Abstract
The effects of challenges with low (0.25%, vol/vol) and high (0.75%) concentrations of butanol on the growth, glucose metabolism, product formation, and transcriptional program of the solvent-tolerant Clostridium acetobutylicum strain 824(pGROE1) and the plasmid control strain 824(pSOS95del) were used to study solvent tolerance and stress response. Strain 824(pGROE1) was generated by groESL overexpression. The growth of 824(pGROE1) was less inhibited than that of 824(pSOS95del), and 824(pGROE1) was able to metabolize glucose over the entire course of the culture (60 h postchallenge) while glucose metabolism in 824(pSOS95del) lasted 24 h. A comparison of their respective DNA array-based transcriptional profiles identified genes with similar expression patterns (these genes are likely to be part of a general butanol stress response) and genes with opposite expression patterns (these genes are likely to be associated with increased tolerance to butanol). Both strains exhibited a butanol dose-dependent increase in expression of all major stress protein genes, including groES, dnaKJ, hsp18, and hsp90; all major solvent formation genes, including aad, ctfA and -B, adc, and bdhA and -B (an unexpected and counterintuitive finding); the butyrate formation genes (ptb and buk); the butyryl coenzyme A biosynthesis operon genes; fructose bisphosphate aldolase; and a gene with homology to Bacillus subtilis kinA. A dose-dependent decrease in expression was observed for the genes of the major fatty acid synthesis operon (also an unexpected and counterintuitive finding), several glycolytic genes, and a few sporulation genes. Genes with opposite expression kinetics included rlpA, artP, and a gene encoding a hemin permease. Taken together, these data suggest that stress, even when it derives from the solvent product itself, triggers the induction of the solvent formation genes.
Metabolically engineered solventogenic clostridia may potentially lead to industrial processes for the production of solvents (such as butanol and acetone), other oxychemicals, and enzymes (30, 35, 41) or for biotransformation (57). In addition, clostridia can degrade a number of toxic chemicals and have great potential for bioremediation applications (15, 16, 25, 46, 54). In all such applications, the ability of cells to withstand “stressful” conditions such as high concentrations of substrates and the accumulation of toxic products without loss of productivity is a most significant goal. The difficulty—but also the intellectual and biotechnological challenge—is that the desirable phenotypic trait may be determined by several genes or a complex regulatory network. Solvents are inherently toxic to bacteria. Solvent tolerance, particularly ethanol tolerance, has been extensively examined in gram-negative organisms (38) but rarely, if at all, in gram-positive organisms. Gram-negative bacteria are generally much more resistant to increasingly polar solvents that gram-positive prokaryotes (27, 28, 51). The initial effect is disruption of membrane fluidity, and cells may attempt to adjust lipid composition to maintain fluidity, a process known as homeoviscous adaptation (45). Growth has been shown to be the most sensitive cellular activity, while glycolysis is more resistant to the effects of solvents (26). As for other solvents, the toxicity of butanol in solventogenic clostridia is attributed to its chaotropic effect on the cell membrane (5, 52). Butanol inhibits active nutrient transport, the membrane-bound ATPase, and glucose uptake (5); partially or completely abolishes the membrane ΔpH (5, 18, 48) and Δψ (48); and lowers the intracellular pH (5, 23, 48) and ATP concentration (5). Lipids from exponential-phase cultures (where low levels of butanol are present) contain approximately 58% saturated acyl chains, while stationary-phase lipids (where high levels of butanol are present) contain approximately 77% saturated chains. This alteration occurs in an attempt to maintain membrane fluidity. Exponential-growth static flask cultures grown in 0.5 or 1.0% (vol/vol) butanol had saturated chain contents of 65 or 73%, respectively, but cultures with 1.5% butanol did not grow at all (37, 52). Finally, moderate increases in butanol concentration, similar to heat shock, have been shown to induce known stress proteins (heat shock proteins [HSPs]) (48).
In order to test the hypothesis that overexpression of HSPs will result in increased cellular resistance to toxic solvents, Tomas et al. constructed (50) a recombinant Clostridium acetobutylicum strain [824(pGROE1)] carrying plasmid pGROE1 for overexpressing the groESL operon genes under the control of the clostridial thiolase promoter. By using this strain, it was shown that GroESL overexpression results in prolonged metabolism and increased butanol production and tolerance. That was the first report to show that HSP overexpression imparts solvent tolerance. In 24-h butanol challenge experiments with exponential-growth-phase cultures of 824(pGROE1), 824(pGROE1) was initially inhibited less than 5% at 0.25 and 0.50% (vol/vol) butanol and was inhibited 7% at 0.75% butanol (t, <8 h), but within 24 h, growth inhibition reached 25% at 0.75% butanol while it remained below 5% with the 0.25% butanol challenge. The plasmid control strain [824(pSOS95del)] was inhibited by as much as 30% initially and more than 37% at 24 h. The objective of this study is to examine the long-term (>24-h) growth characteristics, product formation, and large-scale DNA array-based transcriptional program of these strains when they are subjected to low- and high-dose butanol challenges. We hypothesized that these studies would elucidate standing hypotheses regarding solvent tolerance, sporulation, and solvent production. For example, despite the widely held view that sporulation, solvent formation, and the heat shock response are linked in solvent-producing clostridia, the molecular natures of the common and separate control mechanisms remain unclear (3, 29, 34, 41, 42). The major signal transduction event for the initiation of sporulation in Bacillus subtilis is a phosphorelay mechanism which responds to environmental and metabolic signals by phosphorylating Spo0A (the master regulator of differentiation), thereby activating its function. While an exactly analogous phosphorelay system has not been identified in C. acetobutylicum, Spo0A has been shown to play a major role in differentiation. An activated Spo0A has been recently shown to induce all key solvent formation genes (21, 40), thereby mediating many, if not all, possible triggers of solventogenesis.
MATERIALS AND METHODS
Bacterial strains.
C. acetobutylicum ATCC 824 (American Type Culture Collection, Manassas, Va.) is the wild-type strain. Strain 824(pGROE1) overexpresses the groESL operon and has been described previously (50). Strain 824(pSOS95del) serves as the plasmid control strain (50).
Analytical methods.
Cell growth was measured by using the absorbance at 600 nm (A600) on a Thermo Spectronic (Rochester, N.Y.) BioMate3 spectrophotometer. A mass extinction coefficient of 51 (g of cells)−1 cm−1 was used to convert optical density measurements (A600) into cell concentrations (dry weight), expressed in grams of cells per liter (20). Absorbance readings were kept below 0.40 by diluting samples with the appropriate media. Product (acetate, butyrate, acetone, butanol, ethanol, and acetoin) and glucose concentrations were analyzed by using a Waters (Milford, Mass.) high-performance liquid chromatography system (50). Acetoin concentrations were determined to be less than 2 mM in all cultures.
Growth conditions and maintenance.
C. acetobutylicum strains were grown in an anaerobic chamber (Forma Scientific, Marietta, Ohio) at 37°C. Clostridial growth medium (CGM) was used in liquid cultures (55). Colonies used to inoculate liquid cultures were obtained from agar-solidified 2× YTG plates (55). Colonies were picked from plates at least 4 days old and were heat shocked at 70 to 80°C for 8 min before being used to inoculate cultures. Recombinant strains were grown on 100 μg of erythromycin/ml, except for static flask cultures, where 75 μg of clarithromycin/ml was used.
Butanol challenge experiments.
Static flask cultures containing 650 ml of CGM were grown with strains 824(pSOS95del) and 824(pGROE1). The flasks were inoculated with 6.5 ml of preculture (1/100) at an A600 of 0.60. The large flask was grown to an A600 of 0.8 ± 0.05 and then split into three smaller, 200-ml flasks (with closed lids). Two of the flasks were challenged with 0.25 or 0.75% (vol/vol) butanol, and one flask was left unchallenged. Relative growth was calculated as the ratio of the growth of the butanol-challenged culture to that of the unchallenged culture multiplied by 100 to yield a percentage. Six RNA samples were taken for use in DNA array analysis at 0.25, 1, 3, 6, 12, and 24 h after butanol addition.
Western blot analysis.
Crude cell extracts were prepared, and Western blot analysis was performed, as previously described (50).
RNA sampling, isolation, and purification.
RNA sampling and isolation were performed as previously described (50), with the following modifications. Cell pellets from 2 to 5 ml of culture were used. For isolation, thawed samples were diluted twofold (versus fivefold) in ice-cold TRIzol (Invitrogen, Carlsbad, Calif.).
cDNA labeling and hybridization.
Labeled cDNA was synthesized by random hexamer-primed reverse transcription reactions in the presence of Cy3-dUTP or Cy5-dUTP by using SuperScript II (Invitrogen) reverse transcriptase as previously described (49). Labeled cDNA was hybridized on a targeted cDNA array containing spots representing 1,019 open reading frames (ORFs), approximately one-fourth of the C. acetobutylicum genome (49). Genes included in this generation of arrays include, among others, 169 of 178 pSOL1 ORFs (corrected from a previous publication [49]), 123 DNA replication and repair genes (90% of the total of such genes as identified by the genome annotation [36]), 97 cell division- and sporulation-related genes (92%), 85 carbohydrate and primary metabolism genes (31%), 67 energy production genes (52%), 63 outer membrane and cell envelope genes (36%), 48 lipid metabolism genes (80%), 42 motility and chemotaxis genes (39%), and all previously identified stress response genes. A complete list can be found at http://www.chem-eng.northwestern.edu/Faculty/papou.html.
Microarray data analysis.
The data were normalized, and genes that showed significant differences in expression level were determined by using the methods of Yang et al. (56). All array data were subjected to a prefilter criterion as previously described (49). Average linkage hierarchical clustering was performed using Cluster, and gene clusters were visualized in TreeView (13). Data generated using these targeted cDNA arrays and these data analysis methods have been shown to be highly reliable. Analysis of a degenerate C. acetobutylicum strain (M5 [9]) missing 178 genes properly classified genes with an accuracy of 99.4% (49). Comparison to Northern blot (49) and quantitative reverse transcription-PCR (50, 56) analyses has also shown this method to be accurate in identifying genes as differentially expressed and to be conservative in estimating relative gene expression levels.
RESULTS
824(pGROE1) growth and metabolism displays prolonged tolerance to butanol addition.
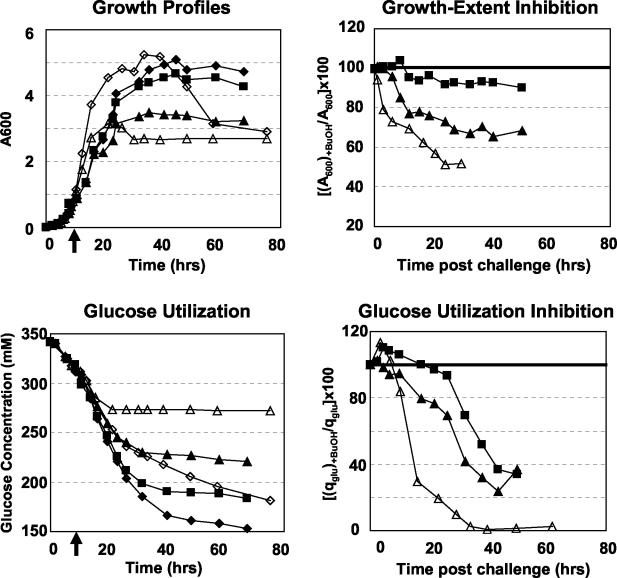
Cultures of 824(pGROE1) and 824(pSOS95del) were subjected to butanol challenges [0.25 and 0.75% (vol/vol) for 824(pGROE1); 0.75% (vol/vol) for 824(pSOS95del)] during mid-exponential growth (A600, 0.8), and the degree of growth inhibition was determined relative to the growth of the unchallenged cultures (Fig. 1). 824(pGROE1) subjected to 0.25% butanol was initially uninhibited (t, <10 h). After 10 h, growth inhibition increased slowly, reaching 15% at 78 h post-butanol challenge. 824(pGROE1) with 0.75% butanol was also initially uninhibited (t, <6 h), but then growth inhibition increased more rapidly (relative to that with 0.25% butanol), finally leveling off at approximately 33%. In contrast to that of 824(pGROE1), the growth of 824(pSOS95del) with 0.75% butanol was inhibited within 1 h and reached A600 levels equal to half those in the unchallenged culture within 24 h, after which no net growth was observed. Glucose utilization was also inhibited by addition of butanol (Fig. 1). In order to assess the effects of butanol on glucose metabolism while accounting for differences in cell densities, the ratio of the specific glucose uptake rate (qglu; expressed in grams of glucose per gram [dry weight] of cells) of the challenged culture to that of the unchallenged culture (multiplied by 100 to yield a percentage) was calculated, and the ratios were plotted (Fig. 1). Specific glucose metabolism was initially higher (t, <15 h) in the 824(pGROE1) culture challenged with a low dose of butanol (glucose utilization, >100%) than in the unchallenged culture, while glucose metabolism was approximately 10% lower in the high-dose butanol-challenged 824(pGROE1) culture. The specific glucose metabolism of 824(pSOS95del) was also initially higher but fell much more rapidly than that of the 824(pGROE1) cultures. Specific glucose metabolism in both of the 824(pGROE1) butanol-challenged cultures fell to less than 40% that of unchallenged cultures by 40 h after butanol challenge, after which all cultures exhibited slow glucose metabolism. Specific glucose metabolism in the 824(pSOS95del) control strain fell to less than 30% within 12 h and to near zero within 24 h.
FIG. 1.
(Left) Growth and glucose utilization profiles for butanol-challenged 824(pGROE1) (solid symbols) and 824(pSOS95del) (open symbols) cultures. Diamonds, unchallenged cultures; squares, 0.25% (vol/vol) butanol [824(pGROE1) only]; triangles, 0.75% (vol/vol) butanol. Arrows along the x axes indicate the time of butanol addition. (Right) Diagrams of growth inhibition and glucose utilization inhibition. Calculations for growth inhibition were carried out up to the point where cell growth and glucose uptake apparently ceased. Specific glucose metabolism (qglu) is calculated as grams of glucose per gram of cells (dry weight).
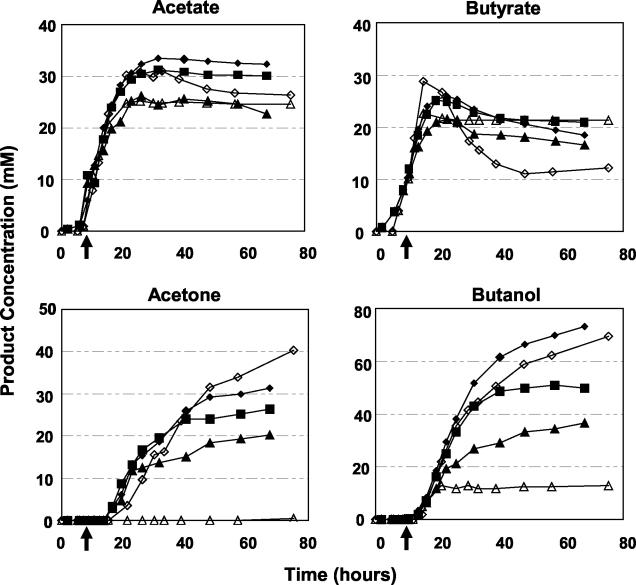
Significant differences in product formation were also observed (Fig. 2). Production and reassimilation of acids, particularly butyrate, were altered in the butanol-challenged cultures. Reductions in peak acetate and butyrate levels in the two strains were very similar (17 and 21% reductions, respectively) after addition of 0.75% butanol. However, butyrate uptake in the butanol-challenged 824(pSOS95del) culture was reduced by 89% by addition of 0.75% butanol, whereas butyrate uptake in 824(pGROE1) was reduced by only 17%. Reassimilation of acids is required for the production of acetone. It is therefore not surprising that acetone production in the butanol-challenged 824(pSOS95del) culture was less than 1 mM (Fig. 2), while the groESL-overexpressing strain was able to produce 20 mM acetone (35% less than the unchallenged culture). Butanol challenge (0.75%) reduced ethanol production (data not shown) by 15% (from 21.7 to 18.5 mM) in 824(pGROE1) compared to 45% (from 15.3 to 8.4 mM) in 824(pSOS95del). Butanol production was reduced in both strains, but to a lesser extent in 824(pGROE1) than in 824(pSOS95del) (a 50 versus an 81% reduction, respectively). Butanol and acetone concentrations were still increasing at the end of the 824(pGROE1) cultures, while butanol production lasted only 9 h in the butanol-stressed 824(pSOS95del) culture. These results suggest that 824(pGROE1) exhibits a prolonged tolerance to butanol addition spanning the entire course of the culture (at least 40 to 60 h after butanol addition).
FIG. 2.
Product formation for 824(pGROE1) (solid symbols) and 824(pSOS95del) (open symbols) at various levels of butanol challenge: diamonds, 0.0% (vol/vol); squares, 0.25% (vol/vol) [824(pGROE1) only]; triangles, 0.75% (vol/vol). Arrows along the x axes indicate the time of butanol addition.
Transcriptional analysis of butanol-stressed 824(pGROE1) cultures.
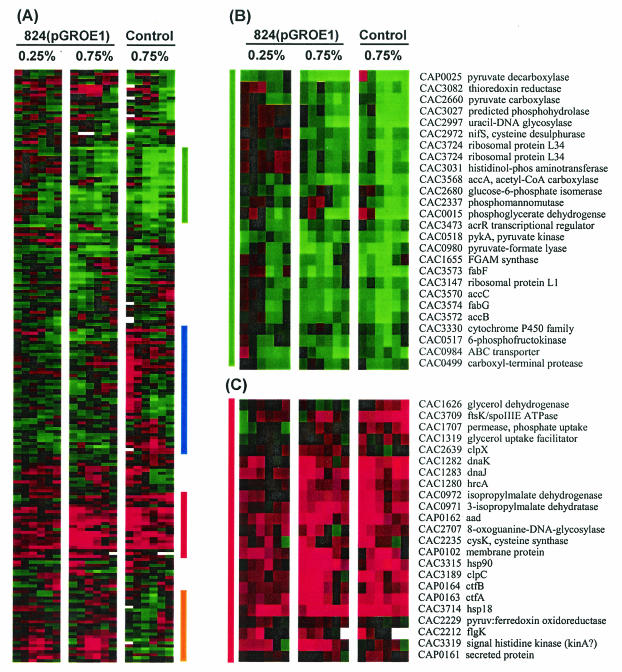
DNA array-based transcriptional analysis of the butanol-challenged 824(pGROE1) and 824(pSOS95del) cultures relative to their respective unchallenged cultures was performed in order to better understand the transcriptional responses of butanol tolerance and butanol stress. Samples for array analysis were taken from each culture at 0.25, 1, 3, 6, 12, and 24 h after butanol addition and were analyzed on duplicate DNA arrays. All duplicate arrays were hybridized with reverse-labeled samples (e.g., challenged-Cy3/unchallenged-Cy5 and challenged-Cy5/unchallenged-Cy3). The resulting array data were normalized, and genes differentially expressed at the 95% confidence level (56) were identified: 152 genes for 824(pGROE1) plus 0.25% butanol, 104 genes for 824(pGROE1) plus 0.75% butanol, and 154 genes for 824(pSOS95del) plus 0.75% butanol. In order to compare the similarities and differences in gene expression for the two butanol challenge levels and the two strains, the three lists of differentially expressed genes were combined into a single list of 199 genes. The complete lists with expression profile data can be found at http://www.chem-eng.northwestern.edu/Faculty/papou.html. The combined gene list was analyzed by average linkage hierarchical clustering (Fig. 3A) (13).
FIG. 3.
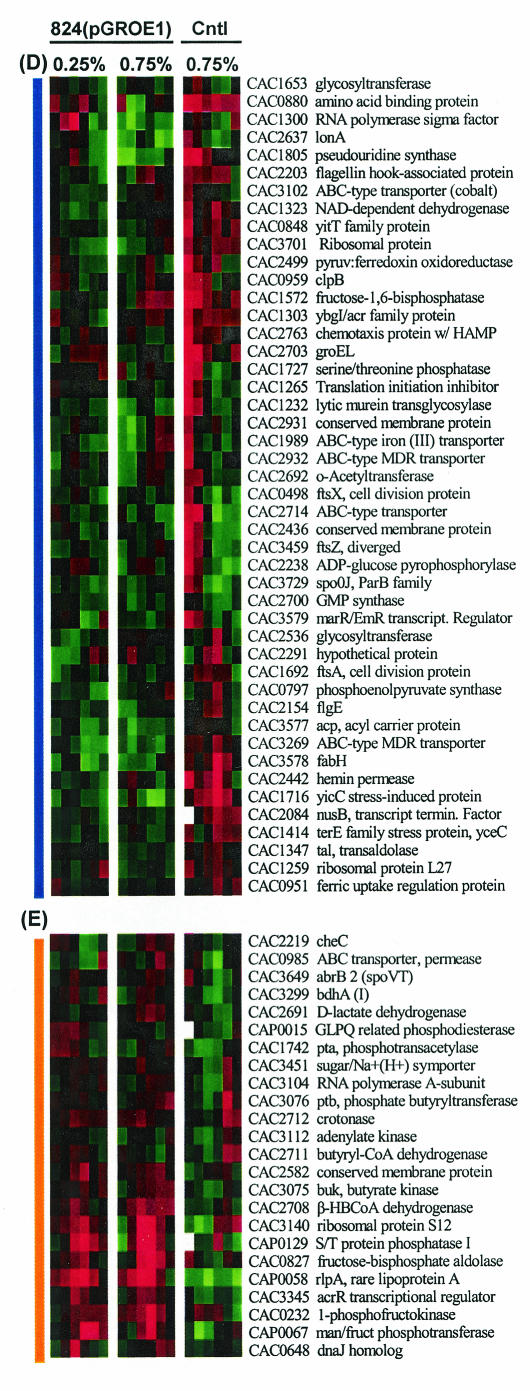
Expression profiles of differentially expressed genes for butanol-challenged 824(pGROE1) and 824(pSOS95del) cultures. (A) Hierarchical gene clustering of the low (0.25%)- and high (0.75%)-dose butanol-challenged 824(pGROE1) and p(SOS95del) cultures. Colored vertical bars on the right indicate clustered regions shown in detail in panels with corresponding bars on the left. (B through E) Detailed views of genes showing generally higher expression (B), generally lower expression (C), lower expression in butanol-challenged 824(pGROE1) cultures but higher expression in butanol-challenged 824(pSOS95del) cultures (D), or higher expression in butanol-challenged 824(pGROE1) cultures but lower expression in butanol-challenged 824(pSOS95del) cultures (E). Red and green indicate higher and lower expression, respectively, in the butanol-challenged cultures relative to the unchallenged culture.
Comparison of the expression profiles (Fig. 3A) for the two 824(pGROE1) butanol challenge experiments (0.25 and 0.75% butanol) reveals that the vast majority of genes identified as differentially expressed showed the same direction (up or down) of differential expression. Several notable exceptions include the thiolase gene (thlA; CAC2873), an acetyl coenzyme A (acetyl-CoA) carboxylase (accD [CAC3569]) belonging to a fatty acid biosynthesis operon, the phosphotransacetylase gene (pta [CAC1742]) involved in acetate production, a phosphotransferase (CAP0066), phosphodiesterase (CAP0015), and a predicted dehydrogenase (CAC1480). Although the direction of differential expression for most genes identified is the same at the two 824(pGROE1) butanol challenge levels for most genes, the fold differences for the 0.75% butanol challenge experiment are generally higher than those for the 0.25% butanol challenge (more-intense red and green in Fig. 3A), suggesting that the transcriptional response to butanol is dose dependent.
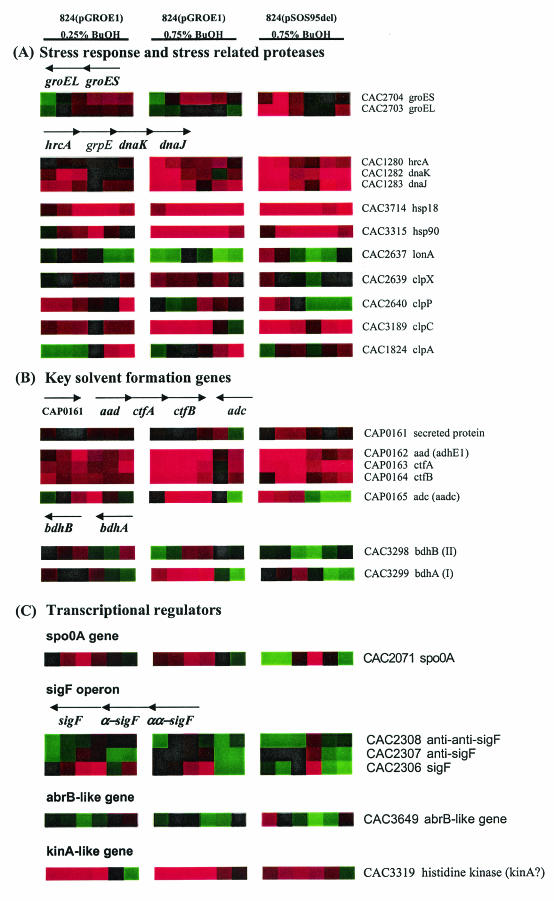
Comparison of all three experiments reveals several areas with very similar gene expression patterns (Fig. 3B and C) and several areas with different expression patterns (Fig. 3D and E). Genes with the same expression patterns for all three experiments are likely to be part of a general butanol stress response. Genes whose expression patterns are opposite for 824(pSOS95del) and 824(pGROE1) are likely to represent candidate genes that have a role in the increased tolerance of 824(pGROE1) to butanol. Among the genes with generally elevated gene expression in all the butanol-challenged cultures (Fig. 3C) are two distinct groups: stress response genes and solvent formation genes. Three major stress response protein families (the dnaKJ operon, hsp18, and hsp90) are highly overexpressed (>2-fold [Fig. 4A]) in all three experiments. All three gene families are overexpressed earlier and more strongly in the 824(pGROE1) high-dose butanol challenge experiment than in the low-dose 824(pGROE1) challenge. The dnaKJ operon is highly overexpressed early in both the 824(pGROE1) and 824(pSOS95del) high-dose experiments. dnaKJ overexpression was less pronounced at 3 and 6 h in both cultures, but it increased back to the initial overexpression level in 824(pSOS95del) but not in 824(pGROE1). Both hsp18 and hsp90 were highly overexpressed throughout the entire course of culture. Expression of groESL was initially (t, 15 min) lower in both butanol-stressed 824(pGROE1) cultures, with slightly higher expression (a <1.5-fold increase) through the remainder of the culture. The lower fold increases in groESL expression relative to those of the other major stress response genes are likely due to overexpression of groESL from the pGROE1 plasmid. Expression of groESL from pGROE1 is driven by the thiolase promoter (50) rather than by its natural promoter, which is negatively regulated by HrcA through a CIRCE element (43). Therefore, expression from the plasmid likely masks differences in expression from the naturally occurring groESL operon. For 824(pSOS95del), groESL expression was initially (t, <1 h) higher in the stressed cultures but had fold ratios near 1 thereafter. This suggests that groESL plays a larger role in the initial response to butanol stress. The proteases of the clp and lon families are another key element of the stress response, responsible for the degradation of misfolded proteins. Differential expression was observed in all three butanol-challenged cultures for several members of the clp family (clpA [CAC1824], clpC [CAC3189], clpP [CAC2640], and clpX [CAC2639]) and for lonA (CAC 2637) (Fig. 4A). Higher expression of clpC was observed in all three experiments. Expression of lonA was lower in the 824(pGROE1) cultures, while it was initially higher in the 824(pSOS95del) butanol-stressed culture.
FIG.4.
Expression profiles of the known stress response genes (A), key solvent formation genes (B), and transcriptional regulators (C). Operon structures are shown above the expression profiles for known operons and gene clusters. Arrows pointing to the right or left indicate transcription from the positive or negative strand, respectively. Red and green indicate higher and lower expression, respectively, in the butanol-challenged cultures relative to the unchallenged culture.
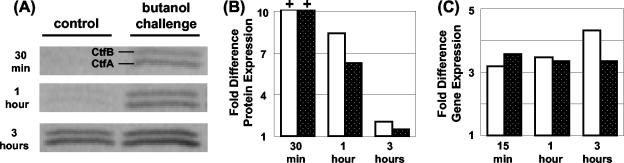
Overexpression of the solvent formation genes in the butanol-challenged cultures is unexpected and counterintuitive but certainly profound (Fig. 4B). Overexpression of the solvent genes was, again, butanol dose dependent. Genes of the sol locus (CAP0161 to CAP0165) exhibited an immediate and high fold increase (>3-fold) in expression at the 0.75% butanol stress level, particularly in the 824(pGROE1) culture. Expression remained highly elevated up to 12 h after butanol addition and showed a slight decrease thereafter. At the lower (0.25%) butanol level, overexpression in 824(pGROE1) was relatively lower (approximately 1.8- to 2.0-fold) over the entire 24 h of analysis. Two genes encoding butanol dehydrogenases (CAC3298 and CAC3299) exhibited a pattern of expression similar to that of adc (CAP0165). The solvent formation genes were highly overexpressed in the butanol-stressed cultures despite the fact that they produced lower levels of solvents. To test whether transcriptional differences in the solvent formation genes result in differences at the protein level, Western blot analyses (n = 2) for the two acetyl-CoA transferase protein units (CtfA and CtfB) were performed on challenged (0.75% butanol) and unchallenged 824(pGROE1) samples at 0.5, 1, and 3 h after butanol addition. Results from the Western blot analysis (Fig. 5) revealed increased levels of CoAT, as predicted by the DNA array transcriptional data. At 30 min post-butanol challenge, a significant band (>29 times background) was present in the butanol-challenged culture, while no detectable band was present in the unchallenged culture. These findings reveal that butanol stress results in increased expression of solvent formation genes and proteins.
FIG. 5.
(A) Western blot for the two units (CtfA and CtfB) of CoAT. Shown is a representative blot for an 824(pGROE1) culture stressed with 0.75% butanol. Times shown are times post-butanol challenge. (B and C) Protein (B) and gene (C) expression levels for a butanol-challenged culture relative to those for an unchallenged control culture. The fold difference in protein or gene expression is calculated as the protein or gene expression level in the butanol-stressed culture divided by that in the control culture without butanol addition. Values are averages from two gels or microarrays. Fold protein differences for the 30-min time point are reported as 10+ because no quantifiable bands were present in the 30-min control sample. Symbols: open bars, CtfA; solid bars, CtfB.
Regulation of the solvent formation genes has been strongly linked to expression and subsequent activation (by phosphorylation) of spo0A (21, 40). The expression pattern of spo0A (Fig. 4C) in the 824(pGROE1) cultures was very similar to that of the solvent formation genes. However, the expression pattern of spo0A in 824(pSOS95del) was lower during the early (t, <1 h) and later (t, 24 h) stages of analysis, with higher expression 6 h after butanol challenge. This is different from the expression pattern of the solvent formation genes. However, the sporulation-specific sigma factor F (sigF [CAC2306]), whose transcription is directed by spo0A, had a gene expression pattern in all three experiments that was very similar to that of spo0A (Fig. 4C). In addition, the expression of abrB (negatively regulated by Spo0A in B. subtilis [14]) was opposite that of spo0A. This further corroborates the spo0A expression data. CAC3319 (annotated as a histidine kinase [36]), which bears a high similarity to B. subtilis kinA (49), was expressed at a significantly higher level (>2-fold) in all three butanol-stressed cultures (Fig. 4C). The KinA protein is a phosphorelay protein that helps initiate sporulation by triggering the cascade which results in increased levels of active Spo0A-P. The expression of other sporulation-related genes was also altered (generally lower expression in all butanol-challenged cultures), including that of spo0J (CAC3729 [Fig. 3D]), spoVR (CAC0581), spoIIP (CAC1276), cotS (CAC2909), and a gene containing a spo0T-type domain (CAC3340).
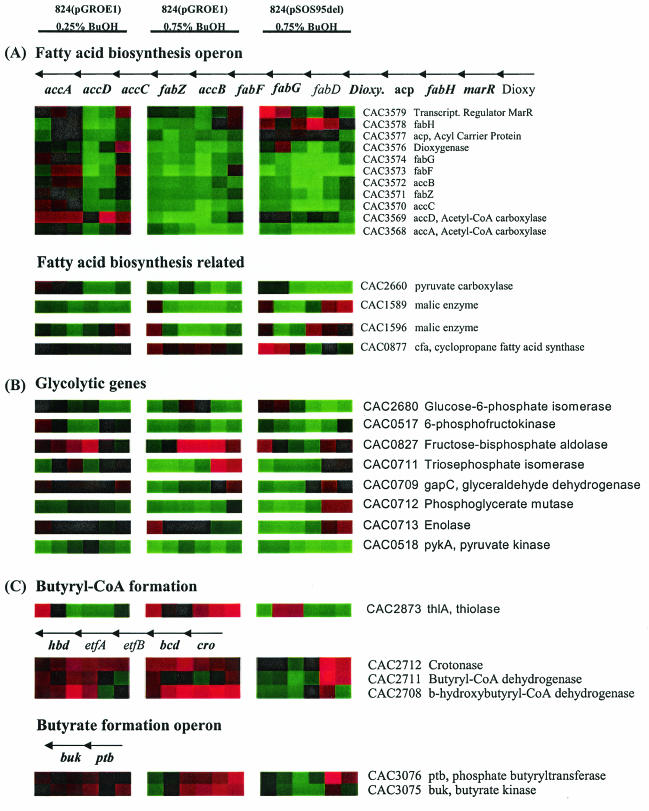
Eleven genes belonging to a fatty acid biosynthesis (fab) operon (CAC3579 to CAC3568 [Fig. 6A]) were identified as having significantly lower expression in the high-dose butanol-stressed cultures. The response to butanol stress in the low-dose 824(pGROE1) culture was initially higher expression for most of the fab genes (up to 3 h) with lower expression thereafter, except for accD (CAC3569). The response was immediate and sustained in both high-dose cultures, with the lowest level of expression in the butanol-challenged 824(pSOS95del) culture. Included in the fatty acid biosynthesis operon are the genes encoding two subunits of an acetyl-CoA carboxylase (CAC3568 and CAC3569). Acetyl-CoA carboxylase plays a critical role in controlling fatty acid metabolism. Expression of the fabH gene, encoding a β-ketoacyl-acyl carrier protein synthase III gene which catalyzes the condensation of malonyl-acyl carrier protein and an acyl-CoA substrate (32), was increased in the less-tolerant 824(pSOS95del) strain and lower in the more-tolerant 824(pGROE1) strain. FabH has been shown to be an important determinant of branched-chain fatty acid synthesis (8) in B. subtilis. In addition, a gene for pyruvate carboxylase (CAC2660 [Fig. 3B]) and two genes encoding a malic enzyme (CAC1589 and CAC1596) had gene expression patterns similar to those of the genes of the fatty acid biosynthesis operon (Fig. 6A). Pyruvate carboxylase and malic enzymes are closely linked to fatty acid biosynthesis through their role in generating NADPH, which is required for fatty acid synthesis. Expression of the malic enzymes in the butanol-challenged 824(pSOS95del) culture was lower than that in the unchallenged culture 6 to 12 h after butanol challenge, the same time at which the dramatic decrease in glucose metabolism occurred and just before cessation of butanol production in 824(pSOS95del). The highly differentially regulated gene expression patterns for the entire fatty acid biosynthesis operon and several related genes provide clear evidence that butanol stress alters the regulation of the fatty acid biosynthesis machinery. Increased expression of the cyclopropane fatty acid synthase gene (cfa [CAC0877]) in C. acetobutylicum has been shown to result in butanol resistance (58). Elevated expression of cfa was observed early in the high-dose 824(pSOS95del) culture, while cfa was <1.7 times higher in the high-dose 824(pGROE1) culture (Fig. 6A). Cfa appears to play a lesser role in response to butanol stress in 824(pGROE1) than in 824(pSOS95del).
FIG. 6.
Expression profiles of the fatty acid biosynthesis operon and related genes (A), glycolytic pathway genes (B), and butyryl-CoA formation genes (C). Operon structures are shown above the expression profiles for known operons. Arrows pointing to the left indicate transcription from the negative strand. Red and green indicate higher and lower expression, respectively, in butanol-challenged cultures relative to the unchallenged culture.
A gene for rare lipoprotein A (CAP0058) is overexpressed in both of the 824(pGROE1) cultures but shows the opposite direction of expression in 824(pSOS95del) (Fig. 3E). Several lipoproteins have been shown to act as membrane chaperones, preventing unproductive interactions with the cell wall (53), and have been shown to be overexpressed in the cyanobacterium Synechocystis sp. after hyperosmotic stress (31). A truncated rlpA gene in Escherichia coli (4) was able to rescue a conditionally lethal mutation in the prc gene (involved in C-terminal processing of penicillin-binding protein-3). prc mutants are sensitive to heat and osmotic stress (19). A conserved membrane protein (CAC2582) is in this same cluster of oppositely regulated genes (Fig. 3E), while two additional conserved membrane proteins (Fig. 3D) had significantly higher expression immediately following butanol addition in 824(pSOS95del) but had slightly lower expression in both 824(pGROE1) experiments. Many genes related to transport system functions displayed this pattern of opposite expression as well. These included an amino acid binding protein (CAC0880), a hemin permease (CAC2442), and several other ABC type transporters (CAC1982, -2714, -2932, -3102, and -3269). The amino acid binding protein (CAC0880) has high homology to artP (formerly yqiX) of B. subtilis, a member of a tricistronic arginine transport operon, which has been shown to be induced during heat shock (22). A BLAST search found two genes with a high degree of homology to the other two members of the B. subtilis arginine transport operon, and these, together with CAC0880, form a predicted operon in C. acetobutylicum (49). The operon has sequences in the promoter region for σA and σF, as well as a Spo0A binding sequence. Hemin permease has been associated with the cellular response of strict anaerobes to oxygen stresses (6). Two genes encoding putative pseudouridine synthase (CAC1805 and CAC1414) also belong to this cluster (Fig. 3D). YceC in E. coli has been shown to modify uridine residues of both 23S rRNA and 16S rRNA (24). Finally, a yicC stress-induced protein (Fig. 3D) required for survival at high temperatures in E. coli (39) also showed an opposite pattern of expression, with higher expression in 824(pSOS95del).
Several glycolytic genes are also included in the cluster containing genes with decreased expression in the butanol-challenged cultures. Included in this group of differentially expressed genes are 6-phosphofructokinase, glucose 6-phosphate isomerase, and pyruvate kinase. Also identified as differentially expressed were phosphoglycerate mutase, triosephosphate isomerase, glyceraldehyde 3-phosphate dehydrogenase, and fructose bisphosphate aldolase. The glycolytic genes are shown together in Fig. 6B. All but fructose bisphosphate aldolase were generally expressed at lower levels in the stressed cultures than in the unstressed culture. Expression of fructose bisphosphate aldolase was elevated in the butanol-stressed 824(pGROE1) cultures between 1 and 12 h. The gene expression patterns correlate well with the observed relative decrease in glucose metabolism (Fig. 1). Although the glycolytic genes responsible for the formation of pyruvate had lower expression in the butanol challenge experiments, the expression of genes responsible for the conversion of pyruvate to butyryl-CoA (Fig. 6C) was significantly higher in the butanol-challenged 824(pGROE1) cultures. Expression of the thiolase gene in the low-dose 824(pGROE1) culture was initially higher, but it was lower than that in the unchallenged culture 3 h after butanol challenge. Butyryl-CoA serves as an intermediate in the production of either butanol or butyrate. The genes encoding the enzymes for production of butyrate (buk [CAC3075] and ptb [CAC3076]) have a pattern very similar to that of the butyryl-CoA pathway genes (Fig. 6C). Finally, several genes encoding ribosomal proteins (CAC3724 [two probes present on the array] and CAC3147 [Fig. 3B]) had lower expression in the butanol-stressed cultures. Two additional genes (CAC1265, a translation initiation inhibitor, and CAC1259 [rpl27]) encoding ribosomal proteins have opposite patterns of expression in 824(pGROE1) versus 824(pSOS95del) (Fig. 3D).
DISCUSSION
Overexpression of groESL in C. acetobutylicum results in long-term (>10 h) butanol resistance compared to that in the plasmid control strain. This finding expands and strengthens previously reported data (50). Experiments with the wild-type strain have produced results similar to those with the plasmid control strain (C. A. Tomas, J. A. Beamish, and E. T. Papoutsakis, unpublished data). While growth is considered to be one of the cellular functions most sensitive to alcohols, glucose metabolism can also be negatively affected (26). 824(pGROE1) was able to metabolize glucose through the entire course of the culture, while glucose metabolism in 824(pSOS95del) ceased 24 h after butanol addition (Fig. 1). The increase in the specific glucose ratio seen in 824(pGROE1) cultures 30 h post-butanol challenge is due to the fact that butanol levels in the unchallenged culture begin to approach butanol levels in the challenged cultures (added plus produced). The growth and glucose metabolism patterns of 824(pGROE1) suggest that increased expression of the GroES and GroEL proteins in 824(pGROE1) serves to stabilize the general biosynthetic machinery. In 824(pGROE1), expression of groESL has been shown to remain elevated, relative to that in the wild type and the plasmid control, throughout the entire course of culture (50), thereby providing extended protection against cellular stress.
Analysis of the transcriptional response of 824(pGROE1) to different levels of butanol challenge (0.25 and 0.75%) and comparison to the transcriptional response of the control strain allow for the identification of genes or groups of genes that are part of a general response to butanol stress, as well as those likely to play a role in the observed solvent tolerance of 824(pGROE1). The increased expression of the stress response genes, solvent formation genes, and many other genes appears to be butanol dose dependent. The strong dependence on the butanol level suggests that C. acetobutylicum has a mechanism for sensing varying levels of butanol and altering gene expression accordingly. Overexpression of the key solvent formation genes upon addition of butanol seems an unlikely response to such a stress. Furthermore, increased expression of the solvent formation genes is not associated with increased solvent production. When viewed in connection with the overexpression of nearly all the known major heat and stress response genes (groESL, dnaKJ, hsp18, hsp90, and several stress response proteases), this result suggests that solvent gene expression is a response to environmental stress (such as butanol addition). Other stresses known to induce solvent production (3, 12, 17) include low pH, high carboxylic acid concentrations, membrane potential uncouplers, and limitations in iron, nitrogen, and phosphate. The stress response in C. acetobutylicum has long been linked to solvent formation and sporulation (3, 42). However, the molecular natures of the common and different control mechanisms are still largely unknown (21). Differential expression of several key transition state regulators and factors responsible for control of many stationary-phase-associated processes upon butanol stress suggests a strong link among the three processes. spo0A, sigF, and abrB are all differentially expressed in butanol-challenged cultures (Fig. 4C). The sigF tricistronic operon, known to be positively regulated by Spo0A in B. subtilis (47), has an expression pattern similar to that of spo0A. Spo0A has also been shown to negatively regulate abrB expression in B. subtilis (47). AbrB is responsible for the regulation of genes involved in sporulation and gene regulation. The expression patterns of sigF and abrB in relation to spo0A suggest a relationship among the three genes identical to that known for B. subtilis (47). In B. subtilis, the sensory transduction kinase KinA activates Spo0A through phosphorylation. The increased expression (under butanol stress) of spo0A and the putative kinA throughout most of the 824(pGROE1) cultures correlates well with increased expression of the solvent formation genes. The level of spo0A expression in 824(pSOS95del) does not correlate as well with the increased expression of the solvent formation genes. However, expression of the putative kinA is higher in the butanol-stressed 824(pSOS95del) culture.
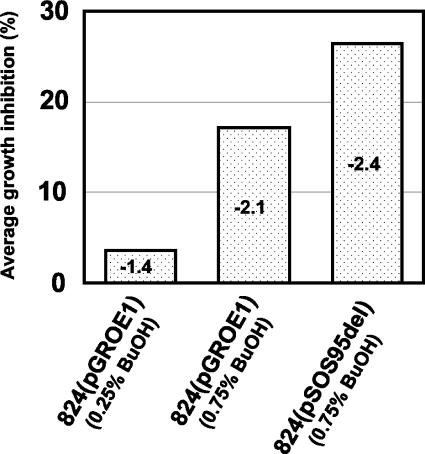
Alterations in cell membrane composition are the most established mechanism which cells utilize to adapt to high levels of solvents (58). The ability of cells to incorporate a higher percentage of transmembrane fatty acids (long-chain C30 fatty acids) in the cell membrane has been shown to increase alcohol tolerance in Thermoanaerobacter ethanolicus (7) from 1.5 to 8% (vol/vol) ethanol. It has been shown that clostridial cells increase the saturated chain content (1, 33, 52) and decrease the fluidity of their lipid membranes (2) in order to adapt to butanol stress. In C. acetobutylicum, butanol stress resulted in decreased expression of a large fatty acid biosynthesis (fab) operon in both 824(pGROE1) and 824(pSOS95del). The organization of the fatty acid biosynthesis genes in a single operon in C. acetobutylicum was similar to that of Streptococcus mutans, Streptococcus pneumoniae, and Streptococcus pyogenes but differed from that of B. subtilis, where the same genes are organized into four operons (10). While the expression patterns for most of the fab operon genes were similar to each other, there were a few notable exceptions. The expression pattern of fabH and marR in the 824(pSOS95del) culture and that of accD in the low-dose 824(pGROE1) culture were opposite that of the other fab operon genes (Fig. 6A) and may be due to an alternative transcriptional or posttranscriptional control mechanism that is not yet known. Despite the unexpected differences in gene expression patterns for these three genes, the degree to which the fab operon had lower relative expression correlates with the level of growth inhibition, i.e., cultures with the lowest average relative expression of fab operon genes had the highest level of growth inhibition (Fig. 7). Large changes in the expression level of a large fatty acid biosynthesis operon are likely to result in membrane composition changes that alter membrane fluidity. The addition of butanol results in changes to fab operon expression that are likely to be detrimental to cell viability in the presence of high levels of butanol.
FIG. 7.
Relationship between fab operon expression and growth inhibition. Numbers inside bars represent the average fab operon expression ratio. Negative expression ratios indicate lower expression in the butanol-challenged cultures. Average fab operon expression ratios were calculated by using the area under the fab operon time course expression profile curves for each strain. Average growth inhibitions were calculated by using the area under the growth inhibition curves for each strain.
Four classes of heat shock genes have been identified in B. subtilis (44). Expression of class I HSP genes, which include the dnaK and groE operons, is HrcA dependent. Class I HSP genes have been identified in C. acetobutylicum; these include the dnaK and groE operons but also the hsp90 gene (50). Class II HSP genes in B. subtilis include a large number of general stress response genes under the control of σB (22), including a large sigB operon (rsbRSTUVW-sigB-rsbX). A search of the C. acetobutylicum genome for a homologous operon resulted in no significant matches, suggesting that a different general stress response mechanism is utilized in C. acetobutylicum. Furthermore, most genes known to be regulated by σB in B. subtilis (22) also lack significant homologies in C. acetobutylicum. Expression of class III HSP genes, which include clpP, clpE, and the four genes of the clpC operon (ctsR, yacH, yacI, and clpC), in B. subtilis is CtsR dependent. A search of the C. acetobutylicum genome (11) for the consensus CtsR operator sequence [(A/G)GTCAAA NAN (A/G)GTCAAA] from B. subtilis yielded an exact match in front of hsp18 and a predicted clpC operon with the exact same arrangement as that in B. subtilis. No CtsR-binding sequence was found upstream of clpP and clpE. The fourth class of heat shock genes includes genes whose expression is responsive to heat shock but whose induction is affected neither by the two repressor proteins HrcA and CtsR nor by σB. Genes with generally higher expression, such as those shown in Fig. 3C (excluding genes classified as class I or III), represent possible class IV genes. In B. subtilis, class I and III genes are considered heat stress specific, while class II and IV genes are considered part of the nonspecific multiple-stress response (44). This differs from what is reported here for C. acetobutylicum, where both class I and class III genes were upregulated upon butanol stress. While an increasing amount of information has recently become available on the stress response in C. acetobutylicum and its possible relation to differentiation and solvent formation, additional studies will be necessary to further elucidate the stress response and its control mechanisms.
Acknowledgments
This work was supported by grants BES-9911231 and BES-0331402 from the National Science Foundation and grant R828562 from the Environmental Protection Agency.
We acknowledge use of the Keck Biophysics Facility and the Center for Genetic Medicine facilities at Northwestern University. We thank Abbott Laboratories for the donation of clarithromycin.
REFERENCES
- 1.Baer, S. H., H. P. Blaschek, and T. L. Smith. 1987. Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol-tolerant Clostridium acetobutylicum. Appl. Environ. Microbiol. 53:2854-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer, S. H., D. L. Bryant, and H. P. Blaschek. 1989. Electron spin resonance analysis of the effect of butanol on the membrane fluidity of intact cells of Clostridium acetobutylicum. Appl. Environ. Microbiol. 55:2729-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahl, H., H. Müller, S. Behrens, H. Joseph, and F. Narberhaus. 1995. Expression of heat shock genes in Clostridium acetobutylicum. FEMS Microbiol. Rev. 17:341-348. [DOI] [PubMed] [Google Scholar]
- 4.Bass, S., Q. Gu, and A. Christen. 1996. Multicopy suppressors of prc mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoAB), DksA, and a truncated RlpA. J. Bacteriol. 178:1154-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowles, L. K., and W. L. Ellefson. 1985. Effects of butanol on Clostridium acetobutylicum. Appl. Environ. Microbiol. 50:1165-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brioukhanov, A. L., R. K. Thauer, and A. I. Netrusov. 2002. Catalase and superoxide dismutase in the cells of strictly anaerobic microorganisms. Microbiology 71:281-285. [PubMed] [Google Scholar]
- 7.Burdette, D. S., S. H. Jung, G. J. Shen, R. I. Hollingsworth, and J. G. Zeikus. 2002. Physiological function of alcohol dehydrogenases and long-chain (C30) fatty acids in alcohol tolerance of Thermoanaerobacter ethanolicus. Appl. Environ. Microbiol. 68:1914-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, K. H., R. J. Heath, and C. O. Rock. 2000. β-Ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J. Bacteriol. 182:365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornillot, E., R. V. Nair, E. T. Papoutsakis, and P. Soucaille. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J. Bacteriol. 179:5442-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Mendoza, D., G. E. Schujman, and P. S. Aguilar. 2002. Biosynthesis and function of membrane lipids, p. 43-55. In A. Sonenshein, J. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 11.Dérre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 12.Dürre, P., M. Bohringer, S. Nakotte, S. Schaffer, K. Thormann, and B. Zickner. 2002. Transcriptional regulation of solventogenesis in Clostridium acetobutylicum. J. Mol. Microbiol. Biotechnol. 4:295-300. [PubMed] [Google Scholar]
- 13.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis, A. J., and C. J. Dodge. 1991. Dissolution of ferrites by Clostridum sp. Geomicrobiol. J. 9:27-40. [Google Scholar]
- 16.Francis, A. J., C. J. Dodge, F. L. Lu, G. P. Halada, and C. R. Clayton. 1994. XPS and Xanes studies of uranium reduction by Clostridium sp. Environ. Sci. Technol. 28:636-639. [DOI] [PubMed] [Google Scholar]
- 17.Girbal, L., and P. Soucaille. 1998. Regulation of solvent production in Clostridium acetobutylicum. Trends Biotechnol. 16:11-16. [Google Scholar]
- 18.Gottwald, M. A., H. Hippe, and G. Gottschalk. 1984. The internal pH of Clostridium acetobutylicum and its effect on the shift from acid to solvent formation. Arch. Microbiol. 143:42-46. [Google Scholar]
- 19.Hara, H., Y. Yamamoto, A. Higashitani, H. Suzuki, and Y. Nishimura. 1991. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J. Bacteriol. 173:4799-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris, L. M., L. Blank, R. P. Desai, N. E. Welker, and E. T. Papoutsakis. 2001. Fermentation characterization and flux analysis of recombinant strains of Clostridium acetobutylicum with an inactivated solR gene. J. Ind. Microbiol. Biotechnol. 27:322-328. [DOI] [PubMed] [Google Scholar]
- 21.Harris, L. M., N. E. Welker, and E. T. Papoutsakis. 2002. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helmann, J. D., M. F. W. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, L., C. W. Forsberg, and L. N. Gibbins. 1986. Influence of external pH and fermentation products on Clostridium acetobutylicum intracellular pH and cellular distribution of fermentation products. Appl. Environ. Microbiol. 51:1230-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, L., J. Ku, M. Pookanjanatavip, X. Gu, D. Wang, P. J. Greene, and D. V. Santi. 1998. Identification of two Escherichia coli pseudouridine synthases that show multisite specificity for 23S RNA. Biochemistry 37:15951-15957. [DOI] [PubMed] [Google Scholar]
- 25.Hughes, J. B., C. Y. Wang, R. Bhadra, A. Richardson, G. N. Bennett, and F. B. Rudolph. 1998. Reduction of 2,4,6-trinitrotoluene by Clostridium acetobutylicum through hydroxylamino-nitrotoluene intermediates. Environ. Toxicol. Chem. 17:343-348. [Google Scholar]
- 26.Ingram, L. O. 1990. Ethanol tolerance in bacteria. Crit. Rev. Biotechnol. 9:305-319. [DOI] [PubMed] [Google Scholar]
- 27.Inoue, A., and K. Horikoshi. 1991. Estimation of solvent-tolerance of bacteria by the solvent parameter LogP. J. Ferment. Bioeng. 71:194-196. [Google Scholar]
- 28.Inoue, A., and K. Horikoshi. 1989. A Pseudomonas thrives in high-concentrations of toluene. Nature 338:264-266. [Google Scholar]
- 29.Jones, D. T., A. V. D. Westhuizen, S. Long, E. R. Allock, S. J. Reid, and D. R. Woods. 1982. Solvent production and morphological changes in Clostridium acetobutylicum. Appl. Environ. Microbiol. 43:1434-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, D. T., and D. R. Woods. 1986. Acetone-butanol fermentation revisited. Microbiol. Rev. 50:484-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanesaki, Y., I. Suzuki, S. I. Allakhverdiev, K. Mikami, and N. Murata. 2002. Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 290:339-348. [DOI] [PubMed] [Google Scholar]
- 32.Khandekar, S. S., D. R. Gentry, G. S. Van Aller, P. Warren, H. Xiang, C. Silverman, M. L. Doyle, P. A. Chambers, A. K. Konstantinidis, M. Brandt, R. A. Daines, and J. T. Lonsdale. 2001. Identification, substrate specificity, and inhibition of the Streptococcus pneumoniae β-ketoacyl-acyl carrier protein synthase III (FabH). J. Biol. Chem. 276:30024-30030. [DOI] [PubMed] [Google Scholar]
- 33.Lepage, C., F. Fayolle, M. Hermann, and J. P. Vandecasteele. 1987. Changes in membrane-lipid composition of Clostridium acetobutylicum during acetone butanol fermentation: effects of solvents, growth temperature and pH. J. Gen. Microbiol. 133:103-110. [Google Scholar]
- 34.Long, S., D. T. Long, and D. R. Woods. 1984. Initiation of solvent production, clostridial stage and endospore formation in Clostridium acetobutylicum P262. Appl. Microbiol. Biotechnol. 20:256-261. [Google Scholar]
- 35.McNeil, B. 1986. The acetone butanol fermentation. Adv. Appl. Microbiol. 31:61-92. [Google Scholar]
- 36.Nölling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. D. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Y. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ounine, K., H. Petitdemange, G. Raval, and R. Gay. 1985. Regulation and butanol inhibition of d-xylose and d-glucose uptake in Clostridium acetobutylicum. Appl. Environ. Microbiol. 49:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piper, P. W. 1995. The heat-shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol. Lett. 134:121-127. [DOI] [PubMed] [Google Scholar]
- 39.Poulsen, P., and K. F. Jensen. 1991. Three genes preceding pyrE on the Escherichia coli chromosome are essential for survival and normal cell morphology in stationary culture and at high temperature. Res. Microbiol. 142:283-288. [DOI] [PubMed] [Google Scholar]
- 40.Ravagnani, A., K. C. Jennert, E. Steiner, R. Grunberg, J. R. Jefferies, S. R. Wilkinson, D. I. Young, E. C. Tidswell, D. P. Brown, P. Youngman, J. G. Morris, and M. Young. 2000. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol. Microbiol. 37:1172-1185. [DOI] [PubMed] [Google Scholar]
- 41.Rogers, P. 1986. Genetics and biochemistry of Clostridium relevant to development of fermentation processes. Adv. Appl. Microbiol. 31:1-60. [Google Scholar]
- 42.Sauer, U., and P. Dürre. 1995. Differential induction of genes related to solvent formation during the shift from acidogenesis to solventogenesis in continuous culture of Clostridium acetobutylicum. FEMS Microbiol. Lett. 125:115-120. [Google Scholar]
- 43.Schulz, A., and W. Schumann. 1996. hrcA, the first gene of the Bacillus subtilis dnaK operon, encodes a negative regulator of class I heat shock genes. J. Bacteriol. 178:1088-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schumann, W., M. Hecker, and T. Msadek. 2002. Regulation and function of heat-inducible genes in Bacillus subtilis, p. 359-368. In A. Sonenshein, J. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 45.Sinensky, M. 1974. Homeoviscous adaptation, a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci. USA 71:522-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spain, J. C. 1995. Biodegradation of nitroaromatic compounds. Annu. Rev. Microbiol. 49:523-555. [DOI] [PubMed] [Google Scholar]
- 47.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 48.Terracciano, J. S., and E. R. Kashket. 1986. Intracellular conditions required for the initiation of solvent production by Clostridium acetobutylicum. Appl. Environ. Microbiol. 52:86-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomas, C. A., K. V. Alsaker, H. P. J. Bonarius, W. T. Hendriksen, H. Yang, J. A. Beamish, C. J. Parades, and E. T. Papoutsakis. 2003. DNA-array based transcriptional analysis of asporogenous, non-solventogenic Clostridium acetobutylicum strains SKO1 and M5. J. Bacteriol. 185:4539-4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomas, C. A., N. E. Welker, and E. T. Papoutsakis. 2003. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell's transcriptional program. Appl. Environ. Microbiol. 69:4951-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vermue, M., J. Sikkema, A. Verheul, R. Bakker, and J. Tramper. 1993. Toxicity of homologous series of organic-solvents for the gram-positive bacteria Arthrobacter and Nocardia sp. and the gram-negative bacteria Acinetobacter and Pseudomonas sp. Biotechnol. Bioeng. 42:747-758. [DOI] [PubMed] [Google Scholar]
- 52.Vollherbst-Schneck, K., J. A. Sands, and B. S. Montenecourt. 1984. Effect of butanol on lipid composition and fluidity of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 47:193-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wahlstrom, E., M. Vitikainen, V. P. Kontinen, and M. Sarvas. 2003. The extracytoplasmic folding factor PrsA is required for protein secretion only in the presence of the cell wall in Bacillus subtilis. Microbiology 149:569-577. [DOI] [PubMed] [Google Scholar]
- 54.Watrous, M. M., S. Clark, R. Kutty, S. Huang, F. B. Rudolph, J. B. Hughes, and G. N. Bennett. 2003. 2,4,6-Trinitrotoluene reduction by an Fe-only hydrogenase in Clostridium acetobutylicum. Appl. Environ. Microbiol. 69:1542-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiesenborn, D. P., F. B. Rudolph, and E. T. Papoutsakis. 1988. Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl. Environ. Microbiol. 54:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, H., H. Haddad, C. Tomas, K. Alsaker, and E. T. Papoutsakis. 2003. A segmental nearest neighbor normalization and gene identification method gives superior results for DNA-array analysis. Proc. Natl. Acad. Sci. USA 100:1122-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young, M., M. E. Collins, J. D. Oultram, and A. Pennock. 1986. Genetic exchange and prospects for cloning in Clostridia, p. 259-281. In A. T. Ganeson and J. A. Hoch (ed.), Bacillus molecular genetics and biotechnology applications. Academic Press, Orlando, Fla.
- 58.Zhao, Y., L. A. Hindorff, A. Chuang, M. Monroe-Augustus, M. Lyristis, M. L. Harrison, F. B. Rudolph, and G. N. Bennett. 2003. Expression of a cloned cyclopropane fatty acid synthase gene reduces solvent formation in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 69:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]