and plasmid-minus colonies, respectively.
Incompatibility assays.
E. coli cells were transformed sequentially with the desired replicons, and the plasmids were maintained by selection on appropriate antibiotics. Survival of the plasmids was then tested by dropping selection by one or both antibiotics and growing the cells in 5 ml of LB for 100 generations, with transfer of approximately 1,000 cells to fresh media at 20-generation intervals. Finally, 50 colonies were replica plated to antibiotic-containing LA plates to score for plasmid retention.
Plasmid curing.
Plasmid curing was based on the procedure developed in the Gerdes laboratory (15). This made use of pKG339, a plasmid that is able to stop the replication of pOU82-based constructs following induction with isopropyl-β-d-thiogalactopyranoside (IPTG). A version of pKG339, called pKGCm, was used in our laboratory (27). Prevention of replication of pOU82-based constructs results in the rapid loss of these plasmids from the cell population. An overnight culture (30°C) was inoculated into fresh medium containing antibiotic selection to ensure that plasmids were present. This 3-h preculture was diluted 1:100 into prewarmed broth minus ampicillin (but containing chloramphenicol for pKGCm selection) and grown in the absence or presence of 2 mM IPTG (added at 3 h). Culture density was monitored at a wavelength of 600 nm. Samples taken throughout the growth period were diluted in saline and plated to LA-chloramphenicol-X-Gal plates to give 100 to 500 colonies per plate. Viable cells were calculated as the total number (blue and white) of CFU per milliliter. Plasmid-containing cells were calculated as blue CFU per milliliter.
RESULTS
Comparative analysis of pas operons of pTC-F14 and pTF-FC2.
The sequence of the pTC-F14 pas operon and flanking regions is shown in Fig. 1. The region between repB and repA of pTC-F14 contains two open reading frames (ORFs) with high sequence homology to pasA and pasB of pTF-FC2 at the levels of both nucleotide sequence (80 and 71%, identity, respectively) and amino acid sequence (81 and 72% identity, respectively). The most obvious difference between the two pas operons is that no region corresponding to the pasC gene of pTF-FC2 was found on pTC-F14. Nucleotide sequence similarity between the two operons terminates at the pTC-F14 pasB stop codon, and for the next 48 bp, there is no obvious similarity between the two plasmids. Clear nucleotide similarity (75%) resumes 68 bp immediately upstream of the repA gene, and there is no space to accommodate a pasC gene. Apart from the absence of pasC, the pTC-F14 pas operon bears striking similarity to that of pTF-FC2. Plasmid pTF-FC2 has a 7-bp duplication (5′-CAGGAGC-3′) in the promoter region of the pas operon, and when the PasA antitoxin was inactivated by a frameshift mutation, a spontaneous deletion occurred between the 7-bp duplications in a small percentage of the plasmids (26). This deletion inactivated the pas operon, and the otherwise sick host cells once again grew strongly. Plasmid pTC-F14 has a 9-bp duplication (5′-AGCAGGAGC-3′) in the same region, which includes the identical 7-bp duplication found in plasmid pTF-FC2. However, pTC-F14 pasA mutants have been even more difficult to construct (see below), and spontaneous deletions in the promoter region have not been detected.
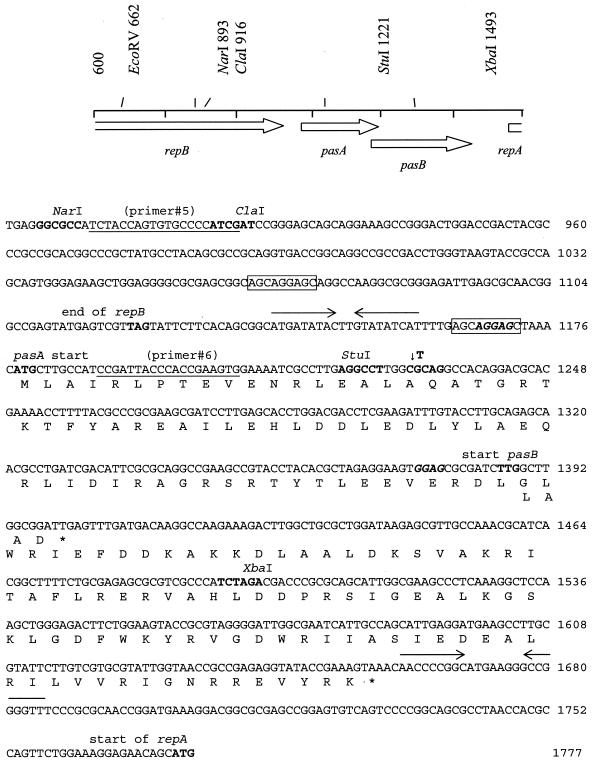
FIG. 1.
Nucleotide sequence of the region of pTC-F14 carrying pasA, pasB, the 3′ end of repB, and the 5′end of repA (accession number AF325537). The amino acid sequences of PasA and PasB are indicated below the nucleotide sequences. Primers 5 and 6 (underlined) were used to make a translational fusion of the 307 bp upstream and the start region of pasA to a lacZ reporter gene. The site of insertion of a T nucleotide into the pasA sequence (↓) to create a frameshift and a PstI site (boldface type) are shown. Repeated sequences similar to those that flank a spontaneous deletion in pTF-FC2 are boxed, and potential ribosome binding sites are in boldface italic type. Two 9-bp inverted repeats immediately upstream of pasA and downstream of pasB are shown by opposing arrows. Restriction enzyme recognition sites are in boldface type.
Both pasA genes have a consensus AGGAG ribosome binding site and an AUG start typical of highly expressed genes. The start codons of both pasB genes (UUG in the case of pTC-F14) are situated within the 3′ end of pasA, and the putative ribosome binding site (GGAG) is less homologous to the consensus sequence than is the case for highly expressed genes. This arrangement of overlapping ORFs is frequently encountered in TA systems, in which the toxin is synthesized in smaller amounts relative to the antidote (e.g., E. coli relBE) (12).
Another pasAB-like operon has been discovered on pAM10.6, a plasmid isolated from Pseudomonas fluorescens that is unrelated to IncQ-like plasmids (20). Interestingly, the amino acid sequence identities of PasA and PasB of pAM10.6 are more closely related to those of pTC-F14 (84 and 74%, respectively) than PasA and PasB of pTF-FC2 are to those of pTC-F14 (81 and 72%, respectively). Like the other pas genes, the genes of the pAM10.6 pas also have overlapping ORFs, but unlike pTF-FC2, no pasC gene is present.
The pas genes of pTC-F14 function as a stability system.
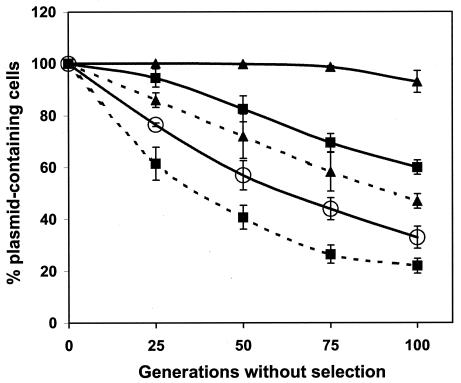
In spite of the high amino acid sequence similarity between PasA and PasB of pTF-FC2 and pTC-F14, it was necessary to demonstrate that the two-component pas-like system of pTC-F14 was able to function as a plasmid stability system. To determine this, the pasAB operon of pTC-F14 was cloned into the unstable, low-copy-number test plasmid pOU82 (10). After 100 generations of growth without selection, the pOU82 control was retained in approximately 33% of the population while plasmids pOU-F14pasAB (plasmid pOU82 containing the pTC-F14 pasAB genes) and pOU-F14pasA (containing the pasA antidote-encoding gene only) were stabilized 60 and 22%, respectively (Fig. 2). By comparison, the three-component pTF-FC2 pas was more effective at plasmid stabilization, and plasmids pOU-FC2pasABC and pOU-FC2pasABC* (pasC inactivated) were retained in 93 and 47% of the population, respectively. Therefore, although the pTC-F14 2-component pas was able to stabilize the pOU82 test plasmid in E. coli, it was less effective than the three-component pTF-FC2 pas.
FIG. 2.
Stabilization of heterologous R1 replicon by the pasAB(C) genes of pTC-F14 and pTF-FC2 in E. coli CSH50 at 30°C. Plasmid stability curves of the tester plasmid pOU82 (○) alone and containing the pasABC (▴, solid line) and pasAB(C) (▴, broken line) genes from pTF-FC2 (constructs pOU-FC2pasABC and pOU-FC2pasABC*, respectively) (Table 1) and the pasAB (▪, solid line) and pasA (▪, broken line) genes from pTC-F14 (constructs pOU-F14pasAB and pOU-F14pasA, respectively) (Table 1). The datum points are averages of the results from three independent determinations with the standard deviations, and values are normalized to 100% at 0 generations.
Difficulty in constructing a pTC-F14 pasA knockout.
We wished to generate a pasA mutant of pTC-F14 to investigate whether this plasmid's PasB toxin could be neutralized by the PasA antitoxin from pTF-FC2. A T was inserted within codon 18 of pasA, creating a frameshift mutation and introducing a PstI site (primer 7) (Fig. 1). The frameshift mutant constructs were transformed into E. coli cells, and transformants were obtained which contained plasmids with the newly introduced PstI site. However, unlike pasA mutants of pTF-FC2, the cells containing this construct were not noticeably sick. On sequencing the construct, it was discovered that a second mutation (G→T) had occurred 5 bp from the stop codon of pasB, resulting in the alteration of a conserved R to an L, which presumably inactivated PasB. To overcome this, the region containing the pasA T-insertion frameshift was recovered by excising an 830-bp EcoRV-XbaI fragment, and an attempt was made to insert (exchange) this into the minimal replicon of pTC-F14. This exchange consistently failed, and when the frameshift-containing insertion was eventually obtained, it was found that a C had been spontaneously deleted, which resulted in the C-terminal 27 amino acids of PasB being out of frame. It was concluded that in the absence of a functional PasA antitoxin, PasB was so toxic to the host cells that a pasA mutant was not viable.
Autoregulation and balanced expression of pasA and pasB is required to control toxicity of the pasAB operon.
Induced expression of the pTC-F14 pasAB genes from the tac promoter was studied in E. coli JM105, as had been done for the pasABC genes of pTF-FC2 (28). Two promoter fusion constructs were used: pTac-F14pasAB, in which the tac promoter was fused to the ribosome binding site and ATG start region of pasAB so as to exclude the natural pas promoter, and pTac-F14Pr-pasAB, in which the tac promoter was fused 276 bp upstream of pasAB and which includes the natural pas promoter. Following induction with IPTG, strong inhibition of growth was caused by expression of pasAB from the tac promoter only when the natural pasAB promoter was not present (results not shown). This confirmed the importance of autoregulatory feedback by PasA-PasB for the balanced production of antitoxin and toxin, as was found in the case of the pTF-FC2 pasABC system (28).
Cross talk between antidote and toxin of pTC-F14 and pTF-FC2.
Unregulated expression caused further problems when we attempted to test cross-complementation of tac-expressed pas genes from pTC-F14 and pTF-FC2. In previous work, site-directed mutagenesis was used to construct a pasA mutant of pTF-FC2. This pasA mutation (pFC2-pasA*BC) resulted in very slow growing host cells, with a small proportion forming large colonies as a result of one of two types of spontaneous pas deletion (26). We wished to test whether the PasB toxin of the pTF-FC2 pasA mutant could be neutralized by the pTC-F14 PasA antitoxin, thereby restoring rapid growth to the host cells. Cells containing the pTC-F14 and pTF-FC2 pasA genes under control of a tac promoter (pTac-F14pasA and pTac-FC2pasA, respectively) as well as cells containing the pKK223-3 vector control were transformed with the pTF-FC2 pasA mutant DNA (pFC2-pasA*BC). Cells were plated on kanamycin and ampicillin, to select for both plasmids, and IPTG to induce pasA expression. However, no transformants were obtained. This lead to the unexpected discovery that the pasA (antitoxin) genes of pTF-FC2 and pTC-F14, when expressed from an IPTG-induced tac promoter, were themselves toxic to host cells. The toxicity of pTac-F14pasA and pTac-FC2pasA alone was evident even in the absence of any other coresident constructs. The degree of toxicity varied slightly from bacterial strain to strain and was more noticeable at 37°C than at 30°C. Attempts to make cells containing the pFC2-pasA*BC construct competent and complement with pTac-F14pasA and pTac-FC2pasA in trans were unsuccessful, as pFC2-pasA*BC made host cells too sick to transform, even when grown at 30°C.
Furthermore, neutralization of pFC2-pasA*BC was not direct evidence of protein-protein interaction, since we could not rule out the influence of PasA binding to the promoter of pFC2-pasA*BC that would also result in a decrease in toxicity. To prove TA interaction at a purely protein level, we attempted to express the toxin from the inducible tac promoter (rather than the natural pas promoter) and then to neutralize the effect of the toxin by placing the antidote in trans. The pTF-FC2 toxin fusion (pTac-FC2pasB) was already available and was not fully lethal in cultures grown at 30°C when uninduced by IPTG. However, the pTC-F14 tac toxin gene fusion (pTac-F14pasB) was much harder to construct, as it appeared to be more toxic than the pTF-FC2 equivalent. pTac-F14pasB was extremely lethal and could not be maintained in cultures, even at 30°C, unless its antidote (on pF14-pasAB [Km]) was supplied in trans. Competent E. coli cells that already contained various plasmids expressing the pTC-F14 or pTF-FC2 antidote were then transformed with these lethal tac-pasB constructs to test which antidote would neutralize which toxin (Table 2).
TABLE 2.
Complementation of lethal expression of PasB toxin by antidote PasA carried in transa
| Resident plasmid in E. coli CSH50-Iq | No. of transformants per ng upon transformation with:
|
|||||
|---|---|---|---|---|---|---|
| pKK223-3 (control)
|
pTac-FC2pasB
|
pTac-F14pasB
|
||||
| −IPTG | +IPTG | −IPTG | +IPTG | −IPTG | +IPTG | |
| pF14-pasAB (Km) | ∼1,000 | ∼1,000 | ∼1,000 | NG | ∼700 | ∼700 |
| pF14-pasΔAB | ∼1,000 | ∼1,000 | ∼1,000 (sick) | NG | NG | NG |
| pFC2-pasABC | ∼1,000 | ∼1,000 | ∼1,000 | ∼1,000 | ∼500 | NG |
| pFC2-pasABC* | ∼1,000 | ∼1,000 | ∼1,000 | ∼1,000 | ∼500 | NG |
| pFC2-ΔpasABC | ∼1,000 | ∼1,000 | ∼1,000 (sick) | NG | NG | NG |
pTC-F14 pasAB and the deletion thereof were carried on plasmids pF14-pasAB (Km) and pF14-pasΔAB, respectively. pTF-FC2 pasABC and pasAB and deletions thereof were carried on plasmids pFC2-pasABC, pFC2-pasABC*, and pFC2-ΔpasABC, respectively. pKK223-3 is the tac promoter fusion vector containing no insert. pTac-FC2pasB contains the pasB of pTF-FC2 under control of the tac promoter. pTac-F14pasB contains the pasB of pTC-F14 under control of the tac promoter. IPTG (1 mM) was used to induce the tac promoter (−, without; +, with). Colony numbers on plates after 24 h of incubation at 30°C are given. NG, no growth.
Without induction by IPTG, it was evident that slight cross-neutralization occurred between the resident plasmid carrying the antidote and the incoming plasmid carrying the toxin in the case of both pTF-FC2 and pTC-F14. The presence of pTac-FC2pasB resulted in healthy colonies in the presence of its own TA system (pFC2-pasABC and pFC2-pasABC*) as well as in the foreign TA system (pF14-pasAB [Km]). However, deletions of either of these TA systems (pF14-pasΔAB or pFC2-ΔpasABC) resulted in sick colonies. Similarly, when not induced by IPTG, pTac-F14pasB gave healthy colonies when in the presence of its own TA system (pF14-pasAB [Km]) as well as in the foreign TA system (pFC2-pasABC and pFC2-pasABC*). In this case, however, deletion of either of these TA systems (pF14-pasΔAB or pFC2-ΔpasABC) resulted in no colonies, illustrating how much more lethal the pTC-F14 tac-pasB fusion was than the pTF-FC2 fusion.
When induction was initiated by IPTG, full neutralization was required between the resident plasmid carrying the antidote and the incoming plasmid carrying the toxin. In this instance, only the related antidote-toxin pairs (pF14-pasAB [Km] + pTac-F14pasB and pFC2-pasABC + pTac-FC2pasB) showed neutralization, whereas the foreign pairs (pF14-pasAB [Km] + pTac-FC2pasB and pFC2-pasABC + pTac-F14pasB) do not cross-react sufficiently to allow cell growth (Table 2). The presence of the extra gene, pasC, on pFC2-pasABC did not appear to affect the neutralization of the toxin by the antidote.
Bearing in mind the lethality of unregulated overexpression of the pas genes, further studies with tac promoter fusions to individual pas genes were dropped in favor of constructs such as pF14-pasAB (Km) (minimal replicon of pTC-F14) and pFC2-pasABC (minimal replicon of pTF-FC2) containing the entire natural promoter region and pasAB(C) operon (Table 1).
Cross-regulation of pas genes.
The pasABC genes of pTF-FC2 were previously shown to be autoregulated, with the PasA antitoxin functioning as the primary regulator (28). We wished to test whether the PasA antitoxins of plasmids pTF-FC2 and pTC-F14 could regulate the pas genes of each other. Translational fusion pasA-lacZ reporter gene constructs were made for both pTF-FC2 [pMC-FC2Pr(pas)] and pTC-F14 [pMC-F14Pr(pas)], containing upstream regions of 298 and 276 bp for the pasA genes of pTF-FC2 and pTC-F14, respectively.
Although β-galactosidase activity varied from experiment to experiment, relative values within individual experiments were highly reproducible. With no other constructs in trans, the pTF-FC2 and pTC-F14 pasA-lacZ reporter genes expressed lacZ at comparable levels (approximately 5,200 Miller units). When the pTF-FC2 and pTC-F14 replicons from which the pas genes had been deleted (pFC2-ΔpasABC and pF14-pasΔAB) were placed in trans, levels of reporter gene expression were reduced slightly and these values were taken as the unregulated level (100% expression) (Table 3). With the natural pasAB genes of pTF-FC2 (pFC2-pasABC) or pTC-F14 (pF14-pasAB [Km]) placed in trans to the pTF-FC2 pasA-lacZ reporter construct, β-galactosidase activity was reduced to 4 and 10% of the unregulated levels of expression, respectively. The pTC-F14 pasA-lacZ fusion was expressed at about 25% of its unregulated level when its own pas genes were placed in trans (pF14-pasAB [Km]), and this level of expression was reduced to 12% when the pas genes of pTF-FC2 were placed in trans (pFC2-pasABC). This indicated that PasABC of pTF-FC2 was twice as efficient at repressing the pas promoter of pTC-F14 than was its own PasAB. Furthermore, pFC2-pasABC was twice as efficient at repressing its own pas promoter [pMC-FC2Pr(pas)] than was pF14-pasAB (Km) (Table 3).
TABLE 3.
Regulation of pas operons of pTC-F14 and pTF-FC2a
| pas-lacZ reporter construct | Coresident plasmid | β-Galactosidase activity (Miller units) ± SD | % Activity |
|---|---|---|---|
| pMC-F14Pr(pas) | pF14-pasΔAB | 5,149 ± 727 | 100 |
| pF14-pasAB (Km) | 1,179 ± 213 | 24.55 ± 1.9 | |
| pFC2-pasABC | 551 ± 188 | 12 ± 2.9 | |
| pFC2-ΔpasABC | 4,763 ± 709 | 93.5 ± 11.6 | |
| pMC-FC2Pr(pas) | pFC2-ΔpasABC | 4,779 ± 1,339 | 100 |
| pFC2-pasABC | 196 ± 10 | 4.25 ± 0.9 | |
| pF14-pasAB (Km) | 510 ± 98 | 10.8 ± 0.98 | |
| pF14-pasΔAB | 4,512 ± 1,048 | 95.05 ± 4.7 |
β-Galactosidase activity was measured in log-phase E. coli strain CSH50-Iq cells containing the pas-lacZ reporter constructs pMC-F14Pr(pas) (pTC-F14 pas promoter fused to lacZ) and pMC-FC2Pr(pas) (pTF-FC2 pas promoter fused to lacZ). The percentage of expression of the promoter regions is calculated as the proportion of β-galactosidase activity measured relative to that in which the inactive pas operons (pF14-pasΔAB and pFC2-ΔpasABC, respectively) are in trans, and it is based on the relative values within the results from each of three experiments. Values represent the averages of the results from three independent experiments with standard deviations.
Therefore, both pas operons could regulate one another, with the three-component pasABC system of pTF-FC2 being the stronger repressor of both promoters. PasC is not responsible for this improved repression, since pFC2-pasABC* (frameshift mutation in pasC) was equally effective as pFC2-pasABC in repressing both its own and pTC-F14's pas operon (results not shown).
It was previously shown that PasA of pTF-FC2 acted as the primary repressor of the pasABC operon and that PasB and PasC played minor and negligible coregulatory roles, respectively. To test whether the PasA of pTF-FC2 could regulate the pTC-F14 pasA-lacZ fusion on its own, a plasmid (pTac-FC2pasA-ACYC) containing the pasA gene of pTF-FC2 under control of the tac promoter was placed in trans to the reporter construct. After 3 h of growth of the host culture, the pasA gene was induced with 2 mM IPTG, and after a further 3 h, the level of reporter gene expression was found to be 13% of the unregulated expression (results not shown). Therefore, PasA is also the major regulator of the pTC-F14 pas operon.
pas-mediated plasmid stability is not dependent on E. coli mazEF.
Aizenman and coworkers (1) presented evidence to suggest that certain PSK systems do not kill the host cells directly but are protein synthesis inhibitors that work through the E. coli mazEF chromosomal TA system. MazEF-mediated cell death can be triggered by several protein synthesis inhibitors, including antibiotics (that are general inhibitors of transcription and/or translation) (24), as well as by the phd-doc TA module from the P1 prophage (13). This prompted us to test the stability of the pas module in the E. coli MC4100 and MC4100 relA/mazEF strains, with the pOU82 assay system. The stability of pOU-FC2pasABC was the same in both MC4100 WT and the relA/mazEF deletion strains over 100 generations without antibiotic selection (results not shown). This indicated that pOU82 stability mediated by the pTF-FC2 pasABC genes was not influenced by the E. coli relA/mazEF operon. Similarly, the stability conferred by the pasAB genes of pTC-F14 on pOU82 was not influenced by the E. coli relA/mazEF operon.
Plasmid curing does not cause cell death.
Since the pOU82-based method of assaying stability is quite different from the methods used by Hazan and colleagues (13) in their E. coli mazEF plasmid stability experiments, a forced plasmid-curing method based on the procedure developed in the Gerdes laboratory (15) was tested. Prevention of replication results in rapid loss of the pOU82-based constructs from a cell population, and if the pas mediated stability was of the PSK type, this would be expected to result in the death of plasmid-minus cells. To test the ability of the pTF-FC2 PasABC to induce cell death, the pOU82-based plasmid constructs were prevented from replicating following induction of CopA inhibitor by plating cells onto LA containing 2 mM IPTG (Table 4). The procedure was effective in curing the cells of plasmids, but no cell death in either the mazEF-positive or -negative hosts was detected, as the number of CFU remained constant for cured and noncured cultures.
TABLE 4.
Ability of pTF-FC2 pasABC system to induce cell death upon curinga
| Strain and plasmid construct | No. of host cells from noncured culture | % Containing plasmid | No. of host cells from cured culture | % Containing plasmid |
|---|---|---|---|---|
| A+M+(pKGcm)(pOU-FC2pasABC) | 255 | 94 | 262 | 0 |
| A+M−(pKGcm)(pOU-FC2pasABC) | 328 | 100 | 326 | 0 |
Colony counts (CFU) of two bacterial cultures diluted and plated onto LA containing X-Gal (blue colonies contained plasmid, white colonies did not contain plasmid) with (cured) and without (noncured) 2 mM IPTG. The E. coli strains used were MC4100 relA+ and mazEF+ (A+M+) and relA+ mazEF-negative (A+M−). The results are representative of those from three separate experiments.
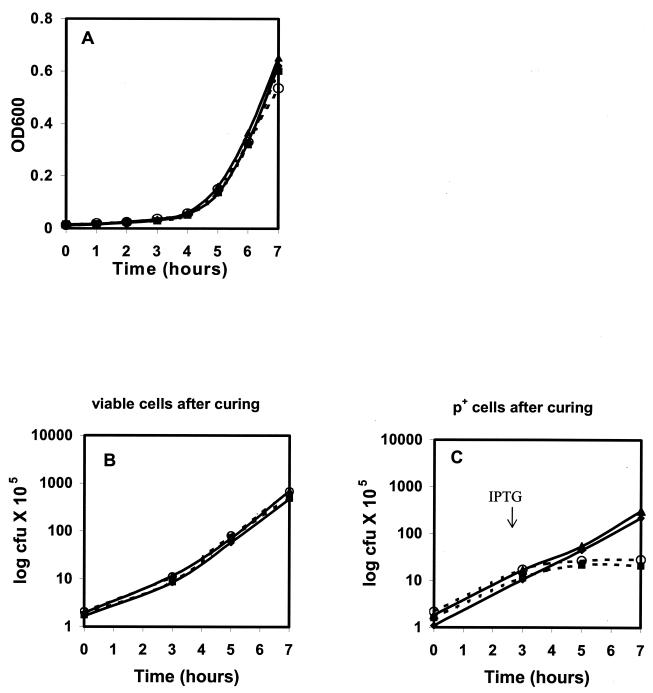
To confirm the result, a similar experiment was set up in which the growth of cells was monitored in liquid medium rather than on plates. A 3-h preculture of E. coli MC4100 relA+ mazEF+ containing pKGCm together with pOU82 or pOU-FC2pasABC was diluted into prewarmed broth without ampicillin and grown for a further 7 h with or without 2 mM IPTG (added at 3 h). At time zero, the inoculum contained approximately 105 cells per ml, of which 94% contained the pOU82-based constructs (Fig. 3).
FIG. 3.
Effect on host cell viability of curing of R1 replicon carrying pTF-FC2 pasABC genes. Turbidity readings (A), total CFU per milliliter (B), and plasmid-containing CFU per milliliter (C) of E. coli MC4100 (relA+ mazEF+) carrying pOU82 (⧫, ▪) and pOU-FC2pasABC (▴, ○) with (broken line) and without (solid line) the addition of IPTG at 3 h. The graphs are representative of the results from three independent experiments.
After 7 h, turbidity readings showed that both cultures had increased in cell mass, with those containing pOU-FC2pasABC (plus IPTG) lagging slightly behind those with pOU82. Viable cell counts likewise increased over 7 h to 4 × 107 to 5 × 107 CFU/ml (for pOU82 plus or minus IPTG) and 5 × 107 to 6 × 107 CFU/ml (for pOU-FC2pasABC plus IPTG). At 0, 3, 5, and 7 h, both cultures were tested for the presence of plasmids by plating onto media containing X-Gal. At the end of 7 h, less than 10% of both types of cells contained plasmids, showing that plasmid curing was successful. Induction of cultures containing pOU-FC2pasABC with IPTG consistently caused a slight lag in the growth rate, as determined by turbidity readings. However, these cultures continued to grow at a constant, if slower, rate and produced high levels of viable plasmid-free cells. This indicated that loss of the pas may have temporarily slowed the growth of plasmid-minus cells but did not cause cell death. Similarly, loss of a plasmid containing a pTF-FC2 pasC mutant (pOU-FC2pasABC*) or a pTC-F14 pasAB-containing plasmid (pOU-F14pasAB) did not cause cell death on curing or the slight lag in growth described for pOU-FC2pasABC (results not shown).
Role of pas genes in competition between replicons of pTF-FC2 and pTC-F14 for maintenance within a bacterial population.
We tested whether the presence or absence of a pas gene conferred a selective advantage to pTF-FC2 or pTC-F14 when both plasmids were coresident in an E. coli cell. E. coli CSH50 cells containing derivatives of plasmid pTF-FC2 or pTC-F14 were grown for approximately 100 generations without antibiotic selection, and the presence of the plasmids was determined by testing 50 colonies for kanamycin or chloramphenicol resistance. Before carrying out these competition experiments, the stability of the replicons with and without functional pas genes was determined. pTF-FC2 replicon-based plasmids pFC2-pasABC (intact pasABC) and pFC2-pasABC* (pasAB with inactive pasC) were stable for 100 generations without antibiotic selection when present on their own in host cells as were pTC-F14 replicon-based plasmids pF14-pasAB (Km) and pF14-pasAB (Cm) (intact pasAB) (Table 5). When the pas genes were deleted, plasmids containing either the pTF-FC2 replicon (pFC2-ΔpasABC) or the pTC-F14 replicon (pF14-pasΔAB) were not as stable (81 and 27% retention, respectively). Frameshift mutations within the pasB gene of the minimal replicon of pTF-FC2 (pFC2-pasAB*C) and pasAB genes of pTC-F14 (pF14-pasA*B*) were also less stable, 60 and 5%, respectively (Table 5).
TABLE 5.
Competitive and noncompetitive plasmid stability in the absence of antibiotic selectiona
| Host cell | pTC-F14-based plasmid and antibiotic marker | pTF-FC2-based plasmid and antibiotic marker | Plasmid retention (%) within cells after 100 generations minus antibiotic selection for:
|
|
|---|---|---|---|---|
| pTC-F14-based plasmids | pTF-FC2-based plasmids | |||
| E. coli CSH50 | pF14-pasAB (Cm) | 99.3 (1.2) | ||
| pF14-pasAB (Km) | 99.7 (0.6) | |||
| pF14-pasΔAB (Km) | 26.6 (29) | |||
| pF14-pasA*B* (Km) | 5.3 (9.2) | |||
| pFC2-pasABC (Km) | 99.0 (1.7) | |||
| pFC2-pasAB*C (Km) | 60.3 (15.4) | |||
| pFC2-pasABC* (Km) | 99.7 (0.6) | |||
| pFC2-ΔpasABC (Km) | 80.7 (14) | |||
| pF14-pasAB (Cm) | pFC2-pasABC (Km) | 1.6 (2.8) | 98.0 (3.5) | |
| pF14-pasAB (Cm) | pFC2-pasAB*C (Km) | 17.0 (5.6) | 91.0 (5.6) | |
| pF14-pasAB (Cm) | pFC2-pasABC* (Km) | 0.6 (1.2) | 89.6 (5.8) | |
| pF14-pasAB (Cm) | pFC2-ΔpasABC (Km) | 75.3 (18) | 89.3 (13.6) | |
All values are normalized to 100% at 0 generations. The percentage of plasmid retention is shown as the average of the results from at least three experiments. Standard deviations are shown in parentheses.
To test whether the pas genes within the minimal replicons of one plasmid affected the stability of the other plasmid, derivatives of plasmids pTF-FC2 and pTC-F14 were cotransformed into E. coli CSH50, which was then subcultured for approximately 100 generations in the absence of selection for either plasmid, and the presence of the individual plasmids within single colonies was determined. When together in a cell, pTF-FC2 (pFC2-pasABC) displaced pTC-F14 (pF14-pasAB [Cm]) and less than 5% of cells retained pF14-pasAB (Cm) (Table 5). A deletion mutant of pTF-FC2 (pFC2-ΔpasABC) that completely lacks the pasABC region was less able to displace pF14-pasAB (Cm) (75% retention). This indicated that the displacement was not due to incompatibility of the replicons and is consistent with previous research in which the iterons of each plasmid cloned on a high-copy-number plasmid would displace the replicon from which the iterons were derived but not the other replicon (8). Plasmids with frameshifts in TF-FC2 pasB (pFC2-pasAB*C) and pasC (pFC2-pasABC*) were able to displace pF14-pasAB (Cm) (17 and 1% retention of pF14-pasAB [Cm], respectively). This indicated that only the pasA region of the A. ferrooxidans plasmid pTF-FC2 was required for the displacement of pF14-pasAB (Cm) from the bacterial population. In contrast, pTC-F14 containing an intact pasAB was not able to displace the pTF-FC2 replicon, even when the pas genes of pTF-FC2 were deleted (pFC2-ΔpasABC) or inactivated (pFC2-pasAB*C).
Noncompetitive and competitive stability of the pTC-F14 and pTF-FC2 derivatives was also studied at 25-generation intervals to determine how rapidly the plasmids were lost from host cells (results not shown). pF14-pasAB (Cm), though highly stable when alone in the host cell, becomes extremely unstable when in the presence of either pFC2-pasABC or pFC2-pasAB*C and begins to be lost from the host population within the first 25 generations.
DISCUSSION
In this work we showed that pasAB TA genes of pTC-F14 were able to improve the stability of an unstable, low-copy-number, heterologous replicon, pOU82. The pasAB of pTC-F14 or pasABC genes of pTF-FC2 were less effective at stabilizing pOU82 than their own replicons (pF14-pasAB [Cm]/pF14-pasAB [Km] or pFC2-pasABC, respectively). The pasABC operon of pTF-FC2 consistently conferred better stability on the pOU82 replicon than the pasAB of pTC-F14. The pasABC operon in which pasC has been inactivated (pOU-FC2pasABC*) conferred plasmid stability to approximately the same level as the pTC-F14 pasAB genes (pOU-F14pasAB). This implied that pasC was responsible for the difference in stability conferred upon pOU82 by the two- and three-component pas, respectively. The role of PasC in the pasABC system is still unclear. If pasC were simply involved in improving the binding of the antidote to the toxin (i.e., more effective neutralization of toxin in cells that inherit the plasmid), one would not expect a decrease in stability when pasC was inactivated. In the absence of PasC, the toxin should be more effective in cells that failed to inherit the plasmid, and this could be expected to increase plasmid stability. The decrease in stability is more consistent with a role for PasC in facilitating the reaction between the PasB toxin and its cellular target. If this were true, an inactive PasC would result in a less effective toxin, which would allow more plasmid-minus cells to escape the effects of the TA module. Although the pas TA is related to the E. coli relBE family (11), the target of the pas TA has yet to be identified.
Using a plasmid replication inhibition system, we showed that the TA systems of both pTF-FC2 and pTC-F14 did not cause cell death upon plasmid loss. Previously, Smith and Rawlings (26) had reported that induction of pasB caused cell death, but we attribute this bactericidal effect to expression of the pasB gene from a heterologous tac promoter. We confirm in this study that autoregulation of the pas operon is essential in both systems for the balanced functioning of the TA locus. Both the pTC-F14 and pTF-FC2 antitoxins (when the pasA genes, alone, were cloned behind the tac promoter) and the toxins (when the pasA antitoxin genes were inactivated) appeared to inhibit E. coli host cells. The toxicity of the antitoxin was less severe in some E. coli strains (such as JM105 and XL1-Blue) but nevertheless restricted our ability to carry out experiments in which either PasA or PasB was to be expressed in the absence of each other or from an IPTG-regulated tac promoter. In spite of the toxicity of the individual products, when both the pasA and pasB genes of pTC-F14 or pTF-FC2 were present and autoregulated, the pas genes did not kill E. coli host cells. Examination of the cultures subjected to plasmid curing revealed a slight lag in growth that was consistent with bacteriostasis rather than cell death.
Because of the report that certain TA PSK systems do not kill host cells directly but are protein synthesis inhibitors that work through the E. coli mazEF chromosomal TA system (13), we tested the ability of the pas genes to stabilize pOU82-based plasmids in mazEF mutants. There was no difference in stability between E. coli mazEF mutant and WT strains using either the pTF-FC2 pasABC or the pTC-F14 pasAB genes. Similarly, the ability of the plasmid addiction systems to cause cell death was not affected by the mazEF status of the host cells.
The effectiveness of plasmid stability conferred by a TA system is correlated to the mode of action of the toxin, i.e., stasis versus death. For example, the parD-pemK locus is responsible for modest stability (approximately 10-fold) and acts by arresting cell division or growth (15). The parDE locus, on the other hand, confers a high degree of stability (approximately 500-fold) by causing cell death of plasmid-free cells. The fairly low level of plasmid stability conferred by the pas genes together with the observation that their function is not affected by the mazEF genes and that the pas genes do not appear to kill host cells, suggests that pas action is most likely the arrest of cell growth or division.
The interpretation of competition experiments between plasmids containing the two TA systems is challenging. The plasmid pTF-FC2 containing the pasABC system clearly displaced the plasmid pTC-F14 containing the pasAB system. This displacement was a function of the pasABC system, as when these genes were deleted, the stability of the pasAB-containing plasmid almost fully recovered. However, the pasAB genes of plasmid pTC-F14 were not able to displace pTF-FC2, even when its pasABC system was deleted. This raises the question of what the mechanism of pas-mediated plasmid displacement is and why it is unidirectional. One possibility is that the pas genes are involved in the regulation of plasmid replication. The pas genes are situated between the repB and repAC genes in a position similar to that of the cac genes of the IncQ plasmid RSF1010, and the cac genes have been shown to be involved in plasmid replication (16). An argument against pas involvement in replication is that when the pasAB genes of pTC-F14 are deleted or the pasABC genes of pTF-FC2 were spontaneously deleted, plasmid replication continued and no change in copy number was detected (26; this study, results not shown). Furthermore, the PasA- and PasB-like proteins of plasmid pAM10.6 are closely related to PasA and PasB of pTF-FC2 and pTC-F14, but pAM10.6 does not have an IncQ-like replicon and the pasAB genes of pAM10.6 do not appear to be part of the pAM10.6 replicon (20).
It is unlikely that the plasmids compete solely via the mechanism of PSK. This would require that the two replicons segregate away from one another at cell division (creating daughter cells lacking one or the other TA system and thus inducing growth inhibition or death). Since the replicons are compatible, this would be unlikely, and if it did occur, one would expect it to give rise to approximately equal numbers of daughter cells containing either pTC-F14 or pTF-FC2. Since we have shown that the non-self toxin-antidote proteins do not completely cross-neutralize one another or are equally effective at partial cross-neutralization, we would expect loss of either plasmid to invoke a PSK effect that would disadvantage or kill the host and, in doing so, eliminate the remaining plasmid. In effect, the two plasmids would be addicted to one another, and the majority of cells would be expected to carry both plasmids. We do not observe this. Instead, pTF-FC2 clearly out-competes pTC-F14, in spite of the observation that the latter seems to have the more virulent toxin.
Using pasA-lacZ reporter gene fusions, it was shown that the proteins of both pas operons could regulate each other, with the three-component pasABC system of pTF-FC2 being the stronger repressor of both, although this stronger repression was not attributable to the presence of pasC. This presents a possible mechanism for how pFC2-pasABC (pTF-FC2) out-competes pF14-pasAB (Cm) (pTC-F14) when together in a cell: the pas promoter of pF14-pasAB (Cm) is twice as effectively repressed by the competitor PasA than it is by its own PasA. Conversely, the pFC2-pasABC pas promoter is repressed less by the competitor PasA than by its native PasA. Presumably, when both plasmid species are present in the same host cell, pF14-pasAB (Cm) is disadvantaged through the action of PasA of pTF-FC2.
The evidence in support of PasA being responsible is that pFC2-pasAB*C (the pTF-FC2 derivative lacking a functional toxin gene) is able to out-compete pTC-F14, presumably through the action of its remaining PasA. Furthermore, pTC-F14 is unable to displace pTF-FC2 lacking its entire pas region (pFC2-ΔpasABC), suggesting that the survival of pFC2-ΔpasABC could be ascribed to its lack of the pas promoter-operator region, thus rendering it immune to any form of interference from the PasA of pTC-F14. It is possible that PasA serves multiple roles. We have shown that it is the primary regulator of the pas operon, causing strong repression of the promoter. Furthermore, when overexpressed, PasA can retard growth or possibly kill the bacterial host cell even in the absence of the PasB toxin. It acts as an antitoxin to the toxin. The role of PasAB(C) in plasmid biology is puzzling, especially as the whole pas of pTF-FC2 can be deleted without an apparent change in plasmid copy number, although there is a fairly modest but clearly detectable decrease in plasmid stability (26). It is possible that pas plays a role only when the copy number of the plasmid falls, whereas the upper limit for plasmid copy number is likely to be controlled by handcuffing in much the same way that it is for other iteron-containing replicons (7). In the same way that relBE (or mazEF) expression is linked to cellular stress (19), the primary role of the pasAB(C) operon may be to integrate plasmid replication into the overall metabolism of the host in which it resides. The pas could function under conditions in which a plasmid that was previously at a normal copy number falls, and hence, the overall expression of the pas operon decreases. By analogy to the mazEF system, failure to express the pas operon sufficiently would result in lower levels of PasA (since it has a shorter half-life) relative to PasB, and this would inhibit cell growth until the pas was again expressed at sufficiently high levels. This inhibition of cell growth would allow the plasmid copy number to catch up and would be expected to confer the relatively low levels of increased plasmid stability observed in the presence of pas without affecting the apparent plasmid copy number under optimal conditions.
Experiments are currently under way to determine whether readthrough from pas results in increased transcription of the downstream repAC genes, thereby ensuring that the plasmid is replicated when the copy number falls. This may not affect the maximum plasmid copy number that is likely to be set by iteron-mediated handcuffing.
Acknowledgments
We thank Kenn Gerdes for the gift of plasmids pOU82 and pKG399, from which pKGCm was constructed, and Hanna Engelberg-Kulka for the E. coli mazEF mutants.
This work was funded by grants provided by BHP-Billiton Johannesburg Technology Centre, the University of Stellenbosch, the National Research Foundation (Pretoria, South Africa), and the Training of Human Resources for Industry Program (THRIP; Pretoria, South Africa).
REFERENCES
- 1.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal addiction module regulated by ppGpp: a model for programmed cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein data base search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 4.Brosius, J., and A. Holy. 1984. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc. Natl. Acad. Sci. USA 81:6929-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadaban, M. J., J. Chou, and S. N. Cohen. 1980. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J. Bacteriol. 142:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifyable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattoraj, D. K. 2000. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 37:467-476. [DOI] [PubMed] [Google Scholar]
- 8.Gardner, M. N., S. M. Deane, and D. E. Rawlings. 2001. Isolation of a new broad host-range IncQ-like plasmid, pTC-F14, from the acidophilic bacterium Acidithiobacillus caldus and analysis of the plasmid replicon. J. Bacteriol. 183:3303-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner, M. N. 2003. Ph.D. thesis. University of Stellenbosch, Stellenbosch, South Africa.
- 10.Gerdes, K., J. E. Larsen, and S. Molin. 1985. Stable inheritance of plasmid R1 requires two different loci. J. Bacteriol. 161:292-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerdes, K. 2000. Toxin-antitoxin molecules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182:561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grønlund, H., and K. Gerdes. 1998. Toxin-antitoxin systems homologous with relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes. J. Mol. Biol. 285:1401-1405. [DOI] [PubMed] [Google Scholar]
- 13.Hazan, R., B. Sat, M. Reches, and H. Engelberg-Kulka. 2001. Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J. Bacteriol. 183:2046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen, R. B., and K. Gerdes. 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 17:205-210. [DOI] [PubMed] [Google Scholar]
- 15.Jensen, R. B., E. Grohmann, H. Schwab, R. Dìaz-Orejas, and K. Gerdes. 1995. Comparison of ccd of F, parDE of RP4, and parD of R1 using a novel conditional replication control system of plasmid R1. Mol. Microbiol. 17:211-220. [DOI] [PubMed] [Google Scholar]
- 16.Maeser, S., P. Scholz, S. Otto, and E. Scherzinger. 1990. Gene F of plasmid RSF1010 codes for a low molecular weight repressor protein that autoregulates expression of the repAC operon. Nucleic Acids Res. 18:6215-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magnuson, R., and M. B. Yarmolinsky. 1998. Corepression of the P1 addiction operon by Phd and Doc. J. Bacteriol. 180:6342-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1983. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Pedersen, K., S. K. Christensen, and K. Gerdes. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45:501-510. [DOI] [PubMed] [Google Scholar]
- 20.Peters, M., E. Jogi, I. Suitso, T. Punnisk, and A. Nurk. 2001. Features of the replicon of plasmid pAM10.6 of Pseudomonas fluorescens. Plasmid 46:25-36. [DOI] [PubMed] [Google Scholar]
- 21.Rawlings, D. E., I.-M. Pretorius, and D. R. Woods. 1984. Expression of a Thiobacillus ferrooxidans origin of replication in Escherichia coli. J. Bacteriol. 158:737-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawlings, D. E. 1999. Proteic toxin-antitoxin, bacterial plasmid addiction systems and their evolution with special reference to the pas system of pTF-FC2. FEMS Microbiol. Lett. 176:269-277. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 183:2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, A. S. G. 1997. Ph.D. thesis. University of Cape Town, Rondebosch, South Africa.
- 26.Smith, A. S. G., and D. E. Rawlings. 1997. The poison antidote stability system of the broad-host-range Thiobacillus ferrooxidans plasmid pTF-FC2. Mol. Microbiol. 26:961-970. [DOI] [PubMed] [Google Scholar]
- 27.Smith, A. S. G., and D. E. Rawlings. 1998. The efficiency of the pTF-FC2 pas poison-antidote stability system in Escherichia coli is affected by the host strain and antidote degradation requires the Lon protease. J. Bacteriol. 180:5458-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, A. S. G., and D. E. Rawlings. 1998. Autoregulation of the pTF-FC2 proteic poison-antidote plasmid addiction system (pas) is essential for plasmid stabilization. J. Bacteriol. 180:5463-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanisch-Perron, C., J. Viera, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]